Abstract
Background
In order for tumors to grow and proliferate, they must avoid recognition by immune cells and subsequent death by apoptosis. Granzyme B, a protease located in natural killer cells, initiates apoptosis in target cells. Inhibition of Granzyme B by PI-9, its natural inhibitor, can prevent apoptosis. Here we investigate whether PI-9 protects prostate cancer cells from apoptosis.
Methods
The expression of PI-9 was quantified by qPCR in several prostate cancer cell lines, and Granzyme B activity was tested in each cell line. PI-9 was overexpressed in LNCaP cells, which lack endogenous PI-9. Apoptosis was induced by natural killer cells in LNCaP cells that either contained or lacked PI-9, and the percent cell death in was quantified. Lastly, PI-9 levels were examined by qPCR and immunohistochemistry in prostate tumor tissue.
Results
Prostate cancer cell lines that expressed PI-9 could inhibit Granzyme B. Overexpression of PI-9 protected LNCaP cells from natural killer cell-mediated apoptosis. Examination of the levels of PI-9 in tissue from prostate tumors showed that PI-9 could be upregulated in low grade tumors and stochastically dysregulated in high grade tumors. Additionally, PI-9 is found consistently in high grade prostatic intraepithelial neoplasia and atrophic lesions.
Conclusions
These results indicate that overexpression of PI-9 can protect prostate cancer cells from apoptosis, and this effect may occur in human prostate tumors. These findings imply that early prostatic inflammation may trigger this increase in PI-9. This suggests that PI-9 upregulation is needed early in tumor progression, before additional protective mechanisms are in place.
Keywords: PI-9, Granzyme B, apoptosis, prostate cancer, immunosurveillanc
Introduction
Immunosurveillance, the process by which the immune system monitors and destroys virally infected or cancerous cells, has emerged as a promising new approach to treating prostate cancer[1]. Cytotoxic lymphocytes (CLs) carry out immunosurveillance by inducing apoptosis in target cells using two pathways: activating death ligand receptors and/or activating granule exocytosis. During granule exocytosis, CLs deliver granules filled with proteases that induce apoptosis, known as granzymes, into aberrant cells. Granzyme B (GrB), a 32 kD serine protease in the S1A family[2], is the main apoptotic initiator. Cleavage of GrB substrates either activates pro-death functions, such as activation of pro-caspases 3, 7[3], and 8[4], or deactivates pro-proliferative functions[5][6]. Granzyme B is the main apoptotic initiator in natural killer cells, and is expressed in all effector CD8+ T cells[7]. GrB is the chief effector of immunosurveillance, and this mechanism must be disrupted for cancer cells to survive. In fact, the ability to evade immunosurveillance has been classified as a defining hallmark of cancer [8].
One way cancer cells could evade immunosurveillance is to prevent the initiation of apoptosis by inhibiting Granzyme B. GrB’s natural inhibitor is PI-9 (serpin B9), a 42 kD clade B serpin which inhibit serine proteases intracelluarly. Serpins irreversibly inhibit their target protease, which can be detected by the formation of an SDS-stable complex with the target [9][10]. PI-9 is abundantly expressed in the cytosol of CLs to protect them from inadvertent exposure to their own GrB[11]. PI-9 is also found in immune-privileged tissues, such as the placenta and the lining of blood vessels, also to protect the cells from nearby GrB[11][12]. Since expression of PI-9 in normal tissue inhibits the apoptotic activities of GrB, overexpression of PI-9 in cancer cells could inhibit GrB-mediated apoptosis. PI-9 expression has been observed in several types of cancer, including breast cancer, cervical cancer, and colon cancer[13]. It has been shown in mice and in HeLa cells that overexpression of PI-9 directly protects cells from apoptosis through GrB inhibition[13][14], and evidence for this protective effect has been observed in breast cancer as well[15].
PI-9 expression may also affect the probability of successful treatment of cancer. PI-9 has been associated with poor clinical prognosis in lymphoma and nasopharyngeal carcinoma[16][17]. PI-9 expression can interfere with hormone therapy in breast cancer[18], and PI-9 expression is correlated with the failure of immunotherapy in melanoma[19]. Immunotherapy is of particular importance in prostate cancer, since the recently approved prostate cancer vaccine, Provenge (sipuleucel-T), uses this approach[20][21]. Taken together, PI-9 has emerged as an important immunoevasive protein in many cancers that has both therapeutic and diagnostic implications.
We hypothesized that PI-9 upregulation occurs in prostate cancer, protecting the cancer cells from GrB-mediated apoptosis. Our data indicates that PI-9 dysregulation may play a protective role early in cancer progression, allowing time for the development of additional protective mechanisms as the tumor grows. This work implies that PI-9 could be a biomarker for early-stage prostate lesions that are resistance to immunotherapy. Elimination of prostate cancer from these patients will require a therapeutic strategy that bypasses PI-9 mediated inhibition of GrB.
