Abstract
Extracellular matrices (ECMs) are complex materials, containing dozens of macromolecules that are assembled together, thus complicating their optimization towards applications in 3D cell culture or tissue engineering. The natural complexity of ECMs has limited cell-matrix investigations predominantly to experiments where only one matrix component is adjusted at a time, making it difficult to uncover interactions between different matrix components or to efficiently determine optimal matrix compositions for specific desired biological responses. Here we have developed modular synthetic ECMs based on peptide self-assembly whose incorporation of multiple different peptide ligands can be adjusted. The peptides can co-assemble in a wide range of combinations to form hydrogels of uniform morphology and consistent mechanical properties, but with precisely varied mixtures of peptide ligands. The modularity of this system in turn enabled multi-factorial experimental designs for investigating interactions between these ligands and for determining a multi-peptide matrix formulation that maximized endothelial cell growth. In cultures of HUVECs, we observed a previously unknown antagonistic interaction between the laminin-derived peptide YIGSR and RGDS-mediated cell attachment and growth. We also identified an optimized combination of self-assembled peptides bearing the ligands RGDS and IKVAV that led to endothelial cell growth equivalent to that on native full-length fibronectin. Both of these findings would have been challenging to uncover using more traditional one-factor-at-a-time analyses.
Introduction
The compositions of extracellular matrices (ECMs) are finely tuned in nature to support the architecture and function of their corresponding cells and tissues. The engineering of ECMs has become a central aspect for many developing technologies, particularly within tissue engineering, regenerative medicine, and 3D cell culture.1, 2 In each of these areas, ECMs must be selected, designed, or optimized to favor a given biological outcome, for example rapid proliferation of a particular cell type. Nevertheless, ECMs are formidable targets for materials design, as their compositions and organization are challenging to measure and specify in a way that would allow their optimization towards specific applications. This challenge arises from their native complexity, as they are physical networks of at least dozens of glycoproteins, proteoglycans, and glycosaminoglycans, and each of these constituents can exhibit additional levels of complexity in the form of modular domains, splice variants, or differential glycosylation.2, 3
Multifactorial experiments, where the concentrations of multiple matrix molecules are systematically varied in precise combinations, would efficiently enable the optimization of ECMs towards specific applications, but they are difficult to implement for currently available ECM materials. Other scientific areas and industries such as the agricultural, chemical, and electronics industries have been transformed by the employment of statistical design of experiments (DOE), including techniques such as Factorial Experimentation, Response Surface Methodology, and Robust Design.4 Although high-throughput techniques have been increasingly employed for biomaterial development over the past decade,5–7, 8 (for review, see Hook et al. and references therein),9 extending these types of analyses to biomaterials presenting combinatorial sets of specific ligands has proven challenging. In ECMs, it is difficult to set the concentration of one factor, ligand, or molecule without inadvertently changing the concentration of another. For example, biologically sourced ECMs such as Matrigel contain multiple different glycoproteins, and independent adjustment of specific molecules within it is not straightforward. Similar difficulties are also encountered in synthetic systems such as co-polymers that bear short biomolecular ligands. In such systems, the composition of a polymer produced from multiple co-monomers may not match the composition of the initial monomer feed, a phenomenon known as compositional drift, which in turn necessitates the serial analysis of each new polymer created from each new monomer mix.10
Previous studies have elegantly described DOE and other high-throughput methods in which multiple ECM proteins are spotted from solution into combinatorial arrays,5, 6, 11 but in these methods the amount of protein that is present on the surface has not specifically been known. Cell responses to the arrays can only be correlated to the amount of each protein in the spotting solution, making it difficult to elucidate how ratios of adsorbed proteins may combine to drive specific cell behaviors. In other techniques utilizing arrays of bound peptide ligands, many different ligands can be explored, but it is likely that combinations of peptides would exhibit compositional drift.12 In this report, we employ modular synthetic ECMs derived from multiple co-assembling peptides that enable straightforward factorial optimization for several different matrix-bound ligands. The approach is based on derivatives of the previously reported peptide Q11 (Ac-QQKFQFQFEQQ-Am), which can be assembled together into fibrillar hydrogels that present short peptide ligands,13 immune epitopes,14 or chemical modifications enabling chemoselective cross-linking.15 Previously, Q11 derivatives were designed that presented the RGDS integrin ligand and the laminin-derived IKVAV ligand within self-assembled fibrillar gels.13 We combined these peptides with two new ligand-bearing Q11 derivatives, REDV-Q11 and YIGSR-Q11 (Table 1), to produce a system where each different peptide could be co-assembled (Figure 1), and where the final amount of each peptide in the resultant composite could be known with a high degree of precision. After characterizing the assembly, fibrillization, and gelation behavior of the new peptides, we exploited the modularity of the system to undertake factorial experiments and response surface methodology (RSM). This series of multifactorial experiments enabled the statistical comparison of how each peptide affected endothelial cell growth individually and in combination, and it permitted the calculation of an optimal formulation. Both of these investigations would have been challenging using previous natural or synthetic ECMs. Moreover, the approach was simple and accessible, not requiring any expensive tools beyond standard culture ware and a source of synthetic peptides.
Table 1.
Peptides investigated within multi-functional matrices.
| Name | Sequence | m/z (calc’d) | m/z (found) |
|---|---|---|---|
| Q11 | Ac-QQKFQFQFEQQ-CONH2 | 1526.7 | 1527.7 |
| RGDS-Q11 | Ac-GGRGDSGGG-(Q11)-CONH2 | 2227.3 | 2227.6 |
| RDGS-Q11 (scrambled) | Ac-GGRDGSGGG-(Q11)-CONH2 | 2227.3 | 2227.0 |
| IKVAV-Q11 | Ac-GGIKVAVGGG-(Q11)-CONH2 | 2322.6 | 2322.4 |
| VAKVI-Q11 (scrambled) | Ac-GGVAKVIGGG-(Q11)-CONH2 | 2322.6 | 2321.9 |
| YIGSR-Q11 | Ac-GGYIGSRGGG-(Q11)-CONH2 | 2388.6 | 2389.1 |
| RYGSI-Q11(scrambled) | Ac-GGRYGSIGGG-(Q11)-CONH2 | 2388.6 | 2387.4 |
| IGSE-Q11 (YIGSR control) | Ac-GGIGSEGGG-(Q11)-CONH2 | 2198.3 | 2199.1 |
| REDV-Q11 | Ac-GGREDVGGG-(Q11)-CONH2 | 2311.5 | 2311.8 |
| EVDR-Q11 (scrambled) | Ac-GGEVDRGGG-(Q11)-CONH2 | 2311.5 | 2311.2 |
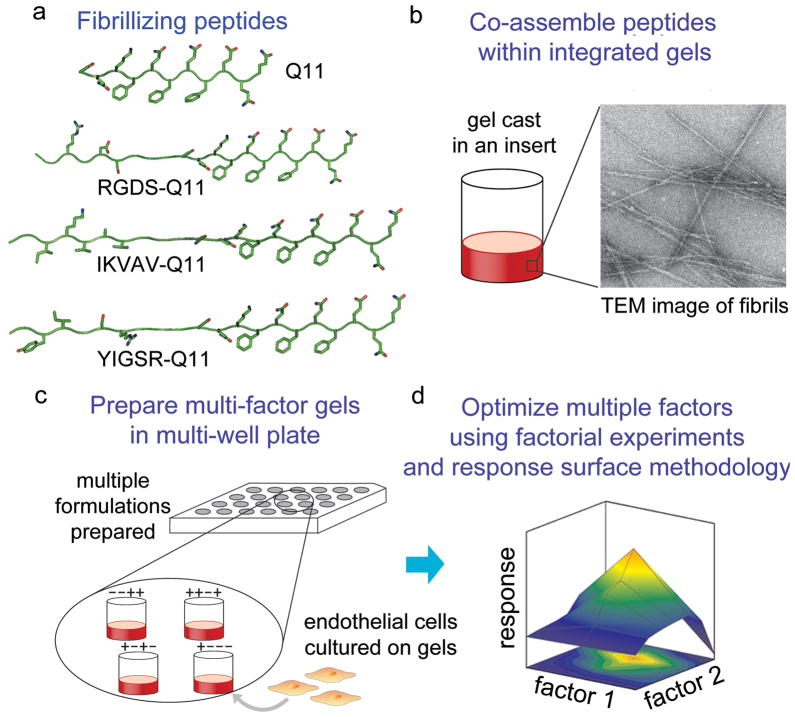
Figure 1.
