Abstract
The expression of endogenous retrotransposable elements including Long Interspersed Nuclear Element-1 (LINE-1 or L1) and Human Endogenous Retrovirus (HERV)-K accompanies neoplastic transformation and infection with viruses such as HIV. The ability to engender immunity safely against such self-antigens would facilitate the development of novel vaccines and immunotherapies. Here we address the safety and immunogenicity of vaccination with these elements. We employed immunohistochemical analysis and literature-precedent to identify potential off-target tissues in humans and establish their translatability in preclinical species to guide safety assessments. Immunization of mice with murine L1 Open Reading Frame-2 (L1O2) induced strong CD8 T cell responses without detectable tissue damage. Similarly, immunization of rhesus macaques with human L1O2 (96% identity with macaque), and Simian ERV (SERV)-K Gag and Env induced polyfunctional T cell responses to all antigens, and antibody responses to SERV-K Env. There were no adverse safety or pathology findings related to vaccination. These studies provide the first evidence that immune responses can be induced safely against this class of self antigens, and pave the way for their investigation as HIV- or tumor-associated targets.
Introduction
Both tumors and tumor cell lines exhibit aberrant expression of L1 (1–5) and HERV-K (6–17). In studies of breast cancer, ovarian clear cell carcinoma, and non-small cell lung cancer, L1 or HERV activity is associated with poor prognosis and tumor progression to a more invasive phenotype (3, 11, 15, 16, 18). Such tumor-associated expression of endogenous retrotransposable element (ERE) antigens makes them potential targets for vaccines and immunotherapies. Naturally arising T and B cell responses to HERV-K Env in breast cancer patients suggest that immune tolerance, if any, of this self-antigen can be overcome even without therapeutic intervention (16). Further, HERV-E-specific T cells mediated tumor regression of a metastatic renal cell carcinoma following an allogenic stem cell transplant (19). These findings suggest that active or passive immunization against ERE antigens could potentially benefit cancer patients whose tumors express such target antigens.
ERE expression is also associated with certain viral infections. EBV infection transactivates expression of HERV-K18 Env, a T cell superantigen, which enables the virus to establish long-term infection of its host (20). HIV-1 infection triggers HERV-K RNA and protein expression since antibody responses to HERV-K are more commonly found in HIV-1 infected persons (21–24). Further, T cell responses have been observed against a variety of HERV peptides in HIV-1-infected patients, with the magnitude and breadth of these responses correlating negatively with viral load (24–26). Indeed, the HERV-K family Human Mouse mammory tumor virus-Like (HML)-2 Gag and Env proteins are detectable in HIV-1-infected cells in vitro (Jones et al., submitted). T cell lines targeting these antigens are capable of killing autologous HIV-1-infected but not uninfected cells independently of the HIV-1 clade (Jones et al., submitted). These studies open the possibility that ERE antigens might be useful surrogate targets for vaccination against HIV-1, which ordinarily escapes immune control by mutation.
The utility of EREs as tumor- or viral infection-associated antigens will be critically dependent on the level of expression in healthy cells, and upon their immunogenicity. Since EREs comprise some 41.3% of the human genome (27, 28) selecting the correct elements will be challenging, especially in the light of T cell responses to degraded open reading frames (ORFs) of HERV-E, H and L (19, 24–26). Indeed, the majority of EREs are inactive after germ-line invasion as a result of inactivating mutations. Only approximately 100 of the half-million genomic copies of L1 are known to be active in modern day humans (2, 29–31). Likewise, no loci of the most recent of the HERV-K subfamily to enter the human genome (HML-2) have proved to be replication competent (32–36).
To define the safety of vaccines and therapeutics targeting EREs, it is critical to first define the expression of L1 and HERV-K(HML-2) in healthy tissues. The reported findings of L1 (37–43) and HERV-K (44) expression underscore the potential for these antigens to be expressed in normal healthy cells in situ. This raises the safety concerns of inducing autoimmune pathology or immune complex disease by ERE-based vaccines and immunotherapies. We sought to investigate the pre-clinical safety and immunogenicity of vaccines based on L1O2, and consensus sequences of Simian ERV-K (SERV-K) Gag and Env in relevant animal models. Here, we probed mouse, human and non-human primate healthy adult tissues with novel or commercially available antibodies to L1O2 and HERV/SERV-K Env and Gag to identify potential target tissues, which together with literature precedent (39, 42, 45), became the focus of our preclinical safety studies. Next we demonstrated that immunization of mice with murine L1O2 (mL1O2) is safe and immunogenic inducing a CD8 T cell response without associated tissue damage, allowing us to proceed to rhesus macaques vaccination studies. Vaccination with human L1O2 (hL1O2) and SERV-K Gag and Env induced T cell responses to all three antigens and antibody responses to SERV-K Env in Indian rhesus macaques without vaccine-related pathology. The safety and immunogenicity findings reported here support the evaluation of ERE-targeting vaccines and immunotherapeutics in relevant disease models.
Materials and Methods
L1O2 antigen constructs
LINE-1.3 (L19088) (46) was used for human constructs (hL1O2). A L1O2 consensus sequence of eight hot L1 elements identified by similarity to L1.3 (47) was used to probe the mouse genome for similar intact L1O2 genes (O88913, O88914, O88915, Q792I9, Q91Z88, Q91Z89, Q9QUI2, Q9QWY0, Q9QWY2 & Q9QWY3), which were used to produce a mL1O2 consensus sequence. Amino acid substitutions D205A in the endonuclease domain, and D702A in the reverse transcriptase (RT) domain were made in both hL1O2 and mL1O2 by site-directed mutagenesis to prevent enzymatic functions. Full-length (FL) L1O2 as well as L1O2 fragment 1 (Fr1) covering amino acids 1–400, including the endonuclease domain; Fr2 covering amino acids 401–800 including the RT domain; and Fr3 covering amino acids 801–1275, were synthesized for both hL1O2 and mL1O2. Alignments of L1O2 were made with the draft rhesus genome sequence, but large gaps were found in the RT domain and the endonuclease domain was missing. Investigation of the translated nucleotide database for rhesus macaques using tBLASTn identified 47 nucleotide sequences for rhesus L1O2. A consensus sequence of these had a predicted amino acid sequence identity of 92% (96% positivity according to the NCBI BLOSUM62 scoring matrix) with hL1O2. We opted to use the hL1O2 sequence in macaque vaccination studies on the grounds of its known provenance and high conservation with the expected rhesus form.
SERV-K antigen constructs
tBLASTn and a library of HERV-K(HML-2) RT peptide sequences were used to screen build 36.3 of the Celera genome assembly. PERL scripts were used to extract, de-fragment and align matches corresponding to proviral insertions containing both LTRs. The alignments constructed were edited manually in Se-Al (http://tree.bio.ed.ac.uk/software/seal/) to construct consensus ORFs. A second round of screening was then performed, in which the consensus gag, pol and env ORFs were used to BLAST search the human genome. Reading frames that approximated the expected size were considered potentially intact and were manually inspected in Se-Al. Phylogenetic screening of the macaque genome identified an ERV family closely related to HML-2 (48). Five intact (or nearly intact) SERV-K (HML-2) gag genes (NC_007858.1, NC_007864.1, NC_007864.1_2, NC_007875.1 and NC_007876.1) and four intact (or nearly intact) env genes (NC_007868, NC_007858, NC_007862 and AC200900_BAC) were identified in the macaque genome, and used to construct consensus SERV-K (HML-2) gag and env sequences.
