Abstract
Purpose
This study was designed to evaluate DNA methylation and the expression of DNA methyltransferases (Dnmt1, Dnmt3a, Dnmt3b and Dnmt3L) in metaphaseII (MII) oocytes and the DNA methylation of pre-implantation embryos during mouse aging to address whether such aging-related changes are associated with decreased reproductive potential in aged mice.
Methods
Oocytes (MII) from 6 to 8 weeks old female mice are referred to as the ‘young group’; oocytes from the same group that were maintained until 35–40 weeks old are referred to as the ‘old group.’ The oocytes were fertilized both in vitro and in vivo to obtain embryos. The DNA methylation levels in the oocytes (MII) and pre-implantation embryos were assessed using fluorescence staining. The expression levels of the Dnmt genes in the oocytes (MII) were assessed using Western blotting.
Results
The DNA methylation levels in the oocytes and pre-implantation embryos (in vivo and in vitro) decreased significantly during the aging of the mice. The expression levels of all of the examined Dnmt proteins in the old group were lower than young group. Both the cleavage and blastocyst rate were significantly lower in the oocytes of the older mice (69.9 % vs. 80.9 %, P < 0.05; 33.9 % vs. 56.4 %, P < 0.05). The pregnancy rate of the old mice was lower than that of the young mice (46.7 % vs. 100 %, P < 0.05). The stillbirth and fetal malformation rate was significantly higher in the old group than in the young group (17.2 % vs. 2.9 %, P < 0.05).
Conclusions
The decreased expression of Dnmt1, Dnmt3a, Dnmt3b and Dnmt3L in oocytes (MII) and the change of genome-wide DNA methylation in oocytes and pre-implantation embryos due to aging may be related to lower reproductive potential in old female mice.
Keywords: Mouse, Aging, Methyltransferase, DNA methylation, Fertility
Introduction
An age-related reduction in female reproductive capacity is common in most mammalian species [1]. Recent changes in lifestyle have led many women to postpone childbearing, and this has been associated with an increased risk of infertility because the reproductive capacity of women declines dramatically beyond the fourth decade of life [2–4]. Although the primary cause for this decline is the gradual depletion of oocytes, it is well recognized that reduced oocyte quality contributes to an overall reduction in fertility and the age-related decline in female fertility [5, 6].
To date, the underlying reasons for the reproductive decline in females remain unclear. It is reported that maternal factors, including neuroendocrine inadequacy [7, 8], uterine apparatus failure [9, 10], and such fetal factors as spotty oocytes or subsequent embryos [11, 12], have been correlated with female reproductive aging. In addition, an increase in the incidence of aneuploidy is well documented with increasing maternal age, particularly in humans [13, 14]. Other studies have indicated that when embryos produced by young women were transferred into older women, the pregnancy rates were consistent with those of the young women [15, 16]. Take together, the quality of the oocyte or embryo could be the primary cause of the decreased reproductive potential in the aging female.
There is a growing perception that epigenetic modifications, such as DNA methylation and histone modification, play important roles in cellular senescence and the aging of an organism [17–19]. DNA methylation is a well-characterized epigenetic modulator and has been shown to play a variety of crucial roles in cell division and proliferation, aging and proper germline and embryo functions [20, 21]. The process of cytosine methylation at C-5 is catalyzed by DNA methyltransferases (Dnmts). Three active DNA methyltransferases—Dnmt1, Dnmt3a and Dnmt3b—and one related protein lacking catalytic activity, Dnmt3L, are present in mammals [22], and DNA methylation patterns change with aging in a complex fashion [23]. For example, age-related demethylation has been reported in the rat brain, liver and small intestine mucosa [24, 25], whereas rat lung genomic DNA does not demethylate as a whole, and the genome-wide DNA methylation content in rat kidneys increases [26]. These results suggest that age-related methylation shows tissue specificity [27–32]. The mammalian genome undergoes profound reprogramming of DNA methylation patterns in germ cells and early preimplantation embryos [23]. Upon fertilization, the gamete methylation patterns from the parents are erased by a genome-wide demethylation event, and during the subsequent implantation, new methylation patterns are established through de novo methylation [33]. These events are important for early embryonic development and the establishment of totipotency or pluripotency [34].
The DNA methylation levels of oocytes and embryos and the expression levels of four key Dnmts were examined in the oocytes from young and old mice. We also investigated the in vivo and in vitro developmental capacities of the oocytes. Our study could lead to a better understanding of the relationship of oocyte DNA methylation and reproductive competence during female aging.
