Abstract
Human pregnane X receptor (hPXR) plays a key role in regulating metabolism and clearance of endogenous and exogenous substances. Identification of novel hPXR activators among commercial drugs may aid in avoiding drug-drug interactions during coadministration. We applied ligand-based computational approaches for virtual screening of a commonly prescribed drug database (SCUT). Bayesian classification models were generated with a training set comprising 177 compounds using Fingerprints and 117 structural descriptors. A cell-based luciferase reporter assay was used for evaluation of chemical-mediated hPXR activation in HepG2 cells. All compounds were tested at 10 μM concentration with rifampicin and dimethyl sulfoxide as positive and negative controls, respectively. The Bayesian models showed specificity and overall prediction accuracy up to 0.92 and 0.69 for test set compounds. Screening the SCUT database with this model retrieved 105 hits and 17 compounds from the top 25 hits were chosen for in vitro testing. The reporter assay confirmed that nine drugs, i.e., fluticasone, nimodipine, nisoldipine, beclomethasone, finasteride, flunisolide, megestrol, secobarbital, and aminoglutethimide, were previously unidentified hPXR activators. Thus, the present study demonstrates that novel hPXR activators can be efficiently identified among U.S. Food and Drug Administration-approved and commonly prescribed drugs, which should lead to detection and prevention of potential drug-drug interactions.
Introduction
Nuclear receptors (NRs) are a class of transcription factors that control gene expression and play a key role in the development, homeostasis, and metabolism of living organisms (di Masi et al., 2009). The pregnane X receptor (PXR) belongs to the NR1I family and regulates enzymes and transporters involved in xenobiotic detoxification as well as maintaining a homeostatic balance of endobiotics, including bile acids, cholesterols, and steroid hormones (Jyrkkärinne et al., 2008). PXR mediates activation of gene sets pertinent to xenobiotic metabolism, such as cytochrome 450 superfamily members CYP1, CYP2B, CYP2C, and CYP3A4 in rodents and humans (Maglich et al., 2002; Plant, 2007; di Masi et al., 2009). A very broad range of substances have been identified as human (h) PXR activators in vitro, including commercial drugs, pesticides, environmental contaminants, and natural products (Timsit and Negishi, 2007).
Because of its vital role in drug metabolism, it is not surprising that human PXR has been found responsible for decreased drug efficacy and increased drug toxicity (Ma et al., 2008; di Masi et al., 2009). For example, coadministration of rifampicin, a hPXR activator used for treatment of tuberculosis (Chrencik et al., 2005) with a variety of drugs [including oral contraceptives (Ma et al., 2008), the anesthetic midazolam (Backman et al., 1996), and HIV protease inhibitors (Ivanovic et al., 2008)], resulted in decreased drug efficacy mainly due to hPXR-mediated increased expression of CYP3A4 (Ivanovic et al., 2008; Ma et al., 2008; di Masi et al., 2009). Thus, identification of novel hPXR activators among commercial drugs is important in predicting hPXR-mediated drug-drug interactions.
Crystal structures of the hPXR ligand-binding domain (LBD) indicate that its binding cavity is much larger than that of other NR family members (Xu et al., 2004; Chrencik et al., 2005; di Masi et al., 2009). Several key amino acid residues are responsible for the high flexibility of its binding site that is critical for recognizing promiscuous ligands of various dimensions and chemical properties (Ekins et al., 2009). Probably because of the flexibility of the hPXR LBD and the limitation of docking algorithms, docking of structurally diverse molecules is a challenge (Ekins et al., 2008, 2009; Khandelwal et al., 2008; Yasuda et al., 2008). Therefore, docking methods have been suggested for use in combination with other computational methods to improve prediction (Khandelwal et al., 2008; Yasuda et al., 2008; Ekins et al., 2009). The flexibility and large size of the hPXR LBD necessitates development of multiple pharmacophores for consensus prediction by considering interactions between a ligand and various binding sites in protein (Yasuda et al., 2008). In a recent study, ligand-based structure-activity relationship approaches, such as machine learning methods (Khandelwal et al., 2008) and Bayesian statistics (Ekins et al., 2009; Zientek et al., 2010), have been applied to generate models by using just binary classification of ligands (e.g., activator and nonactivator) instead of quantitative data while using two-dimensional instead of three-dimensional descriptors.
In the current study, we applied Bayesian models to identify novel hPXR activators by virtual screening of an in-house database of frequently prescribed U.S. Food and Drug Administration-approved drugs (SCUT) (Chang et al., 2006). We confirmed 9 novel hPXR activators of 17 predicted hPXR activators by luciferase reporter assay; this result indicates that ligand-based virtual screening combined with experimental validation assays is a valuable tool for efficient retrieval of novel ligands that interact with hPXR.
Materials and Methods
Principal Component Analysis of SCUT Database Molecules and Training and Test Set Compounds.
Datasets consisting of 177 (Ung et al., 2007; Khandelwal et al., 2008) and 145 (Khandelwal et al., 2008) previously published hPXR activators/nonactivators were used as training and test sets, respectively. In the training set, 98 compounds with EC50 < 100 μM were classified as hPXR activators, and 79 compounds with EC50 > 100 μM were classified as nonactivators. The current test set consisted of 82 activators and 63 nonactivators that were reported previously. A total of 104 independent variables representing molecular size, solubility, flexibility, polarity, charge, surface area, and hydrogen bond features were calculated with the “Calculate Molecular Properties” protocol of Discovery Studio 2.1 (DS2.1; Accelrys, San Diego, CA). The PCA plot is a useful tool to assess similarity among training and test set compounds so as to understand potential outlier prediction (Khandelwal et al., 2007). A PCA plot of training, test, and SCUT database compounds was performed with the protocol “Calculate Principal Components” of DS2.1 by using 3 as the minimum number of components and 0.65 minimum variance explained.
Building and Validation of Bayesian Models.
Bayesian statistics is a classification approach based on a learn-by-example protocol. A Bayesian model is created by estimating the frequencies of features when a hypothesis is true (Xia et al., 2004). To apply the model to a sample, a weight is calculated for each feature using a Laplacian-corrected estimator. The prediction of the likelihood of a sample is made by summing up the weights associated with each feature (Xia et al., 2004; Rogers et al., 2005). Extended connectivity fingerprints of maximum diameter 6 (ECFP_6), functional class fingerprints of maximum diameter 6 (FCFP_6) (Rogers et al., 2005) and another 117 structural descriptors were calculated with DS2.1. Laplacian-corrected Bayesian classification models (Xia et al., 2004; Rogers et al., 2005) were generated using the “Create Bayesian Model” protocol. Leave-one-out cross-validation-based “receiver operator curve” area under the curve (XV ROC AUC) (Zweig and Campbell, 1993) was calculated for the training set compounds as an assessment of predictive capacity of Bayesian models. The Bayesian models were validated with the test set as well. The activities of the test set compounds were predicted by the Calculate Molecular Properties protocol with the Bayesian models.
Virtual Screening of SCUT Database Drugs with Bayesian Models.
Drugs from the SCUT database were virtually screened for prediction of their hPXR activities through the same protocol as for test set compounds by a Bayesian model with the highest ranking predictive performance (see Results).