Vaccines
Genes were synthesized in their native (L1O2) or codon-optimized form (ERV-K), at GeneArt, and cloned into pPJV7563 as described previously (49). DNA vaccines were precipitated onto gold beads as described (50). The control vaccine plasmid was vector backbone only. For rhesus macaque experiments, the antigen plasmids were co-precipitated onto gold beads at a 9:1 ratio together with pPJV7563-encoded rhesus GM-CSF as described (49). For recombinant adenovirus serotype 5 (rAd5) production, the genes were cloned into pShuttle-CMV and recombined with the Ad5 genome using the AdEasy™ system (Q-Biogene Inc, Carlsbad, CA). Control rAd5 vectors encoded eGFP. Final production and purification of rAd5 vaccines was performed by ViraQuest Inc. (North Liberty, IA).
Primary antibodies
Anti-HERV-K Env mouse mAb HERM-1811-5 was obtained from Austral Biologics (San Ramon, CA). Antibodies capable of binding human, macaque and mouse L1O2, or both HERV- and SERV-K Gag, were derived by affinity-purification from peptide-KLH hyper-immunized New Zealand White rabbit serum by Lampire (Pipersville, PA) for hL1O2, and Cambridge Research Biochemicals, (Billingham,UK) for HERV-K Gag. Surface-accessible, immunogen peptides were selected using Protean software from DNASTAR-Lasergene 6. For hL1O2 an anti-RT 781–800 FKENYKPLLKEIKEETNKWK peptide (90% conserved with the predicted macaque sequence) was selected. For HERV-K Gag, peptides were selected from p15: 229–250 ENKTQPPVAYQYWPPAELQYR (92% conserved with the SERV-K p15), and capsid (CA) 337–360 KSFSIKLLKDLKEGVKQYGPNS (96% conserved with the SERV-K CA). Peptide synthesis and conjugations for hL1O2 were performed by New England Peptide; and for HERV-K Gag by Cambridge Research Biochemicals. Antibodies were validated for antigen specificity and cross-species reactivity by ELISA, western blot, and immunoprecipitation followed by in-gel digestion and MALDI-TOF MS to demonstrate antigen specific pull-down from transfected cell lysates.
In-gel digestion and MALDI-TOF MS
Bands of interest in Bis-Tris gels were excised, cut into small pieces and the proteins reduced, alkylated and digested with trypsin (Promega, Madison, WI) overnight at 37°C. The supernatant was removed and two extractions with 60 % acetonitrile, 0.1 % TFA were performed. All three fractions were combined and their volume reduced to 5 µL in a speed-vac. 15 µL of 0.1 % TFA was added to each sample and the peptides were purified using C18 ZipTips (Millipore, Bedford, MA) according to the manufacturer’s instructions. 1 µL of the eluted peptide solution was mixed with 1 µL of a saturated solution of α-cyano-4- hydroxycinnamic acid (in 50 % acetonitrile/ 0.1 % TFA) and spotted onto a stainless steel MALDI plate. Spectra were acquired on a Bruker Ultraflex II MALDI mass spectrometer in reflector mode. After internal calibration using two trypsin autolysis peaks (m/z = 842.50 and 2211.09 Da) experimental peptide masses were matched against mass lists generated by in silico digestion of the antigen sequences.
Tissue Arrays
Tissue arrays were created for humans mice, rhesus and cynomolgus macaques. One or more representative sections from each pivotal organ (with a minimum of one section from each paired organ) were collected and processed from healthy individuals. The tissue list comprised of the following: gastrointestinal tract (tongue, salivary glands, stomach, duodenum, jejunum, ileum, cecum and colon); endocrine organs (thyroid, pancreas, adrenal and thyroid); skeletal muscle (gastrocnemius in mice and quadriceps in macaques); cardiovascular system (heart, aorta and mesenteric arteries); skin; lymphoid organs (tonsil, spleen, lymph node and thymus); bone marrow; central (cerebrum including hippocampus and hypothalamus, cerebellum, medulla, pituitary and spinal cord) and peripheral (sciatic nerve) nervous system; respiratory tract (nasal mucosa, trachea, bronchus and lung); adipose tissue; urinary tract (urinary bladder and kidney); eye (retina); and liver for both sexes. The reproductive tract tissue list was: female (ovary, uterus, cervix, vagina and mammary gland) and male (testis, prostate, seminal vesicle, epididymis and mammary gland).
Processing for Histopathology and Immunohistochemistry
Tissue sections or cell cultures were fixed in 10% neutral buffered formalin, embedded in paraffin, cut into 4 or 5µm sections, and mounted onto Superfrost plus slides (Fisher Scientific, Pittsburg, PA). Immunohistochemistry was performed by loading onto the Ventana XT autostainer (Roche Diagnostics Indianapolis, IN). The system dewaxes the slides, pre-treats them with a Tris/EDTA pH 8.0 antigen retrieval system (Ventana mCC1) for 16 min, blocks endogenous peroxidise and then stains with primary antibody for 1 h at room temperature, and subsequently a biotinylated donkey anti-species (Jackson ImmunoResearch Laboratories Inc, Westgrove, PA) for 30 min before incubation with streptavidin HRP and DAB substrate (Roche Diagnostics). A hematoxylin counterstain (Merck KGaA, Darmstadt, Germany) was applied according to manufacturer’s instructions. After removal of the slides from the Ventana system they were dehydrated, treated with xylene to clear and mounted using DPX. Primary antibodies were validated on fixed eGFP- or antigen-transfected HEK 293 cells. In order to maximize the likelihood of staining at physiological expression levels in tissues, the antibodies were used at the highest concentration that gave isotype control-like staining of the eGFP-transfected cells, but an intense staining of the antigen-transfected positive control cells. The specificity of staining with peptide-specific antibodies was tested by pre-incubation of the antibody with increasing concentrations of the cognate or control irrelevant peptides for 30 min prior to application of the entire mixture to the slide. All images were captured using the NanoZoomer 2.0 (Hamamatsu, Japan) and analyzed by drug safety and ACVP certified pathologists with extensive immunohistochemistry experience. The intensity (scored on a 4-point scale from minimal to marked intensity), distribution (nuclear, cytoplasmic or cell membrane associated), and characteristics (granular, punctuate or diffuse), of the staining as well as the proportion of cells stained (approximate percentage) were recorded. The staining was considered specific to ERE expression if it differed in intensity (usually 3–4), distribution and/or characteristics from the isotype control; and could be inhibited only by the immunogen peptide. Staining consistent with common artefacts (e.g. necrotic cells) was discounted.
Animals
Female Balb/C (18–20g body weight) were purchased from Charles River Canada (Montreal, QC). All mouse experiments were conducted under the guidelines of Pfizer Canada Inc., Animal Care Committees and under the requirements of the Canadian Council on Animal Care (CCAC). Indian rhesus macaques (Macaca mulatta, 16 male and 8 female, median weight 7.78 kg, median age 8 years) were housed at the Wisconsin National Primate Research Center. The University of Wisconsin Institutional Animal Care and Use Committee reviewed and approved all study protocols, which were in accordance with the US Department of Health and Human Services Guide for the Care and Use of Laboratory Animals. Animals used in this study were typed for the MHC-I alleles Mamu-A*01, Mamu-A*02, Mamu-A*08, Mamu-A*11, Mamu-B*01, Mamu-B*03, Mamu-B*04, Mamu-B*08, Mamu-B*17, and Mamu-B*29 using sequence-specific primers (51, 52). We excluded Mamu-B*17+ and Mamu-B*08+ animals from this study because these alleles are associated with spontaneous control of SIV replication.
