Abstract
Systemic lupus erythematosus (SLE) is a complex autoimmune disease with a strong genetic component that determines risk. A common three single-nucleotide polymorphism (SNP) haplotype of the complement receptor 2 (CR2) gene has been associated with increased risk of SLE (Wu et al., 2007) (Douglas et al., 2009), and a less common haplotype consisting of the major allele at SNP1 and minor alleles at SNP2 and 3 confers protection (Douglas et al., 2009). SNP1 (rs3813946), which is located in the 5´ untranslated region (UTR) of the CR2 gene, altered transcriptional activity of a CR2 promoter-luciferase reporter gene construct transiently transfected into a B cell line (Wu et al., 2007) and had an independent effect in the protective haplotype (Douglas et al., 2009). In this study, we show that this SNP alters transcriptional activity in a transiently transfected non B-cell line as well as in stably transfected cell lines, supporting its relevance in vivo. Furthermore, the allele at this SNP affects chromatin accessibility of the surrounding sequence and transcription factor binding. These data confirm the effects of rs3813946 on CR2 transcription, identifying the 5´UTR to be a novel regulatory element for the CR2 gene in which variation may alter gene function and modify the development of lupus.
Keywords: Human, B cells, Systemic Lupus Erythematosus, Gene Regulation, Transcription Factors, Complement Receptor 2
1. Introduction
Complement receptor type 2 (CR2/CD21) is a 145 kDa glycoprotein that binds C3 degradation products generated during complement activation, specifically iC3b, C3dg, and C3d. It has a number of important functions in normal immunity, including cooperating with the B cell receptor to activate B cells, targeting antigen to follicular dendritic cells in secondary lymphoid organs, processing and presenting complement-coated antigens to T cells, and shaping the natural antibody repertoire [reviewed in (Holers, 2005)]. Its critical role in host defense is apparent from studies showing blunted IgG responses in Cr2-deficient mice after immunization with T-dependent or –independent antigens (Ahearn et al., 1996; Croix et al., 1996; Haas et al., 2002; Molina et al., 1996).
CR2 is also believed to participate in the development of autoimmune disease. CR2 levels are decreased by ~50% in patients with systemic lupus erythematosus (SLE) (Marquart et al., 1995; Wilson et al., 1986), and SLE-prone MRL/lpr mice demonstrate a similar phenotype prior to the onset of clinical evidence of disease (Takahashi et al., 1997), suggesting a role in disease pathogenesis. Mice rendered deficient in Cr2 develop either enhanced or diminished symptoms of autoimmunity, depending on the disease model (Del Nagro et al., 2005; Kaya et al., 2001; Prodeus et al., 1998; Wu et al., 2002). Furthermore, Cr2 is a strong candidate gene for disease susceptibility in the NZM2410 murine model of SLE due to gene polymorphisms that alter the structure and function of the CR2 protein product (Boackle et al., 2001). The mechanism by which CR2 participates in the onset and evolution of autoimmune disease is not known.
CR2 expression is restricted primarily to B cells and follicular dendritic cells, and its expression on B cells is tightly regulated, first detected at the late immature/transitional stage, increasing as the cells mature into follicular and marginal zone B cells respectively (Takahashi et al., 1997; Tedder et al.,1984a; Thomas et al., 2006), and disappearing as cells differentiate into plasma cells (Tedder et al., 1984b). The transition of human transitional B cells from CR2low to CR2high is believed to be a checkpoint for the deletion of self-reactivity (Suryani et al., 2010). Surface expression of CR2 is closely linked to the presence of mature mRNA, which is controlled primarily at the level of transcription (Makar et al., 1998a)). Critical elements that regulate basal transcription of the human CR2 gene have been targeted to the region spanning -315 to +75 of the genomic sequence. Although this region of the CR2 promoter is sufficient to drive basal transcription (Ulgiati et al., 2002), the strict developmental regulation and inducible expression of CR2 require additional elements (Makar et al., 1998a; Makar et al., 2001; Tolnay et al., 2002; Vereshchagina et al., 2001). A number of regulatory elements have been identified in the human CR2 gene, including sequences homologous to TATA, SP1, AP-2, AP-1 and Ig enhancer E motif DNA protein binding sites in the proximal promoter (Rayhel et al., 1991; Ulgiati et al., 2002) as well as distal regions that control inducible expression including a NF-κB site (Tolnay et al., 2002). Furthermore, appropriate tissue-specific expression of CR2 requires an intronic silencer element (Makar et al., 1998b). However, the 5´UTR of the CR2 proximal promoter was not known to be relevant in the regulation of CR2 transcription until our previous report, in which we showed that a lupus-associated polymorphism (rs3813946) located in this region altered transcriptional activity of a CR2 promoter-luciferase reporter gene construct that was transiently transfected into a B cell line (Wu et al., 2007).
In our initial study conducted in Caucasian and Chinese lupus simplex families, we demonstrated that the major allele of rs3813946 was transmitted preferentially from heterozygous parents to their affected offspring with a P value of 0.007 and an estimated odds ratio of 1.53 (1.11–2.10) (Wu et al., 2007). In our subsequent case-control study conducted in unrelated Caucasian subjects, we identified a protective CR2 haplotype that contains the major allele of rs3813946 and the minor alleles of 3 other exonic SNPs (haplotype P value 0.003, odds ratio 0.79 [0.68–0.92]), on which the major allele of rs3813946 exhibited an independent genetic effect although it was not associated with increased risk of lupus in single SNP analysis. (Douglas et al., 2009). These data suggest that the ability of the major allele of rs3813946 to alter transcription could modify the effects of the causal SNP(s) in the protective CR2 haplotype. In order to verify that rs3813946 alleles alter gene function and to further characterize the role of the 5´UTR of CR2 in transcriptional regulation, we performed the additional studies outlined in this report. Herein, we show allelic differences of rs3813946 in a second non-CR2 expressing cell line as well as in a stably integrated reporter construct. Furthermore, its variants alter chromatin accessibility and binding of several proteins, which may explain its transcriptional effects. These data confirm a functional effect for this polymorphism and demonstrate the contribution of the 5´UTR in which it is located to the regulation of CR2 transcription, supporting further study of the effects of this region on the complex developmental and tissue-specific expression of CR2.
2. Materials and Methods
2.1. Cell lines and culture conditions
The human Burkitt’s lymphoma cell line Raji and the human erythroleukemic cell line K562 were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cell lines were maintained at 37°C with 5% CO2 in RPMI 1640 with L-glutamine (Gibco BRL, Invitrogen Life Technologies, Melbourne, Australia) supplemented with 10% FBS (Gibco BRL, Invitrogen Life Technologies), 100 µg/ml streptomycin, and 100 IU/ml penicillin (Gibco BRL, Invitrogen Life Technologies).
2.2. Construction of CR2 mutant reporter constructs
A NheI/XhoI fragment of the CR2 promoter containing the −315/+75 of the CR2 promoter was cloned into the luciferase reporter pGL3-basic vector (Clontech Laboratories, Palo Alto, CA, USA) as described previously (Ulgiati et al., 2002). Site-directed mutagenesis was performed using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA, USA), which enabled the incorporation of the minor +21C allele at rs3813946 in the CR2 promoter-luciferase reporter fusion construct. Correct orientation and sequence of constructs was verified by restriction enzyme digestion and nucleotide sequence analysis.
2.3. Transfection and quantitation of promoter activity
Transfections were performed by harvesting cells (Raji or K562) grown to log phase using the Superfect reagent (QIAgen, Valencia, CA, USA) according to the manufacturer’s specifications with plasmid DNA prepared using Endofree Maxiprep-500 columns (QIAgen). Briefly, 4 µg of plasmid DNA and 120 ng of pRL-TK (Renilla) control vector were complexed in combination with Superfect reagent for 10 min at room temperature. The transfection complexes were added to suspensions of cells in 24-well plates to a final density of 5 × 105 cells/ml. Cell lysates from the transfected cells were prepared and assayed for both Firefly and Renilla luciferase using the Dual-Luciferase Reporter Assay system according to the manufacturer’s instructions (Promega, Madison, WI, USA). All transfection data shown are the mean of 3–5 independent transfections, with n values shown in each experiment. Promoter activity is expressed as relative Firefly luciferase activity normalized against Renilla luciferase activity.