Docking of Test Set and SCUT Drugs to hPXR Ligand-Binding Domain with FlexX and Surflex.
Docking programs were applied to evaluate binding between ligands and the LBD of hPXR. In the FlexX docking algorithm (BioSolveIT GmbH, Sankt Augustin, Germany), the protein is kept rigid while the ligand is treated flexibly. The overall strategy of this docking method is the incremental construction algorithm (Rarey et al., 1996), in which a ligand is first split up into fragments. The selection of the base fragment is centered on a recognition technique called pose clustering; subsequently, the ligand is built up incrementally by adding other fragments with a simple greedy strategy. The ranking of fragment placements and estimation of binding energy use terms of the Böhm scoring function with minor changes (Rarey et al., 1996).
Surflex (Tripos, St. Louis, MO) is similar to FlexX in that a molecule is first fragmented, and the conformations of each piece are further explored. Here, a “protomol,” an idealized active site ligand comprising a cluster of molecular fragments featuring the binding pocket surface (Ruppert et al., 1997), is generated and serves as a target for alignment of fragment conformations based on the molecular similarity method (Jain, 2003). An entire molecule is then assembled by the incremental construction approach (Welch et al., 1996) or a genetic algorithm called the “Whole Molecule Algorithm” (Jones et al., 1997). The Hammerhead empirical function is used as a scoring function of putative poses as well as an objective function of local optimization (Jain, 2003).
Previous docking studies indicated that the crystal structure of the hPXR LBD (Protein Data Bank 1NRL) (Watkins et al., 2003) performed better in predicting hPXR activators/nonactivators than three other hPXR LBD crystal structures with FlexX (Khandelwal et al., 2008). Accordingly, the crystal structure 1NRL was selected as the docking target. In FlexX, residues within 6.5 Å of a ligand were defined as the active site. The maximum number of solutions per iteration and fragmentation was 500. The single top docked orientation with the best score (in kilocalories per mole) (Stahl, 2000) of each ligand was generated. In Surflex docking, the protomol was generated based on active site residues, with proto_thresh of 0.5 and proto_bloat of 0. The maximum number of fragment conformations was 20, and the maximum number of poses per ligand was 20. Selected compounds retrieved from Bayesian modeling were tested in vitro to validate their activity against hPXR.
Chemicals and Reagents.
Rifampicin (RIF) was purchased from Sigma-Aldrich (St. Louis, MO). The dual-luciferase reporter assay system was purchased from Promega (Madison, WI). Drugs butorphanol (+)-tartrate salt, megestrol 17-acetate Vetranal, cefoxitin sodium, estramustine sodium phosphate, flunisolide, fludrocortisone acetate, fluticasone propionate, sulfasalazine, secobarbital methanol solution, and triamcinolone were purchased from Sigma-Aldrich. Finasteride, tobramycin, amikacin disulfate, and nimodipine were purchased from Alexis Biochemicals (San Diego, CA). dl-Aminoglutethimide was purchased from LKT Laboratories (Gardena, CA). Oridonin was from ChromaDex (Irvine, CA). Nisoldipine and beclomethasone dipropionate were obtained from the laboratories of Dr. James Polli and Dr. Richard N. Dalby, University of Maryland (Baltimore, MD).
Plasmid Constructs.
The CYP2B6 reporter construct, containing both the phenobarbital-responsive enhancer module and the distal xenobiotic-responsive enhancer module [CYP2B6-2.2 kilobases (kb)], were generated as described previously (Wang et al., 2003; Tolson et al., 2009). The pSG5-hPXR expression plasmid was acquired from Dr. Steve Kliewer (University of Texas, Southwestern Medical Center, Dallas, TX). The pRL-TK Renilla luciferase plasmid used to normalize firefly luciferase activities was purchased from Promega.
Transient Transfection in HepG2 Cells.
HepG2 cells seeded in 24-well plates were transfected with the CYP2B6-2.2 kb reporter construct in the presence of the hPXR expression vector using a FuGENE 6 Transfection Kit (Roche Applied Science, Indianapolis, IN) according to the manufacturer's protocol. Twenty-four hours after transfection, cells were treated with solvent [0.1% dimethyl sulfoxide (DMSO)] or test compounds (including positive control RIF and drugs) at a concentration of 10 μM for another 24 h. In parallel experiments, the highly cytotoxic chemotherapeutic agent, mitomycin, was tested at 0.1, 1, and 10 μM concentrations. Subsequently, all cell lysates were assayed for firefly activities normalized against the activities of Renilla luciferase using a dual-luciferase kit (Promega). Data are presented as means ± S.D. of three individual transfections.
Dose-Dependent Assay.
On the basis of the preliminary screening results, four drugs highly responsive to hPXR activation or deactivation at 10 μM were further tested at the concentrations of 0.1, 1, and 10 μM in HepG2 cells transfected with hPXR expression and CYP2B6 reporter constructs. The dual-luciferase activities of different treatments were measured and calculated as described above.
Cytotoxicity Assay.
To ensure that the observed hPXR activation of mitomycin in HepG2 cells was not confounded by potential cytotoxic effects, an MTT assay was performed in parallel. The cells were dispensed in a 96-well plate at a density of 1.5 × 104 cells/well. After a 24-h incubation, cells were treated with oridonin at 20 and 40 μM (positive control) or mitomycin at various concentrations for 24 h. A 20-μl aliquot of MTT solution (5.0 mg/ml) was added to each well followed by a 4-h incubation, and the resulting crystals were dissolved in 150 μl of DMSO. Absorbance (A) was measured with a microplate reader (Bio-Rad Laboratories, Hercules, CA).
Results
Principal Component Analysis.
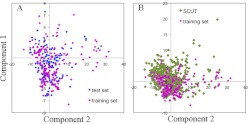
PCA is useful as an estimation of whether molecules occupy different descriptor spaces so as to understand potential outlier predictions of the test set and database compounds. PCA was performed on the SCUT database and the test and training set compounds based on 104 descriptors. The first two and three principal components explained 62 and 68.1% of the variance, respectively, indicating that the plot of the first two components roughly represented the descriptor space occupied by the molecules. Figure 1A demonstrates that the test set accommodated a space similar to that of the training set molecules; the training set drugs covered most of the descriptor space of that occupied by the SCUT database drugs (Fig. 1B).
Fig. 1.
PCA plots among the test set and training set drugs (A) and among training set and SCUT database drugs (B). The first two principal components explained 62% of variance.
Bayesian Model Building and Validation.
The hPXR data for model development (training set n = 177) and validation (test set n = 145) were taken from recent publications (Ung et al., 2007; Khandelwal et al., 2008). A total of 117 structural descriptors including topological variables as well as structural fingerprints ECFP_6 and FCFP_6 (calculated with DS2.1) were applied for model development. Four Bayesian models were generated with combinations of parameters. The first two models, ECFP-1 and FCFP-1 (Table 1), were obtained with 117 descriptors together with ECFP_6 or FCFP_6, respectively. The other two models, ECFP-2 and FCFP-2, were developed by 74 descriptors without topological features as well as ECFP_6 or FCFP_6. The predictive performance of Bayesian models was evaluated by XV ROC AUC based on leave-one-out cross-validation of training set compounds. XV ROC AUC reflects the relationship between sensitivity and specificity, ranging from 0 to 1 with a higher number indicating a better model (Zweig and Campbell, 1993). The models were also validated with an external test set consisting of 145 molecules. The predicted performance of the models was demonstrated by their sensitivity, specificity, overall prediction accuracy (Q), and Matthew's correlation coefficient (C) values calculated from the empirical true positive, true negative, false positive, and false negative (Table 1).