Animal procedures
Mice: DNA vaccination was given using the PMED technology which delivers DNA directly into the cells of the epidermis (53). Mice were randomly assigned to groups (n = 10) and the abdomen was shaved 20 min prior to DNA vaccination. 2 µg pDNA was administered to the abdomen by the PMED ND10 device, or rAd5 (108 virus particles in 50 µL PBS per dose) was i.m. injected to the tibialis anterior. Mice were primed with pDNA followed by boosts with pDNA or rAd5 in 4-week-intervals and examined two weeks following the final boost. At the end of the vaccination phase following termination, the mice underwent necropsy examination. The following tissues based either on biological importance and/or ERE expression were then collected and processed as described above for histopathological evaluation: lungs, heart, kidneys, liver, urinary bladder, pancreas, mesentery, adrenal glands, thymus, lymph node, brain, skeletal muscle (gastrocnemius/soleus block), haired skin and injection site (skeletal muscle). Rhesus macaques: animals were prepared for vaccinations as described (49). A total of six actuations of the PMED ND10 device were given spread bilaterally into the epidermis of the inguinal lymph node regions to vaccine group 1 and the controls at weeks 0, 6 and 12, and to vaccine group 2 at weeks 8, 14 and 20. At week 33 vaccine group 2 was given 12 actuations using the PMED X15 device spread over both inguinal lymph node regions and the lower abdomen as a final boost. The total DNA doses were 3.6 or 7.2 µg of each of three antigen encoding plasmids and 1.8 or 3.6µg of RhGM-CSF (co-formulated onto the same gold beads) per dosing session for the ND10 and X15 immunizations respectively. Four weeks after the final vaccination, the macaques were challenged by the intrarectal route with SIVsmE660 once per week until they had detectable plasma viremia, or by the intravenous route if they remained uninfected, as described (54, 55) (and Sheppard et al manuscript in preparation). 10 to 12 weeks following infection the macaques were euthanized with Beuthanasia-D (up to 177 mg/k IV) (manufactured by Schering-Plough Animal Health Corp., Union NJ) and were necropsied. The following tissues were collected and prepared as described above: cerebrum, cerebellum, brain stem, spinal cord, pituitary gland, stomach, duodenum, jejunum, ileum, cecum, colon, pancreas, liver, gallbladder, lung, kidneys, thyroid glands, trachea, esophagus, ascending aorta, adrenal glands, axillary lymph nodes, inguinal lymph nodes, mesenteric lymph nodes, mandibular salivary glands, spleen, tongue, skeletal muscle, urinary bladder, diaphragm, eyes with optic nerve, bone (stifle), bone marrow, thymus (if present), mammary gland, haired skin, ovary/testis, prostate, seminal vesicles, uterus/cervix, vagina, heart and any lesions noted during gross examination.
Clinical chemistry, haematology and urinalysis
The complete blood count, differential and reticulocyte parameters were measured using whole blood (K2 EDTA) on the Siemens Advia 120 hematology analyzer. Standard clinical chemistry parameters were measured in serum on the Siemens Advia 2400 chemistry analyzer. Serum insulin was measured using the Siemens Advia Centaur automated immunoassay platform. Glucagon was measured in plasma (K2 EDTA and aprotonin) using the BioPlex Luminex suspension array system. Urinalysis was performed using the Clinitek Atlas Chemistry analyzer. Additionally, urine creatinine and N-acetylglucosamide were also performed on the Advia 2400. Microscopic analysis of urine sediment was performed on all urinalysis samples using light microscopy.
Peptides
For mL1O2, peptides were synthesized by New England Peptide Inc (Gardner, MA). Two sets were obtained: 15-mer peptides overlapping by 10 residues; and predicted MHC class I (H-2d) restricted epitope peptides of 9 amino acids generated using the MHC-I processing method at IEDB (ANN). Seventeen predicted peptides were derived from Fr1; 18 from Fr2; and 27 peptides from Fr3. For the macaque studies, 15-mers overlapping by 11 amino acids spanning the entire protein sequence of hL1O2 (JPT, Berlin, Germany), SERV-K Gag and Env (both from Pepscan, Lelystad, The Netherlands) were obtained. 9-mer peptides predicted to bind the Mamu-A*01 and –A*02 MHC-I molecules for the same proteins were predicted using the MHCPathway Macaque algorithm (www.mamu.liai.org) and obtained from Genescript (Piscataway, NJ). Peptides were divided into pools of no more than 10 peptides.
T cell cytotoxicity assay
Ex vivo CTL assay was conducted using Cr- release assay as described previously (56) with modifications. Briefly, splenocytes (3×107 cells) from individual mice were cultured in complete RPMI1640 medium supplemented with recombinant mouse IL-2 (10U/ml) and a pool of predicted 9-mer peptides in the context of H-2d (10 µg/ml each) at 37°C for 5 days ex vivo. Target cells P815 (H-2d) were pulsed with and without peptides and then labeled with sodium chromate (Perkin Elmer, Waltham, MA). Unpulsed target cells were used as negative controls. Various effector: target ratios were incubated for 4 h. Data presented as percentage of specific lysis (mean ± S.D.).
T cell enrichment
Splenocytes (2×107 cells) were enriched for either CD4+ or CD8+ T cells by negative selection using magnetic beads following the manufacturer’s protocol (Milltenyi Biotechnoies Inc., Auburn, CA). Flow through cells were collected and counted. The purity of T cells as assessed by Flow cytometry analysis using FlowJo Software V8.1 (Tree Star Inc., Ashland, OR) was consistently over 93%.
Intracellular cytokine staining
All antibodies were purchased from BD Biosciences (San Jose, CA). We incubated 1 × 106 PBMC with 10 µg of Brefeldin A (Sigma–Aldrich, ST. Louis, MO) per ml for 6 h at 37 °C in 100 µL of R10 [RPMI 1640 containing 10% fetal calf serum, 2 mM l-Glutamine, 10 U/ml Penicillin G, 10 µg/ml Streptomycin, and 0.025 µg/ml Amphotericin B (HYClone, Logan, UT)] to prevent protein transport from the Golgi apparatus with or without peptide stimulation with 10 µM peptide. For experiments with CD107a, we added the anti-CD107a at the beginning of the assay and added Monensin according to the manufacturer’s directions with the BFA. After the incubation period we washed and stained the cells for selected surface markers (CD8 and CD4) and fixed overnight in 2% paraformaldehyde at 4 °C. On the following day, we permeabilized the cells with 0.1% saponin in PBS containing 10% FCS (HyClone) and stained them for intracellular expression of CD3 and the cytokines gamma-interferon (IFN-γ) and TNF-α. After staining, we washed the cells with PBS containing 10% FCS and fixed in 2% paraformaldehyde for at least 30 min at 4 °C. We collected 1–3 × 105 events within the lymphocyte gate with FASCDiva 6.0 software on a custom made BD LSR II flow cytometer (BD Biosciences). We analyzed the data with FlowJo v.8.7.3. (Treestar Inc.).