Materials and Methods
Cell culture
All cell lines were obtained from the ATCC. LNCaP cells were grown in RPMI-1640 containing 10% FBS, 10 mM HEPES, 0.11mg/mL sodium pyruvate, 4.5g/L glucose, 10units/mL penicillin, and 10mcg/mL streptomycin. PC3 cells were grown in F-12K nutrient mixture (Gibco) containing 10% FBS, 10units/mL penicillin, and 10mcg/mL streptomycin. NK-92 cells were grown in Myelocult media (Stem Cell Technologies) containing 200 U/mL interleukin-2 (NCI).
qPCR
RNA was prepared from each cell line using the RNEasy kit (Qiagen). For prostate tissue, scrapings were taken from frozen prostate sections and sonicated in RLT buffer (Qiagen), then RNA was prepared as above. RNA was quantified using a ND-1000 Spectrophotometer (NanoDrop). Equal amounts of RNA were used to synthesized cDNA. The RNA, was combined with RNase-free water, oligo-dT and random decamer primers (Ambion) and heat denatured at 70°C for 5 minutes. M-MLV reverse transcriptase and its buffer (Promega) and RNase inhibitor (Roche) were added to the reaction, which was carried out at 42°C for one hour, followed by 95°C for 10 minutes. PI-9 (SerpinB9Hs00244603_A1), k-alpha tubulin (Hs0074482_sH), and HPRT (433768F) Taqman probe sets were purchased from Applied Biosystems/Ambion. qPCR reactions containing Taqman Universal PCR Master Mix (Applied Biosystems), one set of Taqman probes, and the appropriate cDNA were set up in triplicate. qPCR was performed on the ABI 7300 Real Time PCR system instrument. qPCR raw data (Ct) for each sample was normalized to the reference gene for comparison.
To enable normalization between cell lines, a baseline Ct value of 45 was established at which no expression was observed. This value was assigned to LNCaP cells, which do not express PI-9, and was used to normalize the other cell lines examined to LNCaP cells.
BioPorter delivery
The BioPorter kit was purchased from Genlantis. Granzyme B was expressed and purified as described previously[22]. Briefly, 72 pmole of Granzyme B was incubated with the hydrated BioPorter reagent. The BioPorter/Granzyme B mixture was added to 1×10^5 cells/well in a 6-well plate at a final concentration of 2.1 uM Granzyme B. The cells and protein mix were incubated for 4 hours at 37°C, then lysed in 0.5% NP-40 buffer containing a protease inhibitor cocktail (Roche).
Western blotting
All samples were brought up in SDS loading buffer (Invitrogen) containing 5mM Bond-Breaker TCEP Solution (Thermo Scientific). The mouse anti-Granzyme B antibody was purchased from USBiological and used at 1:1000 in 5% BSA. The polyclonal rabbit anti-PARP antibody was purchased from Cell Signaling and used at 1:1000 in 5% milk. The mouse anti-cMyc antibody was purchased from Santa Cruz Biotechnology and used at 1:1000 in 5% milk. Goat anti-mouse-HRP and goat anti-rabbit-HRP were purchased from Bio-Rad and used at 1:2000 in 5% milk. All blots were developed using the Amersham ECL-Plus Western blotting detection system (GE Healthcare).
Flow cytometry
All flow cytometry was carried out on the FACScaliber instrument from BD Biosciences. For detection of PI-9, cells were fixed and permeabilized using the BD Cytofix/Cytoperm kit (BD) according to the manufacturer’s instructions. For detection of endogenous PI-9, a mouse monoclonal anti-PI-9 antibody (clone 7D8, MBL Medical and Biological Labs) were added at a final concentration of 5.0 μg/ml and incubated for 30 minutes at 4°C. As a control, an untargeted mouse IgG antibody (BD Biosciences) was added at the same concentration. Following the removal of unbound primary antibody, a goat anti-mouse secondary antibody that was conjugated to the dye FITC (BD Biosciences) was added to a final concentration of 10 μg/ml. After incubating for 30 min at 4°C, unbound secondary antibody was removed and cells were analyzed. For detection of myc-tagged PI-9, a similar protocol was followed except an anti-myc antibody directly conjugated to a fluorochrome was used.
For detection of cell surface MIC A and MIC B, LNCaP pcDNA cells were harvested and re-suspended in Stain Buffer FBS (BD Biosciences). A mouse anti-human MIC A/MIC B antibody or its isotype control (BioLegend) were added to approximately 2×105 cells at a final concentration of 20.0 μg/ml and incubated for 30 minutes at 4°C.Following removal of unbound primary, a FITC-conjugated goat anti-mouse secondary antibody (BD Biosciences) was used at a final concentration of 10 μg/ml. Cells were incubated for 30 minutes at 4°C washed to remove unbound secondary antibody, and resuspended in Stain Buffer FBS for flow cytometry analysis.
Transfection
The PI-9 (serpinB9) gene was synthesized with an N-terminal myc tag by GeneArt using human codon bias. After the start Met, the myc tag (EQKLISEEDL) is followed by a GGS linker. The synthesized gene was sub-cloned into pcDNA3.1(+) and the identity of the construct confirmed by HindIII and XhoI digestion and DNA sequencing.
To create the LNCaP-PI9 and LNCaP-pcDNA stables, both the PI-9 construct sub-cloned into pcDNA3.1(+) and empty pcDNA3.1(+) was linearized with PvuI restriction enzyme. Following purification, 2.0 μg of DNA was transfected into LNCaP cells using GenJet in vitro DNA transfection reagent for LNCaP cells, version II (SignaGen Labs) following the manufacturer’s protocol. Approximately 48 h post-transfection, cells were switched into media containing 500 μg/ml G418 (Invitrogen). At this G418 concentration, un-transfected cells were killed in approximately 10 days. Cells that survived the G418 selection were expanded and used for all subsequent assays.