Materials and methods
All of the chemicals and media were purchased from Sigma Chemical Co. (St. Louis, MO, USA), unless otherwise indicated. The animals used in the study were Kunming white mice (Academy of Military Medical Sciences, Beijing, China) and were maintained at 20–22°C under a 14 h (6:00–20:00) light and 10 h (20:00–6:00) dark schedule. The experimental protocols for handling the mice were in accordance with requirements of the Institutional Animal Care and Use Committee of the China Agricultural University.
Natural mating
Young (6–8 weeks old) females were mated with 8–10 weeks old Kunming males in rut. The next morning, those females with a vaginal plug were considered pregnant. The females were allowed to deliver normally, and the litter size and number of stillbirths and fetal malformations were recorded. Following the weaning of their litters, the females were retired from breeding until they reached 35–40 weeks old when they were mated again. Their reproductive performance (the rate of pregnancy and the rate of stillbirths and fetal malformations) was then recorded.
MII oocyte collection
The same group of female mice were divided into two groups, one group of mice at 6–8 weeks old were superovulated using 10 IU (intraperitoneal) of pregnant mare gonadotropin (PMSG; Ningbo Hormone Products Co., Ningbo, Zhejiang, People’s Republic of China, L/N: 20100704), followed by injection of 10 IU of human chorionic gonadotropin (hCG; Ningbo Hormone Products, L/N: 100801, Ningbo, China) 48 h later. The other group was maintained until they were 35–40 weeks old and then superovulated as described above. 14 h after the hCG injection, the ampullae of the oviducts was opened with forceps, and the cumulus–oocyte complexes were recovered in M2 medium supplemented with 4 mg/mL of bovine serum albumin (BSA, albumin fraction V powder; Roche Diagnostics GmbH, Mannheim, Germany). The cumulus cells were removed from the oocytes with hyaluronidase (300 IU/mL) treatment for 3–5 min in M2. Only oocytes with normal morphology were used in the subsequent manipulations.
In vitro fertilization (IVF) and embryo culture
The oocytes collected from the younger and older females are referred to as the ‘young oocyte’ and ‘old oocyte’ groups, respectively. The oocytes in the two groups were individually placed into 75 μL drops of Human Tubal Fluid (HTF) medium under mineral oil, into which a 10-μL spermatozoa suspension with a concentration of 5 × 106 sperm cells per milliliter was added for insemination. Five hours after the IVF, the eggs were removed from the fertilization drop, washed in HTF medium and cultured in 75 μL drops of HTF medium. Then, the 2-cell, 4-cell, 8-cell, morula and blastocyst stage embryos were scored individually at approximately 24 h, 48 h, 60 h, 72 h and 96 h after fertilization. These embryos are referred to as the ‘in vitro pre-implantation embryo’ group.
In vivo embryo collection
Young (6–8 weeks old) and old (35–40 weeks old) mice were mated with 8–10 weeks old Kunming white male mice in rut. Embryos at the 2-cell, 4-cell, 8-cell, morula and blastocyst stages were collected at 44–48 h, 52–56 h, 60–64 h, 68–72 h, 84–96 h after mating. These embryos are referred to as the ‘in vivo pre-implantation embryo’ group.
Immunocytochemical staining of oocytes and pre-implantation embryos
The MII oocytes and in vivo and in vitro pre-implantation embryos used for staining were washed in 0.1 % Tween-20 in PBS, fixed for 30 min in 4 % paraformaldehyde in PBS, permeabilized with 0.5 % Triton X-100 in PBS for 30 min and treated in 2 M HCl for 30 min at 25°C. After extensive washing with 0.1 % Tween-20 in PBS, the cells were blocked for 2 h in 2 % BSA in PBS and incubated with anti-5-MeC antibodies (Epigentek Group Inc., USA) (1:500) at 4°C overnight. The oocytes and embryos were then washed extensively and probed with fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (Santa Cruz Biotechnology, Inc., USA) (1:100) for 1 h at 37°C, and the DNA was counterstained with 10 μg/mL propidium iodide for 10 min. After extensive washing, the oocytes and embryos were incubated in 10 % DABCO (triethylenediamine) in PBS. The material was then mounted on slides, and the fluorescence was detected using a Nikon spectral confocal scanning microscope (Nikon Corporation, Tokyo, Japan) at excitation wavelengths of 488 nm and 543 nm. The system settings were constant for all of the examinations. Each experiment was repeated at least three times, and a minimum of 20 oocytes or embryos was used for each group. The oocytes were treated with 2 % BSA instead of polyclonal primary antibody for the negative control.
The fluorescence intensities were quantified using EZ-C1 Free Viewer software (Nikon) as described by Worrad and Aoki [35, 36], with some modifications. In brief, the pixel value of the fluorescence was measured within a constant area from ten different regions of the nucleus and ten different regions of the cytoplasm, and the average cytoplasmic value was subtracted from the average nuclear value.
