Abstract
UDP-3-O-(R-3-hydroxyacyl)GlcN N-acyltransferase (LpxD) has been shown to be essential to survival of lipid A producing Gram-negative bacteria. In this study, LpxD-binding peptides 12 amino acids in length were identified from a phage-bound random peptide library screen. Three peptides displayed antibacterial activity when expressed intracellularly, one of which (RJPXD33) represented 15% of the total hits. RJPXD33 binds to E. coli LpxD with a Kd of 6 μM and is competitive with R-3-hydroxymyristoyl-ACP binding. RJPXD33 can be C-terminally fused in vivo with thioredoxin or N-terminally modified in vitro with beta-alanyl-fluorescein and maintain LpxD binding. The latter was used to develop an LpxD fluorescent binding assay used to evaluate unlabeled ligands, and is amenable to small molecule library screening. Furthermore, RJPXD33 also binds to and inhibits E. coli UDP-N-acetylglucosamine acyltransferase (LpxA) with a Kd of 20 μM, unearthing the possibility for the development of small molecule, dual-binding LpxA/LpxD inhibitors as novel antimicrobials.
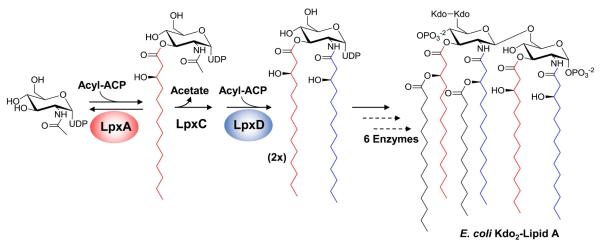
Lipopolysaccharide (LPS) constitutes the environmental facing monolayer of the outer membrane of Gram-negative bacteria. When released from the bacterial membrane during infection, the membrane-anchoring portion of LPS, lipid A, is able to stimulate the host innate immune system, and is responsible for the endotoxic effects associated with Gram-negative sepsis (1, 2). Disruption of early catalytic steps in the biosynthesis of the lipid A, either by chemical or genetic disruption, have proven lethal in Escherichia coli and other Gram-negative bacteria (3-5). Among the essential early pathway enzymes are two acyltransferases (Figure 1), UDP-N-acetylglucosamine acyltransferase (LpxA) and UDP-3-O-(R-3-hydroxyacyl)glucosamine N-acyltransferase (LpxD), which catalyze the first and third steps in the pathway, respectively (4, 6). Structurally, each acyltransferase consist of a left-handed parallel beta-helix (LβH) core structure first discovered through crystallization of LpxA (7-9). Both LpxA and LpxD are Type II acyl carrier protein (ACP)-dependent enzymes and, in E. coli, utilize R-3-hydroxymyristoyl-ACP to catalyze functionally similar reactions (4, 6).
Figure 1.
Kdo2-lipid A biosynthesis in E. coli. Acyltransferases, LpxA and LpxD, catalyze functionally similar acylations of their respective UDP-Glucosamine-based substrates using R-3-hydroxymyristoyl-ACP as the acyl donor.
Both acyltransferases have been touted as novel subcellular targets for antimicrobial development (10, 11), and while in vivo chemical inhibition of the lpxA gene product has been shown to display antimicrobial activity (11), there are no known in vivo active LpxD inhibitors. Of the two acyltransferases, LpxD is thought to be the better target due to the build-up of UDP-3-O-(R-3-hydroxyacyl) glucosamine that occurs in LpxD mutants grown at non-permissive temperatures (6, 10). This compound has detergent-like properties and its accumulation is potentially toxic. In addition to the dearth of bioactive LpxD inhibitors, the historic lack of a non-radioactive enzymatic assay has hampered chemical genetic approaches to finding small molecule inhibitors. Even with the development of a newly describe LpxD fluorescent assay, the scarcity and complex nature of the LpxD substrates required for high throughput screening (HTS) efforts limits the assay’s utility to secondary screening validation.
Peptides that bind to and inhibit essential enzymes have been invaluable as chemical probes used to validate intracellular targets, modulate biosynthetic pathways, develop target-specific binding assays, as well as serve as templates for the development of more pharmacologically relevant peptidomimetic agents (12-14). Therefore, as a prelude to small molecule inhibitor discovery, here we describe the use of a random phage-bound peptide library, combined with ligand-competitive phage elution, to identify bioactive LpxD inhibitory peptides. One bioactive peptide in particular was identified that binds to and inhibits both purified recombinant LpxD and LpxA. A single peptide with the ability to inhibit two steps in early lipid A biosynthesis represents a unique finding and serves as a new example of a dual targeting antimicrobial agent.
Phage display was employed to identify LpxD-binding peptides. LpxD contains a C-terminal helical extension spanning 45 Å from the active site contained within its LβH core structure that proved to be an ideal position to place a biotin tag for biopanning experiments (7, 8). Initial attempts at labeling the C-terminus via cleavage of an LpxD-intein fusion, expressed from pUMGD13, using cysteine-biotin or ethylenediamino-biotin failed to generate biotinylated protein (15). As well, other nucleophiles known to cleave intein protein fusions, such as mercaptoethane sulfonic acid (MESNA) and/or DTT, also failed to cleave the LpxD-intein fusion protein. Therefore, a biotin ligase recognition (BLR) motif coding region was cloned upstream of the intein coding region in pTYB2 to allow for in vivo biotinylation of protein fusions (16, 17). The construct pUMRJ4 contains lpxD cloned upstream of the BLR coding region and upon induction in E. coli BL21-AI/pBirAcm/pUMRJ4, allows for the expression of a 96 kDa protein fusion (LpxD-BLR motif-intein-chitin binding domain). The 96 kDa biotinylated-LpxD intein fusion was bound to chitin resin and the intein linkage cleaved using MESNA, resulting in elution of biotinylated LpxD (38 kDa). The purified protein contained 0.97 moles of biotin per mole of LpxD.