Cell death assay
LNCaP cells were washed in PBS and resuspended at 2×106 cells/ml in PBS. TFL4 dye (OncoImmunin) was added at 1:300,000 and the cells were incubated for 15 minutes at 37°C. LNCaP cells were washed in PBS, resuspended in Myelocult media plus 200U/mL IL-2, then aliquoted in triplicate into wells at 1×105 cells/well. NK-92 cells in Myelocult media plus 200U/mL IL-2 were added to the appropriate wells at ratios of 0:1, 5:1, 10:1 or 30:1 in a final volume of 200ul/well. The plate was spun at 175xG for 5 minutes, then incubated at 37°C for 4 hours. The NK-92/LNCaP cell mixure was brought up in 500μl of PBS containing a final concentration of 2μg/mL propidium iodide (BioVision). Cell death was assessed on a FACSCaliber flow cytometer (BD Biosciences) by counting the number of propidium iodide and TFL4 positive cells.
Immunohistochemistry
Immunohistochemical staining was conducted on paraffin-embedded prostate tissue sections by the UCSF Tissue Core. Tissue sections were deparaffinized with xylene and alcohol, and were then subjected to heat-based antigen retrieval in the microwave for 10 minutes in 10 mM sodium citrate buffer, pH 6.0 followed by cooling at room temperature for 30 minutes. Samples were rinsed in water and PBS and then blocked in normal horse serum for 30 minutes. Tissue sections were then incubated overnight at room temperature with a 1:10 dilution of the PI9-17 monoclonal primary anti-PI-9 antibody (Monosan) in PBS containing 1% bovine serum albumin (BSA). On the following day, slides were rinsed in PBS and incubated for 30 minutes with the secondary biotinylated horse anti-mouse antibody at a 1:200 dilution in PBS containing 1% BSA. The ABC (Vector) stain was then applied at a 1:100 dilution and incubated for 30 minutes. After rinsing again in PBS, slides were subjected to DAB chromogen stain, hematoxylin counterstain, and were dehydrated in alcohol and xylene.
Results
PI-9 is expressed in cell lines derived from prostate cancer
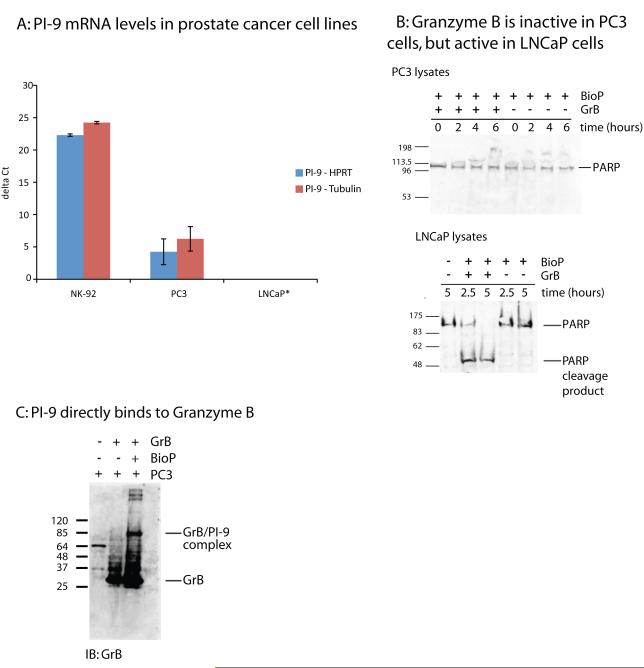
To determine the role PI-9 plays in prostate cancer, we first tested whether PI-9 is expressed in cell lines derived from prostate cancers. qPCR was used to measure the amount of PI-9 mRNA in the highly invasive and androgen independent PC3 cell line, and in the non-invasive and androgen responsive LNCaP cell line. As a positive control, the amount of PI-9 mRNA was also measured in natural killer (NK-92) cells, a type of cytotoxic lymphocyte known to express high levels of PI-9. mRNA was measured using Taqman probes for PI-9 and for two reference genes, HPRT and k-α-tubulin. As shown in Figure 1A, PC3 cells expressed measureable quantities of PI-9, but LNCaP cells did not. As expected, NK-92 cells expressed PI-9 in great abundance, 324-fold more than PC3 cells. PI-9 levels were confirmed at the protein level by flow cytometry (Supplemetary Figure 1). A third cell line, the androgen independent and invasive DU-145 cells, were also found to contain PI-9 (data not shown). However, these cells were resistant to the reagents used in subsequent experiments, so functional studies were continued with the PC3 and LNCaP cell lines. These results show that PI-9 is expressed in PC3 cells but not in LNCaP cells at both the mRNA and protein levels.
Figure 1. PI-9 inhibits Granzyme B when present in prostate cancer cell lines.
1A: PI-9 levels were measured by qPCR. The threshold cycle (Ct) of PI-9 mRNA for each cell line was normalized to the Ct two reference genes. Each cell line was then normalized to LNCaP cells. *LNCaP cells contained no detectable message, and were assigned a Ct value after which no message was observed in any cell line to establish a baseline.
1B: The protein transfection reagent BioPorter (BioP) was used to deliver recombinant GrB to both PC3 and LNCaP cells. Lysates of GrB treated cells were immunoblotted for the GrB substrate PARP to determine if GrB was active in these cell lines. Controls showed that protein delivery into PC3 cells with BioPorter was possible.
1C: PC3 cell lysates containing GrB delivered by BioPorter were blotted with an anti-GrB antibody. A higher molecular weight complex appears which corresponds to PI-9 bound to GrB.