Western blotting
The expression levels of methyltransferases (Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3L) were determined by Western blot analysis. Briefly, an equal amount of protein (100 μg) from young or old MIIoocytes was separated by 10 % sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were transferred onto polyvinyl difluoride (Pierce, 0.4 5 μm) membranes at 300 mA for 3 h. The blots were blocked for 1.5 h at room temperature in 5 % nonfat dry milk in TBST buffer. The blots were then incubated with primary antibodies (for Dnmt1, mouse monoclonal antibody, Epigentek Group Inc., USA; for Dnmt 3a, Dnmt 3b and Dnmt 3L, rabbit polyclonal antibody, Epigentek Group Inc., USA) overnight at 4°C. After washing with TBST, the blots were then incubated with the secondary antibodies (goat anti-rabbit or goat anti-rat IgG conjugated to horseradish peroxidase). Subsequently, the blots were washed with TBST, and the HRP-bound secondary antibody was detected using ECL-Dura (Thermo Scientific). The relative densities of the methyltransferases (Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3L) were analyzed using ImageJ software. The experiment was replicated three times for each protein.
Statistical analysis
All of the data in the present study were analyzed using the Student’s t-test with SPSS (Statistical Package for the Social Sciences) 12.0 software (SPSS, Inc., Chicago, IL, USA); P < 0.05 was the criterion for statistical significance.
Results
In vivo and in vitro developmental potential of oocytes in young and old mice
As shown in Table 1, after natural mating, the pregnancy rate of the young females was higher than that of the old individuals (100 % vs. 46.7 %, P < 0.05). There were no significant differences in the average litter size (the average litter size was compared only for the mice that had successfully given birth at both young and old ages, 13.7 ± 0.4 vs. 12.3 ± 0.5, P > 0.05). However, the stillbirth and fetal malformation rates were significantly lower in the young females (2.9 ± 0.8 vs. 17.2 ± 0.9, respectively, P < 0.05).