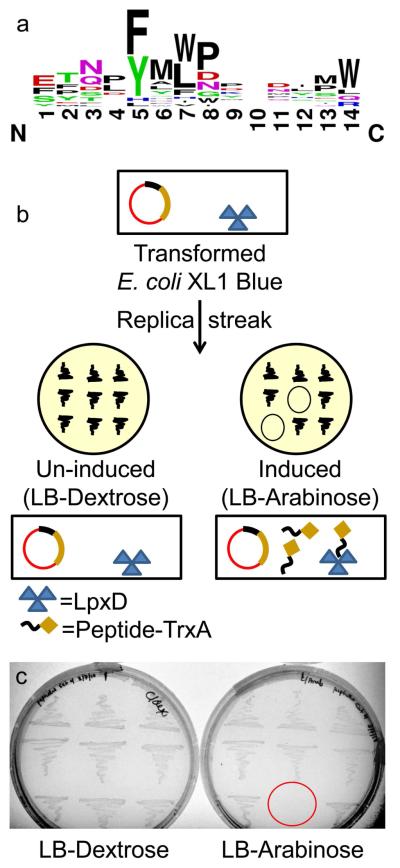
Phage display screening was performed using the Ph.D.-12 random peptide library (NEB), which contains approximately 1.9 billion phage-bound, random 12-amino acid peptides fused to the N-terminus of the M13 phage pIII coat protein. Biopanning experiments were carried out against biotinylated LpxD immobilized on magnetic beads coated with streptavidin (Dynabeads M-270). Bound phage were eluted from immobilized LpxD with either UDP-GlcNAc, previously shown to bind to Chlamydia trachmoatis LpxD (8), or ACP as competitive ligands. Peptide coding sequences from ten randomly selected phage were obtained after rounds three and four of panning. The twenty DNA sequences isolated encoded 17 unique peptides. Peptide sequences were aligned in Clustal W (18) to determine sequence homology (Figure 2, panel a; Supplementary Figure 1). A consensus YMLP motif was identified among a third of the sequences, which is similar to the WMLDP motif located within the LpxA-specific peptide, peptide 920 (P920; SSGWMLDPIAGKWSR) (13). The aromatic-Met-hydrophobic motif is heavily conserved among 50% of the sequences, with variations between tyrosine and phenylalanine in the lead position. Also, the sequences TNLYMLPKWDIP (RJPXD33) and AWWEFNPFAWPY were identified multiple times among the twenty isolated phages.
Figure 2.
Identification and selection of bioactive peptides. a) Sequences of the peptides identified from phage display after rounds 3 and 4 of selection. The figure was generated by WebLogo (weblogo.berkeley.edu) subsequent to alignment in Clustal W. The size of the characters at any given position denotes the frequency of an amino acid, while the height of the stack is indicative of sequence conservation. b) Outline of bioactivity screen for peptides from phage display: E. coli XL1 Blue was transformed with a pool of plasmids coding for the expression of peptides identified from phage display (under the control of the ParaBAD) and evaluated under peptide induction and non-induction conditions. c) Representative replica plates from bioactivity screen: LB-Dextrose (non-induction of peptide), LB-Arabinose (induction of peptide). Red circle indicates no growth of bacteria upon in vivo peptide expression.
The M13 pIII coat protein containing the N-terminal peptide is expressed in the E. coli host with a signal sequence immediately upstream of the peptide library site. This signal sequence targets the modified pIII protein to the E. coli periplasm wherein the signal peptide is cleaved (19). While this compartmentalization away from the cytoplasm is ideal for amplifying and expressing phage displaying inhibitory peptides to essential cytosolic enzymes, bioactivity of such peptides cannot be ascertained within this context (20). Therefore, a general strategy utilized for bioactivity screening of LpxD-binding peptides identified from phage display was worked out (Figure 2, panel b). A low copy, tightly controlled expression construct was employed to promote stringent conditions for bioactivity assays. DNA oligonucleotides, coding for the 17 peptides identified in the phage display screen were cloned into a low-copy (p15A ori) plasmid, pUMRJ100, upstream of the thioredoxin (trxA) coding region (Supplementary Figure 2). The expression of the peptide-TrxA fusions were under the control of the l-arabinose promoter (ParaBAD). The resultant constructs were pooled and transformed into E. coli XL-1 Blue cells and selected on LB-chloramphenicol (LB-cam) plates containing dextrose to suppress basal expression of the peptide fusions from the araBAD promoter. Individual colonies were then re-streaked onto both LB-cam/dextrose (non-induction) and LB-cam/l-arabinose (induction) plates to probe for toxicity (Figure 2, panel c). No toxicity was seen on dextrose containing media. However, individual constructs expressing peptides RJPXD31, RJPXD33, or RJPXD34 displayed in vivo toxicity (no growth) when plated on LB-cam/arabinose media. These three peptides had a more extensive N/QXYMLP motif in common, again with variations in their respective aromatic and hydrophobic amino acids. Plasmids from random colonies not displaying toxicity were sequenced and demonstrated representation of all of the pooled constructs and proper orientation of the peptide coding regions.
