Abstract
Protein C is activated by thrombin with a value of kcat/Km = 0.11mM−1s−1 that increases 1700-fold in the presence of the cofactor thrombomodulin. The molecular origin of this effect triggering an important feedback loop in the coagulation cascade remains elusive. Acidic residues in the activation domain of protein C are thought to electrostatically clash with the active site of thrombin. However, functional and structural data reported here support an alternative scenario. The thrombin precursor prethrombin-2 has R15 at the site of activation in ionic interaction with E14e, D14l, and E18, instead of being exposed to solvent for proteolytic attack. Residues E160, D167, and D172 around the site of activation at R169 of protein C occupy the same positions as E14e, D14l, and E18 in prethrombin-2. Caging of R169 by E160, D167, and D172 is responsible for much of the poor activity of thrombin toward protein C. The E160A/D167A/D172A mutant is activated by thrombin 63-fold faster than wild-type in the absence of thrombomodulin and, over a slower time scale, spontaneously converts to activated protein C. These findings establish a new paradigm for cofactor-assisted reactions in the coagulation cascade.
Introduction
Trypsin-like proteases are responsible for digestion, blood coagulation, fibrinolysis, development, fertilization, apoptosis, and immunity and remain major targets of therapeutic intervention.1 Nearly all members of this family are expressed as inactive zymogens and converted to the mature enzyme by proteolytic cleavage at the conserved residue R15 (chymotrypsinogen numbering). The cleavage generates a new N-terminus that relocates within the protein and orchestrates alignment of the catalytic triad and optimal architecture of the oxyanion hole and primary specificity pocket required for substrate binding and catalysis. A common theme in the blood coagulation and complement cascades involves cofactor-assisted zymogen activation,2,3 where the cofactor corrects defects in the enzyme and promotes efficient activation of zymogen acting as substrate. The well-established conformational selection of the trypsin fold, enabling a switch of the enzyme from the inactive E* form when free to the active E form when bound to cofactors,4 provides a molecular framework for the effect. For example, complement factors B and C2 are mostly inactive until binding of complement factors C3b and C4b enables catalytic activity at the site where amplification of C3 activation leads to formation of the membrane attack complex.3,5,6 Complement factor D assumes an inactive conformation with a distorted catalytic triad7,8 until binding to C3b and factor B promote substrate binding and catalytic activity.9,10 Clotting factor VIIa circulates in the blood as a poorly active protease but acquires full catalytic activity on interaction with tissue factor exposed to the bloodstream on vascular injury.11 Clotting factor Xa requires the action of factor Va on a membrane surface to efficiently convert prothrombin into thrombin.12 Other enzymes challenge the paradigm. Thrombin hydrolyzes synthetic and macromolecular substrates, such as fibrinogen and PAR1 with values of kcat/Km that are nearly diffusion-limited,13 as expected of an enzyme stabilized in the active E form when free. Yet, activity toward the anticoagulant substrate protein C is extremely poor (kcat/Km = 0.11mM−1s−1) and requires the cofactor thrombomodulin to be increased to a physiologically meaningful level (kcat/Km = 190mM−1s−1). The molecular origin of this drastic effect has eluded experimentalists since the discovery of thrombomodulin 3 decades ago.14 How can thrombin work at the diffusion-controlled limit with most substrates yet be such a poor catalyst of protein C?
The value of kcat/Km for the hydrolysis of substrate by a trypsin-like protease is defined as the ratio k1k2/(k-1 + k2) and depends on the rate of diffusion of substrate into the active site, k1, the acylation rate k2 that corresponds to kcat and the rate of dissociation of the enzyme-substrate complex, k−1. Hence, kcat/Km depends on parameters that reflect properties of the enzyme-substrate complex (k−1 and k2), but also of the free enzyme and substrate (k1). A defect in protein C may compromise the value of kcat/Km independent of a defect in thrombin, so it is possible that the cofactor thrombomodulin corrects a defect in the substrate protein C rather than the enzyme thrombin. The proposal that thrombomodulin changes the conformation of the active site of thrombin is supported by spectroscopic15 and mutagenesis16–19 studies. Structural biology has remained inconclusive because the thrombin-thrombomodulin complex has been crystallized with the active site occupied by an inhibitor20 or crystal contacts.21 The alternative view that thrombomodulin induces a conformational change in protein C22,23 enjoys substantial experimental support.18,19,24–27 The cofactor effect of thrombomodulin is more difficult to reproduce with mutations in the enzyme16,17,25,28 than mutations of protein C.18,19,26,27 Of particular importance is previous mutagenesis work on the activation domain of protein C. Mutations of D167 and D172 near the site of cleavage at R169 enhance activation by thrombin 30-fold in the absence of thrombomodulin and generate a protein C variant of potential clinical relevance as a clot inhibitor activatable by thrombin in the arterial circulation independent of thrombomodulin.19 Replacement of R169 with Trp produces a variant that is activated by chymotrypsin with a kcat/Km value comparable to that measured for activation of wild-type protein C by the thrombin-thrombomodulin complex.27 These seminal observations suggest that residues around the site of cleavage at R169 control the mechanism of protein C activation and hold the key to unravel the role of the cofactor thrombomodulin. The case is strengthened by a recent structural observation on the activation domain of prethrombin-2,29 which provides the motivation for the work reported here.
