Abstract
Recent advances in studies of bacterial gene expression have brought the realization that cell-to-cell communication and community behavior are critical for successful interactions with higher organisms. Species-specific cell-to-cell communication is involved in successful pathogenic or symbiotic interactions of a variety of bacteria with plant and animal hosts. One type of cell–cell signaling is acyl-homoserine lactone quorum sensing in Gram-negative bacteria. This type of quorum sensing represents a dedicated communication system that enables a given species to sense when it has reached a critical population density in a host, and to respond by activating expression of genes necessary for continued success in the host. Acyl-homoserine lactone signaling in the opportunistic animal and plant pathogen Pseudomonas aeruginosa is a model for the relationships among quorum sensing, pathogenesis, and community behavior. In the P. aeruginosa model, quorum sensing is required for normal biofilm maturation and for virulence. There are multiple quorum-sensing circuits that control the expression of dozens of specific genes that represent potential virulence loci.
Keywords: bacterial signaling, biofilms, pathogenesis, Pseudomonas aeruginosa, virulence mechanisms
It was once held that most bacteria function only as individuals designed to compete with one another and to multiply rapidly under appropriate conditions. After all, dense cultures of bacteria can be grown from a single cell. This concept has given way to the view that, like other creatures, bacteria can communicate with each other and form communities that represent more than the sum of the individuals (for example, see refs. 1–3). It is now clear that many bacterial species use chemicals to signal each other and to coordinate their activities. Several different types of signals have now been described. Many Gram-positive bacteria use small peptides in signaling one another (4, 5), Gram-negative bacteria appear to use small molecule signals of various sorts (1, 6–9). Perhaps the best-studied signaling system is the acyl-homoserine lactone (acyl-HSL) system used by a large number of Gram-negative bacterial species. This type of bacterial cell-to-cell communication was discovered in the context of microbial ecology, but it is now evident that acyl-HSL signaling is important in plant and animal (including human) diseases. The signaling pathway is an enticing target for the development of antipathogenic therapies. As will be discussed below, acyl-HSL signaling represents a dedicated communication system that is used by bacteria to control specific genes in response to population density. Acyl-HSLs are small molecule signals with no other known function. These chemical signals are produced by specific enzymes, and they are detected by specific receptors. Because acyl-HSL signaling provides a mechanism by which a bacterial species can monitor its own population density, this type of signaling and other signaling systems that achieve the same purpose have been termed quorum-sensing systems (10).
Overview of Acyl-HSL Quorum Sensing
In most described cases, acyl-HSL signals are generated by the activity of a single enzyme that uses as substrates S-adenosylmethionine and an intermediate of fatty acid biosynthesis, acyl-acyl carrier protein (11–15). The enzyme is generally a member of the LuxI family of acyl-HSL synthases. Different LuxI homologs generate different acyl-HSLs. Thus Pseudomonas aeruginosa RhlI primarily catalyzes the synthesis of N-butyryl-HSL (C4-HSL), and P. aeruginosa LasI directs the synthesis of N-(3-oxododecanoyl)-HSL (3OC12-HSL). The acyl side-chain length and the substitutions on the side chain provide signal specificity. Acyl side chains of these signals can be fully saturated, they can have hydroxyls or carbonyls on the third carbon, and they can have lengths of 4 to 16 carbons (ref. 7; A. Schaefer and E.P.G., unpublished data). Short-chain signals such as C4-HSL diffuse freely through the cell membrane (16, 17), and 3OC12-HSL partitions into cells, presumably in the membrane. The 3OC12-HSL signal can diffuse into the surrounding environment but export is enhanced by the mexAB-oprM, and perhaps other, efflux pumps (17, 18). Regardless, the cellular concentration of an acyl-HSL is defined by the environmental concentration, and environmental concentrations can rise only when there is a sufficient population of the signal-producing bacterium.
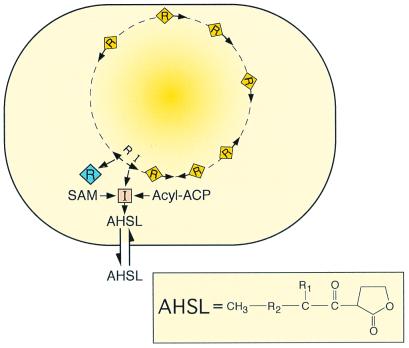
The specific receptors for acyl-HSL signals are members of the LuxR family of transcriptional regulators. LuxR family members have been proposed to consist of two domains, a C-terminal DNA-binding domain, and an N-terminal acyl-HSL-binding domain (for a review see ref. 19). A simple model depicting an acyl-HSL quorum-sensing circuit is shown in Fig. 1. Quite often the two regulatory genes (the R and I genes) are linked, but not always. The orientation of the two genes with respect to each other is variable.