Table 1.
In vivo and in vitro developmental potentials of oocytes collected from the mice at young and old age
| Group | In vivo development | In vitro development | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Individuals (n) | Pregnancies (n, % ± SE) | Total litter size (n) | Average litter size (n ± SE)a | Stillbirth plus fetal malformations (n, % ± SE)b | Individuals (n) | No. of oocytes | 2-cell (% ± SE) | Blastocyst (% ± SE) | |
| Old | 15 | 8 (46.7 ± 6.7)c | 98 | 12.3 ± 0.5c | 17 (17.2 ± 0.9)c | 20 | 212 | 149 (69.9 ± 0.9)c | 72 (33.9 ± 0.3)c |
| Young | 15 | 15 (100 ± 0.0)d | 205 | 13.7 ± 0.4c | 6 (2.9 ± 0.8)d | 9 | 249 | 202 (80.9 ± 1.2)d | 126 (56.4 ± 1.9)d |
aAverage litter size was compared only in the mice that had successfully given birth at both young and old age
bStillbirth and fetal malformation rate is the percentage of total litter size
c,dDifferent superscripts within same column indicate statistically significant difference (P < 0.05)
As shown in Table 1, when the MII oocytes were fertilized in vitro, both the cleavage and blastocysts rates were significantly lower in the old oocyte group than in the young oocyte group (69.9 % vs. 80.9 %, P < 0.05; 33.9 % vs. 56.4 %, P < 0.01).
Genome-wide DNA methylation in oocytes and pre-implantation embryos during maternal aging
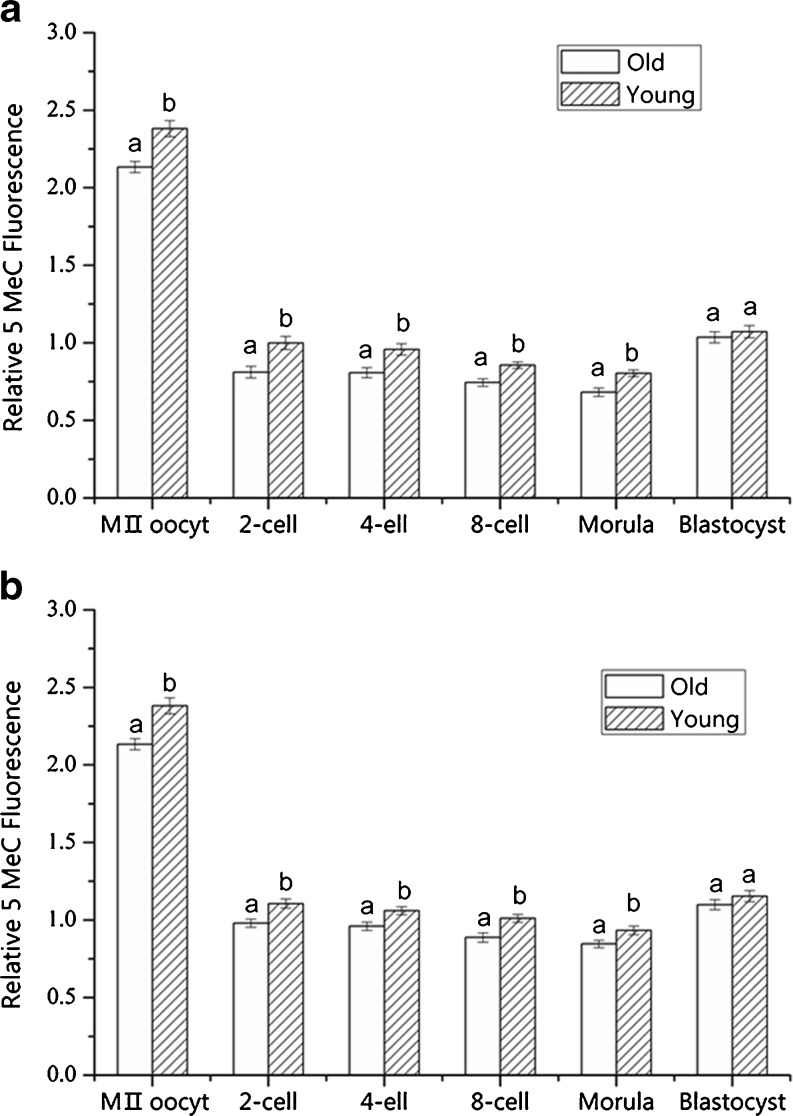
The level of genome-wide DNA methylation was measured by the 5-MeC fluorescence intensity in the MII oocytes and pre-implantation embryos (Figs. 1 and 2). The level of DNA methylation in the old oocyte group was significantly lower than that in the young oocyte group (P < 0.01) (Fig. 2a and b). After in vitro fertilization and natural mating, the level of DNA methylation in the old oocyte group was significantly lower in the 2-cell, 4-cell, 8-cell and morula embryos (P < 0.05) when compared with that in the young oocyte group; however, there was no significant difference in DNA methylation in the blastocysts (P > 0.05) (Fig. 2a and b).
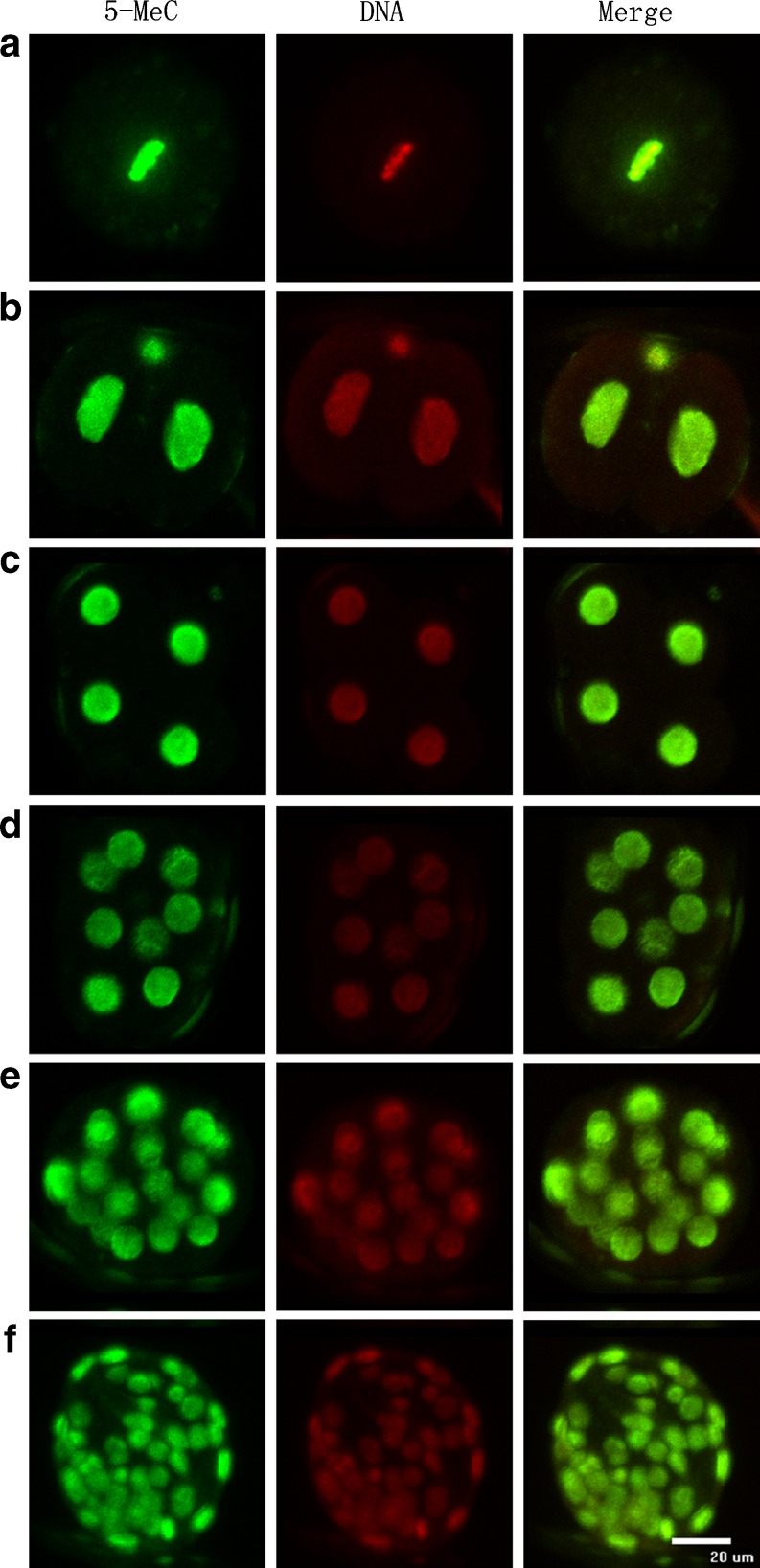
Fig. 1.