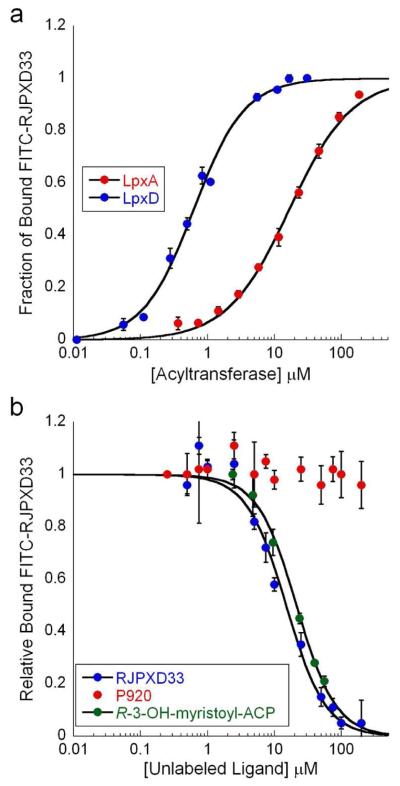
Purified native LpxA and His6-LpxD, which has been shown in literature to exhibit equal activity to wild-type LpxD, were used in peptide binding studies (10). The binding of fluorescein-labeled peptide to acyltransferase was assessed by measuring the changes in fluorescence polarization, which reflects changes in polarization of emitted light due to the differences in mobility between free and protein bound fluorescein-labeled peptide, when excited by polarized light. RJPXD33 was chosen for fluorescent labeling because of its bioactivity and disproportioned representation among the randomly selected phage. Increasing concentrations of acyltransferase, in the presence of fixed [FITC-RJPXD33], resulted in a sigmoidal binding isotherm upon plotting fluorescence polarization (mP) versus Log [acyltransferase]. FITC-RJPXD33 exhibited a Kd = 607 ± 40 nM (Hill coeff = 1) for His6-LpxD with a ΔmP = 295 ± 3 (Figure 3, panel a). While in the presence of LpxA, FITC-RJPXD33 exhibited a Kd = 17 ± 1.6 μM (Hill coeff = 1) and a ΔmP of 254 ± 3. As a negative control experiment, FITC-RJPXD33 was tested against E. coli MurA, UDP-N-acetylglucosamine enolpyruvyl transferase, under the same conditions as with LpxA and LpxD. At 300 μM MurA no significant polarization was observed with FITC-RJPXD33.
Figure 3.
Direct and competetion binding data for bioactive peptides. a) Binding plot showing binding of FITC-RJPXD33 to both LpxD and LpxA. FITC-RJPXD33 was held constant at 20 nM while LpxD or LpxA were titrated at increasing concentrations to determine the binding isotherms. b) A fluorescent, competition LpxD binding assay, in which FITC-RJPXD33 (20 nM) and LpxD (660 nM) were held constant while unlabeled competing ligand (RJPXD33 (blue), P920 (red), or R-3-OH-myristoyl-ACP (green)) was titrated at various concentrations. P920 did not demonstrate binding to LpxD. All data points represent the mean of three individual experiments with error bars representing the standard deviation. The relative amount of bound labeled peptide was normalized by dividing the ΔmP obtained in the presence of competing ligand by the ΔmP obtained in the absence of competing ligand.
Competition binding experiments against unlabeled peptides were also carried out. In these experiments, the ability of labeled peptide to bind to acyltransferase, at varying concentration of unlabeled peptide, was ascertained. RJPXD33 was able to compete against FITC-RJPXD33 for binding to His6-LpxD (Kd = 6.5 ± 0.2 μM; Figure 3, panel b) and against FITC-P920 for binding to LpxA (Kd = 22 ± 2.1 μM; Supplementary Figure 3). RJPXD31 and RJPXD34 were able to displace FITC-RJPXD33 from LpxD as well, with Kds of 41 ± 3 μM and 31 ± 1.5 μM, respectively (Table 1; Supplementary Figure 4). The binding of these two peptides to LpxA were weaker than that of RJPXD33, with RJPXD31 displaying Kd = 119 ± 9 μM, while RJPXD34 did not bind at concentrations up to 300 μM (Supplementary Figure 5). Unlike the LpxD-binding peptides, FITC-P920 showed binding to LpxA, Kd = 187 ± 19 nM, Hill coeff = 1.5, ΔmP = 269 ± 3, but did not bind to His6-LpxD at concentrations up to 50 μM. Unlabeled P920 demonstrated a Kd of 4.7 ± 0.2 μM for LpxA (Supplementary Figure 3), but could not compete against FITC-RJPXD33 for binding to His6-LpxD (Figure 3, panel b), as evidenced by no decrease in polarization at concentrations up to 300 μM of unlabeled P920.
Table 1.
Binding constants of labeled and unlabeled peptides for acyltransferases.
| Peptide Sequence | LpxD Kd (IC50) μM |
LpxA Kd (IC50) μM |
|---|---|---|
| TNLYMLPKWDIP-NH2 (RJPXD33) | 6.5 ± 0.2 (3.5 ± 0.08) | 22 ± 2.1 (19 ± 1.2) |
| FITC-(βa)TNLYMLPKWDIP-NH2 (FITC-RJPXD33) | 0.6 ± 0.04 | 17 ± 1.6 |
| SENNFMLPLLPL-NH2 (RJPXD34) | 31 ± 1.5 (26 ± 3.0) | DNB |
| QHFMVPDINDMQ-NH2 (RJPXD31) | 41 ± 3 | 119 ± 9 |
| SSGWMLDPIAGKWSR (P920) | DNB | 4.7 ± 0.2 |
| FITC-(βa)SSGWMLDPIAGKWSR-NH2 (FITC-P920) | DNB | 0.19 ± 0.02 |
(βa) - β-alanine used for the N-terminal labeling with FITC. DNB – Did Not Bind. Binding constants for FITC labeled peptides were obtained from direct binding experiments as described in Methods. Binding constants for nonlabeled peptides were obtained from competition binding experiments against FITC labeled peptides. IC50 values were obtained using a fluorescent acyltransferase assay as described in Methods.