TABLE 1.
Predictive performance of FlexX docking and Bayesian models with training set (n = 177, leave-one-out cross-validation) and test set (n = 145)
| ECFP-1 | FCFP-1 | ECFP-2 | FCFP-2 | |
|---|---|---|---|---|
| Two-dimensional fingerprints | ECFP_6 | FCEP_6 | ECFP_6 | FCFP_6 |
| No. of descriptors | 117 | 117 | 74 | 74 |
| XV ROC AUC (%)a | 87.0 | 85.2 | 86.4 | 87.7 |
| TP/FP/TN/FNb | 37/8/55/45 | 48/11/52/34 | 29/5/58/53 | 33/8/55/49 |
| SE (%)b | 45.1 | 58.5 | 35.4 | 40.2 |
| SP (%)b | 87.3 | 82.5 | 92.1 | 87.3 |
| Q (%)b | 63.4 | 69.0 | 60.0 | 60.7 |
| C (%)b | 34.7 | 41.4 | 32.1 | 30.3 |
Based on training set compounds.
Predictive performance validation by test set compounds: TP, true positive; TN, true negative; FP, false positive; FN, false negative; SE, sensitivity; SP, specificity; Q, overall prediction accuracy; C, Matthew's correlation coefficient (Ung et al., 2007; Khandelwal et al., 2008). SE = TP/(TP + FN), SP = TN/(TN + FP), Q = (TP + TN)/(TP + TN + FP + FN), C = [(TP *TN) − (FN × FP)]/[(TP + FN) (TP + FN) (TN + FN)(TN + FP)]1/2.
The XV ROC AUC values of ECFP-1, FCFP-1, ECFP-2, and FCFP-2 models revealed good internal prediction in terms of leave-one-out cross-validation of training set compounds (Table 1). Model validation with the external test set (Table 1) revealed a good specificity for all models, but a lower sensitivity. This result indicated that the models identified a low ratio of false positives but a high ratio of false negatives.
The effects of using topological descriptors and different fingerprints, i.e., ECFP_6 and FCFP_6, on the predictive performance of the Bayesian models are discussed in Supplemental Results 1. Compared with previous models developed in our laboratory using machine learning methods (Khandelwal et al., 2008), the Bayesian models performed better in terms of higher accuracy and Matthew's correlation values. Among the four Bayesian models, the ECFP-2 model was deemed superior: although ECFP-2 has lower overall prediction accuracy and Matthew's correlation coefficient than ECFP-1 and FCFP-1, the advantage of this model is its high specificity (92.1%), indicating a lower occurrence of false positives. To possibly increase the chance of a hit during experimental tests, the ECFP-2 model was selected for virtual screening to identify novel hPXR inhibitors.
Virtual Screening of SCUT Database with ECFP-2 Bayesian Model and Docking Programs.
SCUT database compounds were screened with the ECFP-2 model, retrieving 113 hits according to their Bayesian scores. The results of the docking test set and SCUT compounds with hPXR are included in Supplemental Results 2 and Tables S1 and S2. Among the 113 hits obtained by virtual screening with Bayesian model ECFP-2, 8 compounds were removed from the list because of failed docking by FlexX and disfavored binding energies indicated by Surflex. Among the 105 hits left, 28 compounds belonging to the training or test set were discarded. The top 50 hits were used to search against PubMed for previously documented hPXR affinity. Four compounds with recorded studies on their hPXR activity were removed. The top 25 hits without any previously documented hPXR study were checked for the feasibility of using them in experimental assays. Ultimately, 17 compounds were selected for in vitro testing based on their commercial availability (Table 2). Interestingly, four predicted hPXR activators, i.e., beclomethasone, triamcinolone, fludrocortisone, and fluticasone, were docked unsuccessfully by FlexX but had favorable Surflex scores. This divergent result could be attributed to the different docking algorithms between FlexX and Surflex and the lack of protein flexibility in docking.
TABLE 2.
Molecules predicted after SCUT database search with Bayesian model, FlexX, and SurfleX
| No. | Name | Bayesian Score | FlexX Score | Surflex Score (−LogKd) | Category | PXR Luc Activity (Mean ± S.D.) |
|---|---|---|---|---|---|---|
| kcal/mol | ||||||
| 1 | Beclomethasone | 21.201 | xa | 4.47 | Corticosteroid | 3.95 ± 0.25 |
| 2 | Triamcinolone | 19.05 | x | 4.33 | Corticosteroid | 0.8 ± 0.07 |
| 3 | Megestrol acetate | 18.56 | −10.2 | 5.91 | Corticosteroid | 5.61 ± 0.35 |
| 4 | Fludrocortisone | 16.43 | x | 4.05 | Corticosteroid | 0.9 ± 0.08 |
| 5 | Fluticasone propionate | 15.71 | x | 6.25 | Corticosteroid | 18.5 ± 1.46 |
| 6 | Estramustine | 11.65 | −5.8 | 4.53 | 17β-Estradiol | 1.04 ± 0.08 |
| 7 | Finasteride | 10.49 | −15.7 | 6.47 | Antiandrogen | 2.97 ± 0.31 |
| 8 | Nisoldipine | 10.14 | −24.3 | 5.19 | Dihydropyridine | 11.7 ± 2.69 |
| 9 | Flunisolide | 9.85 | −16.8 | 4.8 | Corticosteroid | 2.7 ± 0.14 |
| 10 | Mitomycin | 8.09 | −13.5 | 4.1 | 0.12 ± 0.01 | |
| 11 | Nimodipine | 7.99 | −23.2 | 7.63 | Dihydropyridine | 16.5 ± 0.44 |
| 12 | Cefoxitin | 7.51 | −17 | 5.11 | Cephalosporin | 1.07 ± 0.04 |
| 13 | Amikacin | 7.22 | −15.2 | 7.73 | 0.92 ± 0.03 | |
| 14 | Aminoglutethimide | 7.12 | −20 | 6.63 | 1.42 ± 0.07 | |
| 15 | Tobramycin | 6.43 | −9.6 | 7.78 | 1.13 ± 0.07 | |
| 16 | Sulfasalazine | 6.31 | 6.81 | Mesalazine | 0.8 ± 0.08 | |
| 17 | Secobarbital | 6.1 | −12 | 5.53 | Phenobarbital | 1.57 ± 0.06 |
x indicates unsuccessful dock by FlexX.
hPXR Activation by Predicted Drugs.