schematic diagram of immunofluorescence results on 5-MeC of oocytes and embryos. A-G refers to immunofluorescence results on 5-MeC of MIIoocyte, 2-cell, 4-cell, 8-cell, morula and blastocyst separately (left), PI-stained nuclei (middle), Merged (right). Bar = 20 μm
Fig. 2.
Effect of maternal age on genome-wide DNA methylation levels of MIIoocytes and pre-implantation embryos. A and B refers to fluorescent intensity of MIIoocytes and in vitro pre-implantation embryos (a), in vivo pre-implantation embryos (b) separately. The fluorescent intensity of in vitro 2-cell embryo from the young group was set as 1. a,b different superscript letters identify significant differences (P < 0.05)
Expression of Dnmt1, Dnmt3a, Dnmt3b, Dnmt3L in oocytes during maternal aging
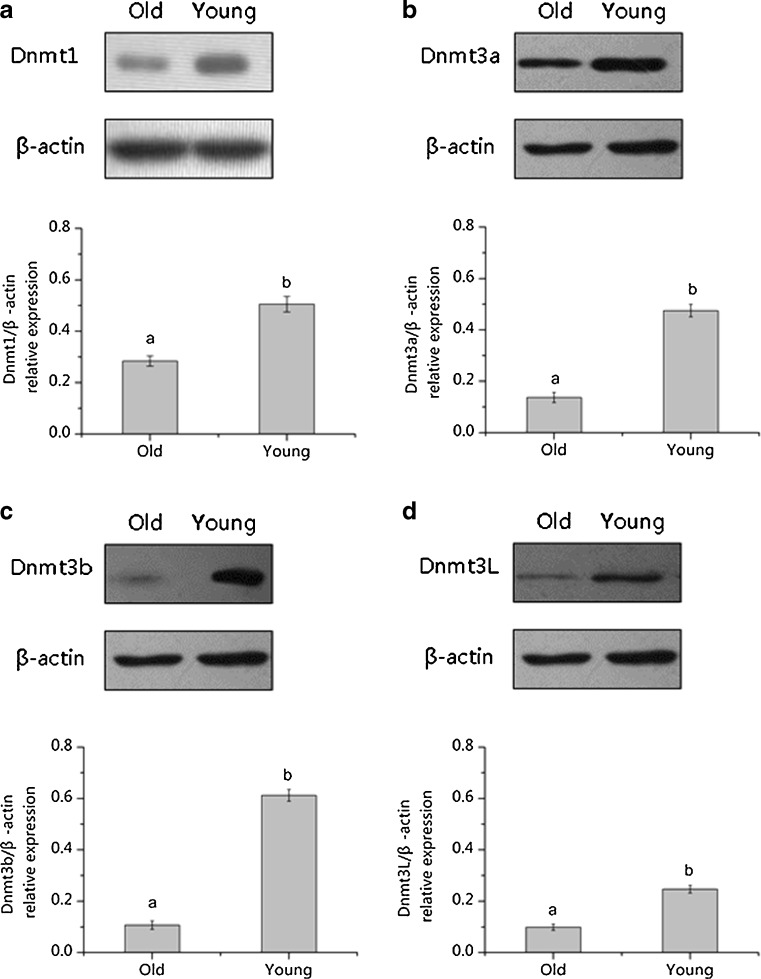
We detected the expression of Dnmt1, Dnmt3a, Dnmt3b and Dnmt3L in the MIIoocytes from young and old mice using Western blotting (Fig. 3), revealing a significant decrease in the expression of Dnmt1 (Fig. 3a), Dnmt3a (Fig. 3b), Dnmt3b (Fig. 3c) and Dnmt3L (Fig. 3d) in the aged MII oocytes.