Figure 1.
Generalized scheme for an acyl-HSL quorum-sensing circuit in a bacterial cell. The orange square indicates an acyl-HSL synthase-LuxI homolog. The diamonds are a LuxR homolog. Yellow diamonds on the bacterial chromosome are the LuxR homolog activated by the acyl-HSL signal. The arrows on the chromosome are qsc genes. The acyl-HSL (AHSL) signal can diffuse in and out of cells. The compound in the box is an acyl-HSL (R1, H, OH, or O; R2, (CH2)2–14, or CH2-CH2-CH⩵CH-CH2-CH2). The substrates for the acyl-HSL synthase are an acylated acyl carrier protein (Acyl-ACP) and S-adenosylmethionine (SAM).
Acyl-HSL quorum sensing is commonly found in Gram-negative bacteria that interact with plant and animal hosts. Quorum sensing was first discovered to control the luminescence of Vibrio fischeri, a bacterium that forms a mutualistic light organ symbiosis with certain marine animals (20, 21). Here quorum sensing is critical to the symbiosis. Acyl-HSL signaling is critical for virulence of the plant pathogen Erwinia carotovora (22) and for virulence of P. aeruginosa in mouse models of lung (23) and burn infections (24), in invertebrates (25–27), and in plants (28). Thus acyl-HSL quorum sensing appears as a common theme in the interaction of several different bacterial species with eukaryotic hosts. We will describe the elements of quorum sensing, and discuss some of the factors controlled by quorum sensing in P. aeruginosa. In this paper P. aeruginosa will serve as a model for the role of bacterial communication in community behaviors important in pathogenesis.
Quorum Sensing in P. aeruginosa
P. aeruginosa can be isolated from soil and water. It is also an opportunistic pathogen of humans, other animals, and plants. One of the reasons P. aeruginosa is a successful opportunistic pathogen is that it produces a battery of secreted virulence factors. These virulence factors include exoproteases, siderophores, exotoxins, and lipases. Many of these virulence factors are regulated by quorum sensing (for reviews see refs. 1, 29, and 30). Of what advantage to P. aeruginosa is quorum sensing control of virulence factors? First, it is economical to produce extracellular factors only after a critical population has been achieved. A mass of cells is required to produce sufficient quantities of these factors to influence the surrounding environment. Furthermore, in the host, timing of the deployment of virulence factors may be critical. The pathogen can amass without displaying its virulence factors, and then the pathogen can mount a surprise attack in which the arsenal of virulence factors is deployed in a coordinated and overwhelming fashion.
Genetic studies have revealed two quorum-sensing systems in P. aeruginosa. Both of these systems, LasR-I and RhlR-I, have linked R and I genes. They are the quorum-sensing systems (31–36). In addition, the recently completed P. aeruginosa genome sequencing project has revealed a third LuxR homolog that is adjacent to a cluster of quorum-sensing-controlled (qsc) genes (37). However, a third LuxI homolog is not evident from the sequence, and the function of the third LuxR homolog is as yet unknown. LasR is a transcriptional regulator that responds primarily to the LasI-generated signal, 3OC12-HSL, and RhlR is a transcriptional regulator that responds best to the RhlI-generated signal, C4-HSL. The current model for quorum sensing in P. aeruginosa is as follows: at low population densities LasI produces a basal level of 3OC12-HSL. As density increases, 3OC12-HSL builds to a critical concentration, at which point it interacts with LasR. This LasR–3OC12-HSL complex then activates transcription of a number of genes. The list of target genes includes lasB, toxA, rhlR, and lasI (29, 32, 36, 38, 39). A curious fact is that different target genes are activated at different 3OC12-HSL concentrations (39). Activation of lasI by LasR creates a positive autoregulatory loop. The activation of rhlR by LasR results in a quorum-sensing regulatory cascade, in which activation of the rhl system requires an active las system. RhlR responds best to the RhlI-generated C4-HSL. RhlR then activates expression of genes required for production of a variety of secondary metabolites such as hydrogen cyanide and pyocyanin (for a review see ref. 29). A DNA sequence with dyad symmetry called a lux-box-like sequence can easily be identified in the promoter regions of many quorum-sensing-controlled (qsc) genes (10, 37, 40, 41). By analogy to other acyl-HSL quorum-sensing systems we deduce that the lux-box-like sequences function as binding sites for LasR and RhlR. It is not yet clear how RhlR and LasR discriminate between their respective binding sites. In fact, many genes show partial activation with either LasR or RhlR and the appropriate acyl-HSL (for example see refs. 30 and 37). One explanation for the partial activation or incomplete specificity is that binding site discrimination is less than perfect and either LasR or RhlR can bind with varying efficiency to any lux-box-like element. However, lux-box-like sequences are not apparent in the promoter regions of all qsc genes. This observation suggests that LasR or RhlR may also bind to identified sequences, or that some qsc genes are controlled by LasR or RhlR indirectly.