Schematic for modular scaffolds enabling multifactorial experimentation. A series of ligand-bearing peptides were synthesized that contained the self-assembling Q11 domain at their C-termini (a). These were co-assembled into fibrillar gels (b). An array of gels possessing different combinations of peptides was produced in multi-well plates, on which HUVECs were cultured (c). Using full factorial experiments and response surface methodology, interactions between immobilized ligands were investigated with respect to their combined ability to modulate cell growth, and optimum formulations that maximized cell growth were identified (d).
Materials and Methods
Peptide design and synthesis
All ligand-bearing peptides possessed the same basic construction as previously reported,13 consisting of two N-terminal Gly residues, the ligand sequence, a triglycine spacer, and a C-terminal Q11 sequence (Table 1). Peptide synthesis reagents were purchased from NovaBiochem. All peptides were synthesized as previously reported on a CS Bio 136 automated peptide synthesizer using standard Fmoc-based solid phase chemistry, HOBt/HBTU activation, and Rink amide AM resin. All peptides were N-terminally acetylated, cleaved using conventional TFA/TIS/H2O cocktails, and collected by precipitation in diethyl ether. Peptides mass was verified with MALDI mass spectrometry, and peptides were stored as lyophilized powders at −20°C until experimentation.13, 15
Gel formation and bulk measurements of peptide entrapment
To produce multi-peptide matrices, stock solutions of each peptide were first made in water with extensive vortexing and bath sonication. Peptide concentrations were verified and adjusted using Phe absorbance as previously reported.13, 15 Stock solutions were then mixed to provide solutions with specified peptide ratios, which were then pipetted into culture inserts (transparent PTFE membrane, 0.4 μm pore size, 1.13 cm2 area, Millipore cat# PICM01250), microfuge tubes, or on glass slides as previously reported.13 For culture experiments, the mixed peptide solutions were additionally incubated overnight at 4 °C in sealed 24-well plates, a step that increased the viscosity of the peptide solution. It was observed that the surface of gels was topographically uniform by bright field microscopy, and it did not appear to change grossly for the different peptide formulations studied. Gelation of the peptide solutions was then achieved by overlaying them with Dulbecco’s phosphate buffered saline (PBS), as previously reported.13
To measure the co-assembly of ligand-bearing peptides in the gels, peptide solutions were prepared that consisted of 20 mM total peptide containing 2–10 mM ligand-bearing peptide and the balance Q11. Mixed peptide solutions were overlaid with PBS, incubated for 1 h, washed three times with PBS for 30 min each, dissolved by mixing 1:2.3 with TFA, and analyzed on a C4 HPLC column as previously reported.13 By comparing peptide ratios in gelled and ungelled samples, any unpredictable incorporation or peptide leaching from the gels could be detected in the bulk material. Gels were analyzed in triplicate.
Transmission electron microscopy
Solutions of 1.5 mM aqueous peptide were mixed 1:2 with PBS, vortexed, sonicated, and allowed to fibrillize overnight at room temperature. Fibrils were then applied to 400 mesh carbon grids, stained with 1% uranyl acetate, and analyzed immediately using transmission electron microscopy TEM (FEI Tecnai F30), similarly to previous reports.13, 14
Oscillating rheometry
A Bohlin Gemini rheometer (Malvern Instruments Ltd., Malvern, UK) with an 8 mm parallel plate configuration was utilized to measure gel viscoelasticity. Gels were produced on glass slides using a filter paper template with an 8 mm diameter hole, as previously reported.13, 15 Removing the template after gelation resulted in circular gels of 8 mm diameter that were adherent to the glass slide. Frequency sweep measurements were performed from 0.01–3.3 Hz at 0.1% strain, and three independent gels were tested at each formulation.
Endothelial cell culture, growth, and attachment
Primary human umbilical vein endothelial cells (HUVECs), Endothelial Growth Medium (EGM)-2, and subculture reagents were purchased from Lonza. HUVECs were maintained in EGM-2 containing 2% fetal bovine serum at 37 °C/5% CO2 and subcultured according to the supplier’s protocols. HUVECs were used for experiments between the 4th and 6th passages. HUVECs were seeded at a density of 8,850 cells/cm2 on top of peptide gels, and cell growth was measured with an MTS-based proliferation assay (Promega cat# G3582) as previously reported.13 To measure attachment, cells were plated in serum-free media. One hour later, the media was removed, and loosely adherent cells were removed with serial PBS washes. Adherent cells were counted directly, as previously reported.13
Statistical Design of Experiments (DOE)
The efficient investigation of multiple co-assembled ligands and the determination of an optimal combination of ligands for HUVEC growth were facilitated by the modular nature of the self-assembling peptide system developed. Co-assembled multi-peptide matrices were subjected to a series of multi-factorial experiments using statistical “Design of Experiments” methods.4 The work proceeded in three stages: 1) investigation of each ligand by itself to determine its independent effect on HUVEC growth, 2) factorial experiments with mixtures of ligands to determine how significantly each ligand affected HUVEC growth in the presence of the other ligands and whether any response to a particular ligand was dependent on the level of the other ligands (i.e. interactions between factors), and 3) determination of an optimized formulation using response surface modeling (RSM). In the first stage, the effect of gels containing controlled amounts of RGDS-Q11, IKVAV-Q11, YIGSR-Q11, and REDV-Q11 on HUVEC growth was independently investigated by dosing one of the ligand-bearing peptides into backgrounds of 30 mM Q11. In these initial dose-response experiments, scrambled peptides were used as controls (Table 1). Maximal responses observed in these dose responses were used to set initial boundary conditions for the second stage, the factorial experiments.
In the second stage, a two-level, four-factor (24) design was utilized, in which each factor (ligand-bearing peptide) was set at a high or low level (concentration), producing the matrix of experimental runs shown in Table 2. This design allowed for the identification of the most significant factors and the identification of interactions between those factors. JMP software (SAS, North Carolina, USA) was utilized to facilitate the generation of this experimental matrix, which consisted of seventeen different gel formulations. Gels were produced and analyzed in triplicate, amounting to 51 total gels. HUVECs were seeded onto the gels, and growth was analyzed at 64 h. This time point was selected because it resulted in large enough cell numbers to be able to make distinctions between groups, but not so many that the cultures became confluent. JMP software was utilized to fit the data, estimate the strength of each factor, determine R2 values, analyze the residuals, measure the lack of fit in the model, conduct ANOVA to determine significance, and identify statistically significant factors and interactions.
Table 2.
Experimental runs for factorial experiments. This model is a four-factor, two-level design (24), with one center point. The runs were randomized and conducted in triplicate (51 total gels).
| Run | Patterna | RGDS (mM) | IKVAV (mM) | YIGSR (mM) | Q11 (mM) |
|---|---|---|---|---|---|
| 1 | −−−− | 0 | 0 | 0 | 2 |
| 2 | −−−+ | 0 | 0 | 0 | 20 |
| 3 | − −+ − | 0 | 0 | 6 | 2 |
| 4 | − −++ | 0 | 0 | 6 | 20 |
| 5 | −+ − − | 0 | 1.5 | 0 | 2 |
| 6 | −+ −+ | 0 | 1.5 | 0 | 20 |
| 7 | −++ − | 0 | 1.5 | 6 | 2 |
| 8 | −+++ | 0 | 1.5 | 6 | 20 |
| 9 | 0000 | 3 | 0.75 | 3 | 11 |
| 10 | + | 6 | 0 | 0 | 2 |
| 11 | +− −+ | 6 | 0 | 0 | 20 |
| 12 | +−+ − | 6 | 0 | 6 | 2 |
| 13 | +−++ | 6 | 0 | 6 | 20 |
| 14 | ++− − | 6 | 1.5 | 0 | 2 |
| 15 | ++−+ | 6 | 1.5 | 0 | 20 |
| 16 | +++− | 6 | 1.5 | 6 | 2 |
| 17 | ++++ | 6 | 1.5 | 6 | 20 |
+ and − indicate high and low levels, respectively. 0 indicates a center point.
