Abstract
Purpose
To describe the identification of a new mutation responsible for causing human severe combined immunodeficiency syndrome (SCID). In a large consanguineous Israeli Arab family, this served as a diagnostic tool and enabled us to carry out preimplantation genetic diagnosis (PGD). We also demonstrated that PGD for homozygosity alleles is feasible.
Methods
We carried out genome-wide screening followed by fine mapping and linkage analysis in order to identify the candidate genes. We then sequenced DCLRE1C in order to find the familial mutation. The family was anxious to avoid the birth of an affected child, and therefore, because of their religious beliefs, PGD was the only option open to them. The embryos were biopsied at day 3, and a single blastomere from each embryo was analyzed by multiplex polymerase chain reaction for the SCID mutation and 5 additional polymorphic markers flanking DCLRE1C.
Results
Linkage analysis revealed linkage to chromosome 10p13, which harbors the DNA Cross-Link Repair Protein 1 C (DCLRE1C) ARTEMIS gene. Sequencing identified an 8 bp insertion in exon 14 (1306ins8) of DCLRE1C in all the affected patients; this causes an alteration in amino acid 330 of the protein from cysteine to a stop codon (p.C330X). One cycle of PGD was performed and two embryos were transferred, one homozygous wild-type and one a heterozygous carrier, and healthy twins were born.
Conclusions
Identifying the familial mutation enabled us to design a reliable and accurate PGD protocol, even in this case of a consanguineous family.
Keywords: Artemis, DCLRE1C, Autosomal recessive, Immunodeficiency, Homozygosity
Introduction
Severe combined immunodeficiency (SCID) is a heterogeneous group of genetic disorders caused by mutations in a number of different genes and characterized by severely impaired humoral and cellular immunity [3, 4]. Criteria for diagnosis of most forms of SCID are the early onset of clinical manifestations such as severe-to-lethal infections, severe impairment of cell-mediated immunity, failure of antibody synthesis, and lymphopenia, mainly at the expense of T-lymphocytes. Most SCID patients have thymic hypoplasia, peripheral CD3+ T-cell counts of 500 cells/mm3 or less (normal range, 3000–6500 cells/mm3) and variable numbers of B and natural killer (NK) lymphocytes, depending on the underlying genetic defect [2, 5]. There are two types of SCID, the commoner of which is caused by an X-linked recessive mutation in IL2RG [7]. The second type is transmitted by autosomal recessive inheritance and can be caused by mutations in various genes such as JAK3, ADA, RAG1, RAG2, IL7R, CD3D, CD3E, ZAP70, and DNA Cross-Link Repair Protein 1 C (DCLRE1C-ARTEMIS) [2, 5, 7, 12].
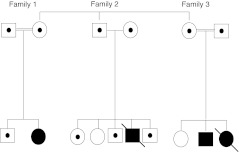
We have identified a large consanguineous Israeli Arab family with several members affected by SCID. The pedigree (Fig. 1) shows that the SCID in this family is transmitted by autosomal recessive inheritance. The family history revealed that two previous affected children had died, one from lymphoma and one after bone marrow transplantation, from a then unknown immunodeficiency. Laboratory study of the cellular profiles of the affected children yielded the following findings: helper T cells—low; CD8+ cells—normal; B cells—normal (2 patients) or reduced (2 patients); NK cells—normal. Response to mutagens was reduced (2 patients) or borderline (2 patients). The laboratory results were unhelpful in indicating which autosomal recessive SCID mutation was responsible for the disease, so we performed a genome wide screen followed by fine mapping.
Fig. 1.
Pedigree of the family
Family 3 (Fig. 1) came to our Genetic Center because they were anxious to have a healthy child, and because of their religious beliefs pregnancy termination was not an option. Performing preimplantation genetic diagnosis (PGD), therefore, was the best approach in this case. In SCID patients bone marrow transplantation from identical human leukocyte antigens (HLA) can sometimes be effective. We offered the family the possibility of finding a suitable HLA match among the tested embryos, but they refused because of their experience of an unsuccessful transplant in their daughter that resulted in her death. Therefore PGD was performed to find healthy/carrier SCID embryos.
We report here the results of a long journey from the identification of a new family mutation to PGD and prenatal diagnosis.