Methods
Thrombin wild-type and mutant S195A were expressed in Escherichia coli, refolded and purified to homogeneity as previously described.29,30 The activation peptide of protein C, 162QEDQVDPR↓LIDGKMTRRGDS181, was synthesized by solid-phase method using Fmoc chemistry on a model PS3 automated synthesizer (Protein Technologies International). The crude peptide was then fractionated by reverse-phase high performance liquid chromatography and finally analyzed by mass spectrometry to an average molecular weight of 2316.5 Da. For crystallization studies, maximum solubility of the peptide occurred when thrombin S195A was dialyzied into 0.1M HEPES, pH 7.0, 150mM NaCl. Preparation of vectors, protein expression, and purification of Gla-domainless protein C wild-type and mutants E160A/D167A/D172A (EDD) and E160A/D167A/D172A/S360A (EDDS) were carried out as described elsewhere with minor modifications.31 Primers used for the EDD mutant were as follows: 5′-CACAGCAGACCAAGAAGACCAAGTAGCTCCGCGGCTCATTGCTGGG-3′ (forward) and 5′-CCCAGCAATGAGCCGCGGAGCTACTTGGTCTTCTTGGTCTGCTGTG-3′ (reverse); and for the EDDS mutant were 5′-TGCCTGCGAGGGCGACGCTGGGGGGCCCATGGTC-3′ (forward) and 5′-GACCATGGGCCCCCCAGCGTCGCCCTCGCAGGCA-3′ (reverse). After validation of the constructs, proteins were expressed in BHK cells in media containing DMEM supplemented with 10% (volume/volume) calf serum and 2mM L-glutamine. Right after collecting cell culture supernatant, benzamidine HCl was added to a 5mM final concentration to prevent proteolysis. Both wild-type and mutants were purified to homogeneity by immunoaffinity chromatography using the Ca2+-dependent monoclonal antibody HPC4. Eluted proteins were concentrated using Vivaspin concentrators (Sartorius Stedim Biotech) and loaded onto a gel filtration Superdex TM 200 column (GE Healthcare; Bio-Sciences AB).
After the gel filtration step, protein concentration was adjusted to 0.8 mg/mL and autoactivation was followed at room temperature for up to 150 hours. To visualize autoactivation in polyacrylamide gels, time reaction aliquots were collected and quenched with reducing SDS protein loading buffer and immediately stored at −80°C. Samples were processed and analyzed by SDS-PAGE electrophoresis, and gels were stained with Coomassie brilliant blue R250. The kinetics of autoactivation were monitored by collecting samples over time and measuring activity toward the chromogenic substrate H-D-Asp-Arg-Arg-p-nitroanilide (DRR) specific for activated protein C32 under experimental conditions of 10mM Tris, pH 7.4, 145mM NaCl, 2mM CaCl2, 0.1% PEG8000 at 37°C, and in the presence of 1μM hirudin as a control to rule out contaminating effects from thrombin. The concentration of protein C was kept at 50nM, and DRR was used at 50μM. Activation of protein C wild-type and mutant EDD by thrombin was carried out as described25,30 in the absence or presence of 50nM thrombomodulin under experimental conditions of 5mM Tris, pH 7.4, 145mM NaCl, 5mM CaCl2, 0.1% PEG 8000 at 37°C. Alternatively, a discontinuous assay was performed33 where protein C wild-type and mutant EDD (0.2μM) were activated by thrombin (30nM) in 20mM Tris, pH 7.4, 145mM NaCl, 5mM CaCl2, 0.1% PEG8000 at 37°C. In the presence of thrombomodulin (50nM), the concentration of thrombin was reduced to 1nM. At different time intervals, 10 μL of the reaction was transferred into a solution containing 150 μL of 1μM hirudin, 200μM S2366, 20mM Tris, pH 7.4, 145mM NaCl, 5mM CaCl2, 0.1% PEG8000 at 25°C. The concentration of activated protein C was monitored at 405 nm by measuring the rate of chromogenic substrate S2366 hydrolysis and quantified from a standard curve prepared by complete activation of either protein C wild-type or mutant EDD (0.5μM) with thrombin (20nM) and thrombomodulin (50nM) at the time of the experiment. The contribution of autoactivation was established in experiments carried out under identical solution conditions in the absence of thrombin.
Crystallization of thrombin S195A in complex with the uncleaved fragment 162QEDQVDPR↓LIDGKMTRRGDS181 of the activation domain of protein C (1:5 molar ratio) was achieved at 22°C by the hanging drop vapor diffusion technique using an Art Robbins Instruments Phoenix liquid handling robot and mixing equal volumes (0.2 μL) of protein (10 mg/mL) and reservoir solution. Optimization of crystal growth was achieved by hanging drop vapor diffusion method mixing 3 μL of protein (11 mg/mL) with equal volumes of reservoir solution (Table 1). Crystals were grown in 0.1M Tris, pH 8.5, 0.2M CH3COONa, and 30% PEG4000 in 2 weeks. Diffraction quality crystals were cryoprotected in paraffin oil at 100°K. X-ray diffraction data were collected with a home source (Rigaku 1.2 kW MMX007 generator with VHF optics) Rigaku Raxis IV++ detector and were indexed, integrated, and scaled with the HKL2000 software package.34 The structure was solved by molecular replacement using MOLREP from the CCP4 suite35 and Protein Data Bank accession code 1SHH as a search model. Refinement and electron density generation were performed with REFMAC5 from the CCP4 suite, and 5% of the reflections were randomly selected as a test set for cross-validation. Model building and analysis of the structures were carried out using COOT.36 In the final stages of refinement, TLS tensors modeling rigid-body anisotropic temperature factors were calculated and applied to the model. Ramachandran plots were calculated using PROCHECK.37 Statistics for data collection and refinement are summarized in Table 1. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (accession code 4DT7).
Table 1.