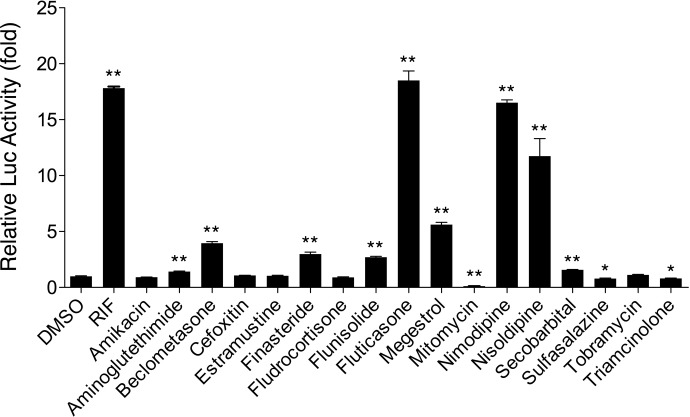
hPXR governs the transactivation of multiple drug-metabolizing genes such as CYP3A4, CYP2B6, and UDP-glucuronosyltransferase 1A1, as well as drug transporters such as multidrug resistance 1. The CYP2B6 reporter construct containing both the proximal phenobarbital-responsive enhancer module and the distal xenobiotic-responsive enhancer module is highly responsive to chemical-mediated hPXR activations (Tolson et al., 2009). Here, we investigated the ability of 17 drugs to activate hPXR-mediated CYP2B6 reporter expression in HepG2 cells. As shown in Fig. 2 and Table 2, compared with vehicle control, nine drugs, i.e., beclomethasone, finasteride, flunisolide, fluticasone propionate, megestrol acetate, nimodipine, nisoldipine, secobarbital, and aminoglutethimide, showed noticeable increases of hPXR-mediated CYP2B6 reporter activities at 10 μM. RIF is an established hPXR activator (Cheng et al., 2009; Messina et al., 2009). Among the active molecules, fluticasone propionate and nimodipine exhibited robust hPXR activation to a level that challenges that of RIF, whereas nisoldipine was not as potent as RIF but still caused a significant increase in hPXR-mediated CYP2B6 reporter activities. Megestrol, beclomethasone, finasteride, and flunisolide caused increased activity by 2.7- to 5.6-fold. Secobarbital and aminoglutethimide demonstrated 1.57- and 1.42-fold higher Luc activity than the vehicle control, indicating their marginal effects on hPXR activation. One interesting observation was that instead of being an hPXR activator, mitomycin had dramatically decreased the CYP2B6 luciferase activity to just 12% of basal level. The drugs amikacin, cefoxitin, estramustine, fludrocortisone, sulfasalazine, tobramycin, and triamcinolone did not have observed enhanced Luc activity (Table 2; Fig. 2).
Fig. 2.
Effects of drugs on hPXR-mediated CYP2B6 reporter gene activation. HepG2 cells were transfected with hPXR expression vectors in the presence of the CYP2B6-2.2 kb reporter construct. Transfected cells were then treated with vehicle or drugs (10 μM) for 24 h. RIF (10 μM) was used as a positive control for hPXR. Luciferase activities were determined and expressed relative to the vehicle control. Data represent the mean ± S.D. (n = 3).
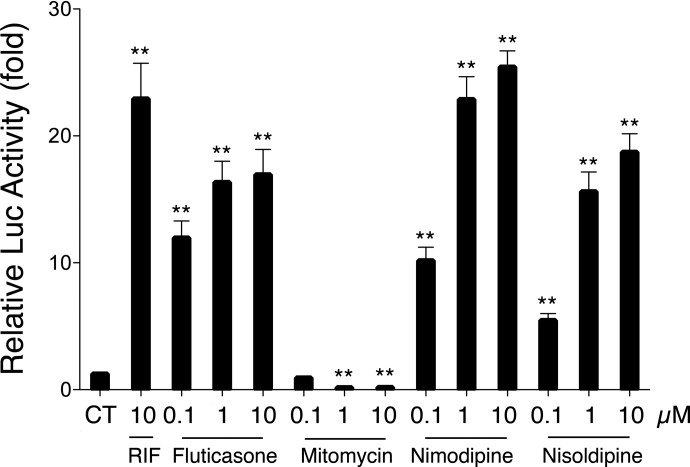
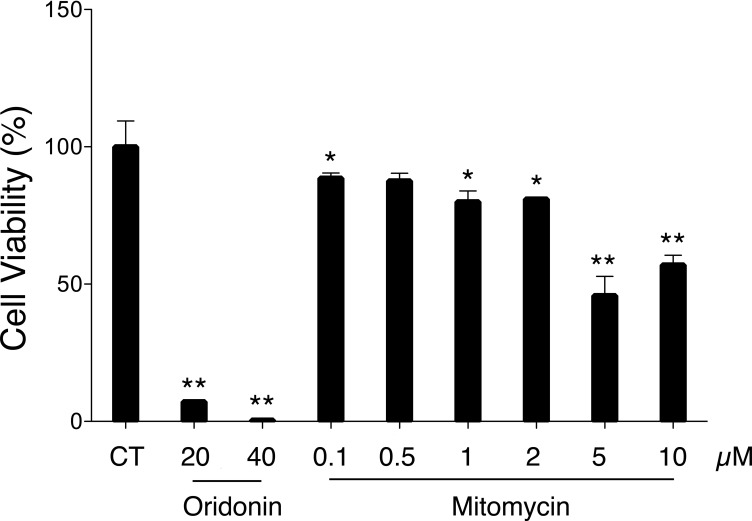
To further confirm their effects on hPXR-mediated Luc activity, a dose-dependent assay of mitomycin and the three compounds with the most potent Luc activity, i.e., fluticasone propionate, nimodipine, and nisoldipine, was performed (Fig. 3). Fluticasone propionate, nimodipine, and nisoldipine significantly enhanced the PXR-mediated CYP2B6 reporter expression in a dose-dependent manner, with the highest activation at the concentration of 10 μM. In contrast, mitomycin (0.1 μM) only moderately decreased CYP2B6 Luc activity compared with the vehicle control. Notably, at the concentrations of 1 and 10 μM, Luc activity decreased by approximately 90% compared with the control. Mitomycin is a chemotherapeutic agent associated with high cytotoxicity (Crowston et al., 2006). To further confirm whether the decreased Luc response with mitomycin was due to the deactivation of hPXR or to its nonspecific cytotoxicity, cell viability testing was performed on mitomycin in HepG2 cells as measured by an MTT assay. At concentrations of 0.1, 0.5, 1, and 2 μM, mitomycin demonstrated minor cytotoxicity, with cell viability between approximately 90 and 80% (Fig. 4). However, at concentrations greater than 5 μM, mitomycin showed clear cytotoxicity with cell viability dropping to approximately 50%. Given that mitomycin decreases PXR-mediated CYP2B6 Luc activity by 90% at concentrations that only cause minor cytotoxicity, mitomycin may represent a novel hPXR deactivator.
Fig. 3.
Dose-dependent assay of fluticasone, nimodipine, nisoldipine, and fluticasone. The compounds were tested for luciferase activity in HepG2 cells at the concentrations of 0.1, 1, and 10 μM. CT indicates the control with 0.1% DMSO). Data represent the mean ± S.D. (n = 3).
Fig. 4.