Fig. 3.
Western blot analysis for Dnmt1(a), Dnmt3a(b), Dnmt3b(c) and Dnmt3L(d) protein expression levels in MII oocytes from old and young mice. a,b different superscripts letters means significant difference (P < 0.05)
Discussion
DNA methylation plays an important role in gene expression and regulation and participates in cell differentiation and proliferation, organism aging, tumor formation and other important cellular activities [37–40]. Previous reports have shown that the process of age-related methylation varies among different tissues [27–32, 41]. Our study showed that the genome-wide DNA methylation in oocytes and 2-cell to morula stage embryos was significantly lower in old mice than in young mice. It has been reported that DNA methylation has important implications on chromatin structure and gene expression [42, 43] and may provide a unique mechanism for organizing local histone deacetylation and generating maintainable epigenetic chromosomal states in higher organisms [44]. Our previous study demonstrated that the changes in the histone acetylation pattern during aging could affect chromosomal structures and the developmental potential of oocytes [45]. Epigenetic chromosomal states due to decreased DNA methylation in oocytes from aged female mice might negatively affect the subsequent development of oocytes after fertilization.
DNA methylation is introduced into DNA by a group of enzymes called DNA methyltransferases [22]. The results of our Western blot experiment indicate that the expression levels of Dnmt1, Dnmt3a, Dnmt3b and Dnmt3L in the oocytes from the old mice were significantly decreased compared to those from the young mice, which could account for the decline of genome-wide DNA methylation in MII oocytes. In addition, several lines of evidence suggest that histone methylation has an important significance to the formation of DNA methylation [46–49]. Manosalva and González reported that a proportion of old MII oocytes exhibit histone demethylation [50], a finding that may be another reason for the decrease of genome-wide DNA methylation in old MII oocytes. A previous study also reported mutations in the Dnmt1 gene caused genome-wide genome demethylation and embryonic lethality [51]. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprinting in mice [52]. Mice lacking either Dnmt3a or Dnmt3b display different defects and die at different stages of development [53]. In this study, we also found that the expression of Dnmt proteins decreased with aging, and this event may be related to the lower pregnancy rates and higher stillbirth and fetal malformation rates in the old female mice.
In our study, we found no difference in the DNA methylation of the in vitro and in vivo blastocysts between the old and young female mice. This result might be due to a repair of the DNA methylation to the normal level at the blastocyst stage, as it has been reported that de novo methylation is performed by Dnmt3a and Dnmt3b at the time of implantation [54]. A previous report indicated that embryos and placentas capable of developing to mid-gestation showed normal methylation at several imprinted genes and in genome-wide methylation levels, most likely because the hypo-methylation of genomic DNA may occur in pre-implantation or late pregnancy [55]. There is limitation of using fluorescence staining to assess the levels of DNA methylation, but it is suggested that this method is invaluable for the large-scale screening of genome-wide methylation and has a high degree of reproducibility, which is essential for the analysis of small numbers of valuable samples and to provide information on the methylation profiles of individual cells and embryos [56].
In conclusion, our results showed that the DNA methyltransferase expression and DNA methylation patterns are abnormal in the oocytes and early embryos of aged mice, observations that could be associated with the age-related decline in reproductive potential in older females.
Acknowledgments
This work was supported by the National Natural Science Foundation Project of China (No. 30972102); China Agricultural University Graduate Scientific Research and Innovation Special Project (No. kycx09027). We thank Professor William Hohenboken, Professor Tiantian Zhang and Dr. QingGang Meng for proofreading the manuscript.
Footnotes
Capsule
Aging caused a significant decrease in the expression of four key Dnmts and genome-wide DNA methylation in oocytes and pre-implantation embryos.
Ming-xing Yue and Xiang-wei Fu contributed equally to this work.