Crystallographic data for the thrombin-protein C fragment complex
| Buffer | 0.1M Tris, pH 8.5, 0.2M CH3COONa |
| PEG | 4000 (30%) |
| PDB ID | 4DT7 |
| Data collection | Raxis IV++ |
| Wavelength, Å | 1.5418 |
| Space group | P21 |
| Unit cell dimensions, Å | a = 46.4, b = 84.3, c = 66.4 β = 94.6° |
| Molecules/asymmetric unit | 2 |
| Resolution range, Å | 40-1.9 |
| Observations | 140 938 |
| Unique observations | 38 884 |
| Completeness, % | 97.8 (81.1) |
| Rsym, % | 8.3 (34.0) |
| I/σ(I) | 13.4 (2.4) |
| Refinement | |
| Resolution, Å | 40-1.9 |
| Rcryst, Rfree | 0.175, 0.218 |
| Reflections (working/test) | 34 903/1953 |
| Protein atoms | 4755 |
| Na+ | 2 |
| Solvent molecules | 326 |
| Rmsd bond lengths, Å | 0.010 |
| Rmsd angles, degrees | 1.2 |
| Rmsd ΔB (Å2), mm/ms/ssb | 2.11/1.32/2.28 |
| protein, Å2 | 30.0 |
| Na+, Å2 | 26.3 |
| solvent, Å2 | 36.9 |
| Ramachandran plot | |
| Most favored, % | 99.2 |
| Generously allowed, % | 0.2 |
| Disallowed, % | 0.6 |
Rmsd indicates root-mean-squared deviation from ideal bond lengths and angles and Rmsd in B-factors of bonded atoms; mm, main chain-main chain; ms, main chain-side chain; and ss, side chain-side chain.
Results
The activation domains of prethrombin-2 and protein C share a striking sequence similarity: prethrombin-2 14dRELLESYIDGR↓IVEG19; and protein C 159TEDQEDQVDPR↓LIDG173. Particular attention should be paid to the residues in bold. In both zymogens, an Arg residue (R15 for prethrombin-2 and R169 for protein C) is present at the site of cleavage (↓) for the conversion to the mature enzyme. The cleavage is carried out by the prothrombinase complex for prethrombin-2 and thrombin for protein C. There is currently no crystal structure of protein C, whether free or bound, but a crystal structure of prethrombin-2 in the free form shows R15 in electrostatic interaction with the side chains of E14e, D14l, and E18, instead of being exposed to solvent as typically observed in other zymogens.29 Mutation of the E14e/D14l/E18 “anionic cage” of R15 produces a triple mutant of prethrombin-2, E14eA/D14lA/E18A, which spontaneously converts to thrombin without the need of prothrombinase or ecarin.29 Exposure of R15 endows prethrombin-2 with properties not present in the wild-type. These recent findings make it difficult to envision how efficient activation of prethrombin-2 can take place without a cofactor-assisted exposure of R15 in the substrate. An alternative possibility therefore exists that some cofactors in the blood coagulation cascade may influence the conformation of the substrate rather than, or in addition to, the conformation of the enzyme. This paradigm-shifting scenario is highly relevant to the mechanism of protein C activation by the thrombin-thrombomodulin complex.
The charged residues E160, D167, and D172 in protein C occur at the same positions as E14e, D14l, and E18 of prethrombin-2. We therefore tested the hypothesis that the anionic cage E160/D167/D172 sequesters R169 of protein C away from the solvent to prevent autoactivation and to reduce the activity of thrombin toward protein C. We also hypothesized that thrombomodulin corrects the molecular defect of protein C by promoting exposure of R169 to solvent for proteolytic attack by thrombin. The triple mutant E160A/D167A/D172A (EDD) of protein C was constructed with the expectation that it would be activated more rapidly by thrombin in the absence, but not the presence, of thrombomodulin and would spontaneously convert to activated protein C.
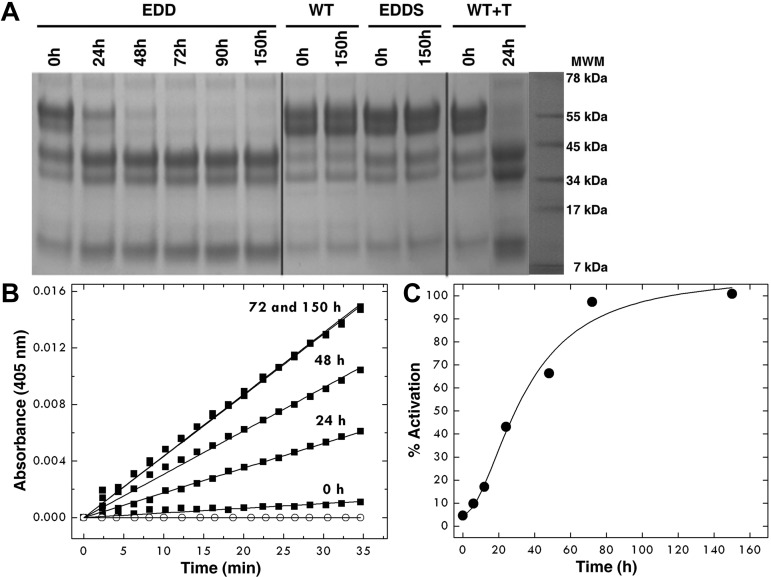
The EDD mutant of protein C shows a significant level of activity toward the chromogenic substrate DRR specific for activated protein C38 that increases over time up to 72 hours (Figure 1). No activity is detected for wild-type up to 150 hours under identical solution conditions. The activity is the result of spontaneous conversion to the mature enzyme, is retained in the presence of saturating amounts (1μM) of the potent thrombin inhibitor hirudin, but is suppressed by mutation of the catalytic S360, indicating that the zymogen protein C itself initiates cleavage at R169. This property, never before reported for wild-type or mutant protein C, is analogous to that of the E14eA/D14lA/E18A autoactivating mutant of prethrombin-229 and is consistent with the minuscule but significant activity present in other zymogens, such as chymotrypsinogen.40
Figure 1.
Autoactivation of the EDD mutant of protein C. After affinity chromatography, the protein concentration was adjusted to 0.8 mg/mL and protein C was left at room temperature for 150 hours. (A) SDS-PAGE electrophoresis documents the spontaneous conversion of the EDD mutant of protein C to activated protein C with disappearance of the band at 55 kDa and thickening of the bands in the 34- to 45-kDa range. The chemical identity of the downshifted bands as the activated product was confirmed by N-terminal sequencing and mapped to the sequence LIDGK corresponding to the new N-terminus of the heavy chain of activated protein C. Double bands reflect the intrinsic heterogeneity of protein C resulting from posttranslational modifications.39 The profile after 72 hours becomes identical to that observed on activation of wild-type protein C with thrombin. No autoactivation is observed with wild-type protein C and the inactive mutant EDDS up to 150 hours. (B) The kinetics of autoactivation was monitored from hydrolysis of DRR, a chromogenic substrate specific for activated protein C, under experimental conditions of 5mM Tris, pH 7.4, 145mM NaCl, 2mM CaCl2, 0.1% PEG8000 at 37°C. ○ represents wild-type protein C. (C) The amount of activation of the protein C mutant EDD was monitored over time from the progress curves in panel B and converted into a percentage. The sigmoidal shape of the curve is indicative of the presence of intermediates along the autoactivation pathway, consistent with an autocatalytic process.