2.4. Chromatin accessibility assays
Chromatin accessibility measured by real-time PCR (CHART-PCR) assays were performed as described previously (Cruickshank et al., 2008; Rao et al., 2001) using 1.25 × 106 nuclei with 5 U MNase/ml (Worthington Biochemicals, Lakewood, NJ, USA). Following digestion of nuclei, DNA was recovered using the QIAamp DNA blood mini-kit (QIAgen) and used in quantitative (Q)-PCR assays with primers targeting the β-actin promoter (forward: 5´-CAGCACCCCAAGGCGGCCAACG-3´; reverse: 5´-GCAACTTTCGGAACGGCGCACGC-3´), PAX7 promoter (forward: 5´-CCGAACCTATCAGATCGCGCTCAC-3´; reverse: 5´-GTCACCCCCTGTCTCCTCCGTCCAG-3´), CR2 upstream region (forward: 5´-GATGTGGATTCGCCTATCCC-3´; reverse: 5´-CCCTGAAGGTAGTGGTGTAAAGC-3´) and CR2- reporter transgene (forward: 5´-GATGTGGATTCGCCTATCCC-3´; reverse 5´-TCTTCCAGCGGATAGAATG-3´). Q-PCR was performed on the CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) using the QuantiTect™ SYBR® Green PCR kit (QIAgen) according to the manufacturers’ instructions with 0.5 µM each primer in 20 µl reaction. Thermal-cycling conditions were as follows: 95°C for 15 min; followed by 40 cycles of 95°C for 15 s, 57°C for 15 s and 72°C for 15 s; followed by a melt-curve cycle (gradual increase in temperature from 60°C up to 99°C). Acquisition of sample fluorescence occurred after 72°C cycles and at 1°C increments during melt-curve analysis. The efficacy of Q-PCR primers was verified by amplification of serially diluted genomic DNA or plasmid DNA to ensure linear detection of template and by agarose gel electrophoresis of PCR products and routine melt-curve analysis to ensure specificity.
2.5. Preparation of nuclear and cytosolic proteins
For preparation of nuclear extract (NE), 4 × 107 cells from Raji and K562 were harvested and prepared essentially as described previously (Li et al., 1991). Fractions were snap-frozen in liquid N2 and stored at −80°C. Determination of protein concentration of the fractions was performed using the Bio-Rad protein assay kit (Bio-Rad) according to the manufacturer’s protocol. For preparation of cytosolic extract (CE), immediately following wash steps, cell pellets were resuspended in 500 µL of Cytoplasmic Extraction Buffer (CEB; 10 mM HEPES [pH 7.9], 3 mM MgCl2, 14 mM KCl, 5 % [v/v] glycerol, 0.2 % [v/v] NP-40) and incubated on ice for 20 min. The nuclei were pelleted and the supernatant containing the CE was removed and snap-frozen in liquid N2 and stored at −80°C.
2.6. EMSA analysis
For EMSA, 10–20 µg NE were preincubated on ice for 10 min together with 1 µg poly(dI-dC) in a binding buffer consisting of 4% Ficoll (Amersham Biosciences, Piscataway, NJ, USA), 20 mM HEPES (pH 7.9), 1 mM EDTA, 1 mM DTT, and 50 mM KCl. When required, competitor oligonucleotides were incubated with NE for 30 min on ice. The NE was then incubated with 80 fmol of 32P-labeled oligonucleotide for 30 min on ice before loading onto a 6% polyacrylamide gel. The gel was electrophoresed at 150 V using 0.25x Tris-taurin-EDTA running buffer. EMSA gels were dried under vacuum and exposed to x-ray film. All double-stranded oligonucleotides were end labeled using γ-[32P]ATP and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA). Probes were designed using the +10 to +37 region of the CR2 promoter for the major (forward: 5´-CTGCTCCAGCCTTGCCCTCCCAGAGCTG-3´; reverse 5´-CAGCTCTGGGAGGGCAAGGCTGGAGCAG-3´) and minor (forward: 5´-CTGCTCCAGCCCTGCCCTCCCAGAGCTG-3´; reverse 5´-CAGCTCTGGGAGGGCAGGGCTGGAGCAG-3´) rs3813946 alleles.
2.7. REMSA analsyis
In vitro transcription reactions were performed according to the manufacturers instructions to generate single-stranded RNA probes using T7 RNA polymerase (Promega) in the presence of α-[32P]UTP (Amersham Pharmacia Biotech AB). For binding reactions, 10 µg of either NE or CE was used along with 5 × 104 cpm radiolabelled probe and CEB in a final volume of 10 µl and incubated at 22°C for 10 min. If required, increasing amounts of tRNA was added prior to incubation to determine sequence specificity. Binding reactions were incubated for a further 10 min with the addition of 1U RNase T1 to degrade unbound RNA following addition of RNA-loading dye. Electrophoresis was carried out on a 6% non-denaturing polyacrylamide mini-gel at 4°C for 25 min at 200 V. Gels were fixed in 10% (v/v) isopropanol/7% (v/v) acetic acid for 10 min, air-dried for 1 hour at 80°C under vacuum and the RNA protein complexes visualized by exposure to a phosphoimaging plate (Molecular Dynamics, USA) overnight and developed with a PhosphorImager 445 SI (Molecular Dynamics, USA). Binding reactions were performed as described above using 1 × 105 cpm radiolabelled RNA oligonucleotide and 20 µg of CE per reaction. Reactions were then irradiated for 2 min using a UV Crosslinker (Stratagene Model 1800, USA). This step degraded all labeled RNA except that which was covalently cross-linked to protein. SDS was added to each reaction which was incubated for 3 min at 80°C then loaded on a 12.5% polyacrylamide gel at 4°C for 20 min at 200 V. The protein-RNA complexes were then visualized as described for REMSA as above.
2.8. Chromatin immunoprecipitation (ChIP)
ChIPs were performed based on the Upstate Biotechnology (Millipore, Lake Placid, NY) ChIP assay kit protocol with modifications (Cruickshank et al., 2009). Briefly, ChIP assays used 107 cells per immunoprecipitation, cross-linking with 1% formaldehyde in PBS at room temperature for 15 min and then adding glycine to 0.125 M for 5 min. After washing with PBS, nuclei were isolated by incubating cells for 15 min in 10 ml NP-40 lysis (10 mM Tris-Cl pH 8.0, 10mM NaCl, 3mM MgCl2, 0.5% NP-40, 0.15 mM spermine and 0.5 mM spermidine) then dounce homogenizing 10 times on ice. Nuclei were collected by centrifugation, lysed in SDS lysis buffer (5×107 cells per ml; 1% SDS, 10 mM EDTA, 50 mM Tris-Cl, pH 8.1) and sonicated 8 times on ice with output set at 3.5. Soluble chromatin was isolated, diluted as described and incubated overnight with 20 µg antibodies; α- C/EBP-β (Santa Cruz, Cat#-150X), α-PAX5 (Santa Cruz, Cat#sc-1974X), α-MAZ (Santa Cruz, Cat#sc-28745X), IgG control (BD Biosciences PharMingen, Cat#18413) or no antibody. An additional untreated aliquot of chromatin was used to monitor DNA shearing by agarose gel electrophoresis and to generate standard curves for ChIP quantitation. Immune complexes were collected with 60 µl Salmon Sperm DNA/Protein A/G-agarose beads (Upstate Biotechnology) for 1 h at 4°C and washed according to the manufacturer’s specifications. Bound complexes were extracted from antibody twice with 250 µl elution buffer (1% SDS, 0.1 M NaHCO3) and cross-links were reversed by adding 25 µl 4 M NaCl and heating to 65°C for 6 h. DNA was extracted by adding 20 µl 1M Tris-HCl (pH 6.4), 20 µl 0.5 M EDTA (pH 8.1) and 2 µl 20 mg/ml Proteinase K and incubating for 10 h at 42°C, before using the QIAamp DNA blood mini-kit (QIAGEN) to elute DNA in 100 µl 0.5 × TE. Q-PCR was performed using 4 µl template, with the CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories) and analyzed using the CFX Manager Software v1.0 (Bio-Rad Laboratories). Primer pairs spanning the +21 SNP (forward: 5`-GCTCACAGCTGCTTGCTGCT-3` and reverse: 5`-GCATCGCGGCACAGAAACTTTC-3`) were used to detect CR2-5`UTR-specific enrichment with an annealing temperature of 55°C.