ELISPOTs
For murine studies, the frequency of IFN-γ-secreting cells in whole splenocytes, or enriched CD4+ or CD8+ T cells was determined using ELISPOT assay as previously described (57) with modifications. Syngenic naive splenocytes were incubated with a pool of 9-mers or two pools of 15-mers derived from designated fraction of mL1O2, at a concentration of 10µg/ml each followed by X-ray (22Gy) irradiation. The irradiated cells were co-cultured with effectors for overnight at 37°C. Following IFN-γ spot development, spots were enumerated on a Zeiss Reader using KS ELISPOT 4.5 software by ZellNet Consulting Inc., (Fort Lee, NJ). For rhesus macaque ELISPOTs fresh peripheral blood mononuclear cells (PBMC) isolated from EDTA-anticoagulated blood were used for the detection of IFN-γ-secreting cells as previously described using both predicted MHC-binding peptides and overlapping 15-mer peptides (55, 58).
ELISA assays
Recombinant SERV-K Gag CA, HERV-K Env transmembrane (TM) and surface unit (SU) protein ELISAs were performed in 384-well plates. Proteins were diluted to 2µg/ml in PBS and the plates coated overnight at 4°C before washing, blocking for 1h with 1% (w/v) BSA/PBS, and application of a eight-point 0.5 Log10 dilution series of rhesus macaque sera diluted from a top concentration of 1:10 (v/v) in 1% (w/v) BSA/PBS. After extensive washing, the binding of antigen-specific IgG was measured using HRP-conjugated anti-human IgG (Southern Biotech, Birmingham, AL). The reciprocal titer was reported as the intercept of the curve with an arbitrary cut-off value of OD450nm = 1 (based on a reader range of 0 – 4), intersecting the linear portion of the curves of positive samples in all cases and excluding weakly- or non-reactive samples.
Protein expression and antigen-lysate preparation
SERV-K CA consensus residues 282–554 (with N-terminal His-tag) was expressed in E. coli (T7 express, Merck KGaA). Soluble protein was purified from cell lysates by affinity chromatography using a 5ml HisTrap FF column on an Äkta Xpress (GE Healthcare, Port Washington, NY). HERV-K Env SU and TM were expressed at 10–25L wave-bag scale in transiently-transfected HEK293 cells (Invitrogen) and were harvested 5 days post-transfection. Secreted protein was purified via N- and C-terminal His-tags by overnight incubation with 5ml resin (HisSelect, Sigma). Lysates containing full-length SERV-K Gag and Env, and hL1O2 Fr2 for use in western blots were expressed by transient transfection of 106 HEK293 cells with 5µg of the appropriate expression plasmid followed by lysis via sonication in lysis buffer (sterile PBS/0.05% Tween 20 plus cOmplete Mini protease inhibitor cocktail [Roche Diagnostics]) at 48 h post transfection.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software (v5.01, GraphPad Software, San Diego, CA) and were vetted by a statistician. Groups were compared using Kruskal-Wallis test followed by two-tailed, unpaired Mann-Whitney tests, except in the case of ELISA titer data, which was log-transformed and analysed by ANOVA and unpaired Student’s t-test.
Results
Target colocalization studies in human, rhesus and cynomolgus macaque and mouse tissues
To predict the potential risks of ERE vaccination in humans and how well these might be modelled in non-human primates (SERV-K and L1O2) and mice (L1O2 only), we first performed immunohistochemistry of mouse, human, and rhesus and cynomolgus macaque healthy tissue arrays. Our reagent antibodies were the commercial HERV-K Env mAb HERM-1811-5, and our own antibodies raised against one hL1O2 peptide and two HERV-K Gag peptides. We validated the antibodies extensively for our purposes demonstrating specificity and cross-species reactivity (Fig S1–3).
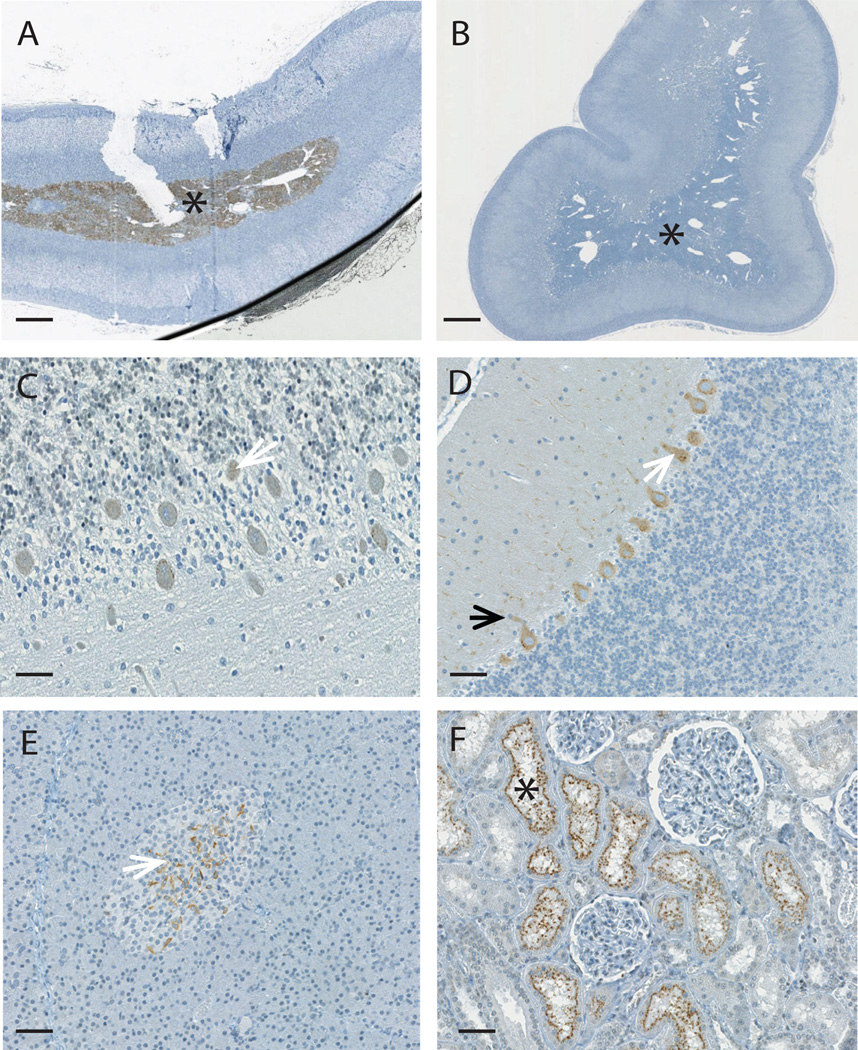
Using unfractionated L1O2 reverse transcriptase (RT) antiserum we identified staining in normal healthy testes similar to that reported previously (39). However, the presence of both the immunogen RT peptide and an irrelevant peptide abrogated staining, calling the specificity of this staining into question (data not shown). Thus, we used the RT peptide affinity-purified anti-hL1O2 RT IgG pAb, which showed an enhanced specificity of staining of L1O2 fragment 2 (Fr2)-transfected HEK 293 cells when compared to the whole unfractionated serum (compare Fig S1. E & H). Indeed, we observed no staining of healthy testes with this validated antibody. Upon probing tissue arrays from the four species with this purified pAb only the medullary region of the adrenal gland demonstrated specific staining (Fig. 1A and B and Table I). In mice, specific L1O2 staining was also seen in the adrenal medulla, and additionally in the β-islets of the pancreas and in the brain (Table I). Within the brain the immunostaining was seen in the pituitary gland (occasional pituitocytes in the pars distalis), cerebellum (purkinje cells) and cerebrum (occasional neurons). These studies suggest macaques would more closely model safety and immunogenicity aspects of L1O2 vaccination in humans, while mice might over-estimate the potential autoimmunity risks and/or show greater immune tolerance of L1O2 due to wider tissue expression.