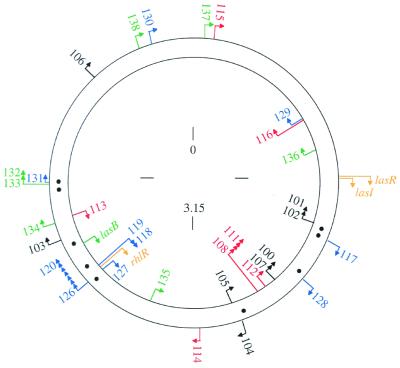
As discussed above, many genes have been reported to come under the control of quorum sensing in P. aeruginosa. For some genes such as lasB there is a considerable amount of evidence in support of this conclusion (36, 38). For other genes, the data are limited, and in many cases the degree of transcriptional control reported is low. A recent study used a random mutagenesis approach to identify 39 genes that were highly regulated (minimum 5-fold induction, maximum 740-fold induction) by quorum sensing (37). The genes were divided into four different classes, two of which respond to 3OC12-HSL, and two of which required both C4-HSL and 3OC12-HSL for maximal induction. The qsc genes map throughout the P. aeruginosa chromosome (Fig. 2), confirming the view that quorum sensing in this bacterium represents a global regulatory system (29). The 39 genes revealed by the random mutagenesis study represent only a subset of the qsc genes in P. aeruginosa. It was estimated that as many as 4% of the roughly 6,000 P. aeruginosa genes are controlled by quorum sensing (37).
Figure 2.
Map of qsc genes or operons on the P. aeruginosa chromosome. Arrowheads indicate the direction of transcription. The different colors indicate different regulatory classes. Black, genes that respond primarily to 3OC12-HSL and that can respond to added signal in early logarithmic phase. Red, genes that respond primarily to 3OC12-HSL, but only after cultures have entered stationary phase (late genes). Blue, genes that respond best to both 3OC12-HSL and C4-HSL, and that can respond to added signal in early logarithmic phase. Green, late genes that require both signals for full expression. The lasR, lasI, and rhlR genes are shown in gold. The locations of lux-box-like elements are shown as black dots between the two DNA strands. These elements were identified in putative promoter regions of some but not all of the qsc genes. The numbers are distance in megabases (Mb) on the approximately 6.3-Mb chromosome (numbering starts at oriC). This figure is from ref. 37.
One report indicates that transcription of rpoS, a gene encoding an RNA polymerase σ subunit involved in expression of stationary-phase factors, is activated by RhlR and C4-HSL (42). This finding raises the possibility that many genes may be controlled indirectly rather than directly by quorum sensing. It is also an enticing hypothesis because it lends itself to the idea that one specific cue that enables a cell to anticipate stationary phase is crowding. Unfortunately, quorum-sensing control of rpoS transcription is an example for which there is limited evidence. It is also an example for which there are low levels of induction (at best 3-fold). In fact, recent investigations suggest that quorum sensing may have no significant influence on rpoS transcription in P. aeruginosa (43).
There are other control elements that affect the quorum-sensing regulatory circuit. The gacA gene product is a transcriptional activator that among other things induces C4-HSL production (44). The rsaL gene is downstream of the lasR gene and it is involved in negatively regulating lasR (45). Vfr is a global regulator that affects a mild activation of lasR (46). The environmental signals that these regulators respond to are unknown. Furthermore, the physiological significance of the observed levels of regulation by these factors remains to be determined. Further complicating the quorum-sensing regulatory scheme is the recent discovery that a specific quinolone produced by P. aeruginosa can serve as an extracellular signal to activate lasB (8). The mechanisms that underlie quinolone signaling remain unknown.