The above results underscore the unique nature of peptides identified herein. Each of the bioactive peptides identified in this study bound to LpxD with dissociation constants in the range of 6-40 μM. Despite the sequence similarities however, our studies show that RJPXD34 is specific for binding to LpxD, showing no binding to LpxA, while RJPXD31 and 33 show affinity for LpxD as well as LpxA. Those peptides having affinity for both acyltransferases bound approximately three times more tightly to LpxD than to LpxA, which is expected since they had been identified from biopanning against LpxD. Interestingly, the FITC-labeled RJPXD33 bound approximately thirty times more tightly to LpxD than to LpxA. While labeled RJPXD33 had the same affinity as the unlabeled peptide to LpxA, the labeled peptide bound ten times tighter than the unlabeled peptide to LpxD. This may indicate that the β-alanyl-fluorescein moiety is making more extensive interactions within LpxD. However, β-alanyl-fluorescein alone shows no significant polarization (ΔmP ~ 26) at up to 180 μM LpxD.
Bioactive peptides were then tested for their abilities to interfere with substrate binding and acyltransferase activity. Inhibition of enzyme activity was determined for our highest affinity dual binding peptide (RJPXD33) and our LpxD-specific peptide (RJPXD34), using a recently described fluorescent acyltransferase assay (Table 1; Supplementary Figure 6). RJPXD33 displayed IC50s of 3.5 ± 0.08 μM and 19 ± 1.2 μM against LpxD and LpxA, respectively, which are consistent with the relative binding affinities of the unlabeled peptide for each of the acyltransferases (above). Likewise, RJPXD34 had an IC50 of 26 ± 3.0 μM against LpxD and did not inhibit LpxA. Next, competition binding experiments against acyl-ACP substrate was carried out. In this experiment the ability of labeled peptide to bind to acyltransferase in the presence of substrate was ascertained. Utilizing FITC-RJPXD33 as our tracer, R-3-hydroxymyristoyl-ACP shows competitive binding with labeled peptide to LpxD, with a dissociation constant of 8.8 ± 0.7 μM (Figure 3, panel b). This binding constant for acyl-ACP was in good accordance with the published Km value for acyl-ACP to LpxD (10). As a control, fluorescein-labeled RJPXD33 was evaluated for binding to 50 μM holo-ACP or acyl-ACP alone and displayed no significant increase in fluorescence polarization, indicating that the peptide was not binding to ACP. Recent kinetic characterization of E. coli LpxD described an ordered mechanism where acyl-ACP binding is the initial event (10). Peptides in this study were also selected to bind to the unbound form of LpxD and our binding studies show that RJPXD33 binds to LpxD in a mutually exclusive manner with R-3-hydroxymyristoyl-ACP, representing a potential basis for its inhibitory activity. Likewise, RJPXD33 binds to LpxA in a competitive fashion with P920, which has previously been shown to be a competitive inhibitor of LpxA with respect to R-3-hydroxymyristoyl-ACP (11). These observations are consistent with other studies in which randomly selected phage-bound peptides have been shown to bind to “hot spots” at protein•protein or protein•ligand interfaces (21, 22). Also, P920 has been shown to bind within the active site groove of LpxA and the dual binding peptides identified in this study may have an overlapping binding site with P920 on LpxA and bind in a similar fashion within the active site of LpxD.
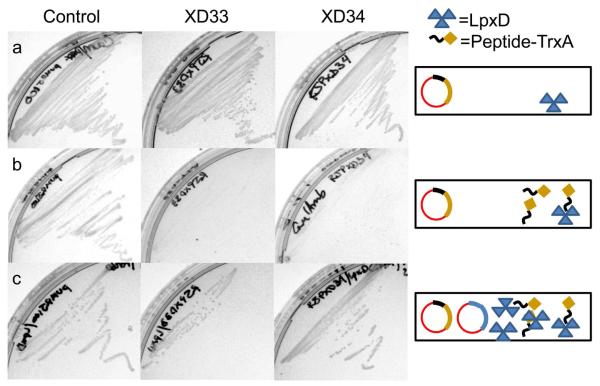
Overexpression of an essential enzyme to suppress the toxicity of an antibacterial agent is a validated strategy used to identify subcellular targets (23-25). This multicopy suppression strategy was adopted to determine whether or not the antibacterial activity displayed by peptides identified in this study were due to inhibition of LpxD in vivo. Again, for this evaluation the most potent dual binding peptide, RJPXD33, as well as the LpxD-specific peptide, RJPXD34 were assessed in cells containing pUC18 or pUC18::lpxD. As seen previously, induction of peptide fusions alone expressing RJPXD33 or RJPXD34 showed no growth. However, as expected, suppression of antibacterial activity was seen upon induction of peptide fusions in cells containing additional extrachromasomally expressed LpxD resulting in bacterial growth (Figure 4). This data is consistent with the in vitro binding and inhibition studies and indicate that the bioactive LpxD-binding peptides primary mode of antibacterial activity is due to inhibition of LpxD. The stronger LpxD affinity along with the LpxA binding ability of RJPXD33 may account for the fewer number of colonies in the LpxD overexpression strain, as compared to RJPXD34.
Figure 4.
Suppression of peptide-mediated antibacterial activity by LpxD overexpression. a) Growth of peptide expression strains on non-induction, LB-Dextrose agar plates: Control = E. coli XL1 Blue/pUMRJ100, XD33 = E. coli XL1 Blue/pUMRJ41, and XD34 = E. coli XL1 Blue/pUMRJ40. b) No growth of peptide expression strains on induction, LB-l-arabinose agar plates: same constructs as in panel a (above). c) Suppression of antibacterial activity of expressed peptide on LB-l-arabinose agar plates, due to overexpression of LpxD from pUMRJ45: same strains as in panels a and b (above) with the addition of pUMRJ45.