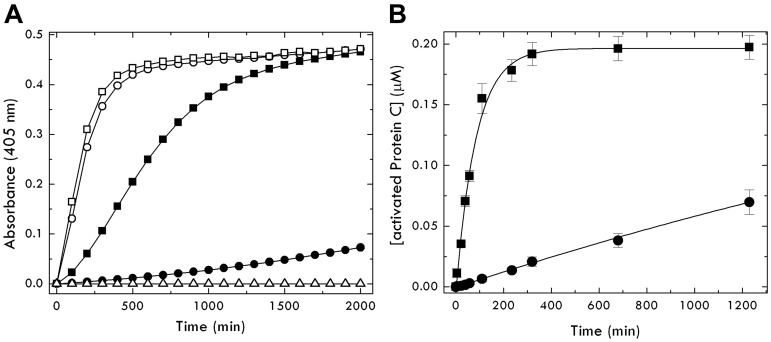
The EDD mutation also affects the rate of conversion to activated protein C by thrombin (Figure 2). The value of kcat/Km = 6.9mM−1s−1 for activation of EDD is 63-fold faster than the value of 0.11mM−1s−1 measured for thrombin activation of wild-type protein C. A significantly smaller effect of the EDD mutation is observed on the kcat/Km value for protein C activation in the presence of thrombomodulin (kcat/Km = 560mM−1s−1 for activation of EDD vs kcat/Km = 190mM−1s−1 for activation of wild-type). Hence, the EDD mutation causes structural changes in the activation domain of protein C that mimic the effect of thrombomodulin. We speculate that this conformational transition is equivalent to a jackknife movement of R169, which is buried inside the E160/D167/D172 anionic cage in the free zymogen but becomes exposed to solvent for proteolytic attack on binding of thrombomodulin. Mutation of the residues of the anionic cage constitutively exposes R169 to solvent and causes enhanced activation by thrombin in the absence of thrombomodulin. This scenario prompts a reevaluation of the role of residues D167 and D172 in protein C activation, previously assumed to cause electrostatic clash with residues E192 and E39 of thrombin.16–19,41
Figure 2.
Activation of the EDD mutant of protein C by thrombin. (A) Shown are progress curves of DRR hydrolysis by activated protein C generated from the zymogen form (circles represent wild-type, and squares, EDD) on interaction with thrombin, in the absence (closed symbols) or presence (open symbols) of 50nM thrombomodulin, under experimental conditions of 5mM Tris, pH 7.4, 145mM NaCl, 5mM CaCl2, 0.1% PEG8000 at 37°C. Analysis of the curves gives the value of kcat/Km for activation of protein C as follows: 0.11 ± 0.01mM−1s−1 (●), 6.9 ± 0.1mM−1s−1 (■), 190 ± 10mM−1s−1 (○), and 560 ± 20mM−1s−1 (□). ▵ represents the inactive EDDS mutant of protein C as a control. The sigmoidal nature of the progress curve, most visible in the case of the EDD mutant, is the result of the continuous nature of the assay32 that measures hydrolysis of DRR after the buildup of activated protein C. Autoactivation of EDD is negligible under the conditions used in the assay because it evolves over a time scale that extends beyond the complete generation of activated protein C by thrombin (Figure 1). (B) Kinetics of protein C wild-type (0.2μM, ●) and mutant EDD (0.2μM, ■) activation by thrombin (30nM) under conditions of pseudo–first order kinetics ([protein C] ≪ Km). Continuous lines were drawn according to a single exponential with values of kobs/[thrombin] = kcat/Km of 0.11 ± 0.02 (wild-type) and 6.7 ± 0.3 (EDD mutant), in excellent agreement with the values determined independently from progress curves of DRR hydrolysis (see A). No lag phase is observed in this discontinuous assay31 because activation of protein C is determined by quenching the thrombin-catalyzed reaction at different times under pseudo–first order kinetics.
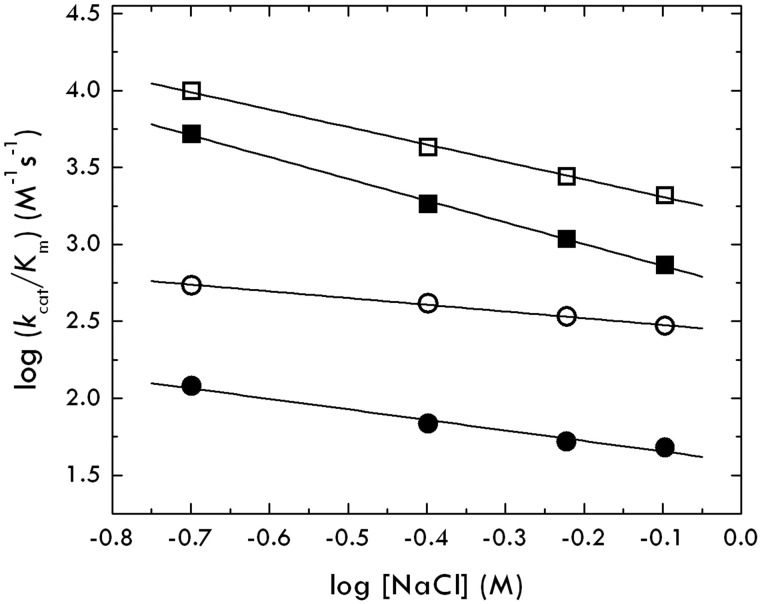
When a binding interaction depends on electrostatics, it is influenced significantly by changes in the salt concentration or ionic strength of the solution. An increase in the salt concentration decreases/increases the strength of interaction depending on whether electrostatic coupling/clash is involved. If the thrombin-protein C interaction is hindered by electrostatic clash between residues D167 and D172 of protein C and residues E192 and E39 of thrombin, then the value of kcat/Km should increase with increasing salt concentration. Contrary to this expectation, the kcat/Km for the thrombin-protein C interaction decreases with increasing salt concentration, implying that electrostatics do not oppose but actually favor the interaction (Figure 3). Importantly, the slope of the plot increases with the EDD mutation of protein C and is independent of the E192Q mutation of thrombin.