Regulation of Virulence by Quorum Sensing in P. aeruginosa
Mutations in elements of the quorum-sensing machinery in P. aeruginosa do not markedly influence the growth of this bacterium in the laboratory. For example, the growth rate of the LasI−, RhlI− double mutant PAO-MW1 in Luria–Bertani broth at 30°C or 37°C is similar to that of the wild-type PAO1 under normal laboratory culture conditions. Yet quorum-sensing mutant strains show severe virulence defects in various mouse models, in invertebrate models, and in a plant model system. We will briefly describe the results of experiments in which colonization of the lungs in a neonatal mouse model by a LasR mutant was compared with colonization by the wild-type parental strain (23). When neonatal BALB/cByJ mice were inoculated intranasally with wild-type P. aeruginosa, the bacteria colonized the lung, causing an acute pneumonia, bacteremia, and death. In contrast, a LasR mutant strain could colonize the lung, but it did not achieve high densities and it did not cause pneumonia, bacteremia, or death. The mutant did not have a growth defect under laboratory conditions, it could invade the lung and survive but it could not cause disease.
Biofilms and Quorum Sensing
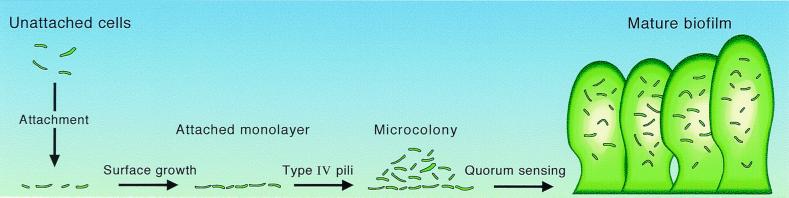
Bacteria often tend to attach to surfaces and form communities enmeshed in a self-produced polymeric matrix. These communities are called a biofilm (2, 47). P. aeruginosa is often found in naturally occurring biofilms. Under the appropriate laboratory conditions, P. aeruginosa forms characteristic biofilms that can be several hundred micrometers thick (Fig. 3). Development of a mature biofilm proceeds through a programmed series of events (for a recent review see ref. 2). After attachment, cells multiply to form a layer on a solid surface. Individuals in the layer then exhibit a surface motility called twitching. Twitching depends on type IV pili. As a result of twitching motility, small groups of P. aeruginosa called microcolonies form. Microcolonies then differentiate to form a mature biofilm. Microcolonies in a mature biofilm have tower- and mushroom-shaped architectures. The cells in these structures are encased in an extracellular polysaccharide matrix. Water channels that allow the flow of nutrients into and waste products out of the biofilm innervate these structures. There is a significant physiological heterogeneity within biofilms. In P. aeruginosa biofilms there is a steep oxygen gradient. Oxygen is present at measurable concentrations mainly at the periphery of the biofilm. Oxygen microelectrode studies have also shown that water channels serve to bring oxygen to deeper areas of the biofilm. Similar gradients may be expected for pH and nutrients. These gradients dictate physiological variability among individual cells in the biofilm, with slower-growing cells present deeper within the biofilm and more actively growing cells at the periphery. This heterogeneity in physiological activity makes studying biofilms with traditional molecular microbiological techniques difficult. Bacteria in these mature biofilms are phenotypically resistant to microbicidal agents, including antibiotics. Thus biofilms cause many different types of chronic or persistent bacterial infections (for a recent review of biofilm infections and biofilm physiology see ref. 2).
Figure 3.
Diagram of the P. aeruginosa biofilm-maturation pathway. Unattached cells that approach a surface may attach. Attachment involves specific functions. Attached cells will proliferate on a surface and use specific functions to actively move into microcolonies. The high-density microcolonies differentiate into mature biofilms by a 3OC12-HSL-dependent mechanism.
Recent studies have linked quorum sensing and biofilm maturation (48). This is a particularly gratifying finding because quorum sensing functions to control gene expression in groups of bacteria, and biofilms are just that, organized groups of bacteria. A mutation in lasI has a dramatic affect on biofilm maturation. LasI mutants are incapable of 3OC12-HSL synthesis, and the development of LasI mutant biofilms is arrested after microcolony formation but before maturation of the microcolonies into thick structured assemblages. Thus LasI mutant biofilms appear flat and undifferentiated. The normal biofilms architecture can be restored to the mutant by addition of the LasI-generated quorum-sensing signal 3OC12-HSL. A RhlI mutant exhibits normal biofilm development and architecture. The 3OC12-HSL-responsive qsc genes involved in biofilm maturation remain unknown. Of interest, the LasI mutant biofilms were susceptible to treatment with the detergent SDS, whereas wild-type biofilms were resistant. LasR− mutants have biofilm phenotypes similar to that of LasI− mutants (Fig. 4). These observations suggests that antimicrobial therapies targeting the quorum-sensing mechanism of P. aeruginosa may result in the formation of abnormal biofilms that are more amenable to treatment. Another study showed that acyl-HSLs could be detected in clinical biofilm isolates and on catheters colonized by P. aeruginosa in mice (49).