PI-9 inhibits Granzyme B in PC3 cells
After demonstrating that PI-9 is made in cell lines derived from some prostate cancers, we next asked whether cellular PI-9 could inhibit its target protease, Granzyme B (GrB). Two cell lines with no detectable PI-9 (LNCaP cells) and measurable PI-9 (PC3 cells) were tested for GrB resistance. GrB was introduced into each cell line using BioPorter, a reagent for protein transfection. The cells were lysed at various time points for up to six hours after the introduction of GrB. GrB activity was measured by monitoring cleavage of PARP, a 113kD substrate of GrB, by Western blot. As shown in Figure 1B, GrB cleaved PARP in LNCaP cells, which lack PI-9. However, GrB did not cleave PARP in PC3 cells, which contain PI-9. GrB, a cationic protein, is known to bind to the surface of cell membranes, though it requires perforin for entry[23]. Granzyme B binding to the membrane can be observed in the absence of BioPorter (Figure 1C, lane 2), however, these lysates were prepared in the present of SDS to ensure this external GrB did not react with internal PI-9. These results indicate that PI-9 inhibits GrB in a cellular context.
Since PARP cleavage suggested that PI-9 inhibited GrB activity, we next set out to directly demonstrate that PI-9 inhibited GrB. To directly test inhibition, GrB was added to PC3 cells using BioPorter, and the cells were lysed after 6 hours. Since GrB has been shown to bind to the plasma membrane of cells, lysates were prepared under denaturing conditions to prevent cell surface GrB from forming a complex with intracellular PI-9. These lysates were processed by Western blot to detect GrB. As shown in Figure 1C, PI-9 formed a stable complex with GrB that appeared at 85kD, and this complex was not observed in the absence of BioPorter. This result indicates that PI-9 binds to GrB and suggests that the GrB inhibition observed in PC3 cells is mediated by PI-9.
PI-9 protects prostate cancer cells from apoptosis
Having shown that PI-9 inhibited GrB in vitro, we next asked whether PI-9 inhibition could prevent GrB-mediated apoptosis in the context of natural killer cell mediated death. To answer this question, NK-92 cells were chosen as effectors because selective inhibition of GrB in this cell line significantly reduced target cell lysis [24][25]. To test the role of PI-9 in apoptosis assays, PI-9 was overexpressed in LNCaP cells. Overexpression was chosen after attempts to knockdown PI-9 expression or remove PI-9 by immunodepletion in PC3 cells were unsuccessful, due to the stability and long half life of PI-9. LNCaP cells were selected as targets for two reasons: LNCaP cells express no detectable PI9, and LNCaP cells express the NK activating ligands MHC class I chain–related molecules (MICs, Supplementary Figure 2)[26], which suggested that NK-92 cells could recognize LNCaP cells.
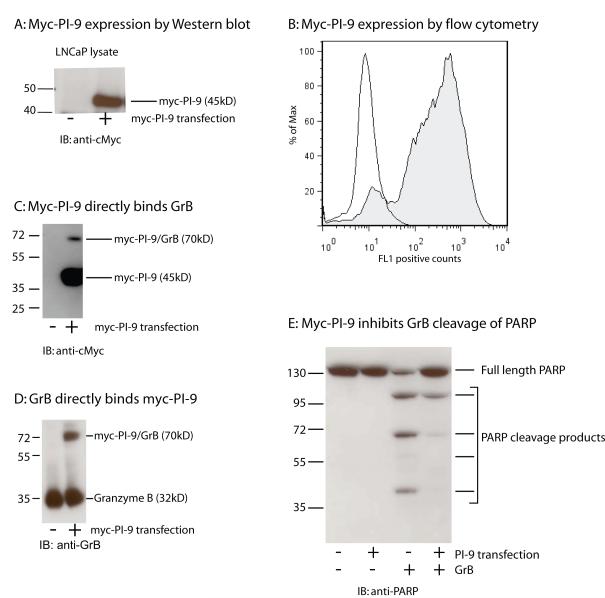
To generate a PI-9 over-expression cell line, a Myc-tagged PI-9 gene or empty pcDNA vector control was transfected and stably integrated into LNCaP cells. These cell lines were referred to as LNCaP PI-9 and LNCaP pcDNA respectively. Myc-PI-9 expression was tested by immunoblotting of lysates using an anti-Myc antibody. A 45-kD band appeared in lysates from LNCaP PI-9 and was absent in lysates from LNCaP pcDNA (Figure 2A). Analysis of myc staining by flow cytometry showed that greater than 80% of LNCaP PI-9 cells expressed Myc-PI-9 relative to LNCaP pcDNA cells (Figure 2B). An identical result was obtained when staining was performed with an anti-PI9 antibody (data not shown). This result indicated successful PI-9 overexpression.
Figure 2. Overexpression of PI-9 in LNCaP cells.
2A: LNCaPs transfected with either Myc-PI-9 (+) or a pcDNA control vector (-) were lysed and blotted with anti-Myc.
2B: myc-PI-9 levels in LNCaP pcDNA (unfilled) and LNCaP PI9 (filled). Identical results were obtained with the anti-myc antibody and the anti-PI9 antibody.
2C: LNCaP lysates transfected with either Myc-PI-9 (+) or a pcDNA control vector (-) were incubated with recombinant GrB and blotted with anti-Myc.
2D: The experiment in 2C was repeated and blotted with anti-GrB
2E: LNCaP lysates transfected with either Myc-PI-9(+) or a pcDNA control vector (-) were incubated with recombinant GrB and PARP cleavage was assessed.