Figure 3.
Salt dependence of the thrombin-protein C interaction. The value of kcat/Km for protein C activation by thrombin under experimental conditions of 50mM Tris, pH 7.4, 5mM CaCl2, 0.1% PEG8000 at 37°C, was measured as a function of salt concentration in the range 200 to 800mM. The slope in the log-log plot, Γ, gives a measure of the electrostatic coupling (ie, charges involved or ions exchanged) on formation of the complex. Symbols refer to protein C wild-type (circles) or mutant EDD (squares) in the presence of thrombin wild-type (closed symbols) or mutant E192Q (open symbols). Values of Γ are as follows: −0.7 ± 0.1 (closed circles, thrombin-protein C), −1.4 ± 0.1 (closed squares, thrombin-protein C EDD), −0.5 ± 0.1 (open circles, thrombin E192Q-protein C), and −1.1 ± 0.1 (open squares, thrombin E192Q-protein C EDD). These values prove that there is no electrostatic clash between thrombin and protein C. The thrombin-protein C interaction is actually favored by electrostatic coupling that increases with the EDD mutation of protein C but is not affected by the E192Q mutation of thrombin.
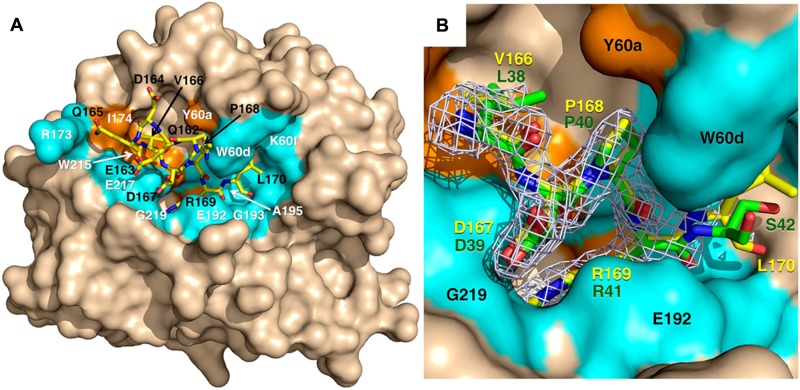
A direct test of the role of D167 of protein C in thrombin recognition comes from the X-ray crystal structure of the thrombin mutant S195A bound to the uncleaved fragment 162QEDQVDPR↓LIDGKMTRRGDS181 of the activation domain of protein C. The relevance of this fragment to understand the mode of interaction of protein C with thrombin is illustrated by the kinetics of hydrolysis. Under physiologic conditions (0.1M Tris, 145mM NaCl, pH 7.4, 5mM CaCl2, 37°C), the fragment is cleaved by thrombin with a value of kcat/Km = 0.16mM−1s−1, which is comparable to the value of 0.11mM−1s−1 measured for the thrombin-protein C interaction in the absence of thrombomodulin. Although the structure was solved at high resolution (Table 1), the fragment is visible in the density map only from Q162 to L170 (Figure 4). The 166VDPR169 segment of the protein C fragment, encompassing the P1 to P4 positions of substrate, binds to thrombin like the 38LDPR41 segment of the thrombin receptor PAR1.42 R169 penetrates the primary specificity pocket and P168 fits snugly against W60d and Y60a, as expected. D167 at the P3 position provides no steric or electrostatic hindrance to binding and actually makes a favorable polar interaction with the backbone N atom of G219, as observed for D39 of PAR1. The enhancement of protein C activation observed with the E192Q mutation of thrombin16 or the D167F mutation of protein C18,19 cannot be the result of removal of unfavorable electrostatic clash. V166 at the P4 position of the activation peptide of protein C penetrates the aryl binding site of thrombin to engage W215 of thrombin and neighbor residues L99 and Y60a as seen for other substrates.41–47 Q165 further upstream of the cleavage site makes a 114-degree turn and drastically changes direction, thereby forcing the peptide to leave the active site. The side chain of Q165 relocates next to T172 of thrombin, but the electron density becomes weak toward the N-terminus of the peptide. The constrained conformations of R169 and D167 that presumably exist in the free form of protein C are removed on formation of the enzyme-substrate complex as documented by the crystal structure. The energetic cost of this conformational change is what reduces the value of kcat/Km in the hydrolysis of protein C by thrombin compared with cleavage of PAR1 that binds to the active site in a similar conformation.
Figure 4.
X-ray crystal structure of thrombin S195A in complex with a fragment of the activation domain of protein C. (A) Thrombin is rendered in surface representation (wheat) with the active site in the center and exosite I on the right. The protein C peptide is rendered in stick representation (yellow). Residues of thrombin interacting with the protein C peptide through molecular contacts within 4 Å are in orange (hydrophobic contacts) or marine (polar contacts). (B) Details of how the P4 to P1′ residues of the protein C peptide (yellow sticks) dock into the active site of thrombin. The 2Fo-Fc electron density map (light green mesh) is contoured at 1 σ. A direct comparison is shown with the P4 to P1′ residues of the thrombin receptor PAR1 (green sticks).42 R169 penetrates the primary specificity pocket and is partially covered in this view by the side chain of E192. P168 at the P2 position makes strong hydrophobic interactions with residues P60b, P60c, and W60d. D167 at the P3 position makes a polar interaction with the backbone N atom of G219 of thrombin, which mimics the interaction of D39 at the P3 position of PAR1. V166 at the P4 position points toward W215 in the aryl binding site. Overall, the binding mode of the P1 to P4 residues of protein C is very similar to that of the P1 to P4 residues of PAR1 and D167 at the P3 position of substrate makes favorable contribution to binding. Values of the B-factor for residues of the protein C peptide are given in parentheses (Å2): Q162 (65), E163 (65), D164 (67), Q165 (68), V166 (46), D167 (37), P168 (34), R169 (32), and L170 (57).