Materials and methods
Finding the family mutation
Whole-genome linkage analysis was performed using 450 highly polymorphic fluorescent-labeled markers from the ABI PRISM Linkage Mapping Set (version 2.5, Applied Biosystems) on an ABI PRISM 3100 Genetic Analyzer. The distance between markers ranged from 2 to 10 centimorgans (cM; equivalent to 1 recombination in 100 meioses) with an average spacing of 5 cM. This analysis revealed linkage to chromosome 10p13, which harbors the DNA Cross-Link Repair Protein 1 C (DCLRE1C) ARTEMIS gene, and therefore a further 10 markers from the Genome Database were included to obtain a higher density of markers on the short arm of chromosome 10. Fine mapping with 4 additional markers, D10S1168, D10S1653, D10S504 and D10S548, confirmed homozygosity in all the affected individuals. Two-point linkage analysis was performed by using the MLINK program. The genetic model used in the analysis was for a fully autosomal recessive trait.
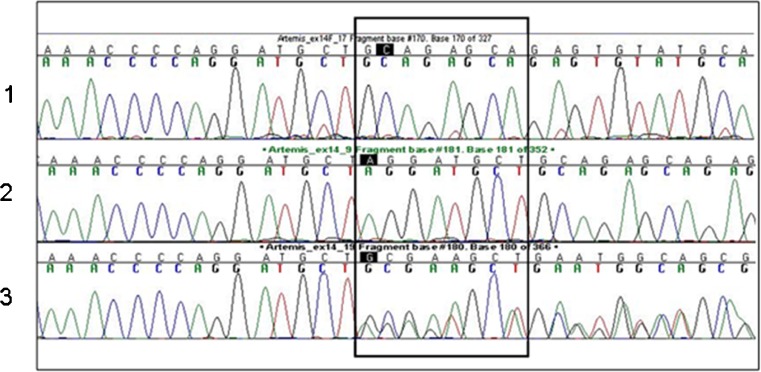
The genes in the candidate locus were identified by means of available databases (NCBI and UCSC Genome Browser, assembly hg18). Sequencing of the candidate genes was performed with primer sets designed using standard software Primer 3. All exons, including exon-intron junctions, were amplified from genomic DNA. Sequence chromatograms were analyzed using SeqScape software version 1.1 (Applied Biosystems). We initially tested one affected individual and one heterozygous parent. Exons 1 to 14 of the Artemis gene isoform α were amplified. The sequence conditions were: initial denaturation at 96°C for 1 min, followed by 26 cycles of 96°C for 10 s, 50°C for 5 s and 60°C for 4 min. We identified a new mutation—an 8 bp insertion—in exon 14 (1306ins8) of DCLRE1C in all the affected patients studied (Fig. 2). The insertion causes an alteration in amino acid 330 of the protein from cysteine to a stop codon (p.C330X).
Fig. 2.
Chromatograph results of a wild-type (1), affected (2) and carrier (3)
Mutation screening of the general population was performed by using genomic DNA extracted from whole blood of 100 individuals of Israeli Arab origin who were unrelated to the family under study. Mutation analysis of the sequence amplified by forward and reverse primers was performed as previously described (Fig. 2) [1].
IVF and blastomere biopsy procedure
The ovarian stimulation procedure was carried out using a standard long follicular protocol, with depot injection of Decapeptyl CR 3.75 mg (Ferring, GmbH, Germany), and Menopur (Ferring, GmbH, Germany). For final oocyte maturation, recombinant hCG (Ovitrelle 250 mcg, Mereck Serono) was used.
Twenty oocytes were recovered, all of which were suitable for ICSI, and 16 were fertilized. The embryos were cultured in P-1 medium supplemented with 10 % synthetic serum substitute (Irvine Scientific, California, USA).
On day 3 eleven embryos that contained at least six cells and ≤50 % fragmentation were biopsied in a biopsy medium (Sydney IVF, Cook, Australia). Zona ablation was carried out using a monocontact laser (Zilos-tk laser, Hamilton Thorne Biosciences, USA). One blastomere from each embryo was removed using a biopsy pipette (Humagen, USA) and placed immediately into sterile 0.2-ml PCR tubes containing 5 μL of alkaline lysis buffer. After biopsy the embryos were placed in blastocyst medium supplemented with 20 % synthetic serum protein substitute (Sage, USA).
On day 4 two embryos, one homozygous wild-type and one a heterozygous carrier, were transferred to the patient. Five blastocysts (four carriers and one homozygous wild-type) were vitrified on days 5 and 6.