In conclusion, the LpxD N-acyltransferase is an essential enzyme of lipid A biosynthesis and has been touted as a novel subcellular target for antimicrobial development (10). The toxicity associated with the expression of LpxD-binding peptides in E. coli in this current study serves as the first example of in vivo chemical inhibition of LpxD activity and further validates LpxD as an antibiotic target. With the continuing rise in resistance to current antimicrobial chemotherapy, much interest exists in finding new, less resistance-prone ways to inhibit bacterial growth. An underexplored, yet promising, niche in the field of antimicrobial development is the targeting of multiple essential proteins with a single antimicrobial agent (26, 27). Such unique agents require an organism to obtain multiple mutations in several essential targets in order to display clinically relevant resistance (27). With the identification of RJPXD33, the targeting of the structurally and functionally similar acyltransferases, LpxA and LpxD may provide another such opportunity. Moreover, protein•ACP interaction inhibitors directed against bacterial Type II ACP-dependent acyltransferases represent an untapped area for the development of unique protein•protein interaction inhibitors as potential antibacterial agents. Finally, this work has identified novel peptides that can serve as surrogate ligands for both LpxA and LpxD. Surrogate ligands such as these can serve as templates for the development of peptidomimetic based compounds more amenable to chemotherapeutic drug delivery. Furthermore, such peptides can be used to develop fluorescent binding assays, such as described above, to facilitate biological screening activities for the identification of small molecule acyltransferase inhibitors.
METHODS
Materials
All DNA primers and Dynabeads M-270 Streptavidin were purchased from Invitrogen. Ph.D.-12 phage peptide library, pTYB2, pTYB3, Chitin beads, and all restriction enzymes were purchased from New England BioLabs (NEB). Plasmid pBirAcm was purchased from Avidity. Bio-Rad Protein Assay and Bio-Gel P2 size-exclusion resin were purchased from Bio-Rad. LB (Lennox) Broth and Agar, and dextrose were purchased from Difco. Peptide synthesis reagents and resins were purchased from Anaspec. Fluorescein isothiocyanate (FITC) was purchased from Acros Organics. ThioGlo1 reagent was purchased from Calbiochem. R-3-hydroxymyristic acid was obtained from Wako Chemicals. All antibiotics and other chemicals were purchased in the highest grade from Fisher Scientific or Sigma-Aldrich. SpectraMax M5 spectrophotometer was from Molecular Devices. DNA sequencing was performed at the University of Michigan DNA Sequencing Core.
Cell culture
A list of all bacteria strains and plasmids are described in Supplementary Table 1. All LB-containing agar plates were incubated at 37 °C unless otherwise indicated. For protein expression and purification, all recombinant E. coli/T7 RNA polymerase promoter constructs were grown in 1 L baffled flask containing 250 mL of LB media supplemented with the appropriate antibiotics. The flasks were shaken (250 rpm) at 37 °C until the cells reached an optical density of OD600 = 0.6. To the cultures were added either 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) (Rosetta (DE3)) or 1 mM IPTG and 0.2% l-arabinose (BL21-AI), and the cultures allowed to incubate at 37 °C for an additional 4 hours. Cells were harvested at 4 °C by centrifugation at 6,000 × g, and lysed by French press. Crude cytosol was prepared by centrifugation of the lysate at 20,000 × g for 30 min at 4 °C.
Biotinylation of E. coli LpxD
The lpxD gene was amplified (pfu DNA polymerase) from E. coli MG1655 (ATCC) genomic DNA by standard PCR protocols with two primers corresponding to the 5′ and 3′ ends of the lpxD open reading frame. The forward primer was CCTTCAATTCGACTGGCTGATTTAGCG and the reverse primer was ATATCTCGAGGTCTTGTTGATTAACCTTGCGCTCAAGCG. The forward primer begins at the second codon of the lpxD reading frame, while the reverse primer contains an XhoI restriction site (underlined) in place of the lpxD stop codon. The purified PCR product was restricted with XhoI and inserted into pTYB3, which had undergone digestion with NcoI, a fill-in reaction with T4 DNA polymerase, and restriction with XhoI. The resulting construct, pUMGD13, contained lpxD with a 3′-intein coding tag.
In a second lpxD-intein construct, a coding region for a 15-amino acid biotin ligase-recognition motif was inserted in frame with, and upstream of, the intein tag coding region in pTYB2 (13, 16, 17). The following complimentary primer pair, 5′-GGGCTGAACGATATTTTTGAAGCGCAGAAAATTGAATGGCATGAACCG-3′ and 5′-CGGTTCATGCCATTCAATTTTCTGCGCTTCAAAAATATCGTTCAGCCC-3′, coding for the biotin ligase-recognition motif (underlined), was blunt-end cloned into the SmaI restriction site of pTYB2 to give pTYB2btc. The XbaI/XhoI restriction fragment from pUMGD13, containing the lpxD open reading frame, was ligated into XbaI/XhoI restricted pTYB2btc to give pUMRJ4 and the insert was confirmed by DNA sequencing. The resulting pUMRJ4 construct contained lpxD with a 3′-biotin ligase recognition motif coding region followed by the intein coding region.
E. coli BL21-AI harboring the plasmids pUMRJ4 and pBirAcm was grown in a 250 mL culture, as indicated above, with d-biotin (12 mg/L) added along with IPTG and l-arabinose during induction. Crude cytosol (10 mL) was applied to a 5 mL Chitin bead column, and the column wash with 10 column volumes of 20 mM Tris-HCl pH 8.0. Immobilized LpxD was cleaved from the intein fusion tag by washing the column with 15 mL of 50 mM mercaptoethane sulfonic acid (MESNA), stopping the flow, and allowing the resin to sit overnight at 4 °C. Fresh buffer was then added to the top of the column and biotinylated LpxD eluted in the first 4 mL. The protein was desalted through a 12 mL Bio-Gel P2 column in 20 mM Tris-HCl pH 8.0. The purified biotinylated LpxD was analyzed by SDS-PAGE and its concentration determined by UV absorbance at 280 nm (ε = 32805 M−1 cm−1). Molar ratio of biotin was determined by 4′-hydroxyazobenzene-2-carboxylic acid (HABA) assay per manufacturer’s protocol (Thermo Scientific).