2.9. Statistical analyses
Statistical significance (*) was determined by Student’s unpaired t-test using a confidence interval of 95% (P<0.05). All values described represent the mean ± SEM; statistics and graphs were generated using GraphPad Prism v4 (GraphPad, San Diego CA).
3. Results
3.1. Allele-specific transcriptional activity at rs3813946
Previous work investigating the effect of the SNP at position +21 in the 5`UTR of the CR2 gene (rs3813946; T+21C) (Figure 1) revealed a 2-fold decrease in transcription associated with the minor C allele as compared to the major T allele (Wu et al., 2007). These data were obtained using +21 biallelic forms of the −315 to +75 proximal CR2 promoter fused with luciferase as a measure of gene expression and involved the transient transfection of these constructs into the CR2 expressing B cell line, Raji. We sought to confirm the relevance of this observation by performing additional studies in CR2 non expressing cell lines. As in the Raji B cell line, the T+21C transition reduced transcription by more than 40% in the transiently transfected CR2−ve erythroleukemic cell line, K562. To clarify whether these effects persisted upon integration of the constructs into the genome, we used a previously generated isogenic K562 host cell line containing a single FRT site (Karimi et al., 2007) to derive stable reporter clones carrying either of the +21 biallelic forms (+21T/C) of the −315 to +75 proximal CR2 promoter (Figure 2B). These reporter cell lines were identical with respect to integration site of reporter constructs, contained a single copy of the reporter gene and differed only at rs3813946 (+21T/C). Using five independently generated reporter cell lines for each allelic variant, we observed a similar level of inhibition (greater than 40%) due to the T+21C transition (Figure 2C). In conclusion, the CR2 promoter-luciferase constructs yielded comparable results across CR2-expressing and non-expressing cell lines, when either transiently expressed or stably integrated, confirming the previously observed effect of rs3813946 on the transcriptional activity of the CR2 promoter.
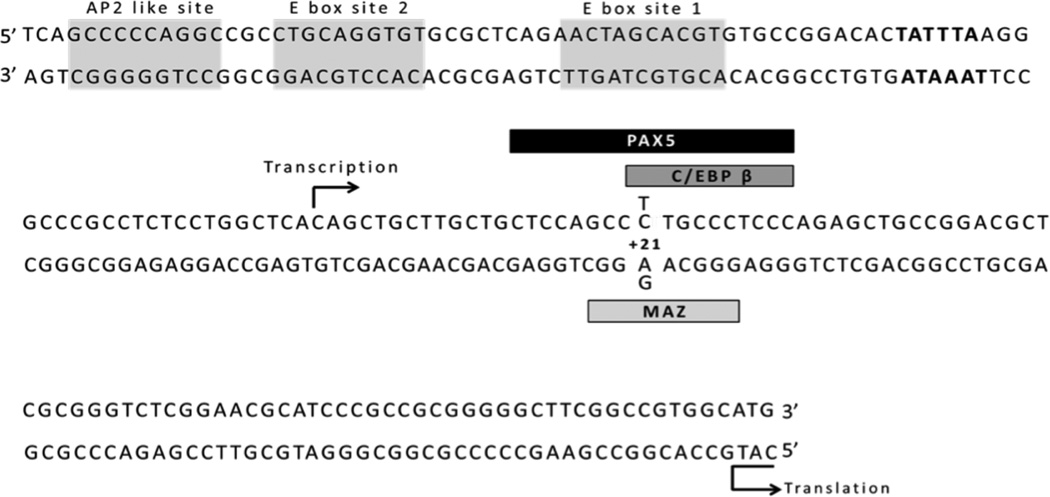
Figure 1. Location of rs3813946 within the proximal promoter region of the human CR2 gene.
The nucleotide sequence from the -133 upstream promoter region to the translation start site of the CR2 gene is shown. Previously identified functional elements, an AP2-like site and the Ebox1 and Ebox2 are shaded. Recently identified transcription factor binding sites for PAX5, C/EBP β and MAZ are shown as boxed regions. The transcription and translation start site is indicated with arrows. The putative TATA box binding site is in bold font. The single-nucleotide polymorphism studied herein (rs3813946) is indicated in the sequence at its nucleotide position (+21T/C).
Figure 2. The minor C allele of rs3813946 decreases proximal promoter activity.
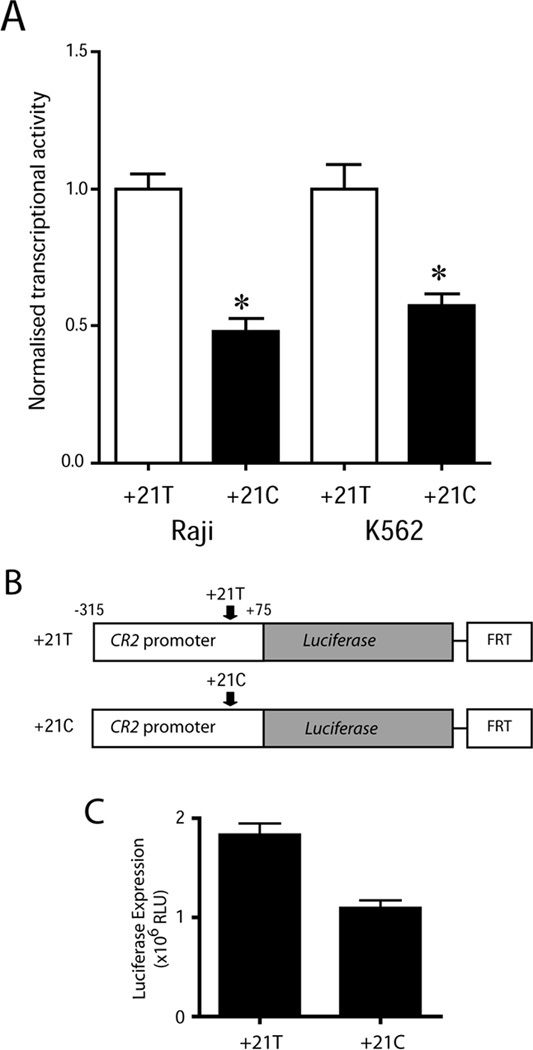
(A) Transcriptional activity of luciferase constructs containing the −315/+75 proximal promoter sequence with the major T (+21T) or minor C (+21C) allele of rs3813946. Constructs were transiently transfected into CR2-negative K562 cells. Results shown represent mean transcriptional activity +SEM (n=6–9) and are expressed as normalized transcriptional activity of each construct relative to the +21T construct. * p < 0.01 when comparing normalized transcriptional activity of the +21C to the +21T construct. (B) Representation of the +21 T/C-luciferase CR2 promoter reporter constructs. Both constructs were integrated into the K562 cell line using FLP recombination. (C) K562 cells containing the stably integrated CR2 promoter constructs were isolated and luciferase activity in cell lysates was assessed. Results show mean luciferase expression ±SEM (n=5).
3.2. Enhanced chromatin accessibility correlates with transcriptional activity due to rs3813946
We next wished to determine if the change in transcriptional activity due to rs3813946 correlated with changes to chromatin structure of reporter gene sequences in stably transfected cells. We performed chromatin accessibility assays using the T/C biallelic stable CR2-reporter K562 cell lines described and assayed the accessibility of a range of endogenous sequences (control genomic regions) in addition to the CR2-luciferase transgene sequences (Figure 3A). Our results show that the β-actin promoter was highly accessible while the PAX7 promoter was protected from MNase digestion in both CR2 reporter cell lines (Figure 3B). Furthermore, an upstream region of the endogenous CR2 promoter (spanning −1215 to -1052) showed similar levels of digestion in both T/C biallelic stable CR2-reporter cell lines (Figure 3B). Therefore, in both CR2-reporter cell lines, CHART-PCR assays revealed differences in accessibility at a broadly expressed promoter (β-actin) compared to a developmentally restricted promoter (PAX7), as well as comparable levels of accessibility at the endogenous CR2 locus. In contrast, chromatin accessibility across the reporter transgene (which included rs3813946) was significantly higher in the cell line carrying the major +21T allele (2B; p<0.005). Thus, inhibition of transcriptional activity due to the minor allele of rs3813946 in CR2-gene reporter cell lines is accompanied by reduced accessibility of promoter sequences to nuclear factors in vivo.