Cytotoxicity of mitomycin in HepG2 cells measured by the MTT assay. The cells were dispensed in 96-well plate at a density of 1.5 × 104 cells/well. After a 24-h incubation, they were treated with oridonin at 20 and 40 μM (positive control) and mitomycin at various concentrations for 24 h. A 20-μl aliquot of MTT solution (5.0 mg/ml) was added to each well followed by a 4-h incubation, and the resulting crystals were dissolved in 150 μl of DMSO. CT indicates the control with 0.1% DMSO. Data represent the mean ± S.D. (n = 3).
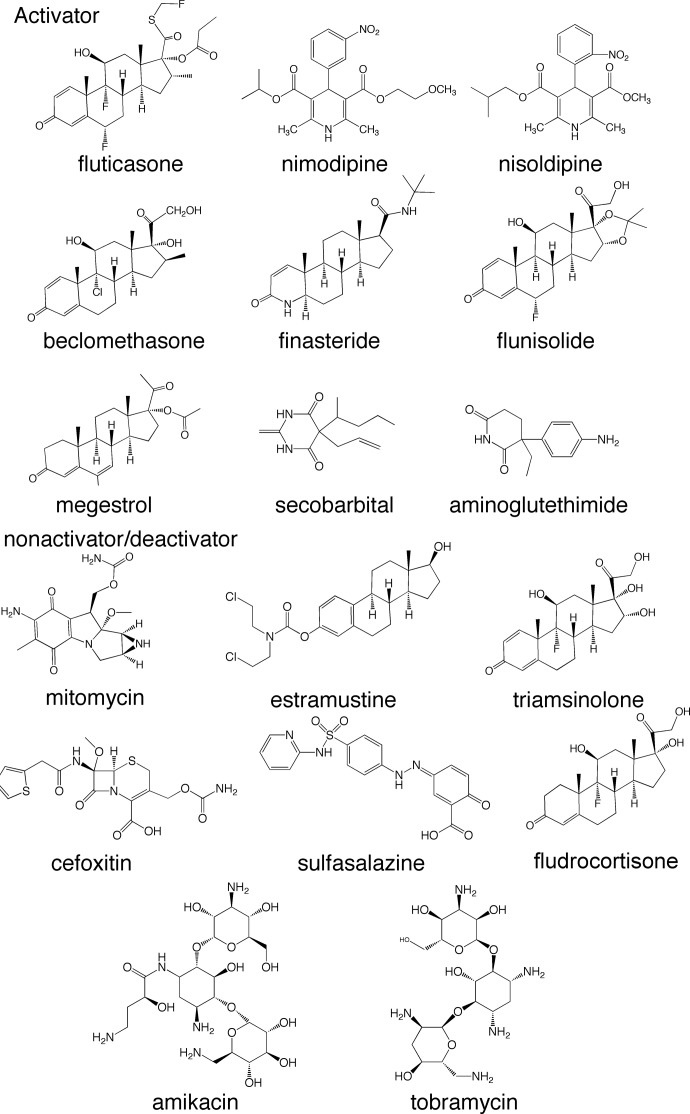
Overall, based on the in vitro tests, with Luc activity 12-fold higher than control, fluticasone propionate, nimodipine, and nisoldipine can be regarded as potent hPXR activators. Megestrol, beclomethasone, finasteride, flunisolide, secobarbital, and aminoglutethimide demonstrated increased Luc activity between 1.57- and 5.6-fold; these compounds can be categorized as weak to moderate hPXR activators (Fig. 5).
Fig. 5.
Chemical structures and therapeutic classifications of newly identified hPXR activators, nonactivators, and a deactivator.
Binding of Newly Identified hPXR Activators with the hPXR LBD.
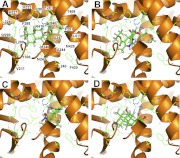
Surflex was used to demonstrate the interactions between the binding pocket of the hPXR LBD and some of the newly identified hPXR activators, i.e., fluticasone, nimodipine, nisoldipine, and beclomethasone (Fig. 6). Fluticasone (Fig. 6A) and nimodipine (Fig. 6B) have hydrogen bond interactions with the side chains of His407 and Ser247, whereas nisoldipine (Fig. 6C) has hydrogen bonds with these residues and with Thr408. Conversely, hPXR LBD docking of beclomethasone reveals hydrogen bonding with residues Asn285 and His407 (Fig. 6D). Docking of the other newly identified hPXR activators is shown in Supplemental Figure S2. In general, we observed that because of the relatively large size of the binding pocket, compounds may move around and interact with different sites within the binding pocket. Similar to the three binding modes of SR12813 in hPXR LBD (Watkins et al., 2001), the side chains of residues His407, Ser247, and Asn285 were predicted to be involved in a hydrogen-bonding interaction with the docked molecules. The compounds beclomethasone, flunisolide, secobarbital, and aminoglutethimide (Fig. 6D; Supplemental Fig. S2, F, H, and I) have hydrogen bonding with a side chain of Asn285. The results are consistent with a previous docking study (Khandelwal et al., 2008), indicating that the side chain NH of Asn285 forms hydrogen bonding interactions with hPXR activators. In addition, the backbone of Leu209 was found to form hydrogen bonds with two drugs, i.e., aminoglutethimide and mitomycin, as shown in Supplemental Fig. S2, I and J. The predicted involvement of Ser247 and Asn285 in the interaction between an activator and hPXR is supported by mutagenesis, in which the mutation of these two residues is responsible for less promiscuity of mouse PXR (di Masi et al., 2009).
Fig. 6.
The Surflex docked (Protein Data Bank 1NRL) conformations of newly identified PXR activators fluticasone (A), nimodipine (B), nisoldipine (C), and beclomethasone (D). The hydrogen-bonding interaction is shown as black dotted lines. The protein backbone is shown as ribbon (orange), amino acid residues are shown in the stick mode, and the PXR activators are shown in the ball and stick mode. The images were created using PyMOL (version 1.3).
Discussion
Need for Integrated Application of Computational and Experimental Approaches.
The integrated use of docking and structure-activity relationship models to determine ligand, substrate, or inhibitor specificity has greatly advanced our understanding of the mechanism of receptors and drug transporters (Khandelwal et al., 2008; Krueger et al., 2009). To date, several combinations of structure- and/or ligand-based methods have been reported to characterize hPXR and activator interactions (Gao et al., 2007; Ekins et al., 2008; Khandelwal et al., 2008; Yasuda et al., 2008), but application of this strategy to identify novel hPXR activators among commercial Food and Drug Administration-approved drugs has not been explored on a large scale.
Previous docking studies on hPXR indicated that directly using the “cutoff score” from docking programs was limited for prediction and classification of hPXR activators and nonactivators (Ekins et al., 2008, 2009; Khandelwal et al., 2008; Yasuda et al., 2008). To overcome this limitation, our present work applied Bayesian models for identification of hPXR activators. The models were generated with known hPXR activators and nonactivators by using different combinations of structural descriptors. The sensitivity, specificity, and overall accuracy evaluated on the basis of external test set compounds served as a means of model validation, and their values indicated that the Bayesian models were able to successfully distinguish hPXR activators from nonactivators. The Bayesian model with the highest specificity was selected for virtual screening of the SCUT database. Although the model has lower sensitivity than other models, the high specificity indicated that this model could result in less false positives, thus increasing the efficiency of experimental tests. The hits were ranked and selected according to their Bayesian scores. Only those with the highest scores and favorable binding energies obtained with docking programs were considered for in vitro testing. The combined computational approach used in this study with experimental assays could be applied in identification of ligands for other proteins.