Once LNCaPs stably expressing PI-9 were generated, the functional ability of myc-PI-9 was tested. LNCaP-pcDNA and LNCaP-PI-9 cell lysates were generated under non-denaturing conditions, then incubated with recombinant GrB. Western blotting for PI-9 using an anti-cMyc antibody showed the appearance of a 70kD band in only the LNCaP-PI-9 samples, which indicated the formation of a PI-9/GrB complex (Figure 2C). The PI-9/GrB complex was also detected using an anti-GrB antibody (Figure 2D). This complex demonstrates that myc-PI-9 is functional through its ability to bind to GrB. To further test the inhibition of GrB by myc-PI-9, the same blots were analyzed for the presence of PARP, a substrate of GrB. In the LNCaP-pcDNA samples, several PARP cleavage products were observed, and there was a 50% reduction in full length PARP (Figure 2E). In the LNCaP-PI-9 samples, very little PARP cleavage was observed, and 97% of the full length PARP band remained. These results indicate that overexpressed myc-PI-9 can successfully inhibit GrB and prevent cleavage of its substrates in LNCaP cells.
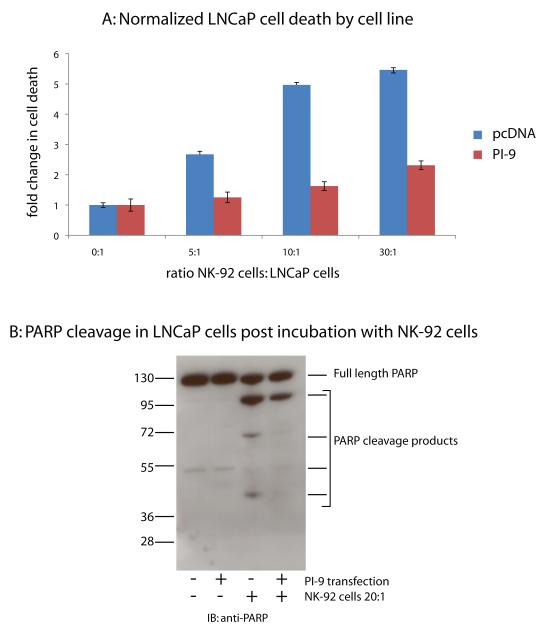
Once these cells lines were generated and PI-9 function verified, they were tested for resistance to NK-92-induced cell death. Fluorescently labeled LNCaP cells were co-incubated with unlabeled NK-92 cells in varying ratios for 4 hours. Cells were then stained with propidium iodide (PI) and target cell death was quantified by the percentage of PI positive, fluorescent cells by flow cytometry. All samples were normalized to LNCaP cells in the absence of NK-92 cells. LNCaP-pcDNA cells were killed by the NK-92 cells at all ratios. (Figure 3A). The LNCaP cells over-expressing PI-9 exhibited less cell death than the LNCaP-pcDNA cells over the entire range of ratios. This result indicates that LNCaP cells expressing PI-9 are resistant to NK-92 cell-induced cell death. The presence of PI-9 can indeed protect prostate cancer cells from NK-92-mediated apoptosis.
Figure 3. LNCaP cells that overexpress PI-9 are resistent to NK-mediated cell death.
3A: LNCaP cells transfected with either Myc-PI-9 (+) or a pcDNA control vector (-) were co-incubated varying ratios of NK-92 cells. Cell death was assayed using flow cytometry by propidium iodide staining. LNCaP-only cell death was used to normalize cell death in the presence of NK cells.
3B: LNCaP cells transfected with either Myc-PI-9 (+) or a pcDNA control vector (-) were co-incubated at 1:20 with NK-92 cells. LNCaPs were isolated and blotted for PARP cleavage
To further confirm the mechanism by which PI-9 prevents cell death, the target cells were examined for the degree of cleavage of the GrB substrate PARP. LNCaP cells were incubated with NK-92 cells in a ratio of 0:1 or 20:1 for 6 hours, lysed, and blotted for PARP. As shown in Figure 3B, PARP was present in both lysates, as indicated by a 113kD band present in both before and after incubation with NK-92 cells. In the LNCaP-pcDNA cells, significant PARP cleavage was observed after incubation with NK-92 cells. These PARP cleavage products were only faintly observed in the LNCaP-PI-9 cells after incubation with NK-92 cells, approximately two fold less than in the LNCaP-pcDNA lysates. These results indicate that PI-9 protected cells from apoptosis by inhibiting GrB.
PI-9 can be detected in prostate tumors
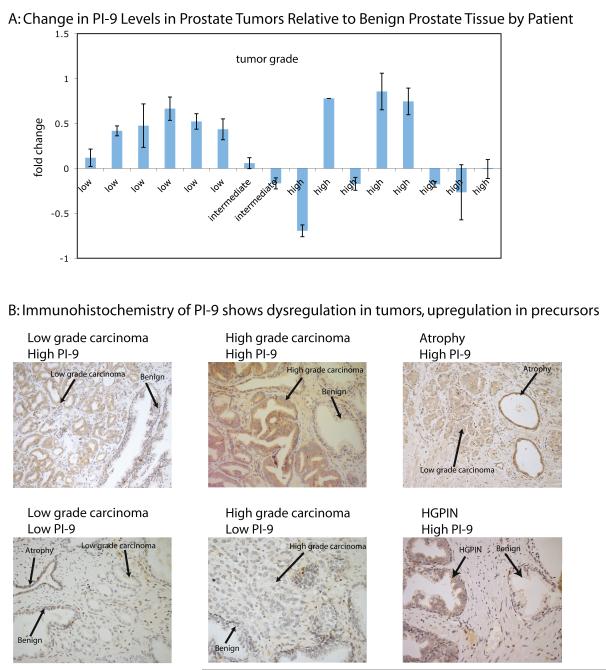
Having shown that PI-9 expression protects prostate cancer cells from NK-92 induced cell death, we undertook a small pilot study to determine whether this effect occurred in prostate tumors. PI-9 mRNA was measured in 32 prostate biopsies containing both cancerous and benign tissue. PI-9 expression was measured by qPCR using the Taqman PI-9 probe and two reference genes, and each tumor sample was normalized to its corresponding benign sample. As shown in Figure 4A, greater PI-9 expression was observed in low grade tumors than in the benign control. However, PI-9 expression in higher grade tumors was stochastic. These results were statistically significant when the high grade tumors were grouped by PI-9 expression using the Mann-Whitney rank sum test (Supplemental Figure 3, P = 0.0043 for low expression of PI-9, P= 0.0238 for high expression of PI-9). These results suggest that PI-9 expression is elevated in low grade tumors but stochastically dysregulated in high grade prostate tumors, implying the function of PI-9 is needed early in cancer progression.