Figure 3. Reduced chromatin accessibility in stably transfected reporter constructs containing the minor C allele at rs3813946 correlates with transcriptional activity.
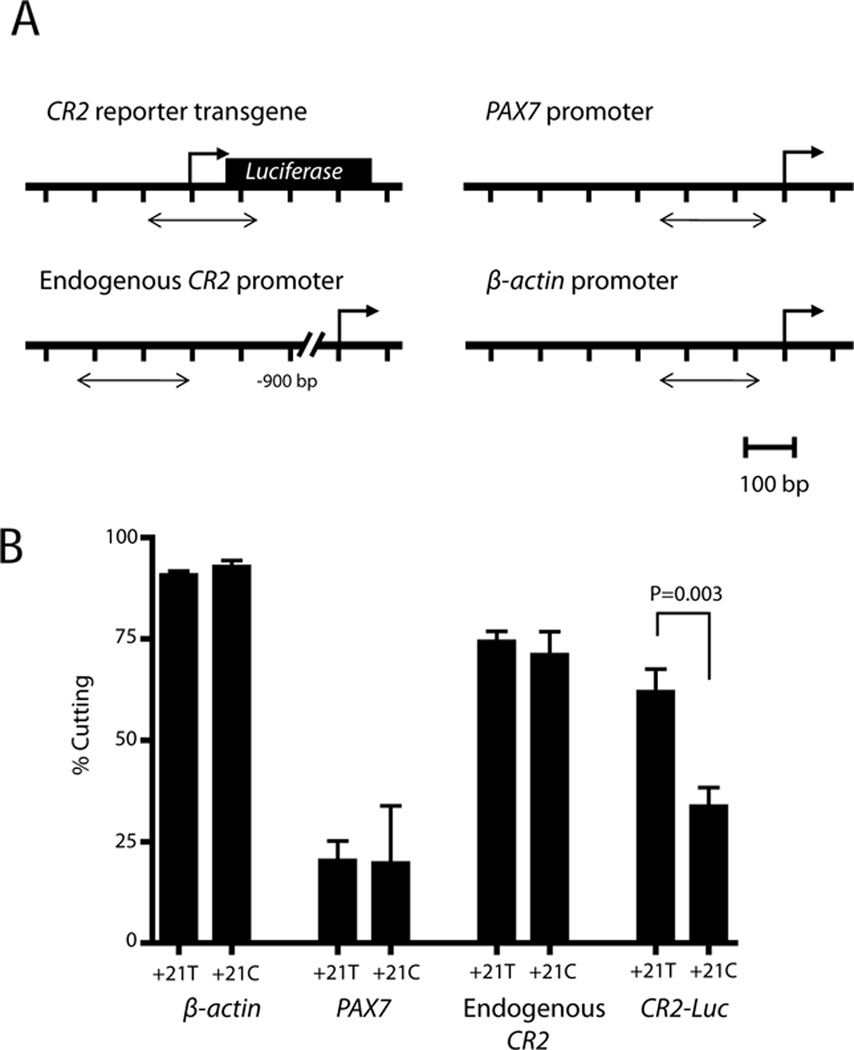
(A) Location of primers used to assess chromatin accessibility by CHART-PCR. (B) Pairs of “Uncut” and “Cut” samples from CHART-PCR assays were used as template in Q-PCR reactions with primer pairs targeting promoter regions of β-ACTIN (−239 to −62), PAX7 (−228 to −64), upstream CR2 (−1215 to −1052), and CR2-reporter transgene sequences (238 bp amplicon including −113 to +75 of the CR2 promoter sequence). MNase chromatin accessibility is expressed as the percentage cutting and plotted as the mean ± SEM from independent pairs of “Uncut” and “Cut” samples (n=6).
3.3. Allele-specific DNA binding factors at rs3813946
Next, we determined whether the allelic variation at rs3813946 altered binding of specific transcription factors. EMSA was employed using Raji nuclear extracts (NE) and dsDNA probes spanning the −5 to +45 region of the 5`UTR containing either the T or C allele at rs3813946 (+21). We observed the presence of five major DNA-binding complexes (Figure 4; complexes A–E) using the probe containing the major T allele at rs3813946. These complexes were specific as they were demonstrated in the presence of scrambled oligonucleotides and their detection was proportionally reduced by the incorporation of increasing amounts of unlabeled oligonucleotide. The EMSA profile using the labeled probe harboring the minor C allele showed distinct changes: complexes A, C, and D were less intense suggesting a difference in binding strength, whereas complexes B and E were completely abolished. These results demonstrate that in vitro, the allele present at rs3813946 alters transcription factor binding strength and influences the formation of distinct DNA-protein complexes, both of which could result in the differences in transcriptional activity associated with this polymorphism.
Figure 4. The T to C transition at rs3813946 alters transcription factor binding in vitro.
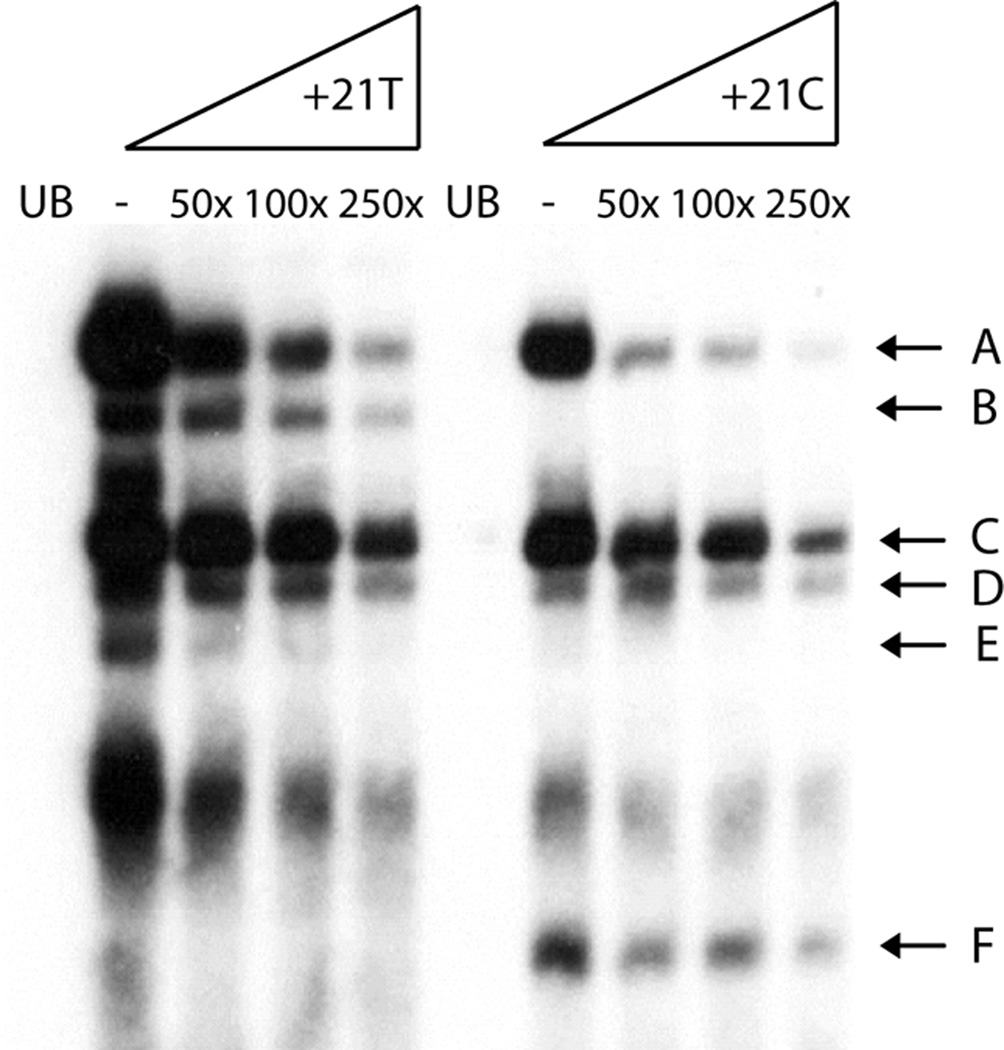
EMSA analysis of the region surrounding rs3813946. Oligonucleotides utilized correspond to +10 to +37 of the proximal promoter sequence and contained either a T (+21T) or a C (+21C) at rs3813946. Nuclear extracts from Raji B cells were pre-incubated with poly(dI-dC) in binding buffer to reduce detection of non-specific DNA-protein interactions. Increasing fold molar excess of unlabeled selfcompetitor oligonucleotide demonstrate the presence of five specific protein-DNA complexes (A–E). Unbound oligonucleotides are represented as UB.