Figure 1. Specific staining with anti-L1O2 RT pAb in the adrenal medulla.
A) At 5 µg/ml the anti-L1O2 RT pAb stained the medullary region (asterisk) of the adrenal gland in mice, human and nonhuman primates (an example from the cynomolgous monkey tissue array is shown). B) The staining could be entirely prevented by peptide competition using 10 µg/ml of the RT peptide. C) At 1µg/ml the anti-HERV-K Gag pAb 4141 stained the cytoplasm of the Purkinje cells (white arrows) in the cerebellum of humans. D) The staining with the anti-HERV-K Gag pAb 4141 was more intense in the nonhuman primates (an example from the rhesus monkey tissue array is shown), and was also seen in the cytoplasm of the axons (black arrow). E) At 1µg/ml the anti-HERV-K Gag pAb 4142 stained occasional cells of the β-islets of the pancreas (white arrow) in human and non-human primates (an example from the cynomolgous monkey tissue array is shown). F) At 1µg/ml the anti-HERV-K Env mAb HERM-1811-5 stained the proximal convoluted tubules (asterisks) of the kidneys of nonhuman primates (an example from the cynomolgous monkey tissue array is shown), but not humans. All images were captured using the NanoZoomer. Magnification is ×20 in parts A and B, with scale bar indicating 500µM and ×200 in parts C–F with scale bar indicating 50µM
Table I. Summary of L1O2 and ERV-K Env and Gag Immunohistochemistry Findings.
In mice only L1O2 immunohistochemistry was conducted as they lack ERV-K. Human and nonhuman primate tissue arrays were probed with all antibodies. Boxes indicate tissues staining by more than one antibody. – indicates a lack of specific staining, as defined by being greater in intensity than the isotype control and reduced upon specific but not control peptide competition.
| Antigen mAb or pAb |
L1O2 | HERV-K Enva,b | HERV-K Gagb | |||
|---|---|---|---|---|---|---|
| RT | HERM-1811-5 | p15 – 4141 | p15 – 4142 | CA – 4143 | CA – 4144 | |
| CNS | Pituitary, Purkinje cellsc cerebrum |
- | Purkinje cells (cerebellum) Neurons Ependymal cells (brain and spinal cord) |
- | ||
| Pancreas | β-isletsc | - | β-islets (endocrine) Ductal epithelial cells Acini (exocrine) |
|||
| Kidneys | - | Tubular epithelial cells (macula densa and proximal convoluted tubules) Medullar (collecting ducts) |
Tubular epithelial cells (cortical convoluted tubules) |
- | - | - |
| Adrenal Glandd |
Medulla (neuroendocrine cells) |
- | Capsule (endocrine and stromal cells) Medulla (neuroendocrine cells) |
- | - | - |
| ♀ tissues | - | Cervical epithelium | Endometrial stroma Glandular epithelum |
- | - | Cervical epithelium |
| ♂ tissues | - | Ductal epithelium (epididymis) | - | - | - | |
| Vasculature | - | - | Smooth muscle (media) Stromal cells (adventitia) |
- | - | - |
| Stomach | - | - | - | Glandular epithelium |
- | - |
| Small intestine |
- | - | - | Brunner glands Enterocytes |
Epithelium (crypts) |
Brunner glands |
| Bone marrow |
- | Multiple lineages (50% of precursors) |
- | Brunner glands Enterocytes |
Multiple lineages |
Multiple lineages Megakaryocytes |
| Liver | - | Periportal hepatocytes |
- | - | - | - |
| Tongue | - | Epithelium | - | - | - | - |
Staining observed in non-human primate but not humans
Not tested on mouse tissues
Staining observed only in mice
Human adrenal gland samples on tissue arrays had insufficient medulla for assessment
We next probed tissue arrays from human and the non-human primate species with four polyclonal antibodies against two HERV-K Gag peptides (p15 and capsid (CA) regions). The tissues demonstrating the most consistently specific staining for all four HERV-K Gag antibodies in one or more species were as followed: central nervous system (neurons, purkinje cells and occasionally ependymal cells) (Fig. 1C & D); endocrine pancreas (few cells in the β-islets of Langerhans) (Fig. 1E) and exocrine pancreas (ductal epithelial cells). The other tissues with specific staining (kidney, adrenal gland, male and female genital tract, vasculature, stomach, small intestine and bone marrow), reacted with one or both antibodies recognizing either just the p15 or the CA regions (Table I). The high degree of inter-species concordance between humans and macaques suggests that vaccination with SERV-K Gag in macaques would be a good model for HERV-K in humans.
For HERV-K Env we used a commercially available mAb, HERM-1811-5 to probe the tissue arrays. HERV-K Env expression was not detected in the human tissue array. However, in non-human primates, SERV-K Env expression was observed in several tissues (Table I), most notably in the kidneys (Fig. 1F). These findings suggest that macaques might over-estimate the risks of HERV-K Env vaccination in humans.
Safety and immunogenicity of vaccination with mL1O2 in Mice
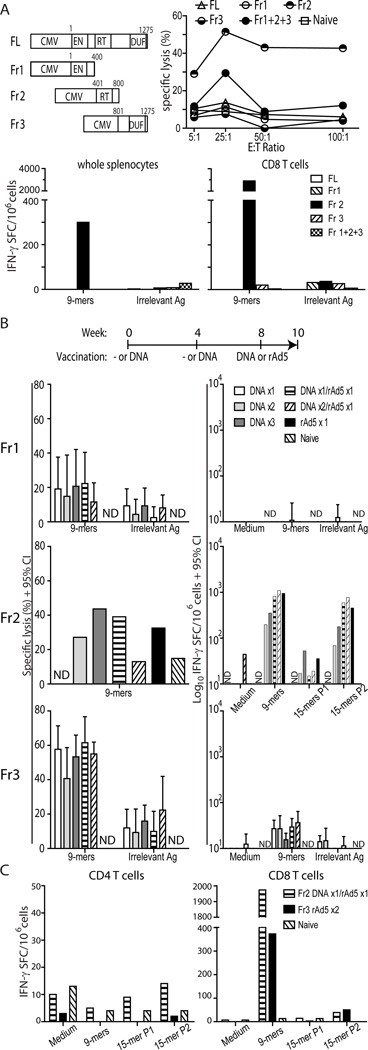
Since L1O2 expression is tightly regulated in vivo, we tested a variety of mL1O2 constructs (Fig. 2A upper left) in initial experiments in order to determine whether self L1O2 immunization could elicit immune responses. Female Balb/C mice were immunized at weeks 0 and 4 with plasmid DNA encoding full-length (FL), or fragment (Fr) 1, Fr2, Fr3 or Fr1+2+3 mL1O2 constructs using Particle Mediated Epidermal Delivery (PMED) (Fig. 2A). Two weeks after the boost, Fr2 elicited antigen-specific killing was superior to the other fractions of mL1O2 (Fig. 2A upper right). Vaccination with the mixture of Fr1+2+3 elicited a relatively lower magnitude CTL response. In contrast, mice immunized with FL, Fr1 or Fr3 mL102 showed background levels of CTL response close to that of naive mice. After an additional boost, the IFN-γ ELISPOT response was assessed in whole splenocytes and enriched CD8 T cells. In accordance with CTL activity, only Fr2 induced an antigen-specific CD8 T cell response, and this response was increased 10-fold by CD8 T cell enrichment (Fig. 2A bottom).