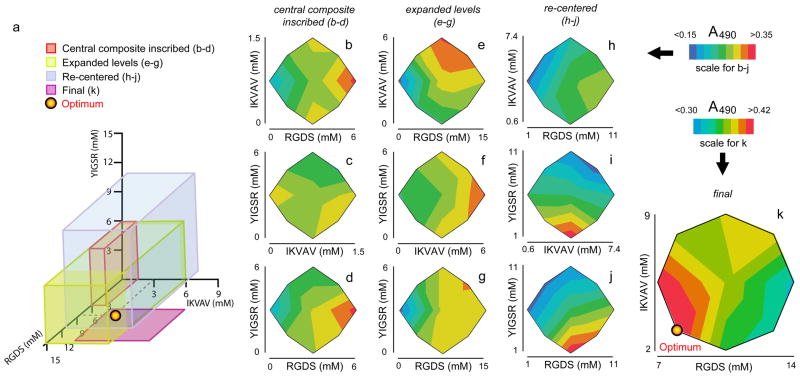
In the third stage, a series of Response Surface Methodology (RSM) experiments were employed to narrow the formulation space to a potential optimal formulation. Four successive rounds of RSM were conducted, each investigating a progressively re-focused range of ligand incorporation in the gels (Figure 6a). The first RSM experiment explored the same range of ligand incorporation as had been investigated in the preceding factorial experiments, utilizing a central composite inscribed design (CCI, Table 3). This design consisted of 16 different combinations of three different factors (RGDS-Q11, IKVAV-Q11, and YIGSR-Q11), in five different levels. Only the three ligand-bearing peptides were controlled as factors in the RSM stage because unmodified Q11 was found to be insignificant in the factorial experiments, and REDV-Q11 was not found to be active in the single-factor experiments. JMP software was again utilized to fit the data into quadratic equations, determine R2 values, analyze the residuals, measure the lack of fit in the model, conduct ANOVA to determine significance, identify statistically significant factors and interactions, and plot the response surface predicted. Each successive round of RSM experiments was used to fine-tune the area of investigation so as to focus on an optimized formulation (experimental designs described in Table 3). This final formulation was then validated with a series of experiments comparing growth and attachment on optimized, unoptimized, and control gels.
Figure 6.
Results from the RSM experiments listed in Table 3. A series of RSM experiments were performed, focusing on sequentially refined areas of formulation space (a). Two-way interaction contour plots are shown for the first central composite inscribed design (b–d), the subsequent design with expanded levels (e–g), the re-centered design (h–j), and the final two-factor RSM experiment (k).
Table 3.
Experimental runs for response surface methodology experiments. All runs were randomized and conducted in triplicate (48 gels for first three rounds, 30 gels for final round).
| Central Composite Inscribed | Expanded Levels | Re-centered | Final | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Run | Patterna | RGDS (mM) | IKVAV (mM) | YIGSR (mM) | Patterna | RGDS (mM) | IKVAV (mM) | YIGSR (mM) | Patterna | RGDS (mM) | IKVAV (mM) | YIGSR (mM) | Patterna | RGDS (mM) | IKVAV (mM) |
| 1 | a00 | 0.0 | 0.75 | 3.0 | a00 | 0.0 | 3.0 | 3.0 | a00 | 1.0 | 4.0 | 6.0 | a0 | 7.0 | 5.5 |
| 2 | 1.2 | 0.3 | 1.2 | 3.0 | 1.2 | 1.2 | 3.0 | 2.0 | 3.0 | −− | 8.0 | 3.0 | |||
| 3 | −−+ | 1.2 | 0.3 | 4.8 | −−+ | 3.0 | 1.2 | 4.8 | −−+ | 3.0 | 2.0 | 9.0 | −+ | 8.0 | 8.0 |
| 4 | −+ − | 1.2 | 1.2 | 1.2 | −+ − | 3.0 | 4.8 | 1.2 | −+ − | 3.0 | 6.0 | 3.0 | 0a | 10.5 | 2.0 |
| 5 | −++ | 1.2 | 1.2 | 4.8 | −++ | 3.0 | 4.8 | 4.8 | −++ | 3.0 | 6.0 | 9.0 | 00 | 10.5 | 5.5 |
| 6 | 0a0 | 3.0 | 0.0 | 3.0 | 0a0 | 7.5 | 0.0 | 3.0 | 0a0 | 6.0 | 0.6 | 6.0 | 00 | 10.5 | 5.5 |
| 7 | 00a | 3.0 | 0.75 | 0.0 | 00a | 7.5 | 3.0 | 0.0 | 00a | 6.0 | 4.0 | 1.0 | 0A | 10.5 | 9.0 |
| 8 | 000 | 3.0 | 0.75 | 3.0 | 000 | 7.5 | 3.0 | 3.0 | 000 | 6.0 | 4.0 | 6.0 | +− | 13.0 | 3.0 |
| 9 | 000 | 3.0 | 0.75 | 3.0 | 000 | 7.5 | 3.0 | 3.0 | 000 | 6.0 | 4.0 | 6.0 | ++ | 13.0 | 8.0 |
| 10 | 00A | 3.0 | 0.75 | 6.0 | 00A | 7.5 | 3.0 | 6.0 | 00A | 6.0 | 4.0 | 11.0 | A0 | 14.0 | 5.5 |
| 11 | 0A0 | 3.0 | 1.5 | 3.0 | 0A0 | 7.5 | 6.0 | 3.0 | 0A0 | 6.0 | 7.4 | 6.0 | |||
| 12 | +−− | 4.8 | 0.3 | 1.2 | +−− | 12.0 | 1.2 | 1.2 | +−− | 9.0 | 2.0 | 3.0 | |||
| 13 | +−+ | 4.8 | 0.3 | 4.8 | +−+ | 12.0 | 1.2 | 4.8 | +−+ | 9.0 | 2.0 | 9.0 | |||
| 14 | ++− | 4.8 | 1.2 | 1.2 | ++− | 12.0 | 4.8 | 1.2 | ++− | 9.0 | 6.0 | 3.0 | |||
| 15 | +++ | 4.8 | 1.2 | 4.8 | +++ | 12.0 | 4.8 | 4.8 | +++ | 9.0 | 6.0 | 9.0 | |||
| 16 | A00 | 6.0 | 0.75 | 3.0 | A00 | 15 | 3.0 | 3.0 | A00 | 11.0 | 4.0 | 6.0 | |||
+ and − indicate high and low levels, respectively. 0 indicates a center point. A and a indicate the highest and lowed axial levels, respectively.
Immunofluorescence Staining
Cultures were fixed with 3.7% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 10% goat serum for 2 hours. The primary antibody was anti-human PECAM-1/CD31 mouse IgG1 (R&D Cat# BBA7), diluted 1:100 in the blocking buffer, and the secondary antibody was AlexaFluor 488 goat anti-mouse IgG1(γ1), diluted 1:200 in the blocking buffer. Nuclei were stained with 10 ng/mL 4′,6-diamidino-2-phenylindole (DAPI).
Results
Overview of matrix design and optimization
A modular strategy was developed for constructing defined multi-ligand culture matrices, and these matrices were utilized to systematically evaluate the influences of precise mixtures of matrix-bound ligands on the growth of HUVECs in culture (Figure 1). The system was based on the co-assembly of multiple ligand-bearing derivatives of the peptide Q11 (Table 1, Figure 1a). Previously, Q11 has been shown to be capable of fibrillizing when appended with short peptide ligands,13, 15 terminal chemical groups for cross-linking,15 or immune epitopes.14 Also previously, TEM with immunogold staining showed that for Q11 derivatives containing N-terminal peptide sequences including RGDS and IKVAV (as shown in Table 1) or longer epitope sequences such as ISQAVHAAHAEINEAGR, the ligands were displayed on the fibrillar surface.13, 14 Here, the set of available ligand-bearing Q11 derivatives was expanded to include two new ligand-bearing peptides, YIGSR-Q11 and REDV-Q11. YIGSR is a peptide ligand from the laminin β1 chain that was originally found to modulate cell attachment, migration, and invasiveness.16, 17 In recent years, YIGSR has been employed as an engineered ligand to modulate the attachment and growth of several cell types on the surface of biomaterials, including endothelial cells.18–22 The peptide REDV, found in the alternatively spliced type-III connecting segment (IIICS) region of fibronectin, is a ligand to which HUVECs can bind through α4β1 integrin.23, 24 While all four short peptide ligands, RGDS, YIGSR, IKVAV, and REDV have been shown to modulate endothelial cell behavior, they have not to our knowledge been explored simultaneously. Here, they were investigated singly and co-assembled together in a broad range of formulations in order to identify interactions occurring between the ligands and to identify optimal matrix formulations that maximized endothelial cell growth. Multifactorial optimization has not previously been utilized to investigate and optimize precise combinations of peptide ligands in this way.