3.4 Sequence specific RNA binding factors at the T+21C SNP show no obvious allelic variation
Since the +21 SNP is located within the 5`UTR we next wished to determine if proteins binding the mRNA encoded by this region differed between the allelic variants. The variant sequences of encoded mRNAs show a substitution of cytosine with uracil due to the T+21C transition (Figure 5A). We asked if there were detectable differences in RNA-protein binding affinity between these allelic forms using REMSA analysis. Single-stranded (ss)RNA probes spanning +6 to +26 (Figure 5A; underlined sequence denotes probes) were generated with either the +21U or +21C and incubated together with either cytoplasmic extracts (CE) or NE. Using increasing amounts of tRNA as a cold competitor, we first determined that CE from Raji cells form three sequence specific RNA-protein complexes (Figure 5B; complexes A–C).
Figure 5. Absence of allele-specific binding of proteins to the 5`UTR of the CR2 mRNA.
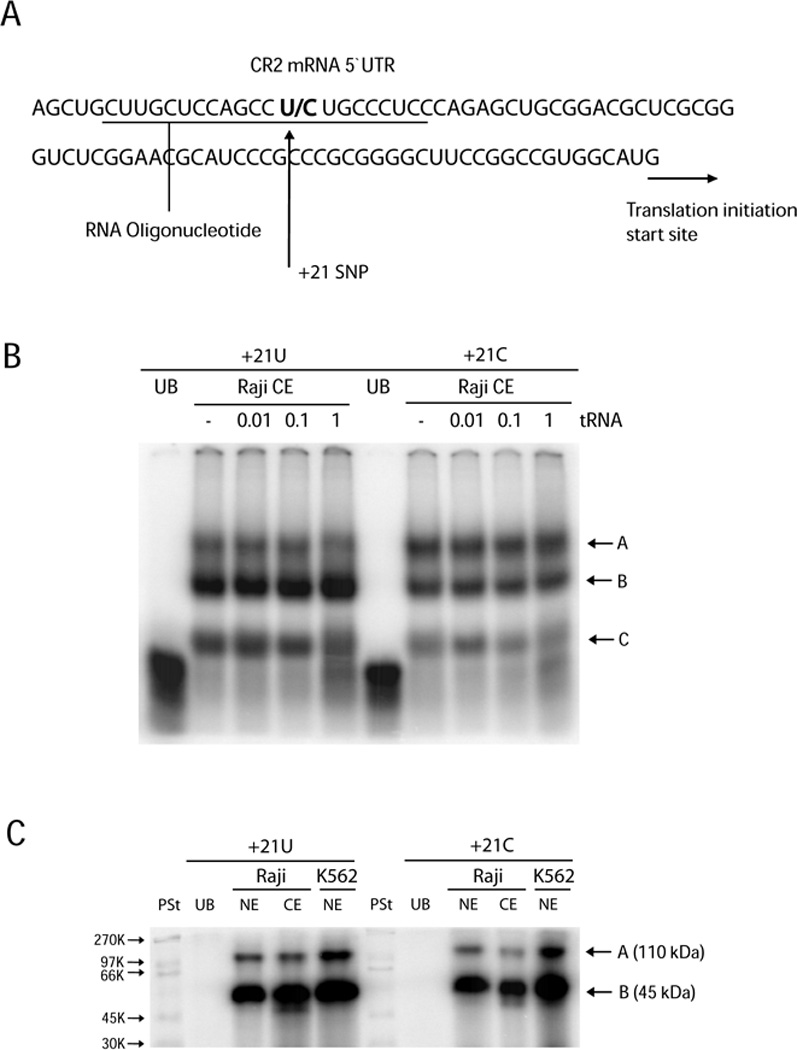
(A) The location of the +21T/C SNP within the transcribed region of the CR2 gene. Single-stranded 20 bp RNA oligonucleotides used for RNA-EMSA (REMSA) corresponded to +6 to +26 of the proximal promoter sequence (underlined). (B) REMSAs were performed using RNA oligonucleotides containing either a U (+21U) or a C (+21C) at position +21 and Raji cytoplasmic extracts (Raji CE). Increasing concentrations of tRNA were used for competition analysis. Unbound oligonucleotides are represented as UB. (C) UV crosslinking with Raji and K562 cytoplasmic (CE) and nuclear (NE) extracts identified two proteins bound to both CR2 transcripts in a sequence-specific manner (A and B). PSt denotes molecular weight protein standard.
As a step towards characterizing proteins which may bind this sequence, ssRNA binding reactions were performed using nuclear and CE from Raji cells (as well as NE from K562 cells) together with the allelic variant +6 to +26 probes that were subsequently treated by UV irradiation to cross-link RNA-protein complexes and degrade RNA that was not covalently bound. The resultant protein-RNA complexes were denatured and analyzed by PAGE. We detected two sequence-specific proteins with approximate molecular weights of 45kDa and 110kDa. These protein-RNA complexes were observed in both nuclear and cytoplasmic fractions of Raji cells using either of the allelic forms (+21 U/C) of the RNA probe (Figure 5C; lanes 3, 4, 8 and 9). The same protein-RNA complexes were observed in K562 NE suggesting that these interactions are not specific to B cells. Therefore, sequence specific RNA-binding proteins bound the region spanning the +21 SNP in a cell-type independent manner; however, these interactions did not show allelic variation.
3.5 In silico analysis of transcription factor binding sites
To determine whether the interval containing rs3813946 contains transcription factor binding motifs, TransFac database searches [v8.3; (Farre et al., 2003; Messeguer et al., 2002)] were performed using the 28-mer oligonucleotide sequences used for EMSA and the default values of TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html). This region contained binding sites for the B cell lineage specific activator PAX5, tumor protein p53, and the alpha isoform of the glucocorticoid receptor (GRalpha) (Figure 1) regardless of the sequence at rs3813946 (Table 1). In addition, binding sites for C/EBP-β and AP2 were identified in the presence of the major T allele and for ETF and the MYC-associated zinc finger protein (MAZ) in the presence of the minor C allele (Table 1). These factors all had threshold scores ≥ 90.9 and were the only factors whose binding site included rs3813946. Both C/EBP-β and an AP2-like element have previously been shown to bind upstream regions of the CR2 promoter (Cruickshank et al., 2009; Ulgiati et al., 2002), whereas ETF and MAZ, (Figure 1) which are typically involved in the initiation of transcription of genes that lack a functional TATA recognition sequence, have not been reported to participate in the regulation of CR2 expression.
Table 1.