Discussion
Previous studies have established that the low specificity of thrombin toward protein C in the absence of thrombomodulin resides in factors that limit the rate of formation of the thrombin-protein C complex.25 Because no such constraints exist when thrombin interacts with PAR130 or chromogenic substrates with the LDPR sequence at the P1 to P4 position,22 the molecular origin of the effect cannot be a defect of the active site of thrombin but rather a nonoptimal conformation of the activation peptide of protein C. Residues D167 and D172 at the P3 and P3′ positions of protein C were originally identified as the major source of the poor activity of thrombin toward protein C because of potential electrostatic clash with E192 and E39 of the enzyme.16–18 The D172N mutation of protein C19 and the E39K mutation of thrombin17 produce only a modest (4-fold and 2.5-fold, respectively) enhancement of protein C activation in the absence of thrombomodulin. The D167F mutation of protein C18,19 and the E192Q mutation of thrombin16 have a more pronounced effect (12-fold and 22-fold, respectively), and the combined mutation D167F/D172N results in 30-fold enhancement of protein C activation.19 However, the crystal structure of thrombin bound to a fragment of the activation domain of protein C reported here shows D167 in favorable interaction with G219 of thrombin (Figure 4), as recently observed for residue D39 of PAR1.42 Furthermore, the salt dependence of the thrombin-protein C interaction is not supportive of electrostatic clash and is not affected by the E192Q mutation of thrombin. The thrombin-protein C interaction is actually favored by electrostatic coupling, which even increases with the EDD mutation of protein C (Figure 3). An alternative explanation for the poor activity of thrombin toward protein C is therefore needed.
We propose that R169 at the site of cleavage of protein C is sequestered by an anionic cage composed of residues E160, D167, and D172. Mutation of these acidic residues compromises the interaction with R169 and facilitates its exposure to solvent for proteolytic attack, thereby explaining the results from previous mutagenesis studies on D167F and D172N.18,19 If the E160/D167/D172 anionic cage of protein C has the same structural architecture as the E14e/D14l/E18 anionic cage of prethrombin-2,29 then it is too small to accommodate a Trp residue at the 169 position. This could explain why the R169W mutation of protein C makes W169 readily available for cleavage by chymotrypsin.27 The EDD mutant of protein C reported here removes all acidic residues in the anionic cage and is activated 63-fold more rapidly by thrombin in the absence, but not the presence of thrombomodulin. This is a very significant enhancement of protein C activation compared with existing mutations of thrombin or protein C.16–18,25,26,28 In energetic terms, the 63-fold enhancement afforded by the EDD mutation accounts for more than half of the 1700-fold total effect of thrombomodulin, thereby making exposure of R169 an important molecular event linked to the thrombomodulin-induced enhancement of protein C by thrombin. The balance of the total effect of thrombomodulin could come from changes in the thrombin active site,15 mapped by mutagenesis to the oxyanion hole, Y60a and D189 in the primary specificity pocket,25 or by an effect of thrombomodulin on the Ca2+ binding site of protein C.26 Hence, thrombomodulin could have a dual cofactor function aimed at optimizing the substrate protein C and the enzyme thrombin.
An unexpected property of the EDD mutant of protein C is its ability to convert spontaneously to activated protein C, without the need of thrombin. This observation provides further support to the hypothesis that R169 is not exposed to solvent, by analogy to R15 in prethrombin-2.29 Furthermore, protein C must be capable of catalytic activity once the site of cleavage at R169 is exposed to solvent, as recently observed for prethrombin-2.29 Other zymogens (eg, chymotrypsinogen) show a minuscule but appreciable level of activity that is the result of the correct architecture of the catalytic triad and open access to the primary specificity pocket.40 The 166VDPR169 sequence in the activation domain of protein C is similar to the 38LDPR41 sequence near the cleavage site of the thrombin receptor PAR1 that activated protein C cuts with a kcat/Km = 1.4mM−1s−1.48 The anionic cages E14e/D14l/E18 of prethrombin-2 and E160/D167/D172 of protein C play the important physiologic role of preventing these zymogens to spontaneously convert to the mature enzyme. If the anionic cage E14e/D14l/E18 is found to play the same role in prothrombin, then a common mechanism emerges where activation is controlled by exposure of a critical Arg residue sequestered within the protein until a trigger ensues. The trigger is provided by physiologic activators: the prothrombinase complex for prothrombin and the thrombin-thrombomodulin complex for protein C. A new paradigm emerges in cofactor-assisted zymogen activation in the blood coagulation cascade, where the cofactor acts on the substrate rather than, or in addition to the enzyme, to promote catalytic conversion of the zymogen to the mature protease.
The ability of prethrombin-2 (or prothrombin) and protein C to autoactivate raises the intriguing possibility that other factors lacking catalytic activity may initiate activation by allosterically exposing the Arg in the activation domain. This could lead to prothrombin conversion to thrombin that bypasses the coagulation cascade, offering an alternative mechanism to that exploited by staphylocoagulase,49 or activation of protein C that does not require thrombomodulin and thrombin. The pathophysiologic implications of prothrombin and protein C variants that spontaneously convert to the mature enzyme could be addressed with mouse models, as recently done for the anticoagulant thrombin mutant W215A/E217A.50 Large-scale production of therapeutically relevant enzymes, such as thrombin and activated protein C, could also be simplified by expressing autoactivating constructs.