Characterization of new self-assembling components: fibrillization, assembly, and mechanics
In previous work, the peptides Q11, IKVAV-Q11, RGDS-Q11, and the corresponding scrambled control peptides were developed as components of co-assembled matrices.13, 14 For the present work, the behavior of the new peptides YIGSR-Q11, REDV-Q11, and their corresponding scrambled peptides were characterized. Like previous Q11-based peptides bearing ligands or epitopes, both YIGSR-Q11 and REDV-Q11 formed fibrils observable by TEM (Figure 2). Using HPLC, it was also determined the extent to which YIGSR-Q11 could be entrapped within Q11 matrices. It was found that in 20mM peptide matrices with at least as high as 40% inclusion of YIGSR-Q11, the ligand was quantitatively incorporated (Figure 2a). No gels tested exhibited ligand incorporation levels that deviated from the ratio of peptides originally mixed in solution. Stated differently, when YIGSR-Q11 was mixed with Q11 in solution and co-assembled by adding PBS, the gel that formed had precisely the same ratio of peptides as the pre-assembled peptide solution. This correspondence between the pre-assembled material and the final gel was also observed previously for RGDS-Q11 and for IKVAV-Q11.13, 14 These results collectively indicated that the system did not exhibit compositional drift upon assembly. REDV-Q11 could not be analyzed with this HPLC method because it co-eluted with Q11 on reverse-phase HPLC columns, but gels containing REDV-Q11 showed no gross indication of gel disruption or destabilization. Inactivity of the REDV-Q11 peptide in initial cell growth experiments and the decision not to use it in the multifactorial optimization steps, described below, also diminished the necessity of fully characterizing it in this context.
Figure 2.
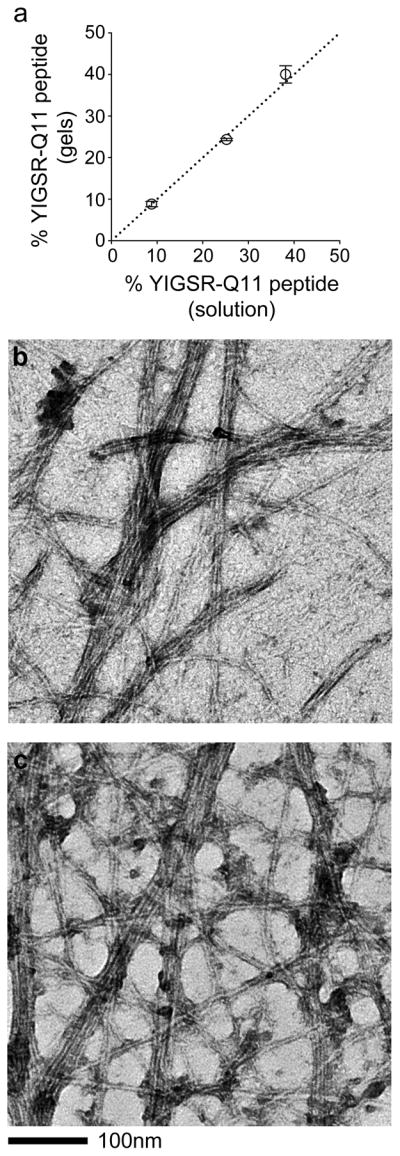
Characterization of newly introduced self-assembled peptides, YIGSR-Q11 and REDV-Q11. YIGSR-Q11, like previous ligand-bearing peptides, quantitatively co-assembled with Q11, as determined by HPLC analysis of the pre-assembled and post-assembled peptide mixtures (a). Both YIGSR-Q11 (b) and REDV-Q11 (c) produced fibrils of similar morphology to those of previously synthesized ligand-bearing peptides.13
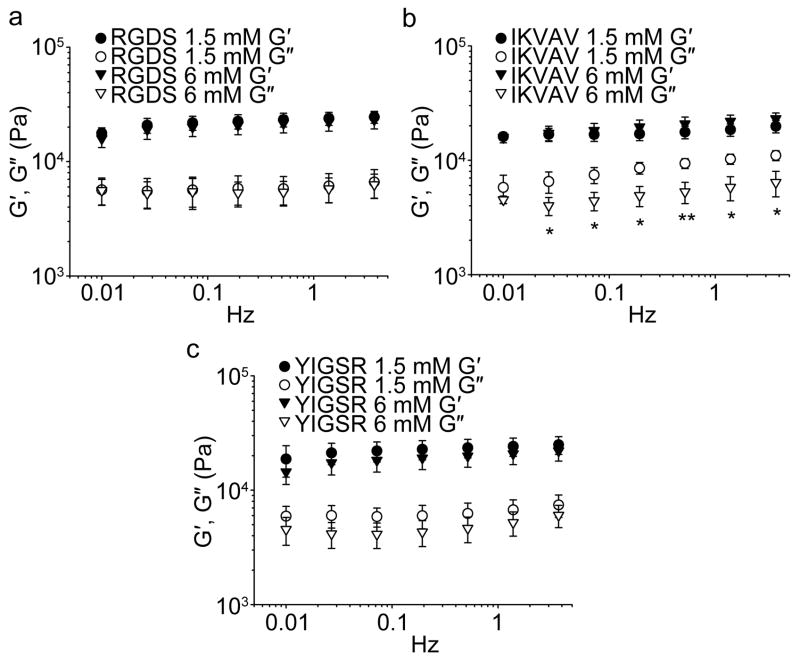
By oscillating rheometry, it was found that the incorporation of RGDS-Q11, IKVAV-Q11, and YIGSR-Q11 did not alter the storage modulus of Q11 gels (Figure 3). The range of formulations evaluated corresponded to the levels initially tested in the first round of full-factorial experiments (0–6 mM ligand bearing peptide). In each case, the storage modulus was about 11 kPa and insensitive to frequency (Figure 3). Owing to the unaffected mechanical properties upon ligand incorporation, the predictable gelation, the uniform incorporation behavior of RGDS-Q11, IKVAV-Q11, and YIGSR-Q11, and the uniform fibrillization of RGDS-Q11, IKVAV-Q11, YIGSR-Q11, and REDV-Q11, it was concluded that each peptide could be reliably incorporated into Q11 gels. This predictable assembly then enabled the multifactorial experiments that followed. It should be noted, however, that an accurate measurement of peptide incorporation does not necessarily mean that the precise amount of peptide that is functionally available to cells is known at this time. For many ligand-bearing hydrogels and biomaterials under development, including this Q11-based system, measurement of the true density of productively available ligands in 3D is a challenging proposition requiring advanced measurement techniques such as FRET labeling of cells and ligands,25 which have not been employed in the present study.
Figure 3.
The stiffness of multi-peptide matrices was independent of the amount of incorporated RGDS-Q11 (a), IKVAV-Q11 (b), or YIGSR-Q11 (c). The total concentration of gels was 30 mM. Closed symbols, storage modulus (G′), open symbols, loss modulus (G″). Loss moduli were slightly lower for gels containing high concentrations of IKVAV-Q11 (*p < 0.05 or **p < 0.01), but G′ was unchanged (ANOVA with Tukey’s HSD post-hoc test, n=3, mean±SD shown for all data).