Figure 4.
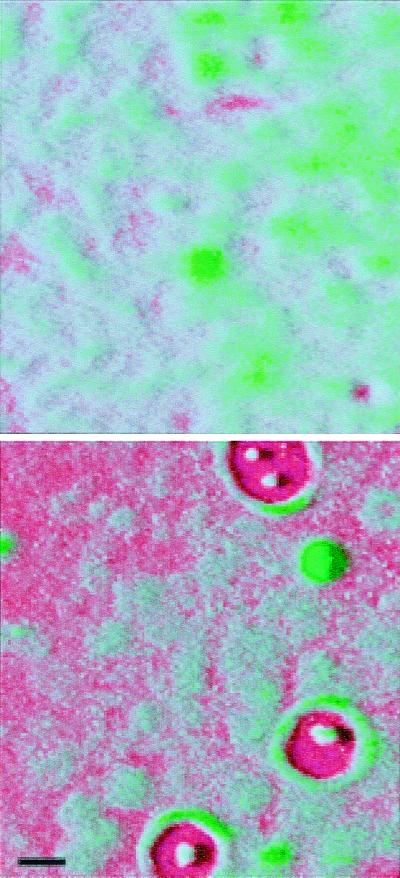
Scanning confocal microscope images of a mature P. aeruginosa wild-type biofilm (Upper) and a quorum-sensing mutant biofilm (Lower). In this case the quorum-sensing mutant was a lasR, rhlR double mutant. The perspective is from above the biofilm on a glass surface. The glass surface is red, and the green is from the green fluorescent protein encoded by the gfp gene in the recombinant P. aeruginosa. The wild-type biofilm consists of thick microcolonies. The immature mutant biofilm appears thinner, and more of the glass surface is exposed. With the lasR, rhlR mutant shown here (but not with lasI, rhlI mutants) zones of clearing around microcolony towers are often observed. Other experiments have shown that these zones are filled with extracellular polysaccharide (M.R.P., unpublished data). The biofilms are in flow-through reaction vessels similar to those described in ref. 48. The colors were applied to the image by computer enhancement with Adobe photoshop 5.0. The black marker bar is 100 μm in length.
Some critical questions remain to be answered regarding the quorum-sensing mechanism in biofilms: What constitutes a quorum in a biofilm? Does a LasR–LasI, RhlR–RhlI regulatory cascade exist within a biofilm? Do acyl-HSL synthesis patterns change within a biofilm? Biofilm and quorum-sensing research has led us to appreciate the fact that P. aeruginosa can behave as a community. Because evidence supports the hypothesis that both biofilms and quorum sensing play integral roles in P. aeruginosa pathogenesis, more studies of the community activities of this bacterium are needed.
Future Challenges
It is now generally appreciated that bacteria possess specific communication systems and that they are capable of organizing into functional communities. We have described one type of signaling system, an acyl-HSL system, in some detail in this article. Other signaling systems are known, but it seems apparent that many more remain to be discovered. Aside from the discovery of different types of chemical communication systems for intraspecies signaling, a challenge for the future is to begin to address the possibility that there is significant interspecies communication. The idea of interspecies communication is supported by a limited body of information. For example, we know that many different bacterial species make a signal to which Vibrio harveyi responds; however, we do not yet know the nature of the signal (for a recent review see ref. 9). Because quorum sensing is required for virulence of P. aeruginosa and other bacteria, the quorum-sensing system is a target for development of new types of therapeutics, antipathogenic agents, agents that do not kill bacterial pathogens but that do interfere with their ability to cause infections. A challenge that faces us is to identify inhibitors of quorum sensing and test their effectiveness in the treatment of infections, particularly persistent biofilm infections. Another challenge is to better understand the network of genes regulated by quorum sensing, and to identify qsc genes involved in normal biofilm maturation and infection by organisms such as P. aeruginosa.