References
- 1.Armstrong DT. Effects of maternal age on oocyte developmental competence. Theriogenology. 2001;55(6):1303–1322. doi: 10.1016/S0093-691X(01)00484-8. [DOI] [PubMed] [Google Scholar]
- 2.Klein J, Sauer MV. Assessing fertility in women of advanced reproductive age. Am J Obstet Gynecol. 2001;185(3):758–770. doi: 10.1067/mob.2001.114689. [DOI] [PubMed] [Google Scholar]
- 3.Assisted reproductive technology in the United States: 1997 results generated from the American Society for Reproductive Medicine/Society for Assisted Reproductive Technology Registry. Fertil Steril. 2000;74(4):641–653; discussion 653–644. [DOI] [PubMed]
- 4.Kooij RJ, Looman CW, Habbema JD, Dorland M, Velde ER. Age-dependent decrease in embryo implantation rate after in vitro fertilization. Fertil Steril. 1996;66(5):769–775. doi: 10.1016/s0015-0282(16)58634-8. [DOI] [PubMed] [Google Scholar]
- 5.Plachot M, Veiga A, Montagut J, et al. Are clinical and biological IVF parameters correlated with chromosomal disorders in early life: a multicentric study. Hum Reprod. 1988;3(5):627–635. doi: 10.1093/oxfordjournals.humrep.a136758. [DOI] [PubMed] [Google Scholar]
- 6.Hamatani T, Falco G, Carter MG, et al. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet. 2004;13(19):2263–2278. doi: 10.1093/hmg/ddh241. [DOI] [PubMed] [Google Scholar]
- 7.Wise PM, Smith MJ, Dubal DB, et al. Neuroendocrine modulation and repercussions of female reproductive aging. Recent Prog Horm Res. 2002;57:235–256. doi: 10.1210/rp.57.1.235. [DOI] [PubMed] [Google Scholar]
- 8.Yeh J, Kim BS, Peresie J. Ovarian vascular endothelial growth factor and vascular endothelial growth factor receptor patterns in reproductive aging. Fertil Steril. 2008;89(5 Suppl):1546–1556. doi: 10.1016/j.fertnstert.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 9.Thorneycroft IH, Soderwall AL. The nature of the litter size loss in senescent hamsters. Anat Rec. 1969;165(3):343–348. doi: 10.1002/ar.1091650303. [DOI] [PubMed] [Google Scholar]
- 10.Finn CA. Reproductive ageing and the menopause. Int J Dev Biol. 2001;45(3):613–617. [PubMed] [Google Scholar]
- 11.Velde ER, Pearson PL. The variability of female reproductive ageing. Hum Reprod Update. 2002;8(2):141–154. doi: 10.1093/humupd/8.2.141. [DOI] [PubMed] [Google Scholar]
- 12.Ito M, Muraki M, Takahashi Y, et al. Glutathione S-transferase theta 1 expressed in granulosa cells as a biomarker for oocyte quality in age-related infertility. Fertil Steril. 2008;90(4):1026–1035. doi: 10.1016/j.fertnstert.2007.07.1389. [DOI] [PubMed] [Google Scholar]
- 13.Pan H, Ma P, Zhu W, Schultz RM. Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev Biol. 2008;316(2):397–407. doi: 10.1016/j.ydbio.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol. 2010;20(17):1522–1528. doi: 10.1016/j.cub.2010.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antinori S, Versaci C, Gholami GH, Panci C, Caffa B. Oocyte donation in menopausal women. Hum Reprod. 1993;8(9):1487–1490. doi: 10.1093/oxfordjournals.humrep.a138284. [DOI] [PubMed] [Google Scholar]
- 16.Sauer MV, Paulson RJ, Lobo RA. Reversing the natural decline in human fertility. An extended clinical trial of oocyte donation to women of advanced reproductive age. JAMA. 1992;268(10):1275–1279. doi: 10.1001/jama.1992.03490100073030. [DOI] [PubMed] [Google Scholar]
- 17.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23(8):413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Fraga MF, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann N Y Acad Sci. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- 19.Munoz-Najar U, Sedivy JM. Epigenetic control of aging. Antioxid Redox Signal. 2011;14(2):241–259. doi: 10.1089/ars.2010.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trasler JM. Gamete imprinting: setting epigenetic patterns for the next generation. Reprod Fertil Dev. 2006;18(1–2):63–69. doi: 10.1071/RD05118. [DOI] [PubMed] [Google Scholar]
- 21.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1(1):11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 22.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. ChemBioChem. 2010;12(2):206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 23.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 24.Golbus J, Palella TD, Richardson BC. Quantitative changes in T cell DNA methylation occur during differentiation and ageing. Eur J Immunol. 1990;20(8):1869–1872. doi: 10.1002/eji.1830200836. [DOI] [PubMed] [Google Scholar]
- 25.Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262(21):9948–9951. [PubMed] [Google Scholar]
- 26.Vanyushin BF, Nemirovsky LE, Klimenko VV, Vasiliev VK, Belozersky AN. The 5-methylcytosine in DNA of rats. Tissue and age specificity and the changes induced by hydrocortisone and other agents. Gerontologia. 1973;19(3):138–152. doi: 10.1159/000211967. [DOI] [PubMed] [Google Scholar]