Molecular analysis for PGD
Genetic testing for the 1306ins8 mutation and microsatellite analysis were performed using fluorescent Polymerase Chain Reaction (PCR) for size discrimination with capillary electrophoresis. DNA was extracted from peripheral blood cells from available family members using the high salt precipitation method [11]. Blastomere analysis was carried out following proteinase K digestion and inactivation at 94°C for 15 min followed by PCR in a 50 μl multiplex PCR reaction containing 0.2 μM dNTPS, 10 % DMSO, 0.1 μM of each primer, and 1.25 U Taq polymerase in a buffer supplied by the manufacture (JMR801, UK). The reaction was thermocycled for 30 cycles using a program of 20 s at 95°C, 1 min at 62°C to 50°C and 30 s at 72°C. The primer sequences are listed in Table 1. From each multiplex reaction, 1.5 μl was used as a template with a hemi-nested primer 5′ fluorescently labeled with 6-FAM or Hex (Metabion, GmbH), with either the forward or reverse unlabeled primer for an additional 35 cycle PCR reaction for each individual marker. Reaction products were diluted and run on an ABI Prism 3100 Avant automated sequencer, and analyzed using GeneMapper software (ABI).
Table 1.
Sequence of the primers used in hemi-nested reaction
| Marker/mutation | Forward primer | Reverse primer | Nested primer |
|---|---|---|---|
| D10S191 | TTTATCCGAAGACCCTGGAA | GTGTTGAGGCTGTCAATCCA | AGGAAATGTATGCCGGGTAGT |
| SCID_TG2 | TGAACCTTGTGGGTTTTGC | CCTGATCCTGTGTGTTTGGA | GCTCTGACAGTACATCCAGGTG |
| SCID_TC1 | AGAGAGACTCGGCCTCCAA | TGCATTTGCAGATGGAAAAA | CAATCGAGAGGTTTCCCTGA |
| SCID_TA2 | TCAAACTCCTCCAAAGAACTGAA | GGGTGTGGTAGAGGGGCTAT | AGACAAAGGCACTGCAAGAAA |
| SCID_MUT | TGATGATCCTCTGCCAATACC | TACATCCCCATCAGCCTTTT | CAAAGTTCCATACCCGGAAA |
| SCID_GA1 | CCATTTTCTCACATTAGTTCATCA | TCCACCACCATTCCATTACTT | AATTGGGATGTCTCCAAGCA |
| SCID_TA3 | CAGGGTTTCACCATGTAGACTG | AGCCTGGGTGACAGAAACTG | TGCTGCTAGTTATTCACTCAACAAA |
| SCID_TA4 | CTGCAGTCAGTCAGGGAAAAA | CGCAGTTCGATCTTGAGGTT | GCAAGTCTCACTTCATGGAACA |
| SCID_CA2 | CCCTCCCTCTCTGCGTCT | GCACATGGTGCAAGAAAGTG | CTCTCCCCCATCCTTCTCTC |
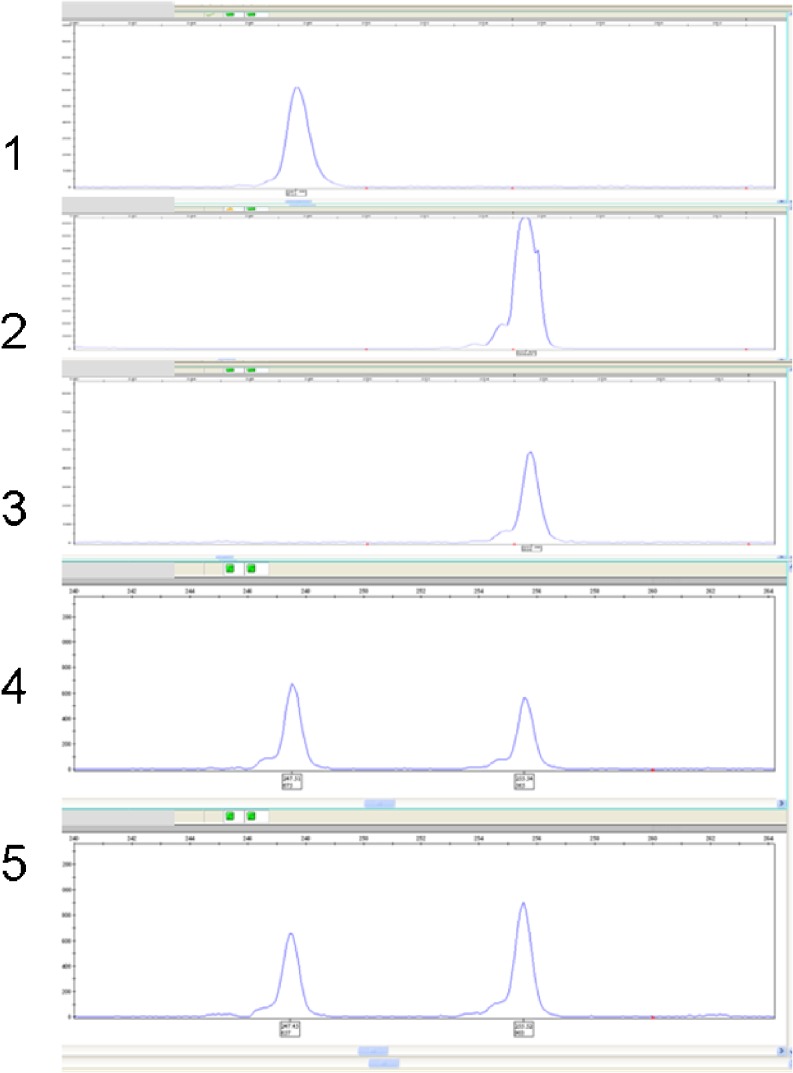
Each multiplex protocol included primers for the family’s specific mutation and 4–8 polymorphic markers flanking the ARTEMIS gene (D10S191, TG2 (chr 10:14786275), TC1 (chr 10: 14849030), TA2 (chr 10: 14919772), GA1 (chr 10: 15047681), TA3 (chr 10: 15194180), TA4 (chr 10: 15221869), CA2 (chr 10: 15516592) (UCSC, assembly hg18), [6] (Fig. 3).
Fig. 3.
Gene-scan results of amplified DNA of the SCID mutation of the family (family 3 in pedigree): 1) The healthy child (homozygous wild-type), 2) The surviving affected child, 3) The deceased affected child, 4) Paternal DNA (healthy heterozygous carrier), 5) Maternal DNA (healthy heterozygous carrier)