Acknowledgments
The authors thank Ms Tracey Baird for her help with illustrations.
This work was supported in part by the National Institutes of Health (research grants HL49413, HL73813, and HL95315).
Footnotes
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: N.P., S.B.-M., and Z.C. performed the research; E.D.C. wrote the manuscript; and all authors designed the research and analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Enrico Di Cera, Department of Biochemistry and Molecular Biology, St Louis University, 1100 Grand Blvd, St Louis, MO 63110; e-mail: enrico@slu.edu.
References
- 1.Page MJ, Di Cera E. Serine peptidases: classification, structure and function. Cell Mol Life Sci. 2008;65(7):1220–1236. doi: 10.1007/s00018-008-7565-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krem MM, Di Cera E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci. 2002;27(2):67–74. doi: 10.1016/s0968-0004(01)02007-2. [DOI] [PubMed] [Google Scholar]
- 3.Gros P, Milder FJ, Janssen BJ. Complement driven by conformational changes. Nat Rev Immunol. 2008;8(1):48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 4.Gohara DW, Di Cera E. Allostery in trypsin-like proteases suggests new therapeutic strategies. Trends Biotechnol. 2011;29(11):577–585. doi: 10.1016/j.tibtech.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arlaud GJ, Barlow PN, Gaboriaud C, Gros P, Narayana SV. Deciphering complement mechanisms: the contributions of structural biology. Mol Immunol. 2007;44(16):3809–3822. doi: 10.1016/j.molimm.2007.06.147. [DOI] [PubMed] [Google Scholar]
- 6.Ponnuraj K, Xu Y, Macon K, Moore D, Volanakis JE, Narayana SV. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol Cell. 2004;14(1):17–28. doi: 10.1016/s1097-2765(04)00160-1. [DOI] [PubMed] [Google Scholar]
- 7.Jing H, Macon KJ, Moore D, DeLucas LJ, Volanakis JE, Narayana SV. Structural basis of profactor D activation: from a highly flexible zymogen to a novel self-inhibited serine protease, complement factor D. EMBO J. 1999;18(4):804–814. doi: 10.1093/emboj/18.4.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narayana SV, Carson M, el-Kabbani O, et al. Structure of human factor D: a complement system protein at 2.0 A resolution. J Mol Biol. 1994;235(2):695–708. doi: 10.1006/jmbi.1994.1021. [DOI] [PubMed] [Google Scholar]
- 9.Forneris F, Ricklin D, Wu J, et al. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science. 2010;330(6012):1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jing H, Babu YS, Moore D, et al. Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity. J Mol Biol. 1998;282(5):1061–1081. doi: 10.1006/jmbi.1998.2089. [DOI] [PubMed] [Google Scholar]
- 11.Banner DW, D'Arcy A, Chene C, et al. The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature. 1996;380(6569):41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 12.Mann KG, Butenas S, Brummel K. The dynamics of thrombin formation. Arterioscler Thromb Vasc Biol. 2003;23(1):17–25. doi: 10.1161/01.atv.0000046238.23903.fc. [DOI] [PubMed] [Google Scholar]
- 13.Di Cera E. Thrombin. Mol Aspects Med. 2008;29(4):203–254. doi: 10.1016/j.mam.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esmon NL, Owen WG, Esmon CT. Isolation of a membrane-bound cofactor for thrombin-catalyzed activation of protein C. J Biol Chem. 1982;257(2):859–864. [PubMed] [Google Scholar]
- 15.Ye J, Esmon NL, Esmon CT, Johnson AE. The active site of thrombin is altered upon binding to thrombomodulin: two distinct structural changes are detected by fluorescence, but only one correlates with protein C activation. J Biol Chem. 1991;266(34):23016–23021. [PubMed] [Google Scholar]
- 16.Le Bonniec BF, Esmon CT. Glu-192–Gln substitution in thrombin mimics the catalytic switch induced by thrombomodulin. Proc Natl Acad Sci U S A. 1991;88(16):7371–7375. doi: 10.1073/pnas.88.16.7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Bonniec BF, MacGillivray RT, Esmon CT. Thrombin Glu-39 restricts the P′3 specificity to nonacidic residues. J Biol Chem. 1991;266(21):13796–13803. [PubMed] [Google Scholar]
- 18.Ehrlich HJ, Grinnell BW, Jaskunas SR, Esmon CT, Yan SB, Bang NU. Recombinant human protein C derivatives: altered response to calcium resulting in enhanced activation by thrombin. EMBO J. 1990;9(8):2367–2373. doi: 10.1002/j.1460-2075.1990.tb07411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson MA, Gerlitz B, Grinnell BW. Enhancing protein C interaction with thrombin results in a clot-activated anticoagulant. Nature. 1992;360(6401):261–264. doi: 10.1038/360261a0. [DOI] [PubMed] [Google Scholar]
- 20.Fuentes-Prior P, Iwanaga Y, Huber R, et al. Structural basis for the anticoagulant activity of the thrombin-thrombomodulin complex. Nature. 2000;404(6777):518–525. doi: 10.1038/35006683. [DOI] [PubMed] [Google Scholar]
- 21.Adams TE, Li W, Huntington JA. Molecular basis of thrombomodulin activation of slow thrombin. J Thromb Haemost. 2009;7(10):1688–1695. doi: 10.1111/j.1538-7836.2009.03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vindigni A, White CE, Komives EA, Di Cera E. Energetics of thrombin-thrombomodulin interaction. Biochemistry. 1997;36(22):6674–6681. doi: 10.1021/bi962766a. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi T, Zushi M, Yamamoto S, Suzuki K. Further localization of binding sites for thrombin and protein C in human thrombomodulin. J Biol Chem. 1990;265(33):20156–20159. [PubMed] [Google Scholar]