Figure 4. PI-9 is found in several stages in prostate tissue.
4A: PI-9 is upregulated in low grade tumors, but dysregulated in high grade tumors. PI-9 levels were measured in tumor and benign patient tissue samples. PI-9 levels in tumors were then normalized to the benign levels to calculate the fold change.
4B: Immunohistochemistry for PI-9 was performed on slices of prostate tissue. PI-9 staining was heterogeneous in both benign and carcinoma regions and consistently positive in atrophy and PIN regions.
To confirm this observation at the protein level, 54 slices from the prostate biopsies of 24 patients were tested for the presence of PI-9 by immunohistochemistry against PI-9. As shown in Figure 4B, PI-9 was found in both low and high grade tumors. The tumors showed a similar pattern as the mRNA data, though fewer tumors overall stained positively than in the qPCR study, where 45% of the low grade tumors had more PI-9 than the benign tissue, but only 25% of the high grade tumors had more PI-9. Interestingly, PI-9 staining intensity was observed most consistently in high grade prostatic intraepithelial neoplasia (HGPIN, 76%), a precursor to prostate cancer, as well as in regions of atrophy (76%). Taken together, this pilot study indicates that PI-9 is present in pre-cancerous states and remains in some tumors.
Discussion
Here we report that the protease inhibitor PI-9 can protect prostate cancer cells from NK-92 cell- induced apoptosis by inhibiting Granzyme B (GrB). We also provide tantalizing preliminary data that this protective mechanism operates early in the progression of prostate cancer. In our experiments, PI-9 expressed by prostate cancer cells inhibited GrB. LNCaP cells that overexpressed PI-9 were resistant to apoptosis mediated by NK -92 cells, but LNCaP cells that lacked PI-9 were sensitive. This observation shows that PI-9 can protect prostate cells from NK cell-mediated apoptosis, one arm of immunosurveillance. Additionally, immunohistochemistry showed that PI-9 was present in HGPIN tumors, one of the earliest forms of prostate cancer, as well as in atrophic lesions. Our results suggest that PI-9 protects prostate tumors from immunosurveillance early in cancer progression by blocking the apoptotic response, while additional mechanisms protect tumors later in their progression through blocking recognition.
Why would PI-9 initially become upregulated in prostate tumors? As shown in Figure 4B, not only is PI-9 upregulated in early tumors, PI-9 is also abundantly expressed during atrophy, including prostatic inflammatory atrophy (PIA, Supplemental Figure 3). Atrophy is a known hallmark of an inflamed prostate, and evidence has shown it can precede the development of PIN[27][28]. A correlation between inflammation and prostate cancer has long been observed, and PI-9 could be the molecular mechanism that connects the two pathologies. PI-9 is often upregulated in response to inflammation to protect bystander cells from inadvertently introduced GrB[12], and PI-9 expression can be induced by pro-inflammatory molecules like IL-1B and TNF-α[29]. Therefore, prostatic inflammation may provide a trigger for PI-9 upregulation. In a subset of inflamed cells, this PI-9 upregulation may become permanent, creating a cancer-prone population of cells that are resistant to immunosurveillance.
While PI-9 allows cancer cells to block the apoptotic response of immunosurveillance, prostate cancer cells have also been shown to block recognition by CLs. through the process of MIC shedding. MICs, or MHC class I chain-related molecules, are expressed on the surface of cancerous cells and target these cells for destruction. MICs bind to the NKG2D receptor on NK cells[26] which initiates granule exocytosis, killing the MIC-expressing cell[30][31]. MIC is expressed in HGPIN and low grade prostate tumors, however, membrane bound MIC cleaved by a metalloprotease in high grade tumors. This allows cancerous cells to evade detection by NK cells[24[26][32]. Early expression of PI-9 could allow prostate cancer cells to survive while MIC is present.
We hypothesize that PI-9 expression may protect prostate cancer tumors early in cancer progression, when MIC is still present on the surface of cells. We find that PI-9 is expressed in the early HGPIN stage, a stage in which MIC is also expressed, but stochastically expressed in advanced tumors (Figure 4B). These results imply that PI-9 is important early in cancer progression, when MIC is still present on the surface of cells. PI-9 may not be important in the later stages, after MIC is shed and cells are in less danger of NK cell-induced apoptosis. Therefore, PI-9 protects prostate cancer cells early in their progression, when surface MIC is expressed, and PI-9 is then dysregulated in later stages when MIC is shed. Future studies beyond cell lines could further elucidate the role of PI-9 and MIC early in prostate cancer progression, as most cell lines and patient samples are derived from late-stage tumors.