The amplification, allele dropout (ADO) and contamination rates obtained during a preclinical test on unrelated single fibroblasts were within the limits set by ESHRE PGD consortium guidelines [14]. Markers exhibiting low amplification rates or high ADO rates were excluded from the PGD assays.
Results
A single 4.85 Mb region of shared homozygosity was found in the affected children. The minimal interval contains 28 known or hypothetical genes. Some of the genes in the candidate region were interesting possibilities for the causative gene in our patients. These include the DNA Cross-Link Repair Protein 1 C (DCLRE1C-ARTEMIS), which maps to chromosome 10p13 (14988878–15036100) and which contains 14 exons that range in size from 52 to 1,160 bp. We identified a homozygous insertion of 8 nucleotides in exon 14 in the affected children, while the parents were heterozygous carriers of the mutation (Fig. 3), seen as a frameshift in the carriers (Fig. 2). The insertion causes an alteration in amino acid 330 of the protein from cysteine to a stop codon in the homozygous affected children (p.C330X), which is predicted to be pathologic.
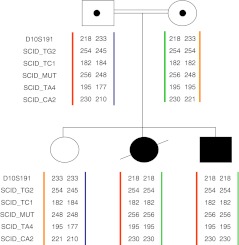
On the basis of these data we designed a PGD program. Eight polymorphic markers flanking DCLRE1C were selected for haplotype analysis as well as the family mutation detected. The husband was found to be heterozygous for five of the eight markers (D10S191, SCID-TG2, SCID-TC1, SCID-TA4, SCID-CA2, Table 1), and all five were used (Fig. 4). The wife and the affected children were each haplotyped with these markers, and this allowed the identification of the individual alleles in the family (mutated alleles and wild-type). Because of the consanguineous marriage and a founder mutation both parents shared the same markers, which is why they are not fully or even highly informative. It is important in PGD to find highly informative markers in order to avoid ADO. In this family we were unable to distinguish between ADO and homozygosity, but we solved the problem by using the 5 markers noted above, and when we rechecked the affected embryos to confirm our results the outcome was the same. In recessive inheritance the finding of one healthy allele in the embryo tested is enough, because even if the result is erroneously reported as wild-type/wild-type, at worst the embryo might be a carrier (wild-type/mutation). The couple agreed to the transfer of both healthy embryos (wild-type/wild-type) and carriers (wild-type/mutation).
Fig. 4.