Newly Identified hPXR Activators, Antagonist, and Nonactivators.
Among the 17 tested drugs, 9 were confirmed as hPXR activators. Mitomycin, although predicted as a hPXR activator, turned out to be a newly identified hPXR antagonist that significantly decreased luciferase activity by 88%. Five of the novel hPXR activators (Table 2), i.e., beclomethasone, finasteride, flunisolide, fluticasone, and megestrol, belong to the corticosteroid/glucocorticoid family, two drugs, i.e., nimodipine, and nisoldipine, are dihydropyridine analogs, and secobarbital is a phenobarbital analog. Although there are no published interactions between any of the nine drugs and PXR, there are quite a few corticosteroid analogs, including dexamethasone, progesterone, 17α-hydroxyprogesterone, 5β-pregnane-3,20-dione, and budesonide (Bhadhprasit et al., 2007; Zimmermann et al., 2009), that are hPXR activators. Dihydropyridines such as nifedipine, nicardipine, and isradipine (Drocourt et al., 2001) have also been reported as PXR activators, whereas a secobarbital analog, phenobarbital, has been identified as a moderate hPXR activator (Lemaire et al., 2004). No aminoglutethimide analog has been shown previously to have an interaction with hPXR. Therefore, the Bayesian model successfully identified nine novel hPXR activators with no previously recorded hPXR interaction, one of which belongs to a novel therapeutic class not previously known to affect PXR activity.
Two tested corticosteroids, triamcinolone and fludrocortisone, did not improve hPXR-mediated CYP2B expression as indicated by the luciferase reporter assay. The structures and logP values of these two drugs are shown in Fig. 5 and Table 3. It can be seen that triamcinolone and fludrocortisone have lower logP values than other corticosteroids that activate hPXR. It is widely known that PXR activators are very hydrophobic (Ekins et al., 2009). The more hydroxyl groups on triamcinolone contribute to its higher hydrophilicity than other corticosteroid analogs, therefore possibly compromising its activity to hPXR. The hydroxyl groups may also represent unfavorable hydrogen bonding interactions. Estramustine is an estradiol derivative and is indicated as a hPXR nonactivator in the current study. 17β-Estradiol is a well established moderate hPXR activator (Xue et al., 2007). As shown in Fig. 3, hPXR activation of estramustine is probably lost because of the attachment of a nitrogen mustard carbamate ester to the phenyl hydroxyl of estradiol. Previous studies showed that the carbamic ester in estramustine is hydrolyzed in vivo by liver, prostate, and intestine (Gunnarsson et al., 1983). As a result, one would expect that estramustine could have in vivo hPXR activity obtained from its metabolite estradiol.
TABLE 3.
Empirical or predicted physicochemical properties of newly identified hPXR activators, an antagonist, and nonactivators
| Name | Molecular Weight | LogP | Solubility | Bioavailability |
|---|---|---|---|---|
| % | ||||
| Fluticasone propionate | 500.6 | 3.4 | Insoluble | 0.51 (intranasal) |
| Nimodipine | 418.4 | 2.7 | 1.20 × 10−2 mg/mla | 100 (intravenous) |
| 13 (oral) | ||||
| Nisoldipine | 388.4 | 3.1 | 5.7 × 10−3 mg/mla | 5 |
| Beclomethasone dipropionate | 408.9 | 1.3 | 49.39 mg/l | 2 |
| Finasteride | 372.5 | 4.7 | 11.7 mg/l | 63 |
| Flunisolide | 434.5 | 1.1 | Insoluble | 6.7 |
| Megestrol | 342.5 | 3.2 | 2 μg/ml | Well absorbed |
| Mitomycin C | 334.3 | −1.6 | Soluble | N.A. |
| Triamcinolone | 394.4 | 0.2 | 80 mg/l | N.A.a |
| Secobarbital | 238.3 | 2.3 | 550 mg/ml | N.A. |
| Aminoglutethimide | 232.3 | 1.3 | Practically insoluble | >95 |
| Fludrocortisone | 380.5 | 0.3 | 140 mg/l | N.A. |
| Estramustine | 440.4 | 5.7 | 3.85 × 10−4 mg/mla | N.A. |
| Cefoxitin | 427.5 | −0.02 | 1.95 × 10−1 mg/mla | 5 |
| Amikacin | 585.6 | −7.4 | 1.85 × 105 mg/ml | 53 |
| Tobramycin | 467.3 | −5.8 | 1 × 103 mg/ml | 11.7 |
| Sulfasalazine | 398.4 | 2.5 | 1.65 × 10−2 mg/ml | <15 |
N.A., not available.
Predicted solubility by DrugBank (http://www.drugbank.ca/).
Clinical Pharmacokinetic Implications for hPXR-Mediated Drug-Drug Interaction.
Because of the critical role of hPXR in regulation of genes involved in metabolism of xenobiotics, identification of novel hPXR activators/nonactivators among commercial drugs could benefit from a priori identification of drug-drug interactions. The empirical or predicted physicochemical properties of newly identified hPXR activators, a suggested antagonist, and nonactivators are included in Table 3. Megestrol acetate can be regarded as a moderate hPXR activator with 5.61-fold increased luciferase activity compared with the vehicle. It has been used for treatment of weight loss in patients with cancer and HIV infection/AIDS (Mulligan et al., 2007). A previous clinical study demonstrated when the anti-HIV drug indinavir was coadministered with megestrol acetate, its efficacy decreased significantly in terms of pharmacokinetic parameters (∼36% for Cmax and ∼28% for AUC) (all clinical pharmacokinetic data in this section were retrieved from http://www.rxlist.com). Indinavir is metabolized mainly by CYP3A in liver (Chiba et al., 1997). Considering that hPXR is a regulator of the cytochrome P450 superfamily and megestrol is an activator of hPXR, one would expect that the drug-drug interaction between megestrol estate and indinavir could be hPXR-mediated.
Nimodipine and nisoldipine are moderate hPXR activators according to the current study. The oral bioavailability of both drugs is relatively low (13 and 5%, respectively), because of the high first-pass metabolism in the liver and gut wall. Drug-drug interactions have been observed between cimetidine and nimodipine or nisoldipine. However, we are not aware of recorded decreases in drug efficacy attributable to coadministration with nimodipine or nisoldipine. Because the available studies of nimodipine/nisoldipine-mediated drug interactions are quite limited, more clinical tests are required. Fluticasone is a potent hPXR activator according to the current study. However, significant involvement of fluticasone in drug interactions is not expected because of its low plasma concentration after extensive first-class metabolism with inhaled dosing.
Beclomethasone, finasteride, and flunisolide were identified as moderate hPXR activators. Beclomethasone dipropionate is metabolized rapidly by high-capacity esterases widely distributed in tissues. Possibly because of the fast clearance of beclomethasone and its major metabolite B-17-MP, no beclomethasone-mediated drug interaction has been observed. Finasteride has been tested with a few clinical drugs, including antipyrine, digoxin, propranolol, and theophylline. Possibly because of its low affinity to hPXR, it did not seem to influence the P450 system, and no meaningful drug interactions have been identified. Flunisolide can be used as nasal spray, and there has been no drug interaction identified.