Predicted transcription factor binding sites in 5´ UTR flanking rs3813946
| rs3813946 | |
|---|---|
| T | C |
| Pax5 P53 GR-Alpha AP2 |
Pax5 P53 GR-Alpha MAZ |
| C/EBP | ETF |
3.6 ChIP analysis of transcription factors binding to the 5` UTR
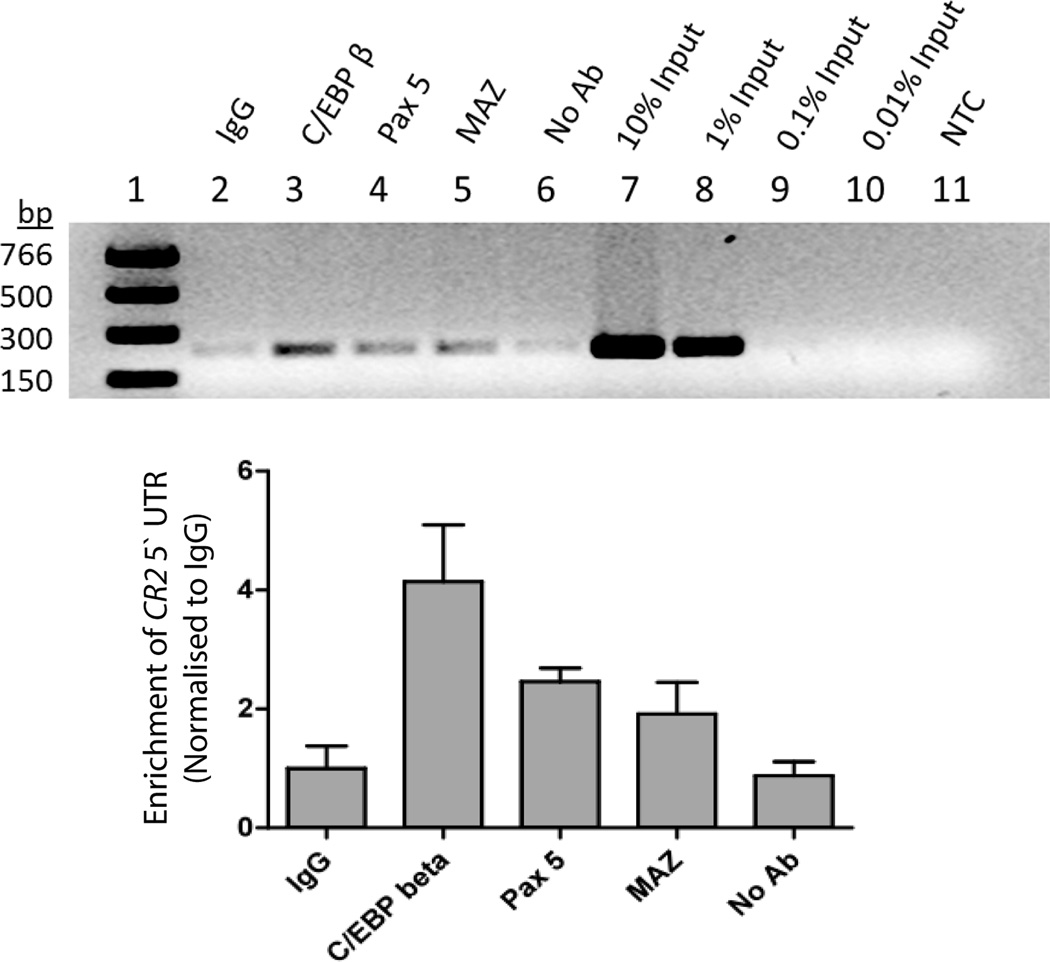
To test the functional relevance of transcription factors identified as containing consensus sites across the sequence containing rs3813946, we performed chromatin immunoprecipitation (ChIP) in Raji cells, hich are homozygous for the major allele at this SNP, using antibodies to C/EBP-β, PAX5, and MAZ. The sequence flanking rs3813946 was enriched in ChIP assays that included each of these antibodies (Figure 6). As a positive control for C/EBP-β specificity, the BCL-2 promoter sequence was also enriched (data not shown) (Heckman et al., 2000; Heckman et al., 2003) and as a positive control for PAX5 specificity, the CD19 promoter was enriched (data not shown) (Bougel et al., 2010). These data confirm the in silico analyses, and provide further support for a role for the 5´UTR in the transcriptional regulation of the CR2 gene.
Figure 6. Evidence that C/EBP β, PAX5 and MAZ interacts with the CR2 5` UTR in Raji cells.
ChIP assays demonstrate specific enrichment of CR2 5` UTR sequences in Raji cells using antibodies targeting C/EBP β, Pax5 and MAZ compared to IgG and no antibody controls. (A) Gel analysis of QPCR products from a representative ChIP assay showing PCR marker (lane 1), IgG (lane 2), C/EBP β (lane 3), PAX5 (lane 4), MAZ (lane 5), no antibody (lane 6), serially diluted inputs (lanes 7–10) and no template control (lane 11). (B) Enrichment at specific promoters was quantified using a standard curve constructed with serially diluted input samples. ChIP was performed at least three times for each antibody and the enrichment (as a percentage of input samples) at target promoters was then normalized to the background level of enrichment generated by a non-specific IgG control.
4. Discussion
The major allele for rs3813946, located in the 5´UTR of the human CR2 gene, has an independent effect in a protective haplotype for lupus, whereas the minor allele, present in a genetically similar haplotype, does not alter risk (Douglas et al., 2009). Our previous analysis of this naturally occurring polymorphism suggested that it affects the transcriptional regulation of the CR2 gene (Wu et al., 2007). We performed additional experiments to evaluate the effect of variants of rs3813946 on the transcriptional activity of this gene locus in order to assess its contribution to the lupus-associated haplotypes. Alteration of the sequence in this region due to this common single-nucleotide polymorphism affects chromatin structure and transcription factor binding, consistent with its effects on transcription.
In this report we demonstrate that the minor C allele of rs3813946 reduced transcription of reporter genes in CR2-nonexpressing erythroleukemia cells as it did in CR2-expressing B cells in our previous experiments (Wu et al., 2007), providing further support for an effect of this polymorphism on transcriptional activity. Although the absolute levels of transcriptional activity were different between the erythroleukemic and the B cell lines, the relative changes within each cell line showed similar patterns, suggesting that broadly expressed transcription factors mediate the allele-specific effects. We extended this observation to an in vivo context by demonstrating that stable isogenic reporter cell lines showed similar differences, indicating that the effect of this SNP was reproducible within native chromatin in a system in which the integration site of each construct was identical. Furthermore, analyses of these unique CR2-reporter cell lines revealed that chromatin accessibility surrounding the transcriptional start site was reduced as a consequence of the T+21C transition at rs3813946, which correlates with the reduced transcriptional activity associated with this allele. Together, these results reveal the functional effects of rs3813946 in vivo and suggest that the transcription factors involved in regulating transcriptional activation via the +21 site may also play a role in effecting changes to chromatin structure that modulate CR2 gene expression.
Our in vitro experiments examining factors binding the region surrounding rs3813946 suggest that this polymorphism affects DNA-protein interactions, consistent with a direct effect of this SNP on RNA transcription. Functional transcriptional regulatory elements have been reported within the 5´UTR of other genes in diverse systems (Landa et al., 2009; Southam et al., 2007) and the 5´UTR has been shown to regulate gene transcription in various species, including human (Brasset et al., 2007; Chen et al., 2011; Greco et al., 2007). Using bioinformatics, PAX5, p53, and GRalpha were predicted to bind the region surrounding rs3813946 regardless of the allele at this position, whereas loss of binding of C/EBP-β and AP2 and gain of binding of ETF and MAZ were predicted by the T+21C transition at this SNP. ChIP assays in Raji B-cells, which are homozygous for the major allele at rs3813946, confirmed that PAX5, C/EBP-β and MAZ bound the CR2 5´UTR sequence containing rs3813946. Interestingly, the greatest level of enrichment was observed using the C/EBP-β antibody. This transcription factor was determined using bioinformatics to bind both alleles at this SNP equally. Conversely, PAX5 and MAZ, which were shown using bioinformatics approaches to preferentially bind the minor allele over the major allele showed a much lower level of enrichment. These differences in enrichment observed experimentally in the ChIP assays could reflect the decreased affinity of these two transcription factors to the homozygous major T allele at rs3813946, present in the Raji cell line. Whether the minor C allele at rs3813946 alters the binding of these or other factors has not been determined in this study as individuals homozygous minor for this allelic variant are rare and currently cell lines are not available.
Since rs3813946 is located within the 5`UTR, we considered whether it might have posttranscriptional effects. Proteins from nuclear and cytosolic extracts bound ssRNA probes containing the allelic variants at rs3813946 equivalently, suggesting that rs3813946 does not alter cis-acting sequence motifs that interact with specific RNA-binding proteins. Nevertheless, additional experiments are planned to directly evaluate allele-specific differences in mRNA stability or translation efficiency since these may be influenced by other factors such as microRNA binding.
The downstream functional effects of the SNP at rs3813946 are currently under active investigation. Under basal conditions, primary B cells from individuals homozygous or heterozygous for the minor allele at rs3813946 showed a trend towards decreased levels of CR2 RNA transcripts as measured by quantitative RT-PCR (Wu et al, 2007 and data not shown) but there was no difference in CR2 surface protein expression as analyzed by flow cytometry (data not shown). However, the transcriptional effects of rs3813946 may only become apparent under conditions that mimic the disease state, as was the case for a rheumatoid arthritis-associated SNP in the RANKL promoter (Tan et al., 2010). Alternatively, rs3813946 may only alter expression of CR2 at certain stages of B cell development, such as the tolerance checkpoint at which transitional B cells progress from CR2low to CR2high, or in specific cell types, such as follicular dendritic cells, which have been implicated to either drive autoimmunity (Victoratos and Kollias, 2009) or minimize it (Kranich et al., 2008).