Figure 2. The immunogenicity of mL1O2 in Balb/c mice.
A) Balb/C mice (n= 10 per group) were administered with 2 µg plasmid DNA encoding various mL1O2 constructs (upper left panel: CMV – promoter; EN – endonuclease; RT – reverse transcriptase; DUF – domain of unknown function 1725 [commonly found in eukaryotic proteins]) by PMED at weeks 0, 4 and 8. At week 6, antigen-specific cytotoxic activity of immune cells was determined by 51Cr- release assay (upper right) for 5 mice per group, whereas at week 10, IFN-γ production in whole splenocytes (bottom left) and enriched CD8+ T cells (bottom right) was determined for the remaining 5 mice. Targets and APCs pulsed with a pool of 57 peptides (9-mers, H2d) derived from full-length mL1O2 were used to determine CTL activity and IFN-γ spot forming cells, respectively. B) Balb/C mice (n= 5 per group) were immunized with truncated form of mL1O2 Fr1 (top), Fr2 (middle) or Fr3 (bottom) according to the indicated schedule using heterologous prime-boost regimens. At week 6, CTL activity (left panel- at an E:T ratio of 50:1) and IFN-γ production (right panel) were determined. Splenocytes were pooled per group for Fr2 and kept separate for Fr1 and Fr3. C) Balb/C mice (n=5 per group) were immunized with mL1O2 Fr2 by DNA prime- rAd5 boost and for Fr3 by rAd5 prime-boost regimen. At week 6, IFN-γ spot forming cells were quantified in enriched either CD4+ or CD8+ T cells. Cells in B) and C) were stimulated with a pool of 9-mers and two pools of 15-mers (56–57 peptides per pool) at 10 µg.ml−1 per peptide, derived from appropriate fragments. Bars indicate the mean + 95% CI., N.D. indicates that a group was not done.
To further our initial observations, we next conducted a series of experiments testing the mL1O2 fragments separately in the context of the following homologous and heterologous prime-boost regimens: DNA or rAd5 alone; DNA then rAd5; DNA twice, DNA twice then rAd5; or three DNA immunizations (Fig. 2B). To enhance sensitivity of our readouts, we used mL1O2 fragment-specific peptide pools, rather than pools comprising all 57 predicted peptides. Fr1 was poorly immunogenic irrespective of the vector or regimen. Fr2 induced modest levels of CTL activity but consistently strong IFN-γ ELISPOT responses against a pool of predicted 9-mer peptides and to a lesser degree to pools of overlapping 15-mers. Regimens including rAd5 alone or preceded by PMED DNA were the most immunogenic in terms of IFN-γ responses. We fractionated the pooled splenocytes from this group and found CD8 T cell IFN-γ responses to the 9-mers but little or no response to the 15-mers from either T cell subset (data not shown). We conclude that: no CD4 T cell IFN- γ responses were elicited; that the15-mers were inefficient at stimulating purified CD8 T cells; and that the response measured in whole splenocytes with 15-mers might be largely of NK cell origin as has been observed elsewhere (59), therefore we only tested fractionated CD4 and CD8 T cells in subsequent mouse experiments. Using our fragment-specific pools, Fr3 induced strong CTL activity but minimal IFN-γ production. No clear pattern of immunogenicity emerged from the different prime-boost regimens with Fr3 (P = 0.1607 by Kruskal-Wallis analysis). Overall, these studies demonstrated that mL1O2 Fr2 and Fr3 could elicit strong CD8+ T cell-dominant immune responses with CTL activity more potent against Fr3 and IFN- γ production much stronger against Fr2.
Having established that Fr2 and Fr3 were immunogenic in Balb/C mice we next conducted an additional immunization study using a PMED-prime rAd5-boost regimen for Fr2, and a homologous rAd5 prime-boost regimen for Fr3. As before, Fr2 elicited much more potent IFN-γ ELISPOT responses than Fr3 (Fig. 2C). Fractionation of the T cells demonstrated that for both Fr2 and Fr3 the response was entirely CD8+ T cell mediated.
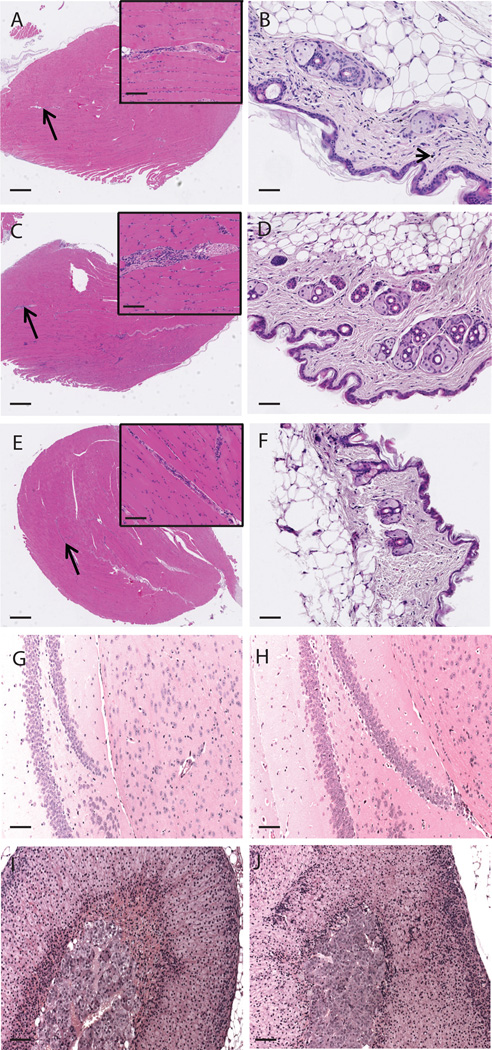
We conducted full histopathological evaluations using mice from the experiments represented in Fig. 2B and 2C. These studies revealed minimal to mild, multifocal aggregates of lymphocytes and monocytes at the injection site (skeletal muscle) in all groups receiving intramuscular injection of rAd5, but not in mice vaccinated exclusively with the PMED device or naive mice (Fig. 3A–F). These aggregates were often found around small arteries and adjacent connective tissue (perimysium) (Fig. 3C). The nature of this histopathological change (almost a pure population of lymphocytes and monocytes), its predilection for the rAd5 vaccinated groups, and its tendency to be less evident the more time had elapsed between rAd5 vaccination and necropsy, indicated that it was not consistent with an autoimmune response to mL1O2; rather, the finding was most likely the result of an immune response against the rAd5 vector itself. Inflammation of the superficial dermis was noted in the skin of mice vaccinated with the PMED device (Fig. 3B). This finding is commonly observed in vaccine studies using this mode of antigen delivery (60). Since mice immunized using PMED achieved similar levels of CD8 T cell responses to mL1O2 as those that received rAd5, the absence of perivascular mononuclear cell infiltrates in the muscle of mice vaccinated with PMED supports the vector-related association of this finding. As we detected mL1O2 expression in the adrenal medulla, and others have documented expression in the mouse hippocampus (42), we scrutinized these tissues for histopathology associated with mL1O2 expression. No signs of inflammatory infiltrates or tissue damage were evident in the hippocampus (Fig 3G & H) or in the adrenal medulla (Fig. 3I & J) when naive and mL1O2 Fr2-immunized mice were compared. Further we did not observe mL1O2 Fr2 or Fr3 antigen-related microscopic findings suggestive of an autoimmune response against mL1O2 antigen in any of the other collected tissues suggesting that vaccination with L1O2 is not likely to result in autoimmune pathology due to low or absent levels of mL1O2 expression in healthy mice tissues.