Single-factor experiments for setting initial bounds for multifactorial experiments
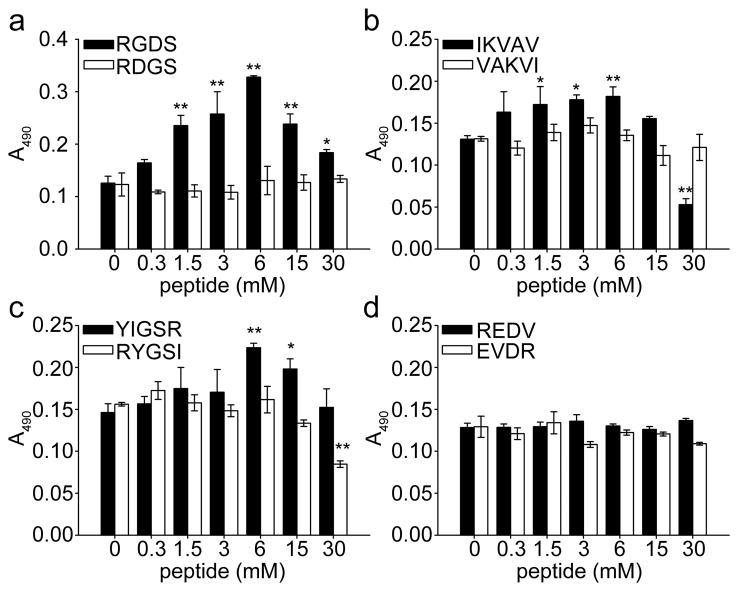
To focus the initial multifactorial experiments on interesting regions of formulation space, the effect of each ligand was first investigated independently using one-factor-at-a-time analysis. Gels were produced containing 30 mM total peptide, with the amount of ligand-bearing peptide varying from 0–100%. HUVECs were cultured on these gels, and their growth was measured at 64 h using an MTS assay, which measures cell metabolic activity. Although the total metabolic activity of a culture often correlates closely with the number of cells present, it is possible that these aspects may become decoupled. Nevertheless, the simplicity and amenability of the assay to multi-well formats led us to use it as a measure of cell growth over time. Figure 4 shows the response of cell growth on gels with varying levels of ligand incorporation. The most striking effect on cell growth was provided by RGDS-Q11, not surprising given the well-known influence of this promiscuous integrin ligand on the attachment and growth of many cell types.2, 26 Gels with immobilized RGDS showed a clear maximum of cell growth at 6 mM Q11, corresponding to one out of every five Q11 peptides in the gel being conjugated to the ligand (Figure 4a). Scrambled control RDGS-Q11 peptides showed no evidence of this increased growth, indicating that the cells interacted specifically with the ligand, presumably through integrins, which for HUVECs could include the β1 or β3 subfamilies of integrins, as well as various other integrins.2, 27 IKVAV-Q11 and YIGSR-Q11 showed similar ligand concentration-dependent effects on cell growth, but to a smaller degree than RGDS-Q11 (Figure 4b,c). IKVAV-Q11 improved cell growth best in a range between 1.5–6 mM ligand, and the response to YIGSR-Q11 was most pronounced for gels containing 6 mM ligand. Scrambled controls for both IKVAV and for YIGSR abolished the effect of the ligands, indicating specific interactions with the ligands. Interestingly, REDV-Q11 showed no influence on HUVEC growth (Figure 4d), despite the fact that REDV has previously been shown to facilitate HUVEC attachment and spreading when immobilized on glass substrates.23 It is possible that the context of this peptide with the spacer sequence, Q11 sequence, and fibrillar conformation disrupted its activity.28 For this reason, REDV-Q11 was not considered further in the multifactorial experiments.
Figure 4.
HUVEC growth as a function of individual ligand incorporation, as determined by MTS signal at 64 h. The total concentration of gels was 30 mM. ■ represents biologically active sequences, □ corresponding scrambled sequences. ** p<0.01 and * p<0.05 compared to Q11 by ANOVA with Tukey’s HSD post-hoc test, n=3, mean±SD.
Factorial experiments for identifying main effects and interactions between ligands
In order to identify the most significant effects in the context of the others, and to indicate any ligands that may interact with each other in their promotion or inhibition of HUVEC growth, experiments were conducted in a two-level, four factor (24) design that included a center point (Table 2). In this factorial design, high and low values were selected for each factor (in this case, Q11, RGDS-Q11, IKVAV-Q11, and YIGSR-Q11). To select these levels, the responses from the single-factor experiments were referenced (Figure 4), leading to the selection of 0 and 6 mM for RGDS-Q11; 0 and 1.5 mM for IKVAV-Q11; 0 and 6 mM for YIGSR-Q11; 2 and 20 mM for Q11. These levels were organized into 16 gel formulations to be investigated (Table 2). One extra “center point” (3 mM RGDS-Q11, 0.75 mM IKVAV-Q11, 3 mM YIGSR-Q11, 11 mM Q11; run 9 in Table 2) was added because the response between the low and high levels for each factor was not expected to be linear, owing to the non-linear responses shown by the single-factor experiments (Figure 4). The 17 experimental runs were conducted in triplicate, amounting to 51 gels. After responses (MTS absorbance, A490) were obtained from each gel, the absorbance values were fit into a least-squares polynomial regression to estimate the impact of single and multiple ligands on HUVEC growth.
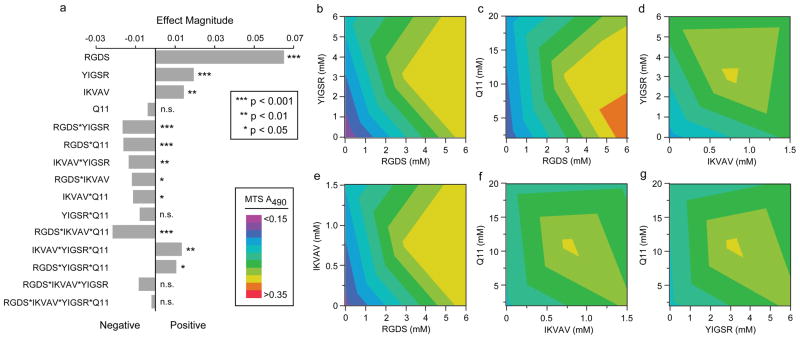
Results from the two-level factorial design indicated that each ligand-bearing peptide had a significant and positive effect on HUVEC growth; in contrast, Q11 did not (Figure 5a). An R2 value of 0.904, a normal distribution of the residuals, and an ANOVA significance level of <0.0001 supported the validity and significance of the model, shown for the single factors and two-factor interactions in Eq. 1,
Figure 5.
Results from the Factorial Experiments described in Table 2. (a) Effect magnitudes of all main factors, two-factor interactions, three-factor interactions, and the four-factor interaction. All ligand-bearing Q11 peptides were determined to be significant main effects, and several two-factor and three-factor interactions were also significant (a). RGDS-Q11 was the strongest main effect, and the combination of RGDS-Q11 and YIGSR-Q11 was the strongest two-factor interaction. All of the two-factor interaction contour plots are shown in (b–g).
| (Eq. 1) |
where the MTS absorbance is measured as A490, the concentration of each peptide is measured in millimolar, and each peptide is represented by the first letter of its name (fit parameters can be found in the Supporting Information). In agreement with the single-factor experiments, RGDS-Q11 showed the most positive effect in the context of the other ligands, followed by YIGSR-Q11 and IKVAV-Q11, as evidenced by their significant factor estimates (Figure 5a). It was interesting that Q11 was not found to be a significant factor, even though its concentration was varied 10-fold in the factorial experiments. This was a surprising finding given the generally appreciated importance of cell behavior on matrix stiffness, which for Q11 is a function of peptide concentration [15]. However, because Q11 concentration was not found to be a statistically significant factor in the present factorial experiments, at least in the context of the other ligand-bearing peptides, it was not adjusted further in the subsequent search for optimal formulations by response surface methodology (RSM). Several interaction effects were observed between the ligands. The strongest of these was the interaction between RGDS-Q11 and YIGSR-Q11 (Figure 5a, b). At low and high concentrations of RGDS-Q11, YIGSR-Q11 had a negligible effect on HUVEC growth, but at intermediate levels of RGDS-Q11 (around 3 mM), YIGSR-Q11 had an optimal concentration of about 3 mM towards promoting HUVEC growth (Figure 5b), above and below which the growth of HUVECs was diminished. The second most prominent two-factor interaction was between Q11 and RGDS-Q11, where RGDS-Q11 incorporation had a larger effect on HUVEC growth at small Q11 concentrations than at larger ones (Figure 5c), followed the interaction between YIGSR-Q11 and IKVAV-Q11 (Figure 5d), between RGDS-Q11 and IKVAV-Q11 (Figure 5e), and between Q11 and IKVAV-Q11 (Figure 5f). The only two-factor interaction that was not found to be statistically significant was between YIGSR-Q11 and Q11 (Figure 5g). Collectively, the factorial experiments indicated that all three of the incorporated ligands significantly modulated HUVEC growth, and in several cases, particularly for RGDS and YIGSR, the strength of the effect of one ligand depended on the concentration of at least one other. These interactions would have been challenging to uncover for materials requiring serial analysis of their composition.