Variants in regulatory domains can also affect molecular processes besides protein expression that can contribute to disease susceptibility. For example, removal of introns from pre-mRNA is physically and temporally linked to RNA transcription, so that differences in the structure of the promoter or the rate of transcription can determine splice site selection (reviewed in Kornblihtt, Nat Struct Mol Biol 13: 5–7, 2006). This may explain our previous observation that the major allele of rs3813946 appeared to amplify the protective effect of CR2 exonic SNPs that modified alternative splicing (Douglas et al., 2009). This effect would be manifested primarily in the follicular dendritic cell, where the alternatively spliced CR2 isoform that includes exon 11 is preferentially expressed (Liu et al., 1997).
SLE is a complex disease that is likely to involve the interaction of many functional variants in addition to environmental influences. The data shown herein demonstrate an effect of rs3813946 on gene transcription that is accompanied by alterations in chromatin accessibility and transcription factor binding. Elucidating how this SNP interacts with other genetic variants that comprise the CR2 haplotypes conferring lupus risk or protection will advance our understanding of the mechanisms that underlie lupus susceptibility and may assist in the development of novel therapeutics to treat this disease.
Highlights.
Common three SNP haplotype of the CR2 gene is associated with increased risk of SLE.
The 5´ UTR SNP shows altered transcriptional activity of CR2 promoter reporter gene.
This SNP also alters transcriptional activity in stably transfected cells.
The 5’UTR SNP affects chromatin accessibility of the surrounding sequence.
rs3813946 affects transcription factor binding.
Binding of previously unidentified transcription factors to rs3813946 shown in vivo.
Identifies the 5´UTR to be a novel regulatory element.
Acknowledgements
Other support for this work included grants from the National Institutes of Health (NIH) [R01 AI070983 to S.A.B., B.P.T. and D.U.], the Lupus Research Institute [S.A.B.], and the Alliance for Lupus Research [S.A.B., B.P.T. and D.U.].
Abbreviations
- b
biotinylated
- B6
C57BL/6
- CR2
Complement Receptor type 2
- NE
nuclear extract
- Q-PCR
Quantitative PCR
- SLE
systemic lupus erythematosus
- SNP
single-nucleotide polymorphism
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare no conflict of interest.
References
- Ahearn JM, Fischer MB, Croix D, Goerg S, Ma M, Xia J, Zhou X, Howard RG, Rothstein TL, Carroll MC. Disruption of the Cr2 locus results in a reduction in B-1a cells and in an impaired B cell response to T-dependent antigen. Immunity. 1996;4:251–262. doi: 10.1016/s1074-7613(00)80433-1. [DOI] [PubMed] [Google Scholar]
- Boackle SA, Holers VM, Chen X, Szakonyi G, Karp DR, Wakeland EK, Morel L. Cr2, a candidate gene in the murine Sle1c lupus susceptibility locus, encodes a dysfunctional protein. Immunity. 2001;15:775–785. doi: 10.1016/s1074-7613(01)00228-x. [DOI] [PubMed] [Google Scholar]
- Bougel S, Renaud S, Braunschweig R, Loukinov D, Morse HC, Bosman FT, Lobanenkov V, Benhattar J. PAX5 activates the transcription of the human telomerase reverse transcriptase gene in B cells. J Pathol. 2010;220:87–96. doi: 10.1002/path.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasset E, Bantignies F, Court F, Cheresiz S, Conte C, Vaury C. Idefix insulator activity can be modulated by nearby regulatory elements. Nucleic acids research. 2007;35:2661–2670. doi: 10.1093/nar/gkm140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dong E, Grayson DR. Analysis of the GAD1 promoter: trans-acting factors and DNA methylation converge on the 5' untranslated region. Neuropharmacology. 2011;60:1075–1087. doi: 10.1016/j.neuropharm.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Croix DA, Ahearn JM, Rosengard AM, Han S, Kelsoe G, Ma M, Carroll MC. Antibody response to a T-dependent antigen requires B cell expression of complement receptors. J. Exp. Med. 1996;183:1857–1864. doi: 10.1084/jem.183.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank M, Fenwick E, Abraham LJ, Ulgiati D. Quantitative differences in chromatin accessibility across regulatory regions can be directly compared in distinct cell-types. Biochem Biophys Res Commun. 2008;367:349–355. doi: 10.1016/j.bbrc.2007.12.121. [DOI] [PubMed] [Google Scholar]
- Cruickshank MN, Fenwick E, Karimi M, Abraham LJ, Ulgiati D. Cell- and stage-specific chromatin structure across the Complement receptor 2 (CR2/CD21) promoter coincide with CBF1 and C/EBP-beta binding in B cells. Mol Immunol. 2009;46:2613–2622. doi: 10.1016/j.molimm.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Del Nagro CJ, Kolla RV, Rickert RC. A critical role for complement C3d and the B cell coreceptor (CD19/CD21) complex in the initiation of inflammatory arthritis. J Immunol. 2005;175:5379–5389. doi: 10.4049/jimmunol.175.8.5379. [DOI] [PubMed] [Google Scholar]
- Douglas KB, Windels DC, Zhao J, Gadeliya AV, Wu H, Kaufman KM, Harley JB, Merrill J, Kimberly RP, Alarcon GS, Brown EE, Edberg JC, Ramsey-Goldman R, Petri M, Reveille JD, Vila LM, Gaffney PM, James JA, Moser KL, Alarcon-Riquelme ME, Vyse TJ, Gilkeson GS, Jacob CO, Ziegler JT, Langefeld CD, Ulgiati D, Tsao BP, Boackle SA. Complement receptor 2 polymorphisms associated with systemic lupus erythematosus modulate alternative splicing. Genes Immun. 2009;10:457–469. doi: 10.1038/gene.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco SJ, Smirnov SV, Murthy RG, Rameshwar P. Synergy between the RE-1 silencer of transcription and NFkappaB in the repression of the neurotransmitter gene TAC1 in human mesenchymal stem cells. The Journal of biological chemistry. 2007;282:30039–30050. doi: 10.1074/jbc.M703026200. [DOI] [PubMed] [Google Scholar]
- Haas KM, Hasegawa M, Steeber DA, Poe JC, Zabel MD, Bock CB, Karp DR, Briles DE, Weis JH, Tedder TF. Complement receptors CD21/35 link innate and protective immunity during Streptococcus pneumoniae infection by regulating IgG3 antibody responses. Immunity. 2002;17:713–723. doi: 10.1016/s1074-7613(02)00483-1. [DOI] [PubMed] [Google Scholar]
- Holers VM. Complement receptors and the shaping of the natural antibody repertoire. Springer Semin Immun. 2005;26:405–423. doi: 10.1007/s00281-004-0186-y. [DOI] [PubMed] [Google Scholar]
- Karimi M, Goldie LC, Ulgiati D, Abraham LJ. Integration site-specific transcriptional reporter gene analysis using Flp recombinase targeted cell lines. Biotechniques. 2007;42:217–224. doi: 10.2144/000112317. [DOI] [PubMed] [Google Scholar]
- Kaya Z, Afanasyeva M, Wang Y, Dohmen KM, Schlichting J, Tretter T, Fairweather D, Holers VM, Rose NR. Contribution of the innate immune system to autoimmune myocarditis: a role for complement. Nature Immunol. 2001;2:739–745. doi: 10.1038/90686. [DOI] [PubMed] [Google Scholar]