Phage Display
Streptavidin-conjugated Dynabeads (10 μL) were washed three times with 100 μL of 50 mM Tris-HCl, pH 7.5, 150 mM NaCl (TBS). Biotinylated LpxD (4 pmol) in 100 μL of TBS was added to Dynabeads (10 μL) and the mixture was allowed to incubate at room temperature for 30 min. The beads were magnetically separated from the supernatant and washed five times with 100 μL TBS. To the affinity captured biotinylated LpxD, 100 μM biotin (100 μL) was added and incubated at room temperature for 10 min to block any excess streptavidin sites. The beads were again washed five times with 100 μL TBS. Phage-bound peptide library (Ph.D.-12), at a concentration of 1 × 109 – 1 × 1010 plaque forming units (pfu), was added to the immobilized LpxD in a total volume of 100 μL TBS. This suspension was allowed to incubate for 45 min at room temperature with gentle mixing. Following the incubation period the supernatant was removed and the beads were washed ten times with 100 μL TBS. Bound phage were eluted by incubating for 15 min with gentle mixing with either 50 μM acyl carrier protein (ACP) or 500 μM UDP-GlcNAc in 100 μL TBS buffer. The phage were then amplified by direct infection with E. coli XL1 Blue, isolated and tittered as previously described, and used in the next cycle (28). In subsequent rounds of panning 0.1% Tween 20 was added to all TBS buffer containing solutions. Following rounds three and four, individual plaques were selected at random, phage purified, and the DNA from each was sequenced.
Bioactivity Assay
pACYC184, isolated from E. coli JM110 (dam−), was restricted with HindIII/BclI and the fragment containing the p15A origin of replication and chloramphenicol resistance marker was isolated from a 1% agarose gel. The araC and thioredoxin genes were individually amplified using pBAD-thio as the template in two separate PCR reactions. For the araC gene 5′-GCGCTGATCATTATGACAACTTGACGGCTACATCATTC-3′ was used as the forward primer and 5′-GCGCCATATGTATGTATATCTCCTTCTTAAAGTTAAAC-3′ was used as the reverse primer. The forward primer contains a BclI restriction site (underlined) and the reverse primer contains an NdeI restriction site (underlined). The thioredoxin gene was cloned using 5′-GCGCCATATGGGGCCCGGATCTGATAAAATTATTCATCTGACTG-3′ as the forward primer (NdeI site underlined) and 5′-CCGAGGAGAGGGTTAGGGATAGGC-3′ as the reverse primer. The araC PCR product was restricted with BclI and NdeI, while the thioredoxin containing PCR fragment was restricted with NdeI and HindIII. These two inserts were ligated together with the BclI/HindIII restricted pACYC184 fragment, resulting in the formation of pUMRJ100.
Complementary DNA oligonucleotides coding for peptides identified from phage display were purchased in the following format, NdeI-(dodecapeptide coding region)-ApaI. Complementary oligos were mixed in equal molar ratios, heated to 94 °C for 2 min, and allowed to cool to room temperature over several minutes. The hybridized oligos were restricted with NdeI and ApaI, pooled, and ligated into pUMRJ100 restricted with the same enzymes. The ligation mixture was used to transform competent E. coli XL1 Blue and the transformed cells selected on LB agar containing 1% dextrose and 10 μg/mL chloramphenicol (LB-cam/dex) at 37 °C overnight. Seventy-two random colonies were selected and streaked onto both LB-cam/dex and LB-cam/0.2% l-arabinose (8 replica plates). Colonies which grew on the dextrose containing plates, but not on the arabinose containing plates were indicative of bioactive peptide expression. Plasmids were isolated from colonies on the LB-cam/dex plates having the desired phenotype and the peptide coding regions determined by DNA sequencing. Empty plasmid vector was used as a negative control and showed no toxicity.
E. coli LpxA and LpxD expression constructs
E. coli His6-LpxD and LpxA were expressed and purified as previously described (29). The pUC18::lpxD construct was made from a previously described wild-type pET expression construct (29). The lpxD gene was excised from the pET construct by XhoI restriction/T4 DNA polymerase fill-in, followed by XbaI restriction. The insert was then ligated into pUC18 which had been restricted with HindIII, filled-in with T4 DNA polymerase, and restricted with XbaI. The resulting lpxD pUC18 construct was designated pUMRJ45. All constructs were verified by DNA sequencing.
Acytransferase substrates
R-3-hydroxymyristoyl-ACP and UDP-3-O-(R-3-hydroxymyristoyl) glucosamine were synthesized and purified as previously described (29).