Secobarbital is a barbiturate derivative. Although there have been few drug interactions observed with secobarbital, its analog phenobarbital (a well recognized hPXR activator) was found to decrease drug efficacy when it was administered concurrently with drugs including anticoagulants, corticosteroids, doxycycline, and estradiol. The phenobarbital-mediated decrease in drug efficacy is attributed to increased metabolism induced by cytochrome P450 enzymes in liver. Because secobarbital is suggested as a weak hPXR activator, its involvement in drug-drug interactions is yet to be determined. Aminoglutethimide was reported to accelerate dexamethasone metabolism and abolish the effects of coumarin and warfarin possibly due to aminoglutethimide-promoted induction of hepatic microsomal enzymes. The confirmation of aminoglutethimide being a weak hPXR activator is consistent with these previously observed drug interactions.
Mitomycin is used as a chemotherapy agent and was identified as a novel hPXR antagonist. Because of the serious side effects caused by its cytotoxicity (Crowston et al., 2006), the application of this drug is tightly restricted. Although an hPXR antagonist could be used with other drugs to increase efficacy, the low basal activity of hPXR and the high toxicity of mitomycin to normal tissues prevent it from being a practical coadministered drug for therapeutic purposes (Pagano et al., 2001). However, it may suggest that we look at other less cytotoxic analogs to understand the activity.
In conclusion, the combined ligand-based screening and experimental assays outlined in this study could present a method to identify potential therapeutic hPXR activators and nonactivators confirmed by in vitro testing. We have shown in this study that Bayesian models generated with available hPXR activators and nonactivators can be used to suggest new potential drug candidates for experimental testing as well as to identify other known activators that are not included for model generation. Identification of hPXR inhibitors provides insights for understanding hPXR mediated drug-drug interactions. Clinical pharmacokinetics and drug interactions pertinent to newly recognized hPXR activators have been investigated, and possible hPXR-mediated drug interactions were discussed. Thus, the application of ligand-based virtual screening in combination with in vitro testing represents a means to rationally identify and subsequently validate commercial drugs as activators for a protein of interest.
Supplementary Material
Acknowledgments
We thank Dr. Alex MacKerell (University of Maryland Baltimore) for making Discovery Studio available to Y.P. We also thank Accelrys for making Discovery Studio available to S.E.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.035808.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- NR
- nuclear receptor
- PXR
- pregnane X receptor
- h
- human
- HIV
- human immunodeficiency virus
- LBD
- ligand binding domain
- PCA
- principal components analysis
- ECFP
- extended connectivity fingerprint
- ECFP_6
- ECFP of maximum diameter 6
- FCFP
- functional class fingerprint
- FCFP_6
- FCFP of maximum diameter 6
- XV ROC AUC
- leave-one-out cross-validation based “receiver operator curve” area under the curve
- RIF
- rifampin
- kb
- kilobase
- DMSO
- dimethyl sulfoxide
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- Luc
- luciferase
- SR12813
- tetraethyl 2-(3,5-di-tert-butyl-4-hydroxyphenyl) ethenyl-1,1-bisphosphonate.
Authorship Contributions
Participated in research design: Pan, Ekins, Wang, and Swaan.
Conducted experiments: Pan, Li, and Kim.
Contributed new reagents or analytic tools: Wang
Performed data analysis: Pan and Swaan.
Wrote or contributed to the writing of the manuscript: Pan, Ekins, Swaan, and Wang.
References
- Backman JT, Olkkola KT, Neuvonen PJ. (1996) Rifampin drastically reduces plasma concentrations and effects of oral midazolam. Clin Pharmacol Ther 59:7–13 [DOI] [PubMed] [Google Scholar]
- Bhadhprasit W, Sakuma T, Hatakeyama N, Fuwa M, Kitajima K, Nemoto N. (2007) Involvement of glucocorticoid receptor and pregnane X receptor in the regulation of mouse CYP3A44 female-predominant expression by glucocorticoid hormone. Drug Metab Dispos 35:1880–1885 [DOI] [PubMed] [Google Scholar]
- Chang C, Bahadduri PM, Polli JE, Swaan PW, Ekins S. (2006) Rapid identification of P-glycoprotein substrates and inhibitors. Drug Metab Dispos 34:1976–1984 [DOI] [PubMed] [Google Scholar]
- Cheng J, Ma X, Krausz KW, Idle JR, Gonzalez FJ. (2009) Rifampicin-activated human pregnane X receptor and CYP3A4 induction enhance acetaminophen-induced toxicity. Drug Metab Dispos 37:1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba M, Hensleigh M, Lin JH. (1997) Hepatic and intestinal metabolism of indinavir, an HIV protease inhibitor, in rat and human microsomes. Major role of CYP3A. Biochem Pharmacol 53:1187–1195 [DOI] [PubMed] [Google Scholar]
- Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, Wisely GB, Lambert MH, Kliewer SA, Redinbo MR. (2005) Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol Endocrinol 19:1125–1134 [DOI] [PubMed] [Google Scholar]
- Crowston JG, Wang XY, Khaw PT, Zoellner H, Healey PR. (2006) Human serum reduces mitomycin-C cytotoxicity in human tenon's fibroblasts. Invest Ophthalmol Vis Sci 47:946–952 [DOI] [PubMed] [Google Scholar]
- di Masi A, De Marinis E, Ascenzi P, Marino M. (2009) Nuclear receptors CAR and PXR: molecular, functional, and biomedical aspects. Mol Aspects Med 30:297–343 [DOI] [PubMed] [Google Scholar]
- Drocourt L, Pascussi JM, Assenat E, Fabre JM, Maurel P, Vilarem MJ. (2001) Calcium channel modulators of the dihydropyridine family are human pregnane X receptor activators and inducers of CYP3A, CYP2B, and CYP2C in human hepatocytes. Drug Metab Dispos 29:1325–1331 [PubMed] [Google Scholar]
- Ekins S, Kholodovych V, Ai N, Sinz M, Gal J, Gera L, Welsh WJ, Bachmann K, Mani S. (2008) Computational discovery of novel low micromolar human pregnane X receptor antagonists. Mol Pharmacol 74:662–672 [DOI] [PubMed] [Google Scholar]
- Ekins S, Kortagere S, Iyer M, Reschly EJ, Lill MA, Redinbo MR, Krasowski MD. (2009) Challenges predicting ligand-receptor interactions of promiscuous proteins: the nuclear receptor PXR. PLoS Comput Biol 5:e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YD, Olson SH, Balkovec JM, Zhu Y, Royo I, Yabut J, Evers R, Tan EY, Tang W, Hartley DP, et al. (2007) Attenuating pregnane X receptor (PXR) activation: a molecular modelling approach. Xenobiotica 37:124–138 [DOI] [PubMed] [Google Scholar]
- Gunnarsson O, Andersson SB, Johansson SA. (1983) The hydrolysis of estramustine phosphate; in vitro studies. Eur J Drug Metab Pharmacokinet 8:395–402 [DOI] [PubMed] [Google Scholar]