- Kranich J, Krautler NJ, Heinen E, Polymenidou M, Bridel C, Schildknecht A, Huber C, Kosco-Vilbois MH, Zinkernagel R, Miele G, Aguzzi A. Follicular dendritic cells control engulfment of apoptotic bodies by secreting Mfge8. J Exp Med. 2008;205:1293–1302. doi: 10.1084/jem.20071019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa I, Ruiz-Llorente S, Montero-Conde C, Inglada-Perez L, Schiavi F, Leskela S, Pita G, Milne R, Maravall J, Ramos I, Andia V, Rodriguez-Poyo P, Jara-Albarran A, Meoro A, del Peso C, Arribas L, Iglesias P, Caballero J, Serrano J, Pico A, Pomares F, Gimenez G, Lopez-Mondejar P, Castello R, Merante-Boschin I, Pelizzo MR, Mauricio D, Opocher G, Rodriguez-Antona C, Gonzalez-Neira A, Matias-Guiu X, Santisteban P, Robledo M. The variant rs1867277 in FOXE1 gene confers thyroid cancer susceptibility through the recruitment of USF1/USF2 transcription factors. PLoS Genet. 2009;5:e1000637. doi: 10.1371/journal.pgen.1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Ross J, Scheppler JA, Franza BR., Jr. An in vitro transcription analysis of early responses of the human immunodeficiency virus type 1 long terminal repeat to different transcriptional activators. Mol Cell Biol. 1991;11:1883–1893. doi: 10.1128/mcb.11.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Xu J, de Bouteiller O, Parham CL, Grouard G, Djossou O, de Saint-Vis B, Lebecque S, Banchereau J, Moore KW. Follicular dendritic cells specifically express the long CR2/CD21 isoform. J Exp Med. 1997;185:165–170. doi: 10.1084/jem.185.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makar KW, Pham CT, Dehoff MH, O'Connor SM, Jacobi SM, Holers VM. An intronic silencer regulates B lymphocyte cell- and stage-specific expression of the human complement receptor type 2 (CR2, CD21) gene. J Immunol. 1998a;160:1268–1278. [PubMed] [Google Scholar]
- Makar KW, Pham CTN, Dehoff MH, O'Connor SM, Jacobi SM, Holers VM. An intronic silencer regulates B lymphocyte cell- and stage-specific expression of the human complement receptor type 2 (CR2, CD21) gene. J. Immunol. 1998b;160:1268–1278. [PubMed] [Google Scholar]
- Makar KW, Ulgiati D, Hagman J, Holers VM. A site in the complement receptor 2 (CR2/CD21) silencer is necessary for lineage specific transcriptional regulation. Int Immunol. 2001;13:657–664. doi: 10.1093/intimm/13.5.657. [DOI] [PubMed] [Google Scholar]
- Marquart HV, Svendsen A, Rasmussen JM, Nielsen CH, Junker P, Svehag S-E, Leslie RGQ. Complement receptor expression and activation of the complement cascade on B lymphocytes from patients with systemic lupus erythematosus (SLE) Clin. Exp. Immunol. 1995;101:60–65. doi: 10.1111/j.1365-2249.1995.tb02277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- Molina H, Holers VM, Li B, Fang Y-F, Mariathasan S, Goellner J, Strauss-Schoenberger J, Karr RW, Chaplin DD. Markedly impaired humoral immune response in mice deficient in complement receptors 1 and 2. Proc. Natl. Acad. Sci. USA. 1996;93:3357–3361. doi: 10.1073/pnas.93.8.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodeus AP, Georg S, Shen L-M, Pozdnyakova OO, Chu L, Alicot EM, Goodnow CC, Carroll MC. A critical role for complement in the maintenance of self-tolerance. Immunity. 1998;9:721–731. doi: 10.1016/s1074-7613(00)80669-x. [DOI] [PubMed] [Google Scholar]
- Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- Rayhel EJ, Dehoff MH, Holers VM. Characterization of the human complement receptor 2 (CR2, CD21) promoter reveals sequences shared with regulatory regions of other developmentally restricted B cell proteins. J. Immunol. 1991;146:2021–2026. [PubMed] [Google Scholar]
- Southam L, Rodriguez-Lopez J, Wilkins JM, Pombo-Suarez M, Snelling S, Gomez-Reino JJ, Chapman K, Gonzalez A, Loughlin J. An SNP in the 5'-UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum Mol Genet. 2007;16:2226–2232. doi: 10.1093/hmg/ddm174. [DOI] [PubMed] [Google Scholar]
- Suryani S, Fulcher DA, Santner-Nanan B, Nanan R, Wong M, Shaw PJ, Gibson J, Williams A, Tangye SG. Differential expression of CD21 identifies developmentally and functionally distinct subsets of human transitional B cells. Blood. 2010;115:519–529. doi: 10.1182/blood-2009-07-234799. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Kozono Y, Waldschmidt TJ, Quigg RJ, Baron A, Holers VM. Mouse complement receptors type 1 (CR1; CD35) and type 2 (CR2; CD21): expression on normal B cell subpopulations and decreased levels during development of autoimmunity in MRL/lpr mice. J. Immunol. 1997;159:1557–1569. [PubMed] [Google Scholar]
- Tan W, Wu H, Zhao J, Derber LA, Lee DM, Shadick NA, Conn DL, Smith EA, Gersuk VH, Nepom GT, Moreland LW, Furst DE, Thompson SD, Jonas BL, Holers VM, Glass DN, Chen PP, Bridges SL, Jr., Weinblatt ME, Paulus HE, Tsao BP. A functional RANKL polymorphism associated with younger age at onset of rheumatoid arthritis. Arthritis Rheum. 2010;62:2864–2875. doi: 10.1002/art.27589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedder TF, Clement LT, Cooper MD. Expression of C3d receptors during human B cell differentiation: immunofluorescence analysis with the HB-5 monoclonal antibody. J Immunol. 1984a;133:678–683. [PubMed] [Google Scholar]
- Tedder TF, Clement LT, Cooper MD. Expression of C3d receptors during human B cell differentiation: immunofluorescence analysis with the HB-5 monoclonal antibody. J.Immunol. 1984b;133:678–683. [PubMed] [Google Scholar]
- Thomas MD, Srivastava B, Allman D. Regulation of peripheral B cell maturation. Cell Immunol. 2006;239:92–102. doi: 10.1016/j.cellimm.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Tolnay M, Vereshchagina LA, Tsokos GC. NF-kappaB regulates the expression of the human complement receptor 2 gene. J Immunol. 2002;169:6236–6243. doi: 10.4049/jimmunol.169.11.6236. [DOI] [PubMed] [Google Scholar]
- Ulgiati D, Pham C, Holers VM. Functional analysis of the human complement receptor 2 (CR2/CD21) promoter: characterization of basal transcriptional mechanisms. J Immunol. 2002;168:6279–6285. doi: 10.4049/jimmunol.168.12.6279. [DOI] [PubMed] [Google Scholar]
- Vereshchagina LA, Tolnay M, Tsokos GC. Multiple transcription factors regulate the inducible expression of the human complement receptor 2 promoter. J Immunol. 2001;166:6156–6163. doi: 10.4049/jimmunol.166.10.6156. [DOI] [PubMed] [Google Scholar]
- Victoratos P, Kollias G. Induction of autoantibody-mediated spontaneous arthritis critically depends on follicular dendritic cells. Immunity. 2009;30:130–142. doi: 10.1016/j.immuni.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Wilson JG, Ratnoff WD, Schur PH, Fearon DT. Decreased expression of the C3b/C4b receptor (CR1) and the C3d receptor (CR2) on B lymphocytes and of CR1 on neutrophils of patients with systemic lupus erythematosus. Arth. Rheum. 1986;29:739–747. doi: 10.1002/art.1780290606. [DOI] [PubMed] [Google Scholar]
- Wu H, Boackle SA, Hanvivadhanakul P, Ulgiati D, Grossman JM, Lee Y, Shen N, Abraham LJ, Mercer TR, Park E, Hebert LA, Rovin BH, Birmingham DJ, Chang D-M, Chen CJ, McCurdy D, Badsha HM, Thong BYH, Chng HH, Arnett FC, Wallace DJ, Yu CY, Hahn BH, Cantor RM, Tsao BP. Association of a common complement receptor 2 haplotype with increased risk of systemic lupus erythematosus. Proc Natl Acad Sci USA. 2007;104:3961–3966. doi: 10.1073/pnas.0609101104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Jiang N, Deppong C, Singh J, Dolecki G, Mao D, Morel L, Molina HD. A role for the Cr2 gene in modifying autoantibody production in systemic lupus erythematosus. J. Immunol. 2002;169:1587–1592. doi: 10.4049/jimmunol.169.3.1587. [DOI] [PubMed] [Google Scholar]