Fluorescence polarization protein binding assay
Peptides FITC-RJPXD33 (FITC-(βA)TNLYMLPKWDIP-NH2), RJPXD33 (TNLYMLPKWDIP-NH2), RJPXD31 (QHFMVPDINDMQ-NH2), RJPXD34 (SENNFMLPLLPL-NH2), and FITC-P920 (FITC-(βA)SSGWMLDPIAGKWSR-NH2) were synthesized using rink-amide resin (0.4 mmol/g capacity) by established peptide coupling procedures. Labeled peptides contained an N-terminal β-alanine (βA) linker coupled to fluorescein using fluorescein isothiocyanate (FITC). Peptides were purified via RP-HPLC and analyzed by electrospray ionization mass spectrometry (ESI-MS) at the University of Michigan Chemistry Mass Spectrometry Services. Purified peptides were suspended in DMSO and diluted to a concentration of 200 nM in 20 mM HEPES, pH 8.0 (~0.1% DMSO). LpxA and His6-LpxD were serially diluted to appropriate concentrations in 20 mM HEPES, pH 8.0 and 45 μL of the dilutions were added to 354-well black Costar plates. To this, 5 μL of labeled peptide was added for a final concentration of 20 nM FITC-peptide. The wells were gently mixed and incubated at 30 °C for 15 min. Fluorescence polarization was measured on a SpectraMax M5 plate reader with data points obtained in triplicate. The data for FITC-RJPXD33 binding to LpxD was normalized to the experimentally determined polarization of the fully bound peptide (mPb), and the resulting binding curves fit to Hill equation (1) using KaleidaGraph software.
| (1) |
Where α is the fraction of FITC-peptide bound, h is the Hill coefficient, [P] is the concentration of protein, and Kd is the dissociation constant of protein•peptide complex. For RJPXD33 binding to LpxA the polarization values of the fully bound labeled peptide (mPb) and Kd were determined by nonlinear fit of the polarization versus [Acyltransferase] plots to Eq. (2). Where mP is the experimentally determined polarization, mPf is the polarization of free FITC-RJPXD33, P is the total amount of acyltransferase, and Kd is the dissociation constant of the protein•peptide complex. The calculated mPb was then used to normalize the experimental data and the resulting binding curves fit to Eq. (1).
| (2) |
For competition binding assays, 220-660 nM of acyltransferase was incubated in the presence of varying concentrations of unlabeled peptides or acyl-ACP for 10 min at 30 °C. FITC-labeled peptide (20 nM) was added and incubated in the dark at 30 °C for 15 min before reading. The [I]50 was determined from the competition binding curve and the dissociation constant of the unlabeled ligand was calculated as previously described using Eq. (3) (30):
| (3) |
Where [I]50 is the free unlabeled ligand (inhibitor) concentration at 50% inhibition, [L]50 is the free ligand (fluorescent tracer) concentration at 50% inhibition, and [P]0 is the free protein concentration at 0% inhibition, Kd is the dissociation constant of the ligand (fluorescent tracer), and Ki is the calculated dissociation constant for the unlabeled ligand.
Acyltransferase inhibition assays
The acyltransferase reaction was observed in the forward direction using a continuous fluorescent assay. The fluorescent assay detects holo-ACP liberation via conjugation with ThioGlo1 reagent, which could be monitored as an increase in fluorescence from excitation and emission wavelengths set to λex = 379 nm and λem = 513 nm. Assays were performed on a SpectraMax M5 microplate reader (auto cutoff set to λ = 495 nm, PMT set to low and precision set to 40 scans per well) using Corning black, 96-well half-area plates. The assay mixture in a final volume of 100 μL contained 20 mM HEPES pH 7.5, 10 μM ThioGlo, 600 μM UDP-GlcNAc (LpxA assay) or 4.5 μM UDP-3-O-(R-3-hyrdoxymyristoyl)-GlcN (LpxD assay), 9 μM R-3-hydroxymyristoyl-ACP, 10 nM acyltransferase and varying concentrations of peptide (200 μM – 1 μM). Assays contained a final concentration of 2% (v/v) DMSO from peptide stock solutions. All components with the exception of enzyme were mixed and incubated in the dark at 30°C for 5 min. To initiate the reaction, a fresh dilution of acyltransferase was added and the reaction was monitored for 10 min. Assays were run in triplicate with the average IC50 being reported. A negative control containing no inhibitor and a positive control containing no acyltransferase were used for 100% and 0% relative activity, respectively.
Multicopy suppression
Competent E. coli XL1 Blue/pUMRJ45 were transformed separately with pUMRJ40 (pUMRJ100::rjpxd34), pUMRJ41 (pUMRJ100::rjpxd33), or pUMRJ100 (control). The transformed cells were grown overnight at 37 °C on LB agar containing 1% dextrose, 100 μg/mL ampicillin, and 10 μg/mL chloramphenicol (LB-amp/cam/dex). Single colonies were selected and streaked onto both LB-amp/cam/dex and LB-amp/cam/0.2% l-arabinose plates. These plates were incubated overnight at 37 °C.
Supplementary Material
Acknowledgement
This research was supported in part by a Valteich Research Award administered by the College of Pharmacy, University of Michigan and by a Pre-Doctoral Research Grant administered by the University of Michigan Rackham Graduate School. R.J.J was supported in part by the University of Michigan Chemistry-Biology Interface (CBI) training program (NIH), grant number 5T32GM008597-14 and an American Foundation for Pharmaceutical Education fellowship. The authors would like to thank the Peptide Synthesis Facility at the Division of Structural Biochemistry of the Research Center Borstel, Germany, for the gift of unlabeled P920.