Linkage analysis of the 1306ins8 mutation in DCLRE1C and 5 microsatellite markers flanking the gene. The PGD analyses were based on these haplotypes
Before the first cycle the PGD protocol was tested using all 5 markers and the familial mutation in a multiplex PCR in single fibroblasts from an unrelated individual. The couple went through one cycle of IVF-PGD using all 5 markers and the family mutation. Amplification was observed in all reactions. ADO observed for 11 blastomeres in the PGD cases was 0–2 events per marker and the amplification rate was 97 %. After the cycle two embryos (wild-type/wild-type and wild-type/mutation) were transferred and a twin pregnancy ensued. The results of the blastomere analysis are summarized in Table 2.
Table 2.
Results of blastomere analysis
| Embryo # | D10S191 | SCID_TG2 | SCID_TC1 | SCID_MUT | SCID_TA4 | SCID_CA2 | Blastomere Result |
|---|---|---|---|---|---|---|---|
| 1 | 233/233 | 245/254 | 182/184 | 247/247 | 195/177 | 210/221 | WT/WT |
| 2 | 218/218 | 254/254 | 182/182 | 255/255 | 195/195 | 230/230 | MUT/MUT |
| 3 | 218/233 | 254/254 | 182/182 | 247/255 | 195/195 | 221/230 | WT/MUT |
| 4 | 233/233 | 254/?* | 182/184 | 247/247 | 177/?* | 210/221 | WT/WT |
| 5 | 218/233 | 254/254 | 182/182 | 247/255 | 195/195 | 221/230 | WT/MUT |
| 6 | 217/233 | 254/254 | NR** | 247/255 | 195/195 | 221/230 | WT/MUT |
| 7 | 218/233 | 254/254 | 182/182 | 247/255 | 195/195 | 221/230 | WT/MUT |
| 8 | 218/233 | 245/254 | 182/184 | 247/255 | 177/?* | 210/230 | WT/MUT |
| 9 | 218/218 | 254/254 | 182/182 | 255/255 | NR** | 230/230 | MUT/MUT |
| 10 | 233/?* | 254/254 | 182/182 | 247/255 | 195/195 | 221/230 | WT/MUT |
| 11 | 218/233 | 245/254 | 182/184 | 247/255 | 177/?* | 210/230 | WT/MUT |
* ? =Allele drop-out ** NR = No reaction
Discussion
In this report we describe the identification of a new mutation responsible for causing SCID; this served as a diagnostic tool and enabled us to carry out PGD. We also demonstrated that PGD for homozygosity alleles is feasible.
The family reported here is the first in Israel known to have SCID and a predisposition for lymphoma caused by the 1306ins8 mutation in DCLRE1C. The nuclease ARTEMIS, encoded by DCLRE1C, is an essential factor in the process of V(D)J recombination and an important component of the nonhomologous end joining (NHEJ) DNA double-strand break (DSB) repair pathway [9, 10].
Finding the mutation in this family was a very important step toward prenatal diagnosis and PGD because the affected children did not show the classical clinical features of SCID and the illness is known to involve several genes in different loci. In order to carry out PGD it is essential to know the precise diagnosis, even though PGD can be done for haplotypes by analyzing the polymorphic markers flanking the region of homozygosity.
However, since the linked markers are not the disease-causing mutation, misdiagnosis is possible if recombination events separate the mutated allele and the linked marker. Even though loci of 1 cM apart are expected to show only approximately 1 % recombination, crossover events between markers and a specific mutation cannot be completely ruled out even for very closely linked markers [8]. In our study the markers are separated by 3 cM, therefore identifying the mutation and associating it with SCID was a necessary step.
Once the familial mutation was found, the family could choose whether to have prenatal diagnosis by either chorionic villus sampling (CVS) or amniocentesis, or to undergo PGD. The couple chose IVF-PGD in order to avoid pregnancy termination in the case of an affected embryo.
In PGD, genetic analysis is performed on single cells, in this case on blastomeres. Molecular testing is done by single cell PCR, which is prone to ADO [13], resulting in failure of amplification of one of the alleles. In blastomeres, simultaneous testing of linked markers reduces the rate of misdiagnosis from 20–30 % to 3 % [15]. ADO is defined as failure of amplification of one locus. ADO of an entire allele may represent aneuploidy for the tested chromosome [1]. In consanguineous marriages with a founder mutation, homozygosity for the mutant alleles, representing affected embryos, can resemble aneuploidy. The question arises, therefore, as to how it is possible to distinguish between aneuploidy and an affected embryo. In our case, if all 5 of the markers together with the mutation gave rise to a single signal, we assumed it was an affected embryo. We tested our assumption by rechecking the affected embryos (8–10 cells) on day 4, and the results confirmed our first diagnosis. We can therefore conclude that PGD for consanguineous families is feasible and reliable.