Response surface methodology experiments for identifying optimal formulations
The objective of the next step in the design of the co-assembled matrices was to approach the maximal growth response possible by finding an optimized combination of matrix-bound ligands. To accomplish this, four rounds of response surface methodology (RSM) were utilized (Experiments outlined in Table 3). In the RSM experiments, RGDS-Q11, IKVAV-Q11, and YIGSR-Q11 were selected as the three factors to be controlled initially. Recall that at the outset the study additionally included REDV-Q11 and Q11 as factors, but REDV-Q11 was eliminated after it was determined that it did not significantly modulate HUVEC growth by itself (Figure 4d). Q11 was kept constant at 2 mM, since it was not found to be a significant main effect in the factorial experiments (Figure 5a). The four rounds of RSM experiments that followed consisted of an initial central composite inscribed (CCI) design, followed by a second CCI design with expanded ranges for all factors, followed by a re-centered central composite circumscribed (CCC) design that focused more directly on a range of formulation space that likely contained the optimum, and culminating in a final CCC design that focused on the two most significant and productive factors, RGDS and IKVAV (Table 3, ranges of formulation space for each RSM experiment shown in Figure 6a). The rationale for each successive step followed from the results of the prior experiment, as described below.
The first RSM experiment investigated the same range of formulation space as the factorial experiments had investigated (red box in Figure 6a). The inscribed design was selected instead of a traditional central composite circumscribed design owing to the fact that the lowest concentrations were at 0 mM, and a circumscribed design would have required physically impossible negative concentration values. It was found that RGDS again was the most significant factor, with the HUVEC growth response increasing significantly as RGDS concentration was increased (Figure 6b, d). Interestingly, in this design, YIGSR was found to have a significantly negative effect on HUVEC growth (p < 0.0001). This was the first indication of this effect, which was subsequently reaffirmed in later RSM experiments as well as confirmatory control experiments, described below. The negative effect of YIGSR can be visualized in its interaction plots with IKVAV and RGDS (Figure 6c–d). Again, a weak but statistically significant interaction was found between YIGSR and IKVAV. The fact that the highest growth response was seen at the highest amount of RGDS tested also suggested that the optimum lay beyond the range studied in this experiment. Therefore, the second round of RSM investigated an expanded range of formulations (Table 3).
The second round of RSM experiments, with expanded levels (green box in Figure 6a) indicated significantly positive effects for both IKVAV and RGDS (p < 0.0001 for both), whereas in this design, YIGSR was not found to be significant. The positive effects of IKVAV and RGDS and the relative ineffectiveness of YIGSR can be visualized in the interaction plots between the three factors (Figure 6e–g). Because of the relative flatness of the response to RGDS-Q11 at higher concentrations, and because the “low” values were zero for all factors, the RSM was then “re-centered” in a new round of experiments, shown in Table 3 and in the blue box in Figure 6a. Here, the significant negative impact of YIGSR was confirmed (p < 0.0001, shown starkly in the interaction plots involving YIGSR, Figure 6i, j). That is, in the range of RGDS-Q11’s maximal effectiveness, around 6–8 mM, YIGSR-Q11 was particularly disruptive.
Owing to the negative influence of YIGSR-Q11 on HUVEC growth, particularly in the range of maximal RGDS-Q11 effectiveness, it was dropped as a factor in the final optimization experiment (pink square in Figure 6a). Using a final two-factor RSM design (Table 3, right column), RGDS-Q11 and IKVAV-Q11 were varied together. The results of this experiment indicated that a good region of formulation space had been reached (note the elevated MTS absorbance scale for the contour plot shown in Figure 6k, as well as the large area covered by high absorbance values). An “optimum” was identified at 8 mM RGDS-Q11, with 3 mM IKVAV-Q11.
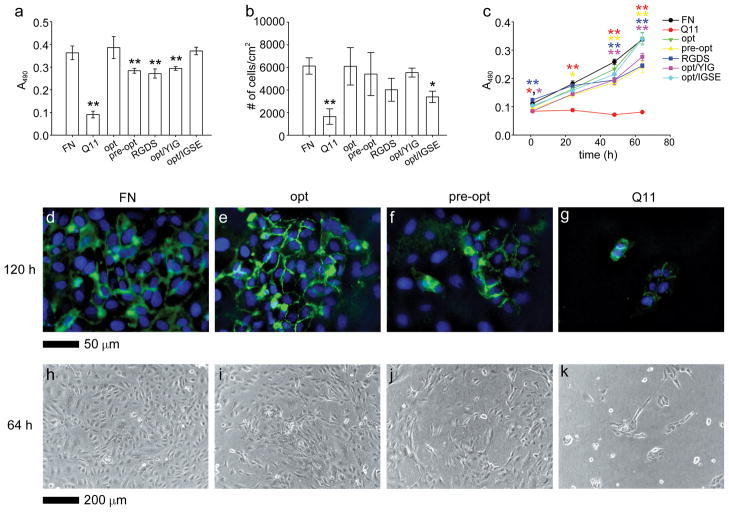
Because the “optimal” formulation was a predicted value, confirmatory experiments were conducted to determine whether this formulation indeed led to increased HUVEC growth. These experiments were also utilized to gauge the benefit realized by undertaking the factorial and RSM experiments instead of more traditional one-factor-at-a-time analysis. HUVECs were cultured on the optimized formulation (8 mM RGDS-Q11, 3 mM IKVAV-Q11) and compared with a “pre-optimized” formulation representing a combination of the best individual ligand concentrations identified before the factorial experiments (6 mM RGDS-Q11, 1.5 mM IKVAV-Q11, 6 mM YIGSR-Q11). It was found that, indeed, the optimized formulation led to HUVEC growth that was significantly greater than the pre-optimized formulation (Figure 7a, this experiment was repeated three times with similar results). Growth on optimized Q11-based gels was indistinguishable from growth on fibronectin-coated gels, whereas pre-optimized gels showed significantly inferior growth. This sub-optimal growth was similar for gels containing 8 mM RGDS-Q11 only and lacking any IKVAV-Q11, indicating that both peptides were necessary for the highest level of growth. The deleterious effect of YIGSR-Q11 on the optimal formulation was also confirmed by adding 6 mM YIGSR-Q11 to the optimized formulation. These gels exhibited sub-optimal growth similar to pre-optimized gels or gels lacking IKVAV-Q11 (Figure 7a). In addition, this YIGSR antagonism could be abolished by substituting a negative control peptide for YIGSR-Q11, 6 mM IGSE-Q11 (Figure 7a, sequence in Table 1),29 which restored HUVEC growth to the same level as the optimized formulation.
Figure 7.
Validation of optimized gel formulations. HUVEC growth and attachment were compared on optimized gels (opt), pre-optimized gels (pre-opt), gels containing only the RGDS ligand (RGDS), optimized gels containing additional YIGSR-Q11 (opt/YIG), optimized gels containing additional IGSE-Q11 (opt/IGSE), and Q11 gels with adsorbed fibronectin (FN). Growth was compared using MTS assays at 64 h (a) and over time (c), and attachment assays were conducted at 1 h (b). **p<0.01 and *p<0.05 compared to “opt” formulation by ANOVA with Tukey’s post-doc test (mean±SD); n=5 (a), n=4 (b), and n=3 (c). Expression of PECAM-1/CD31 (d–g, green, with DAPI counterstain) and phase images (h–k) were compared for FN (d, h), opt (e, i), pre-opt (f, j), and Q11 (g, k) gels.
To determine whether the difference in cell number after 3 days in culture between the optimized and pre-optimized gels was a result of a difference in initial attachment or a difference in growth rate, an attachment assay was performed, and MTS measurements were collected at intermediate time points (Figure 7b, c, respectively). In the attachment assay, optimized and pre-optimized gels were similar, and the YIGSR antagonism was absent (Figure 7b). Only bare Q11 gels and gels containing the IGSE peptide showed statistically inferior cell attachment compared to the optimized formulation. In monitoring the growth of HUVECs over time using the MTS assay, it was found that at short time points (1 h), RGDS-only gels actually supported significantly improved cell numbers, but this advantage dwindled over time, so that by 48 h the RGDS-only gels were significantly inferior to the optimized gels (Figure 7c, blue line). The YIGSR-containing gels also showed inferior MTS readings throughout the course of the experiment, from 1 to 64 h (Figure 7c, purple line). It is not clear why the early-timepoint MTS measurements and attachment assays did not agree with each other for the RGDS-only and YIGSR-containing gels, but it could have arisen from the different stringencies of the washing steps between the two assays, or the influence of trypsinization on the attachment assay. The MTS measurements also were more repeatable than the measurements of attachment, reflecting the relative precisions of these two assays. At 64 h, the improved growth rate on the optimized gels corresponded to a qualitatively improved expression of PECAM-1/CD31 at cell-cell junctions, as well as qualitatively higher degrees of confluence (Figure 7d–k). Quantification of PECAM-1/CD31 levels was challenged by difficulties in recovering enough protein to reliably perform Western blotting. Collectively, these confirmatory experiments indicated that the optimized formulation supported HUVEC growth at the same level as fibronectin-coated gels, that the presence of IKVAV and the lack of YIGSR were both important for maximizing cell growth, that the factorial and RSM experiments achieved an optimized formulation that was superior to the ligand combination suggested by single-factor experiments, and that this optimization produced superior growth rates.