Acknowledgments
We thank M. Hentzer, A. Heydorn, M. Givskov, and S. Mølin for help with the Biofilm experiments. Work by the authors was supported by grants from the National Institutes of Health (GM59026), the National Science Foundation (MCB 9808308), and the Cystic Fibrosis Foundation.
Abbreviations
- acyl-HSL
acyl-homoserine lactone
- C4-HSL
N-butyryl-HSL
- 3OC12-HSL
N-(3-oxododecanoyl)-HSL
Footnotes
This paper was presented at the National Academy of Sciences colloquium “Virulence and Defense in Host–Pathogen Interactions: Common Features Between Plants and Animals,” held December 9–11, 1999, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Fuqua C, Greenberg E P. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 2.Costerton J W, Stewart P, Greenberg E. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 3.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 4.Dunny G M, Leonard B A. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 5.Novick R P, Muir T W. Curr Opin Microbiol. 1999;2:40–45. doi: 10.1016/s1369-5274(99)80007-1. [DOI] [PubMed] [Google Scholar]
- 6.Fuqua W C, Winans S C. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuqua W C, Winans S C, Greenberg E P. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 8.Pesci E C, Milbank J B, Pearson J P, Kende A S, Greenberg E P, Iglewski B H. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassler B L. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua W C, Winans S C, Greenberg E P. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Jr, Greenberg E P. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsek M R, Val D L, Hanzelka B L, Cronan J E, Jr, Greenberg E P. Proc Natl Acad Sci USA. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moré M I, Finger D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 14.Hanzelka B, Parsek M R, Val D L, Dunlap P V, Cronan J E, Jr, Greenberg E P. J Bacteriol. 1999;181:5766–5770. doi: 10.1128/jb.181.18.5766-5770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoang T T, Ma Y, Stern R J, McNeil M R, Schweizer H P. Gene. 1999;237:361–371. doi: 10.1016/s0378-1119(99)00331-5. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan H B, Greenberg E P. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson J P, Van Delden C, Iglewski B H. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans K, Passador L, Srikumar R, Tsang E, Nezezou J, Poole K. J Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens A M, Greenberg E P. In: Cell-Cell Signaling in Bacteria. Dunny G, Winans S C, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 231–242. [Google Scholar]
- 20.Nealson K H, Hastings J W. Microbiol Rev. 1979;43:469–518. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruby E G. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 22.Pirhonnen M, Flego D, Heikiheimo R, Palva E T. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang H B, DiMango E, Bryan R, Gambello M, Iglewski B H, Goldberg J B, Prince A. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rumbaugh K P, Griswold J A, Hamood A N. J Burn Care Rehabil. 1999;20:42–49. doi: 10.1097/00004630-199901001-00008. [DOI] [PubMed] [Google Scholar]
- 25.Tan M W, Rahme L G, Sternberg J A, Tompkins R G, Ausubel F M. Proc Natl Acad Sci USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan M W, Mahajan-Miklos S, Ausubel F M. Proc Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahajan-Miklos S, Tan M W, Rahme L G, Ausubel F M. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 28.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 29.Pesci E C, Iglewski B H. Trends Microbiol. 1997;5:132–135. doi: 10.1016/S0966-842X(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 30.Van Delden C, Iglewski B H. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brint J M, Ohman D E. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gambello M J, Kaye S, Iglewski B H. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latifi A, Winson K M, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Mol Microbiol. 1995;17:333–344. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 34.Ochsner U A, Koch A K, Fiechter A, Reiser J. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochsner U A, Reiser J. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 37.Whiteley M, Lee K M, Greenberg E P. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gambello M J, Iglewski B H. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seed P C, Passador L, Iglewski B H. J Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson J P, Pesci E C, Iglewski B H. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pesci E C, Pearson J P, Seed P C, Iglewski B H. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 43.Whiteley, M., Parsek, M. & Greenberg, E. P. (2000) J. Bacteriol. 182, in press. [DOI] [PMC free article] [PubMed]
- 44.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Hass D. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 45.de Kievit T, Seed P C, Nezezon J, Passador L, Iglewski B H. J Bacteriol. 1999;181:2175–2184. doi: 10.1128/jb.181.7.2175-2184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albus A M, Pesci E C, Runyen-Janecky L J, West S E, Iglewski B H. J Bacteriol. 1997;179:3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 48.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 49.Stickler D J, Morris N S, McLean R J, Fuqua C. Appl Environ Microbiol. 1998;64:3486–3490. doi: 10.1128/aem.64.9.3486-3490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]