The couple underwent one cycle of PGD. The distribution of the genotypes of the embryos was consistent with Mendelian inheritance (18 % homozygous wild-type, 18 % affected, and 64 % heterozygous carriers). Two embryos (one homozygous wild-type and one heterozygous carrier) were transferred, resulting in the delivery of healthy twins at 32 weeks. Their genotypes were retested after birth and the original results were confirmed.
This case demonstrates the importance of finding the mutation causing the disease in individuals, even if the mutation is unique to that family. The efforts expended to identify the mutation are worthwhile in order to design more accurate and reliable diagnostic tests of the fetus and, where relevant, PGD.
Acknowledgments
The authors thank Dr. Gabrielle J. Halpern for her help with editing the manuscript. This study was supported by the Adler Chair in Pediatric Cardiology—Tel Aviv University.
Footnotes
Capsule
Identifying the familial mutation in DCLRE1C was an important step for designing a reliable protocol even in the case of a consanguineous family.
References
- 1.Altarescu G, Eldar Geva T, Brooks B, Margalioth E, Levy-Lahad E, Renbaum P. PGD on a recombinant allele: crossover between the TSC2 gene and “linked” markers impairs accurate diagnosis. Prenat Diagn. 2008;28:929–33. doi: 10.1002/pd.2070. [DOI] [PubMed] [Google Scholar]
- 2.Berthet F, Deist F, Duliege AM, Griscelli C, Fischer A. Clinical consequences and treatment of primary immunodeficiency syndromes characterized by functional T and B lymphocyte anomalies (combined immune deficiency) Pediatrics. 1994;93:265–70. [PubMed] [Google Scholar]
- 3.Buckley RH, Schiff RI, Schiff SE, Markert ML, Williams LW, Harville TO, Roberts JL, Puck JM. Human severe combined immunodeficiency: genetic, phenotypic, and functional diversity in one hundred eight infants. J Pediatr. 1997;130:378–87. doi: 10.1016/S0022-3476(97)70199-9. [DOI] [PubMed] [Google Scholar]
- 4.Buckley RH. Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution. Annu Rev Immunol. 2004;22:625–55. doi: 10.1146/annurev.immunol.22.012703.104614. [DOI] [PubMed] [Google Scholar]
- 5.Elder ME. T-cell immunodeficiencies. Pediatr Clin North Am. 2000;47:1253–74. doi: 10.1016/S0031-3955(05)70270-4. [DOI] [PubMed] [Google Scholar]
- 6.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–4. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard WJ. Cytokines and immunodeficiency diseases. Nat Rev Immunol. 2001;1:200–8. doi: 10.1038/35105066. [DOI] [PubMed] [Google Scholar]
- 8.Lewis R. Human Genetics: Concepts and Applications. 6. New York: McGraw Hill; 2005. pp. 150–4. [Google Scholar]
- 9.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–94. doi: 10.1016/S0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 10.Ma Y, Schwarz K, Lieber MR. The Artemis: DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–51. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pesu M, Candotti F, Husa M, Hofmann SR, Notarangelo LD, O’Shea JJ. Jak3, severe combined immunodeficiency, and a new class of immunosuppressive drugs. Immunol Rev. 2005;203:127–42. doi: 10.1111/j.0105-2896.2005.00220.x. [DOI] [PubMed] [Google Scholar]
- 13.Rechitsky S, Strom C, Verlinsky O, Amet T, Ivakhnenko V, Kukharenko V, Kuliev A, Verlinsky Y. Allele dropout in polar bodies and blastomeres. J Assist Reprod Genet. 1998;15:253–7. doi: 10.1023/A:1022532108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thornhill AR, McGrath JA, Eady RA, Braude PR, Handyside AH. A comparison of different lysis buffers to assess allele dropout from single cells for preimplantation genetic diagnosis. Prenat Diagn. 2001;21:490–7. doi: 10.1002/pd.109. [DOI] [PubMed] [Google Scholar]
- 15.Verlinsky Y, Kuliev A. Micromanipulation and biopsy of polar bodies and blastomeres. In: Verlinsky Y, Kuliev A, editors. Atlas of preimplantation genetic diagnosis. 2. Oxon: Taylor and Francis, Abingdon; 2005. pp. 15–22. [Google Scholar]