- 27.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7(4):536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 28.Eads CA, Lord RV, Wickramasinghe K, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61(8):3410–3418. [PubMed] [Google Scholar]
- 29.Kang GH, Lee HJ, Hwang KS, Lee S, Kim JH, Kim JS. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 2003;163(4):1551–1556. doi: 10.1016/S0002-9440(10)63511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Ahuja N, Shen Y, et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst. 2002;94(10):755–761. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- 31.Tsuchiya T, Tamura G, Sato K, et al. Distinct methylation patterns of two APC gene promoters in normal and cancerous gastric epithelia. Oncogene. 2000;19(32):3642–3646. doi: 10.1038/sj.onc.1203704. [DOI] [PubMed] [Google Scholar]
- 32.Waki T, Tamura G, Sato M, Motoyama T. Age-related methylation of tumor suppressor and tumor-related genes: an analysis of autopsy samples. Oncogene. 2003;22(26):4128–4133. doi: 10.1038/sj.onc.1206651. [DOI] [PubMed] [Google Scholar]
- 33.Tang WY, Ho SM. Epigenetic reprogramming and imprinting in origins of disease. Rev Endocr Metab Disord. 2007;8(2):173–182. doi: 10.1007/s11154-007-9042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 35.Worrad DM, Ram PT, Schultz RM. Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development. 1994;120(8):2347–2357. doi: 10.1242/dev.120.8.2347. [DOI] [PubMed] [Google Scholar]
- 36.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181(2):296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 37.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9(6):765–775. doi: 10.1016/S1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 38.Richardson B. Impact of aging on DNA methylation. Ageing Res Rev. 2003;2(3):245–261. doi: 10.1016/S1568-1637(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 39.Feinberg AP. DNA methylation, genomic imprinting and cancer. Curr Top Microbiol Immunol. 2000;249:87–99. doi: 10.1007/978-3-642-59696-4_6. [DOI] [PubMed] [Google Scholar]
- 40.Feinberg AP, Cui H, Ohlsson R. DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Semin Cancer Biol. 2002;12(5):389–398. doi: 10.1016/S1044-579X(02)00059-7. [DOI] [PubMed] [Google Scholar]
- 41.Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220(4601):1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 42.Cedar H, Razin A. DNA methylation and development. Biochim Biophys Acta. 1990;1049(1):1–8. doi: 10.1016/0167-4781(90)90076-E. [DOI] [PubMed] [Google Scholar]
- 43.Dahl C, Guldberg P. DNA methylation analysis techniques. Biogerontology. 2003;4(4):233–250. doi: 10.1023/A:1025103319328. [DOI] [PubMed] [Google Scholar]
- 44.Eden S, Hashimshony T, Keshet I, Cedar H, Thorne AW. DNA methylation models histone acetylation. Nature. 1998;394(6696):842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 45.Suo L, Meng QG, Pei Y, et al. Changes in acetylation on lysine 12 of histone H4 (acH4K12) of murine oocytes during maternal aging may affect fertilization and subsequent embryo development. Fertil Steril. 2010;93(3):945–951. doi: 10.1016/j.fertnstert.2008.12.128. [DOI] [PubMed] [Google Scholar]
- 46.Bachman KE, Park BH, Rhee I, et al. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Canc Cell. 2003;3(1):89–95. doi: 10.1016/S1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- 47.Lehnertz B, Ueda Y, Derijck AA, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13(14):1192–1200. doi: 10.1016/S0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 48.Strunnikova M, Schagdarsurengin U, Kehlen A, Garbe JC, Stampfer MR, Dammann R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol Cell Biol. 2005;25(10):3923–3933. doi: 10.1128/MCB.25.10.3923-3933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414(6861):277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 50.Manosalva I, Gonzalez A. Aging changes the chromatin configuration and histone methylation of mouse oocytes at germinal vesicle stage. Theriogenology. 2010;74(9):1539–1547. doi: 10.1016/j.theriogenology.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 51.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 52.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129(8):1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 53.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 54.Feil R, Khosla S. Genomic imprinting in mammals: an interplay between chromatin and DNA methylation? Trends Genet. 1999;15(11):431–435. doi: 10.1016/S0168-9525(99)01822-3. [DOI] [PubMed] [Google Scholar]
- 55.Lopes FL, Fortier AL, Darricarrere N, Chan D, Arnold DR, Trasler JM. Reproductive and epigenetic outcomes associated with aging mouse oocytes. Hum Mol Genet. 2009;18(11):2032–2044. doi: 10.1093/hmg/ddp127. [DOI] [PubMed] [Google Scholar]
- 56.Santos F, Dean W. Using immunofluorescence to observe methylation changes in mammalian preimplantation embryos. Meth Mol Biol. 2006;325:129–137. doi: 10.1385/1-59745-005-7:129. [DOI] [PubMed] [Google Scholar]