Figure 3. Inflammatory infiltrate finding at injection site is only present in mice immunized with rAd5 and otherwise no pathology is evident.
A) the anterior tibialis of mice that underwent prime-boost-boost immunization with mL1O2 Fr2 by PMED did not show infiltrates. B) Neither were infiltrates seen at the PMED actuation site, where occassional gold particles (arrowhead) were evident in the dermis. C) Conversely, mice immunized with rAd5-containing regimens displayed an inflammatory infiltrate at the injection site (anterior tibialis) but, D), no gold particles were left in the skin by 6 weeks post the last PMED immunization. As a comparison, representative E) anterior tibialis and F) skin of naive mice are shown. No pathological changes are seen in tissues with potential L1O2 expression when naive and mL1O2-immunized mice are compared. G) and H) The hippocampus, and I) and J) adrenal gland are shown for G) and I) naive and H) and J) mL1O2 Fr2-immunized mice. Black arrows indicate the area shown at higher magnification in the insets in A, C and E. A Black arrowhead indicates a gold particle. All images were captured using the NanoZoomer. Magnification is ×20 in A, C and E with scale bar indicating 500µm except for the inserts where the magnification is ×200 and the scale bar indicates 50µm. Magnification is ×200 in B, D, F with scale bar indicating 50µm. Magnification is ×100 in G – J with scale bar indicating 100µm.
Immunogenicity of hL1O2, SERV-K Gag and Env in Indian rhesus macaques
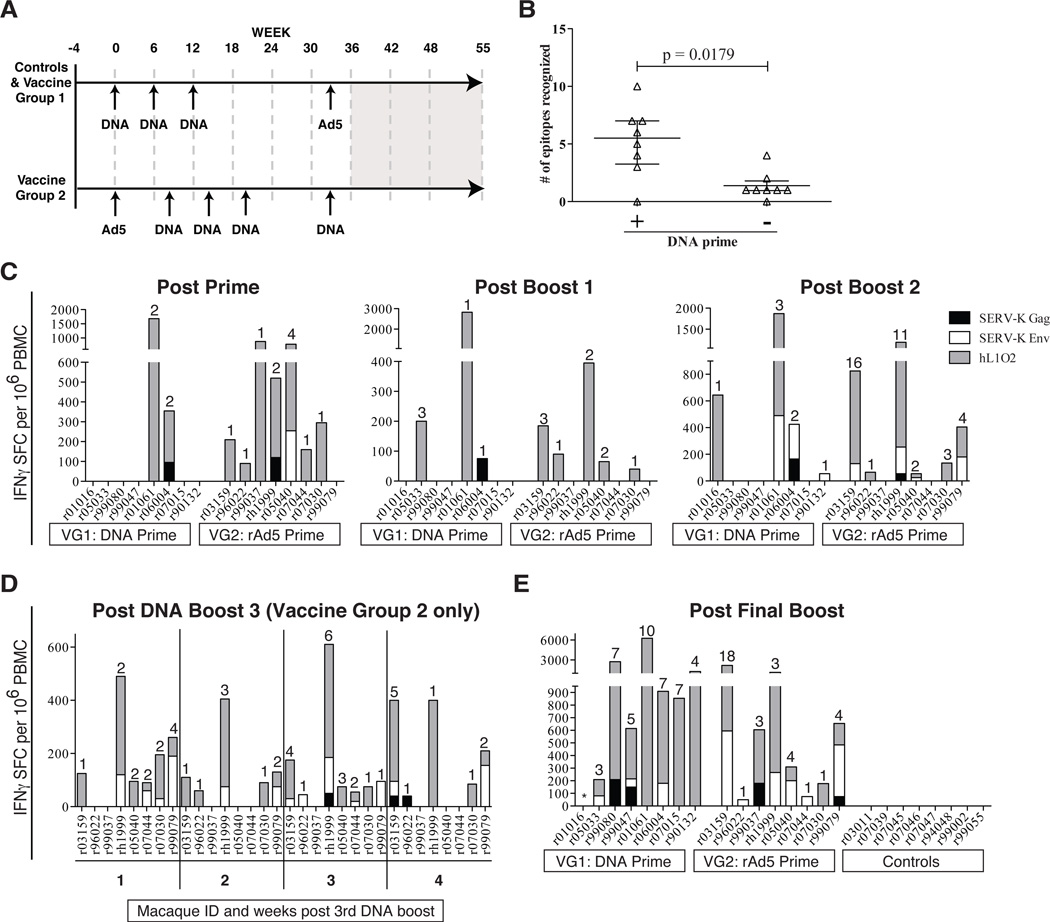
We next expanded our study to the more clinically-relevant non-human primate species of rhesus macaque. Twenty-four Indian rhesus macaques were randomly assigned into three groups of 8 animals. The control group received an empty plasmid PMED DNA prime followed by rAd5 encoding enhanced green fluorescent protein (eGFP). Vaccine group 1 consisted of a PMED DNA prime, rAd5 boost regimen and vaccine group 2 received a reversed modality consisting of a rAd5 prime, PMED DNA boost regimen (Fig. 4A). Both vaccine groups received hL1O2, SERV-K Gag, and SERV-K Env as antigens encoded by separate constructs. PMED vaccinations were given at six weekly intervals with at least 8 weeks between PMED and rAd5. Vaccine group 2 was modified to include an additional PMED DNA booster (four in total versus three PMED immunizations in vaccine group 1).
Figure 4. Vaccination with ERE is immunogenic in Indian rhesus macaques.