Discussion
In the work reported here, we utilized DOE methods to optimize co-assembled peptide matrices towards supporting rapid endothelial cell growth, providing materials that are potentially useful in future in vitro studies of endothelial cell behavior as well as for the development of blood-contacting biomaterials.20, 21 Although only simple metrics were employed (cell attachment and growth), the approach could be directed towards measuring other more specific biological behaviors either singly or in combination. In most practical applications of ligand-modified biomaterials, there are often multiple design goals that run counter to each other, for example the need to maximize endothelial cell proliferation while minimizing thrombogenicity for blood-contacting devices. In contexts such as these, multifactorial optimization and modular matrices may be useful for efficiently understanding the formulations with which such dual objectives could be satisfied. Moreover, the output measured in this study was a broad one, cell growth. It will be interesting to apply these methods to more specific or refined cell behaviors such as differentiation. The approach we describe could be implemented to optimize matrices that favor specific differentiation pathways in cultures of stem cells, for example.
One unexpected result in the study reported here was the apparent antagonistic relationship between the YIGSR and RGDS ligands in terms of their combined ability to support endothelial cell growth. The peptide YIGSR was originally identified as a short synthetic peptide that significantly modulated cell attachment, migration, and invasiveness.16 Its major known cellular receptor is a 67 kDa non-integrin laminin-binding protein, which has been shown to co-localize with the focal adhesion proteins α-actinin and vinculin upon YIGSR binding.17 In recent years, YIGSR has been employed as an engineered ligand to modulate the attachment and behavior of several cell types on the surface of biomaterials, having been attached to a range of different surfaces including polyurethaneureas,21, 30 polyethylene terephthalate,31 peptide amphiphiles,19, 32 glass,17, 33 PEG hydrogels,20 and self-assembling peptides.18 These materials have been constructed in efforts to control the attachment, spreading, growth, and migration of a variety of cell types on these materials, including endothelial cells.18–22 Nevertheless, almost all of these previous studies have evaluated surfaces presenting YIGSR by itself, rather than in combination with other peptide ligands.17–19, 21, 31, 32, 34 RGD, IKVAV, and YIGSR have been investigated in parallel previously,35 but not in combination. Consistent with previous findings, in the present study we have also found that immobilized YIGSR, by itself, can specifically increase HUVEC growth (Figure 4c, Figure 5a). However, by being able to finely adjust both the concentration of YIGSR and RGDS in the co-assembled matrices, we have also observed that in combination with RGDS, YIGSR has an inhibitory effect on HUVEC growth (especially pronounced in Figure 6h–j, confirmed in Figure 7a, c). This effect appeared to be particularly large in the RGD concentration regimes that most effectively supported endothelial cell growth. Previously, two studies have investigated immobilized YIGSR in combination with RGD,20, 22 and in each of these previous studies, only one specific combination of peptides has been evaluated, roughly corresponding to equimolar mixtures of RGD and YIGSR. In one study, the behavior of human microvascular endothelial cells (HMVEC) was studied on PEG-acrylate hydrogels derivatized with YIGSR and RGD peptides, and it was found that the YIGSR/RGD combination selectively enhanced the migration of HMVEC over gels functionalized with RGD alone.20 Spreading and adhesion of HMVECs was not affected by the presence of YIGSR in addition to RGD, but the adhesion of human vascular smooth muscle cells (HVSMC) was increased. In the other study, HUVEC proliferation was found to be enhanced on a mixture of YIGSR and RGD immobilized to polyurethane,22 but again, only one roughly equimolar ratio of the two peptides was studied. The fact that only one formulation was investigated in each case most likely points to the technical challenges associated with precisely preparing surfaces with independently controlled amounts of each peptide, and the approach described in the present work offers a route to overcome such issues. Using the modular materials described here, we were able to ascertain that in some combinations of the two peptide ligands, RGDS and YIGSR did not provide improved HUVEC growth; rather, they inhibited growth. At present the mechanism of this interaction is not clear, but owing to the previous finding that YIGSR does not act through integrins,17 it does not seem likely that YIGSR directly inhibited integrins that would otherwise bind RGDS. It is possible that YIGSR somehow physically occluded the RGDS ligands when the two peptides were co-assembled together within fibrils, but the fact that the IGSE control peptide did not inhibit RGDS (Figure 7a, c) diminishes this possibility to an extent. Given that the 67 kDa non-integrin receptor for YIGSR co-localizes with focal adhesion proteins,17 a more complex mode of interaction within the focal adhesion complex may be possible. The details of such an interaction could be investigated using blocking antibodies or knockout/knock-in models.
One shortcoming of the technique described here is that it has been performed within culture inserts in multi-well plates, resulting in a reduced throughput compared to array-type analyses. However, it may be possible to devise strategies whereby microscale spots of self-assembled peptide gels are arrayed on surfaces or within microwells.8, 36 This would not only enable more extensive variation and replication of gel formulations, but it would also open up possibilities for employing microfluidic techniques, for example to investigate soluble factors in combination with insoluble factors.6 Combination of the self-assembled matrices with newly introduced techniques for producing libraries of suspended microgels would also provide exciting possibilities for exploring a wide range of matrix formulations in more three-dimensional culture systems.37
Conclusions
Statistical design of experiments (DOE) is a powerful technique that has been minimally employed in studying cell-ECM interactions. One of the primary reasons for its low utilization in this context has been the prevailing difficulty of independently adjusting the concentrations of multiple ECM components within traditional protein-based or polymer-based scaffolds. As an alternative, in the work reported here, we utilized a series of co-assembling peptides as chemically defined surrogates of the ECM. We utilized this modular system to conduct multifactorial experiments aimed at identifying a combination of peptides that optimally promoted HUVEC growth on the gels, and we identified a previously unreported antagonism between YIGSR and RGDS ligands that occurred within specific peptide concentration ranges and did not occur for irrelevant control peptides. Both findings would have been challenging to uncover for matrix materials that do not similarly facilitate multifactorial experiments. The materials reported here should be widely applicable for other studies aimed at understanding the combined effects of matrix-bound ligands in various biological contexts.
Acknowledgments
We thank Margaret Gardel for use of the rheometer, and Doris Osei-Afriyie and Katelyn Bird for assistance with peptide synthesis. This research was supported by the National Institutes of Health (NIBIB, 7R21EB007335 and 1R01EB009701), the National Science Foundation (CHE-0802286), and the American Heart Association (JPJ, predoctoral fellowship 09PRE2080000). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and BioEngineering or the National Institutes of Health. TEM was performed at the University of Chicago Electron Microscopy Facility.