In summary, prostate cancer uses both PI-9 upregulation and MIC shedding at consecutive stages in its progression to evade immunosurveillance. Treatment for prostate cancer must take into account both mechanisms, as PI-9 has been shown to affect hormone therapy in breast cancer[18] and immunotherapy in melanoma[19]. MIC shedding could be counteracted by inhibiting matrix metalloprotease-14[32]. PI-9 is a substrate of Granzyme M, so Granzyme M upregulation could alleviate the resistance to cell death caused by PI-9[33]. Targeting both MIC shedding and PI-9 expression could lead to an effective treatment strategy that enhances the immune response to cancer cells.
Supplementary Material
Acknowledgements
We would like to thank Dr. Peter Carroll for performing the prostatectomies, Kevin Chew for sectioning the prostate samples, Sarah Elmes for training and use of the flow cytometer, and the lab of Dr. Keith Yamamoto for the generous use of their qPCR instruments and reagents. We would also like to thank Dr. Aaron Lebeau, Dr. Marc Shuman, and Dr. Lawrence Fong for helpful discussions. This work was funded by the National Institute of Health (R01CA128765)
Financial Support: This work was supported by the National Institute of Health grant # NIH R01CA128765.
Footnotes
None of the authors listed have any significant or perceived conflicts of interest relating to the publishing of this manuscript.
References
- 1.Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 2002;20:323–70. doi: 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed] [Google Scholar]
- 2.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36(Database issue):D320–5. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X, Stennicke HR, Wang B, Green DR, Jänicke RU, Srinivasan A, et al. Granzyme B mimics apical caspases. Description of a unified pathway for trans-activation of executioner caspase-3 and -7. J. Biol. Chem. 1998;273(51):34278–83. doi: 10.1074/jbc.273.51.34278. [DOI] [PubMed] [Google Scholar]
- 4.Medema JP, Toes RE, Scaffidi C, Zheng TS, Flavell RA, Melief CJ, et al. Cleavage of FLICE (caspase-8) by granzyme B during cytotoxic T lymphocyte-induced apoptosis. Eur. J. Immunol. 1997;27(12):3492–8. doi: 10.1002/eji.1830271250. [DOI] [PubMed] [Google Scholar]
- 5.Anthony DA, Andrews DM, Watt SV, Trapani JA, Smyth MJ. Functional dissection of the granzyme family: cell death and inflammation. Immunological Reviews. 2010;235(1):73–92. doi: 10.1111/j.0105-2896.2010.00907.x. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003;3(5):361–70. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 7.Lefrançois L, Obar JJ. Once a killer, always a killer: from cytotoxic T cell to memory cell. Immunol. Rev. 2010;235(1):206–18. doi: 10.1111/j.0105-2896.2010.00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Kaiserman D, Bird PI. Control of granzymes by serpins. Cell Death Differ. 2010;17(4):586–95. doi: 10.1038/cdd.2009.169. [DOI] [PubMed] [Google Scholar]
- 10.Bird PI. Serpins and regulation of cell death. Results Probl Cell Differ. 1998;24:63–89. doi: 10.1007/978-3-540-69185-3_4. [DOI] [PubMed] [Google Scholar]
- 11.Bladergroen BA, Strik MCM, Bovenschen N, van Berkum O, Scheffer GL, Meijer CJLM, et al. The Granzyme B Inhibitor, Protease Inhibitor 9, Is Mainly Expressed by Dendritic Cells and at Immune-Privileged Sites. The Journal of Immunology. 2001;166(5):3218–25. doi: 10.4049/jimmunol.166.5.3218. [DOI] [PubMed] [Google Scholar]
- 12.Buzza MS, Hirst CE, Bird CH, Hosking P, McKendrick J, Bird PI. The Granzyme B Inhibitor, PI-9, Is Present in Endothelial and Mesothelial Cells, Suggesting That It Protects Bystander Cells during Immune Responses. Cellular Immunology. 2001;210(1):21–9. doi: 10.1006/cimm.2001.1806. [DOI] [PubMed] [Google Scholar]
- 13.Medema JP, de Jong J, Peltenburg LT, Verdegaal EM, Gorter A, Bres SA, et al. Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proc. Natl. Acad. Sci. U.S.A. 2001;98(20):11515–20. doi: 10.1073/pnas.201398198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham TD, Jiang X, Shapiro DJ. Expression of high levels of human proteinase inhibitor 9 blocks both perforin/granzyme and Fas/Fas ligand-mediated cytotoxicity. Cellular Immunology. 2007;245(1):32–41. doi: 10.1016/j.cellimm.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang X, Orr BA, Kranz DM, Shapiro DJ. Estrogen induction of the granzyme B inhibitor, proteinase inhibitor 9, protects cells against apoptosis mediated by cytotoxic T lymphocytes and natural killer cells. Endocrinology. 2006;147(3):1419–26. doi: 10.1210/en.2005-0996. [DOI] [PubMed] [Google Scholar]
- 16.ten Berge RL, Oudejans JJ, Ossenkoppele GJ, Meijer CJLM. ALK-negative systemic anaplastic large cell lymphoma: differential diagnostic and prognostic aspects--a review. J. Pathol. 2003;200(1):4–15. doi: 10.1002/path.1331. [DOI] [PubMed] [Google Scholar]