Footnotes
Associated Content
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Meredith TC, Aggarwal P, Mamat U, Lindner B, Woodard RW. Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem. Biol. 2006;1:33–42. doi: 10.1021/cb0500015. [DOI] [PubMed] [Google Scholar]
- 2.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vuorio R, Harkonen T, Tolvanen M, Vaara M. The novel hexapeptide motif found in the acyltransferases LpxA and LpxD of lipid A biosynthesis is conserved in various bacteria. FEBS Lett. 1994;337:289–92. doi: 10.1016/0014-5793(94)80211-4. [DOI] [PubMed] [Google Scholar]
- 4.Anderson MS, Bull HG, Galloway SM, Kelly TM, Mohan S, Radika K, Raetz CR. UDP-N-acetylglucosamine acyltransferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavorable. J Biol Chem. 1993;268:19858–65. [PubMed] [Google Scholar]
- 5.Galloway SM, Raetz CR. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J Biol Chem. 1990;265:6394–6402. [PubMed] [Google Scholar]
- 6.Kelly TM, Stachula SA, Raetz CR, Anderson MS. The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase. The third step of endotoxin biosynthesis. J Biol Chem. 1993;268:19866–19874. [PubMed] [Google Scholar]
- 7.Bartling CM, Raetz CR. Crystal structure and acyl chain selectivity of Escherichia coli LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry. 2009;48:8672–8683. doi: 10.1021/bi901025v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buetow L, Smith TK, Dawson A, Fyffe S, Hunter WN. Structure and reactivity of LpxD, the N-acyltransferase of lipid A biosynthesis. Proc Natl Acad Sci U S A. 2007;104:4321–4326. doi: 10.1073/pnas.0606356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raetz CR, Roderick SL. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 10.Bartling CM, Raetz CR. Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry. 2008;47:5290–5302. doi: 10.1021/bi800240r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams AH, Immormino RM, Gewirth DT, Raetz CR. Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide. Proc Natl Acad Sci U S A. 2006;103:10877–10882. doi: 10.1073/pnas.0604465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaffer ML, Deshayes K, Nakamura G, Sidhu S, Skelton NJ. Complex with a phage display-derived peptide provides insight into the function of insulin-like growth factor I. Biochemistry. 2003;42:9324–9334. doi: 10.1021/bi034386c. [DOI] [PubMed] [Google Scholar]
- 13.Benson RE, Gottlin EB, Christensen DJ, Hamilton PT. Intracellular expression of Peptide fusions for demonstration of protein essentiality in bacteria. Antimicrob Agents Chemother. 2003;47:2875–2881. doi: 10.1128/AAC.47.9.2875-2881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toogood PL. Inhibition of protein-protein association by small molecules: approaches and progress. J Med Chem. 2002;45:1543–1558. doi: 10.1021/jm010468s. [DOI] [PubMed] [Google Scholar]
- 15.Lesaicherre ML, Lue RY, Chen GY, Zhu Q, Yao SQ. Intein-mediated biotinylation of proteins and its application in a protein microarray. J Am Chem Soc. 2002;124:8768–9. doi: 10.1021/ja0265963. [DOI] [PubMed] [Google Scholar]
- 16.Kwon K, Beckett D. Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Sci. 2000;9:1530–1539. doi: 10.1110/ps.9.8.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 19.Wickner W. Mechanisms of membrane assembly: general lessons from the study of M13 coat protein and Escherichia coli leader peptidase. Biochemistry. 1988;27:1081–1086. doi: 10.1021/bi00404a001. [DOI] [PubMed] [Google Scholar]
- 20.Hyde-DeRuyscher R, Paige LA, Christensen DJ, Hyde-DeRuyscher N, Lim A, Fredericks ZL, Kranz J, Gallant P, Zhang J, Rocklage SM, Fowlkes DM, Wendler PA, Hamilton PT. Detection of small-molecule enzyme inhibitors with peptides isolated from phage-displayed combinatorial peptide libraries. Chem Biol. 2000;7:17–25. doi: 10.1016/s1074-5521(00)00062-4. [DOI] [PubMed] [Google Scholar]
- 21.Sidhu SS, Fairbrother WJ, Deshayes K. Exploring protein-protein interactions with phage display. Chembiochem. 2003;4:14–25. doi: 10.1002/cbic.200390008. [DOI] [PubMed] [Google Scholar]
- 22.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 23.Xu HH, Real L, Bailey MW. An array of Escherichia coli clones over-expressing essential proteins: a new strategy of identifying cellular targets of potent antibacterial compounds. Biochem Biophys Res Commun. 2006;349:1250–1257. doi: 10.1016/j.bbrc.2006.08.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Zolli-Juran M, Cechetto JD, Daigle DM, Wright GD, Brown ED. Multicopy suppressors for novel antibacterial compounds reveal targets and drug efflux susceptibility. Chem Biol. 2004;11:1423–1430. doi: 10.1016/j.chembiol.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 25.Chopra I. Over-expression of target genes as a mechanism of antibiotic resistance in bacteria. J Antimicrob Chemother. 1998;41:584–588. doi: 10.1093/jac/41.6.584. [DOI] [PubMed] [Google Scholar]
- 26.Pucci MJ, Podos SD, Thanassi JA, Leggio MJ, Bradbury BJ, Deshpande M. In vitro and in vivo profiles of ACH-702, an isothiazoloquinolone, against bacterial pathogens. Antimicrob Agents Chemother. 2011;55:2860–2871. doi: 10.1128/AAC.01666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charifson PS, Grillot AL, Grossman TH, Parsons JD, Badia M, Bellon S, Deininger DD, Drumm JE, Gross CH, LeTiran A, Liao Y, Mani N, Nicolau DP, Perola E, Ronkin S, Shannon D, Swenson LL, Tang Q, Tessier PR, Tian SK, Trudeau M, Wang T, Wei Y, Zhang H, Stamos D. Novel dual-targeting benzimidazole urea inhibitors of DNA gyrase and topoisomerase IV possessing potent antibacterial activity: intelligent design and evolution through the judicious use of structure-guided design and structure-activity relationships. J Med Chem. 2008;51:5243–5263. doi: 10.1021/jm800318d. [DOI] [PubMed] [Google Scholar]
- 28.Sparks AB, Adey NB, Cwirla S, Kay BK. Screening phage-displayed random peptide libraries. Phage display of peptides and proteins: A laboratory manual. 1996:227–253. [Google Scholar]
- 29.Jenkins RJ, Dotson GD. A continuous fluorescent enzyme assay for early steps of lipid A biosynthesis. Analytical Biochemistry. 2012 doi: 10.1016/j.ab.2012.02.027. doi: 10.1016/j.ab.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.