References
- 1.Collier JH, Rudra JS, Gasiorowski JZ, Jung JP. Chem Soc Rev. 2010;39:3413–3424. doi: 10.1039/b914337h. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Carson AE, Barker TH. Regener Med. 2009;4:593–600. doi: 10.2217/rme.09.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pankov R, Yamada KM. J Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]; Hynes RO. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Box GEP, Hunter JS, Hunter WG. Statistics for Experimenters: Design, Innovation, and Discovery. 2. Wiley; Hoboken, NJ: 2005. [Google Scholar]; Montgomery DC. Design and Analysis of Experiments. 7. Wiley; Hoboken, NJ: 2009. [Google Scholar]; Lazic Z. Design of Experiments in Chemical Engineering. Wiley-VCH; Weinheim, Germany: 2004. [Google Scholar]
- 5.Flaim CJ, Chien S, Bhatia SN. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 6.Flaim CJ, Teng D, Chien S, Bhatia SN. Stem Cells Dev. 2008;17:29–39. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 7.Chung EH, Gilbert M, Virdi AS, Sena K, Sumner DR, Healy KE. J Biomed Mater Res, Part A. 2006;79:815–826. doi: 10.1002/jbm.a.30809. [DOI] [PubMed] [Google Scholar]; Nirmalanandhan VS, Shearn JT, Juncosa-Melvin N, Rao M, Gooch C, Jain A, Bradica G, Butler DL. Tissue Eng, Part A. 2008;14:1883–1891. doi: 10.1089/ten.tea.2007.0125. [DOI] [PubMed] [Google Scholar]; Nirmalanandhan VS, Juncosa-Melvin N, Shearn JT, Boivin GP, Galloway MT, Gooch C, Bradica G, Butler DL. Tissue Eng, Part A. 2009;15:2103–2111. doi: 10.1089/ten.tea.2008.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zapata P, Su J, Garcia AJ, Meredith JC. Biomacromolecules. 2007;8:1907–1917. doi: 10.1021/bm061134t. [DOI] [PubMed] [Google Scholar]; Smith JR, Seyda A, Weber N, Knight D, Abramson S, Kohn J. Macromol Rapid Commun. 2004;25:127–140. [Google Scholar]; Urquhart AJ, Taylor M, Anderson DG, Langer R, Davies MC, Alexander MR. Anal Chem. 2008;80:135–142. doi: 10.1021/ac071560k. [DOI] [PubMed] [Google Scholar]; Mei Y, Saha K, Bogatyrev SR, Yang J, Hook AL, Kalcioglu ZI, Cho SW, Mitalipova M, Pyzocha N, Rojas F, Van Vliet KJ, Davies MC, Alexander MR, Langer R, Jaenisch R, Anderson DG. Nat Mater. 2010;9:768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fernandes TG, Kwon SJ, Bale SS, Lee MY, Diogo MM, Clark DS, Cabral JMS, Dordick JS. Biotechnol Bioeng. 2010;106:106–118. doi: 10.1002/bit.22661. [DOI] [PMC free article] [PubMed] [Google Scholar]; Acharya AP, Clare-Salzler MJ, Keselowsky BG. Biomaterials. 2009;30:4168–4177. doi: 10.1016/j.biomaterials.2009.04.032. [DOI] [PubMed] [Google Scholar]; Bailey SN, Sabatini DM, Stockwell BR. Proc Natl Acad Sci U S A. 2004;101:16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khademhosseini A, Yeh J, Eng G, Karp J, Kaji H, Borenstein J, Farokhzad OC, Langer R. Lab Chip. 2005;5:1380–1386. doi: 10.1039/b508096g. [DOI] [PubMed] [Google Scholar]
- 9.Hook AL, Anderson DG, Langer R, Williams P, Davies MC, Alexander MR. Biomaterials. 2010;31:187–198. doi: 10.1016/j.biomaterials.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 10.Apostolovic B, Klok HA. Biomacromolecules. 2010;11:1891–1895. doi: 10.1021/bm100533g. [DOI] [PubMed] [Google Scholar]
- 11.LaBarge MA, Nelson CM, Villadsen R, Fridriksdottir A, Ruth JR, Stampfer MR, Petersen OW, Bissell MJ. Integr Biol. 2009;1:70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brafman DA, Minicis Sd, Seki E, Shah KD, Teng D, Brenner D, Willert K, Chien S. Integr Biol. 2009;1:513–524. doi: 10.1039/b912926j. [DOI] [PubMed] [Google Scholar]
- 12.Falsey JR, Renil M, Park S, Li SJ, Lam KS. Bioconjugate Chem. 2001;12:346–353. doi: 10.1021/bc000141q. [DOI] [PubMed] [Google Scholar]
- 13.Jung JP, Nagaraj AK, Fox EK, Rudra JS, Devgun JM, Collier JH. Biomaterials. 2009;30:2400–2410. doi: 10.1016/j.biomaterials.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudra JS, Tian YF, Jung JP, Collier JH. Proc Natl Acad Sci U S A. 2010;107:622–627. doi: 10.1073/pnas.0912124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung JP, Jones JL, Cronier SA, Collier JH. Biomaterials. 2008;29:2143–2151. doi: 10.1016/j.biomaterials.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graf J, Iwamoto Y, Sasaki M, Martin GR, Kleinman HK, Robey FA, Yamada Y. Cell. 1987;48:989–996. doi: 10.1016/0092-8674(87)90707-0. [DOI] [PubMed] [Google Scholar]; Iwamoto Y, Robey FA, Graf J, Sasaki M, Kleinman HK, Yamada Y, Martin GR. Science. 1987;238:1132–1134. doi: 10.1126/science.2961059. [DOI] [PubMed] [Google Scholar]
- 17.Massia SP, Rao SS, Hubbell JA. J Biol Chem. 1993;268:8053–8059. [PubMed] [Google Scholar]
- 18.Genove E, Shen C, Zhang S, Semino CE. Biomaterials. 2005;26:3341–3351. doi: 10.1016/j.biomaterials.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Andukuri A, Minor WP, Kushwaha M, Anderson JM, Jun HW. Nanomedicine (N Y, NY, U S) 2010;6:289–297. doi: 10.1016/j.nano.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fittkau MH, Zilla P, Bezuidenhout D, Lutolf MP, Human P, Hubbell JA, Davies N. Biomaterials. 2005;26:167–174. doi: 10.1016/j.biomaterials.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Jun HW, West J. J Biomater Sci, Polym Ed. 2004;15:73–94. doi: 10.1163/156856204322752246. [DOI] [PubMed] [Google Scholar]
- 22.Choi WS, Bae JW, Joung YK, Park KD, Lee MH, Park JC, Kwon IK. Macromol Res. 2009;17:458–463. [Google Scholar]
- 23.Massia SP, Hubbell JA. J Biol Chem. 1992;267:14019–14026. [PubMed] [Google Scholar]
- 24.Heilshorn SC, DiZio KA, Welsh ER, Tirrell DA. Biomaterials. 2003;24:4245–4252. doi: 10.1016/s0142-9612(03)00294-1. [DOI] [PubMed] [Google Scholar]
- 25.Kong HJ, Polte TR, Alsberg E, Mooney DJ. Proc Natl Acad Sci U S A. 2005;102:4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]; Ruoslahti E. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 27.Hall H, Djonov V, Ehrbar M, Hoechli M, Hubbell JA. Angiogenesis. 2004;7:213–223. doi: 10.1007/s10456-004-1328-5. [DOI] [PubMed] [Google Scholar]
- 28.Heilshorn SC, Liu JC, Tirrell DA. Biomacromolecules. 2005;6:318–323. doi: 10.1021/bm049627q. [DOI] [PubMed] [Google Scholar]
- 29.Graf J, Ogle RC, Robey FA, Sasaki M, Martin GR, Yamada Y, Kleinman HK. Biochemistry. 1987;26:6896–6900. doi: 10.1021/bi00396a004. [DOI] [PubMed] [Google Scholar]
- 30.Jun HW, West JL. J Biomed Mater Res, Part B. 2005;72:131–139. doi: 10.1002/jbm.b.30135. [DOI] [PubMed] [Google Scholar]
- 31.Massia SP, Hubbell JA. J Biomed Mater Res. 1991;25:223–242. doi: 10.1002/jbm.820250209. [DOI] [PubMed] [Google Scholar]
- 32.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 33.Massia SP, Hubbell JA. Anal Biochem. 1990;187:292–301. doi: 10.1016/0003-2697(90)90459-m. [DOI] [PubMed] [Google Scholar]
- 34.Boateng SY, Lateef SS, Mosley W, Hartman TJ, Hanley L, Russell B. Am J Physiol Cell Physiol. 2005;288:C30–38. doi: 10.1152/ajpcell.00199.2004. [DOI] [PubMed] [Google Scholar]; Gu X, Masters KS. J Biomed Mater Res, Part A. 2010;93:1620–1630. doi: 10.1002/jbm.a.32660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santiago LY, Nowak RW, Peter Rubin J, Marra KG. Biomaterials. 2006;27:2962–2969. doi: 10.1016/j.biomaterials.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Charnley M, Textor M, Khademhosseini A, Lutolf MP. Integr Biol. 2009;1:625–634. doi: 10.1039/b918172p. [DOI] [PubMed] [Google Scholar]; Jongpaiboonkit L, King WJ, Lyons GE, Paguirigan AL, Warrick JW, Beebe DJ, Murphy WL. Biomaterials. 2008;29:3346–3356. doi: 10.1016/j.biomaterials.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proc Natl Acad Sci U S A. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]