- 17.Oudejans JJ, Harijadi H, Kummer JA, Tan IB, Bloemena E, Middeldorp JM, et al. High numbers of granzyme B/CD8-positive tumour-infiltrating lymphocytes in nasopharyngeal carcinoma biopsies predict rapid fatal outcome in patients treated with curative intent. J. Pathol. 2002;198(4):468–75. doi: 10.1002/path.1236. [DOI] [PubMed] [Google Scholar]
- 18.Jiang X, Ellison SJ, Alarid ET, Shapiro DJ. Interplay between the levels of estrogen and estrogen receptor controls the level of the granzyme inhibitor, proteinase inhibitor 9 and susceptibility to immune surveillance by natural killer cells. Oncogene. 2007;26(28):4106–14. doi: 10.1038/sj.onc.1210197. [DOI] [PubMed] [Google Scholar]
- 19.van Houdt IS, Oudejans JJ, van den Eertwegh AJM, Baars A, Vos W, Bladergroen BA, et al. Expression of the Apoptosis Inhibitor Protease Inhibitor 9 Predicts Clinical Outcome in Vaccinated Patients with Stage III and IV Melanoma. Clinical Cancer Research. 2005;11(17):6400–7. doi: 10.1158/1078-0432.CCR-05-0306. [DOI] [PubMed] [Google Scholar]
- 20.Harzstark AL, Small EJ. Immunotherapy for prostate cancer using antigen-loaded antigen-presenting cells: APC8015 (Provenge) Expert Opin Biol Ther. 2007;7(8):1275–80. doi: 10.1517/14712598.7.8.1275. [DOI] [PubMed] [Google Scholar]
- 21.Small EJ, Schellhammer PF, Higano CS, Redfern CH, Nemunaitis JJ, Valone FH, et al. Placebo-Controlled Phase III Trial of Immunologic Therapy with Sipuleucel-T (APC8015) in Patients with Metastatic, Asymptomatic Hormone Refractory Prostate Cancer. Journal of Clinical Oncology. 2006;24(19):3089–94. doi: 10.1200/JCO.2005.04.5252. [DOI] [PubMed] [Google Scholar]
- 22.Harris JL, Peterson EP, Hudig D, Thornberry NA, Craik CS. Definition and Redesign of the Extended Substrate Specificity of Granzyme B. Journal of Biological Chemistry. 1998;273(42):27364–73. doi: 10.1074/jbc.273.42.27364. [DOI] [PubMed] [Google Scholar]
- 23.Shi L, Keefe D, Durand E, Feng H, Zhang D, Lieberman J. Granzyme B binds to target cells mostly by charge and must be added at the same time as perforin to trigger apoptosis. J. Immunol. 2005;174(9):5456–61. doi: 10.4049/jimmunol.174.9.5456. [DOI] [PubMed] [Google Scholar]
- 24.Willoughby CA, Bull HG, Garcia-Calvo M, Jiang J, Chapman KT, Thornberry NA. Discovery of potent, selective human granzyme B inhibitors that inhibit CTL mediated apoptosis. Bioorg. Med. Chem. Lett. 2002;12(16):2197–200. doi: 10.1016/s0960-894x(02)00363-3. [DOI] [PubMed] [Google Scholar]
- 25.Mahrus S, Craik CS. Selective chemical functional probes of granzymes A and B reveal granzyme B is a major effector of natural killer cell-mediated lysis of target cells. Chem. Biol. 2005;12(5):567–77. doi: 10.1016/j.chembiol.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Wu JD, Higgins LM, Steinle A, Cosman D, Haugk K, Plymate SR. Prevalent expression of the immunostimulatory MHC class I chain-related molecule is counteracted by shedding in prostate cancer. J. Clin. Invest. 2004;114(4):560–8. doi: 10.1172/JCI22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Nunzio C, Kramer G, Marberger M, Montironi R, Nelson W, Schröder F, et al. The controversial relationship between benign prostatic hyperplasia and prostate cancer: the role of inflammation. Eur. Urol. 2011;60(1):106–17. doi: 10.1016/j.eururo.2011.03.055. [DOI] [PubMed] [Google Scholar]
- 28.Borowsky AD, Dingley KH, Ubick E, Turteltaub KW, Cardiff RD, DeVere-White R. Inflammation and Atrophy Precede Prostatic Neoplasia in a PhIP-Induced Rat Model. Neoplasia. 2006;8(9):708–15. doi: 10.1593/neo.06373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kannan-Thulasiraman P, Shapiro DJ. Modulators of Inflammation Use Nuclear Factor-κB and Activator Protein-1 Sites to Induce the Caspase-1 and Granzyme B Inhibitor, Proteinase Inhibitor 9. Journal of Biological Chemistry. 2002;277(43):41230–9. doi: 10.1074/jbc.M200379200. [DOI] [PubMed] [Google Scholar]
- 30.Lanier LL. On guard[mdash]activating NK cell receptors. Nat Immunol. 2001;2(1):23–7. doi: 10.1038/83130. [DOI] [PubMed] [Google Scholar]
- 31.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3(10):781–90. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 32.Liu G, Atteridge CL, Wang X, Lundgren AD, Wu JD. The membrane type matrix metalloproteinase MMP14 mediates constitutive shedding of MHC class I chain-related molecule A independent of A disintegrin and metalloproteinases. J. Immunol. 2010;184(7):3346–50. doi: 10.4049/jimmunol.0903789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahrus S, Kisiel W, Craik CS. Granzyme M is a regulatory protease that inactivates proteinase inhibitor 9, an endogenous inhibitor of granzyme B. J. Biol. Chem. 2004;279(52):54275–82. doi: 10.1074/jbc.M411482200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.