- 24.Gerlitz B, Grinnell BW. Mutation of protease domain residues Lys37-39 in human protein C inhibits activation by the thrombomodulin-thrombin complex without affecting activation by free thrombin. J Biol Chem. 1996;271(37):22285–22288. doi: 10.1074/jbc.271.37.22285. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, Bush LA, Pineda AO, Caccia S, Di Cera E. Thrombomodulin changes the molecular surface of interaction and the rate of complex formation between thrombin and protein C. J Biol Chem. 2005;280(9):7956–7961. doi: 10.1074/jbc.M412869200. [DOI] [PubMed] [Google Scholar]
- 26.Yang L, Manithody C, Rezaie AR. Activation of protein C by the thrombin-thrombomodulin complex: cooperative roles of Arg-35 of thrombin and Arg-67 of protein C. Proc Natl Acad Sci U S A. 2006;103(4):879–884. doi: 10.1073/pnas.0507700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Prasad S, Di Cera E, Rezaie AR. The conformation of the activation peptide of protein C is influenced by Ca2+ and Na+ binding. J Biol Chem. 2004;279(37):38519–38524. doi: 10.1074/jbc.M407304200. [DOI] [PubMed] [Google Scholar]
- 28.Wu QY, Sheehan JP, Tsiang M, Lentz SR, Birktoft JJ, Sadler JE. Single amino acid substitutions dissociate fibrinogen-clotting and thrombomodulin-binding activities of human thrombin. Proc Natl Acad Sci U S A. 1991;88(15):6775–6779. doi: 10.1073/pnas.88.15.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pozzi N, Chen Z, Zapata F, Pelc LA, Barranco-Medina S, Di Cera E. Crystal structures of prethrombin-2 reveal alternative conformations under identical solution conditions and the mechanism of zymogen activation. Biochemistry. 2011;50(47):10195–10202. doi: 10.1021/bi2015019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marino F, Pelc LA, Vogt A, Gandhi PS, Di Cera E. Engineering thrombin for selective specificity toward protein C and PAR1. J Biol Chem. 2010;285(25):19145–19152. doi: 10.1074/jbc.M110.119875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rezaie AR, Esmon CT. The function of calcium in protein C activation by thrombin and the thrombin-thrombomodulin complex can be distinguished by mutational analysis of protein C derivatives. J Biol Chem. 1992;267(36):26104–26109. [PubMed] [Google Scholar]
- 32.Dang OD, Vindigni A, Di Cera E. An allosteric switch controls the procoagulant and anticoagulant activities of thrombin. Proc Natl Acad Sci U S A. 1995;92(13):5977–5981. doi: 10.1073/pnas.92.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rezaie AR, Yang L. Thrombomodulin allosterically modulates the activity of the anticoagulant thrombin. Proc Natl Acad Sci U S A. 2003;100(21):12051–12056. doi: 10.1073/pnas.2135346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected by oscillation methods. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Bailey S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12(4):345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 38.Dang QD, Di Cera E. Chromogenic substrates selective for activated protein C. Blood. 1997;89(6):2220–2222. [PubMed] [Google Scholar]
- 39.Miletich JP, Broze GJ., Jr Beta protein C is not glycosylated at asparagine 329: the rate of translation may influence the frequency of usage at asparagine-X-cysteine sites. J Biol Chem. 1990;265(19):11397–11404. [PubMed] [Google Scholar]
- 40.Wang D, Bode W, Huber R. Bovine chymotrypsinogen A X-ray crystal structure analysis and refinement of a new crystal form at 1.8 A resolution. J Mol Biol. 1985;185(3):595–624. doi: 10.1016/0022-2836(85)90074-9. [DOI] [PubMed] [Google Scholar]
- 41.Bode W, Turk D, Karshikov A. The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1992;1(4):426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gandhi PS, Chen Z, Di Cera E. Crystal structure of thrombin bound to the uncleaved extracellular fragment of PAR1. J Biol Chem. 2010;285(20):15393–15398. doi: 10.1074/jbc.M110.115337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stubbs MT, Oschkinat H, Mayr I, et al. The interaction of thrombin with fibrinogen: a structural basis for its specificity. Eur J Biochem. 1992;206(1):187–195. doi: 10.1111/j.1432-1033.1992.tb16916.x. [DOI] [PubMed] [Google Scholar]
- 44.Sadasivan C, Yee VC. Interaction of the factor XIII activation peptide with alpha-thrombin: crystal structure of its enzyme-substrate analog complex. J Biol Chem. 2000;275(47):36942–36948. doi: 10.1074/jbc.M006076200. [DOI] [PubMed] [Google Scholar]
- 45.Gandhi PS, Chen Z, Appelbaum E, Zapata F, Di Cera E. Structural basis of thrombin-PAR interactions. IUBMB Life. 2011;63(6):375–382. doi: 10.1002/iub.461. [DOI] [PubMed] [Google Scholar]
- 46.Pineda AO, Carrell CJ, Bush LA, et al. Molecular dissection of Na+ binding to thrombin. J Biol Chem. 2004;279(30):31842–31853. doi: 10.1074/jbc.M401756200. [DOI] [PubMed] [Google Scholar]
- 47.Cleary DB, Trumbo TA, Maurer MC. Protease-activated receptor 4-like peptides bind to thrombin through an optimized interaction with the enzyme active site surface. Arch Biochem Biophys. 2002;403(2):179–188. doi: 10.1016/s0003-9861(02)00220-5. [DOI] [PubMed] [Google Scholar]
- 48.Di Cera E. Know your APC. Blood. 2009;113(23):5699–5700. doi: 10.1182/blood-2009-03-211292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedrich R, Panizzi P, Fuentes-Prior P, et al. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003;425(6957):535–539. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 50.Flick MJ, Chauhan AK, Frederick M, et al. The development of inflammatory joint disease is attenuated in mice expressing the anticoagulant prothrombin mutant W215A/E217A. Blood. 2011;117(23):6326–6337. doi: 10.1182/blood-2010-08-304915. [DOI] [PMC free article] [PubMed] [Google Scholar]