- Ivanovic J, Jelena I, Nicastri E, Emanuele N, Ascenzi P, Paolo A, Bellagamba R, Rita B, De Marinis E, Elisabetta de M, et al. (2008) Therapeutic drug monitoring in the management of HIV-infected patients. Curr Med Chem 15:1925–1939 [DOI] [PubMed] [Google Scholar]
- Jain AN. (2003) Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem 46:499–511 [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748 [DOI] [PubMed] [Google Scholar]
- Jyrkkärinne J, Windshügel B, Rönkkö T, Tervo AJ, Küblbeck J, Lahtela-Kakkonen M, Sippl W, Poso A, Honkakoski P. (2008) Insights into ligand-elicited activation of human constitutive androstane receptor based on novel agonists and three-dimensional quantitative structure-activity relationship. J Med Chem 51:7181–7192 [DOI] [PubMed] [Google Scholar]
- Khandelwal A, Bahadduri PM, Chang C, Polli JE, Swaan PW, Ekins S. (2007) Computational models to assign biopharmaceutics drug disposition classification from molecular structure. Pharm Res 24:2249–2262 [DOI] [PubMed] [Google Scholar]
- Khandelwal A, Krasowski MD, Reschly EJ, Sinz MW, Swaan PW, Ekins S. (2008) Machine learning methods and docking for predicting human pregnane X receptor activation. Chem Res Toxicol 21:1457–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger BA, Weil T, Schneider G. (2009) Comparative virtual screening and novelty detection for NMDA-GlycineB antagonists. J Comput Aided Mol Des 23:869–881 [DOI] [PubMed] [Google Scholar]
- Lemaire G, de Sousa G, Rahmani R. (2004) A PXR reporter gene assay in a stable cell culture system: CYP3A4 and CYP2B6 induction by pesticides. Biochem Pharmacol 68:2347–2358 [DOI] [PubMed] [Google Scholar]
- Ma X, Idle JR, Gonzalez FJ. (2008) The pregnane X receptor: from bench to bedside. Expert Opin Drug Metab Toxicol 4:895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. (2002) Nuclear pregnane X receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol 62:638–646 [DOI] [PubMed] [Google Scholar]
- Messina A, Nannelli A, Fiorio R, Longo V, Gervasi PG. (2009) Expression and inducibility of CYP1A1, 1A2, 1B1 by β-naphthoflavone and CYP2B22, 3A22, 3A29, 3A46 by rifampicin in the respiratory and olfactory mucosa of pig. Toxicology 260:47–52 [DOI] [PubMed] [Google Scholar]
- Mulligan K, Zackin R, Von Roenn JH, Chesney MA, Egorin MJ, Sattler FR, Benson CA, Liu T, Umbleja T, Shriver S, et al. (2007) Testosterone supplementation of megestrol therapy does not enhance lean tissue accrual in men with human immunodeficiency virus-associated weight loss: a randomized, double-blind, placebo-controlled, multicenter trial. J Clin Endocrinol Metab 92:563–570 [DOI] [PubMed] [Google Scholar]
- Pagano G, Degan P, De Biase A, Iaccarino M, Warnau M. (2001) Diepoxybutane and mitomycin C toxicity is associated with the induction of oxidative DNA damage in sea urchin embryos. Hum Exp Toxicol 20:651–655 [DOI] [PubMed] [Google Scholar]
- Plant N. (2007) The human cytochrome P450 sub-family: transcriptional regulation, inter-individual variation and interaction networks. Biochim Biophys Acta 1770:478–488 [DOI] [PubMed] [Google Scholar]
- Rarey M, Kramer B, Lengauer T, Klebe G. (1996) A fast flexible docking method using an incremental construction algorithm. J Mol Biol 261:470–489 [DOI] [PubMed] [Google Scholar]
- Rogers D, Brown RD, Hahn M. (2005) Using extended-connectivity fingerprints with Laplacian-modified Bayesian analysis in high-throughput screening follow-up. J Biomol Screen 10:682–686 [DOI] [PubMed] [Google Scholar]
- Ruppert J, Welch W, Jain AN. (1997) Automatic identification and representation of protein binding sites for molecular docking. Protein Sci 6:524–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. (2000) Modifications of the scoring function in FlexX for virtual screening applications. Perspect Drug Discov Des 20:83–98 [Google Scholar]
- Timsit YE, Negishi M. (2007) CAR and PXR: the xenobiotic-sensing receptors. Steroids 72:231–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolson AH, Li H, Eddington ND, Wang H. (2009) Methadone induces the expression of hepatic drug-metabolizing enzymes through the activation of pregnane X receptor and constitutive androstane receptor. Drug Metab Dispos 37:1887–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ung CY, Li H, Yap CW, Chen YZ. (2007) In silico prediction of pregnane X receptor activators by machine learning approaches. Mol Pharmacol 71:158–168 [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. (2003) A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem 278:14146–14152 [DOI] [PubMed] [Google Scholar]
- Watkins RE, Davis-Searles PR, Lambert MH, Redinbo MR. (2003) Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J Mol Biol 331:815–828 [DOI] [PubMed] [Google Scholar]
- Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR. (2001) The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science 292:2329–2333 [DOI] [PubMed] [Google Scholar]
- Welch W, Ruppert J, Jain AN. (1996) Hammerhead: fast, fully automated docking of flexible ligands to protein binding sites. Chem Biol 3:449–462 [DOI] [PubMed] [Google Scholar]
- Xia X, Maliski EG, Gallant P, Rogers D. (2004) Classification of kinase inhibitors using a Bayesian model. J Med Chem 47:4463–4470 [DOI] [PubMed] [Google Scholar]
- Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, et al. (2004) A structural basis for constitutive activity in the human CAR/RXRα heterodimer. Mol Cell 16:919–928 [DOI] [PubMed] [Google Scholar]
- Xue Y, Moore LB, Orans J, Peng L, Bencharit S, Kliewer SA, Redinbo MR. (2007) Crystal structure of the pregnane X receptor-estradiol complex provides insights into endobiotic recognition. Mol Endocrinol 21:1028–1038 [DOI] [PubMed] [Google Scholar]
- Yasuda K, Ranade A, Venkataramanan R, Strom S, Chupka J, Ekins S, Schuetz E, Bachmann K. (2008) A comprehensive in vitro and in silico analysis of antibiotics that activate pregnane X receptor and induce CYP3A4 in liver and intestine. Drug Metab Dispos 36:1689–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientek M, Stoner C, Ayscue R, Klug-McLeod J, Jiang Y, West M, Collins C, Ekins S. (2010) Integrated in silico-in vitro strategy for addressing cytochrome P450 3A4 time-dependent inhibition. Chem Res Toxicol 23:664–676 [DOI] [PubMed] [Google Scholar]
- Zimmermann C, van Waterschoot RA, Harmsen S, Maier A, Gutmann H, Schinkel AH. (2009) PXR-mediated induction of human CYP3A4 and mouse Cyp3a11 by the glucocorticoid budesonide. Eur J Pharm Sci 36:565–571 [DOI] [PubMed] [Google Scholar]
- Zweig MH, Campbell G. (1993) Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39:561–577 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.