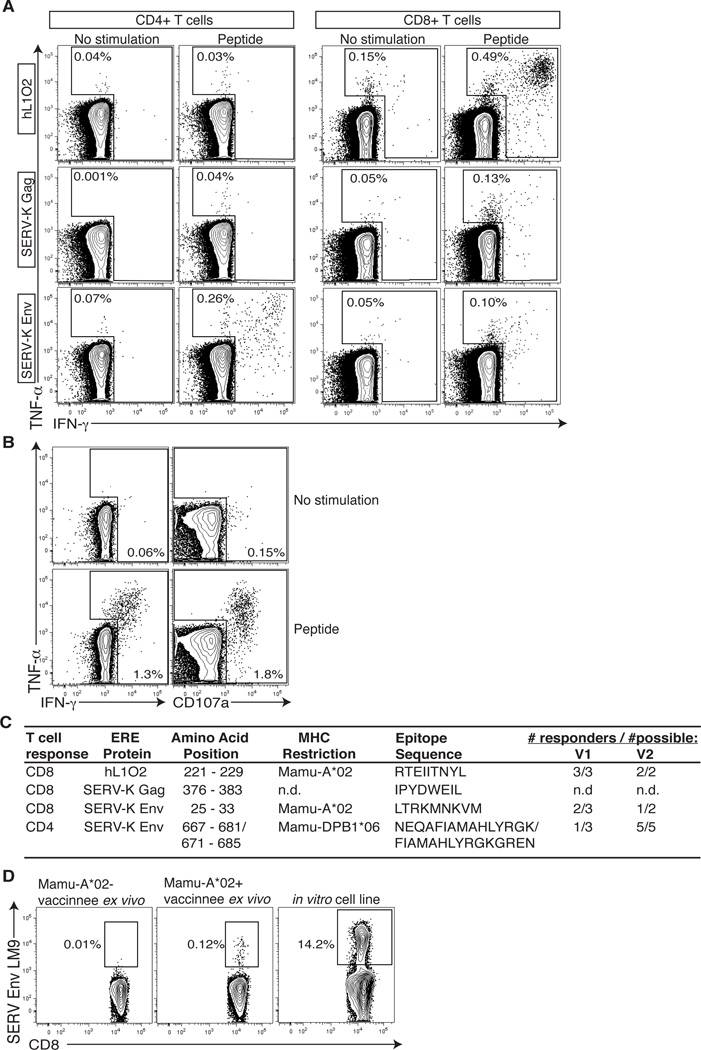
A) Vaccine timeline for study. Controls and Vaccine Group 1 received three DNA primes followed by a rAd5 boost. Vaccine group 2 received a rAd5 prime followed by four DNA boosts. Grey box from study week 36 to 55 indicates SIVsmE660 infection and necropsy. B) Comparison of the number of epitopes 2 weeks following the rAd5 with and without DNA priming beforehand. The p value was determined using a two-tailed, unpaired Mann-Whitney test. C) Immune responses following the prime and first two boosts in both vaccine groups as measured by IFN-γ ELISPOT using both predicted MHC-binding and 15mer peptides. Responses against a predicted MHC-binding peptide contained within a 15-mer peptide also eliciting a response were counted as a single response. Numbers indicate number of peptide pools recognized. Responses are shown as number of IFN-γ spot forming cells (SFCs) per 1 million PBMC. D) Longitudinal analysis of immune responses in vaccine group 2 following the third DNA PMED boost. Individual animal IDs are listed. E) Vaccine-induced responses following final boost in all groups. Asterisk indicates an animal with high spontaneous IFN-γbackground.
DNA priming substantially improves the immune response to foreign antigens encoded by rAd5 vaccines. Whether DNA priming prior to rAd5 vaccination would improve the immune response to self-antigens is unclear. To determine whether PMED DNA immunizations affected the breadth of responses made following rAd5 vaccination with self antigens, we compared the number of T cell epitopes recognized in both vaccine groups at two weeks post rAd5 vaccination (group one at week 35 versus vaccine group 2 at week two) and found a statistically significant 5.5-fold increase in the median number of epitopes recognized (Fig. 4B, P = 0.0176). Thus, we first confirmed the utility of DNA priming prior to rAd5 immunization and extended this observation to include vaccination with self-antigen.
To assess the cellular immune response to vaccination with self-antigens, we performed IFN-γ ELISPOT using pools of both overlapping 15-mer peptides and minimal peptide T cell epitopes predicted to bind to the common MHC class I alleles Mamu-A*01 or -A*02. Both vaccine regimens were immunogenic and induced T cell responses following either a single DNA or rAd5 prime and throughout the vaccine phase (Fig. 4C). Interestingly, a distinct hierarchy emerged in the magnitude with which each antigen was targeted throughout the vaccine phase. hL1O2-specific responses dominated the cellular immune response with SERV-K Env recognized less frequently and SERV-K Gag being the least targeted of the three antigens (Fig. 4C). To further investigate the kinetics of the T cell response to vaccination with these self-antigens, we performed a longitudinal weekly analysis of vaccine group 2 following the third DNA boost. We observed anamnestic T cell responses at one week following the third DNA boost and these responses mostly persisted over the four week monitoring period, although the number of peptide pools recognized varied from week-to-week peaking with 7/8 animals responding to a total of 18 pools at week three (Fig. 4D).
Following the final DNA or rAd5 boost, we observed expansion of anamnestic T cells in 7/8 animals that received a DNA prime (vaccine group 1) and all of the animals that received a rAd5 prime (vaccine group 2) (Fig. 4E). Of note, the single non-responder in vaccine group 1 consistently had extremely high background levels of IFN-γ, making it difficult to identify positive responses. Although this animal did not make a clear response following the rAd5 boost, we observed a strong IFN-γ T cell response following the third DNA prime (Fig. 4C, post boost 2). Therefore, every vaccinated animal in our study mounted at least one T cell response to the vaccine encoding self-antigen while no responses were observed in animals that received only empty vectors (Fig. 4E). The total magnitude and breadth of T cell responses did not vary significantly between the two regimens at the end of the vaccination phase (P = 0.1853 and 0.2477 respectively). However, vaccine group 2 included an additional higher-dose DNA boost using a next-generation PMED platform. This boost increased the magnitude 10-fold and breadth 4.4-fold although these increases were not quite significant (P = 0.0547 and 0.0625 respectively). If vaccine group 1 is compared to vaccine group 2 following the same number of total vaccinations (i.e. week 35 versus week 22), it had superior magnitude and breadth (P = 0.0111 and 0.0093 respectively). Heterologous prime-boost regimens are clearly affected by dose, timing and platform with each component requiring optimization.
To further investigate the T cell response to vaccination with self-antigens, we next selected the strongest T cell response against L1O2, SERV-K Gag, and SERV-K Env and performed intracellular cytokine staining (ICS) assays using 15-mer peptides. The ICS results mirrored the ELISPOT assay as L1O2 was again targeted with the highest magnitude (Fig. 5A). We found that the majority of the responses were CD8+ T cell mediated, with the exception of SERV-K Env, which was also highly targeted by CD4+ T cells (Fig. 5A). The self-antigen specific CD8+ T cells displayed no functional deficiencies, as they were able to produce multiple cytokines (IFN-γ and TNF-α) and degranulate (CD107a) in response to cognate antigen (Fig. 5B). Next, we further explored the characteristics of our self-antigen vaccine-induced T cell responses by mapping their minimal T cell epitope and restricting MHC molecules (Fig. 5C). Knowing the restricting allele allowed us to determine if vaccinated animals expressing these alleles would commonly target the same epitope. Indeed, vaccinees sharing an MHC molecule often mounted the same T cell response as 5/5 and 3/5 Mamu-A*02+ animals targeted the mapped Mamu-A*02 restricted L1O2 and SERV-K Env CD8+ T cell epitopes, respectively. Furthermore, 6/8 vaccinated animals expressing the MHC-II molecule Mamu-DPB1*06 mounted the same CD4+ T cell response against SERV-K Env (Fig. 5D). Finally, knowledge of the minimal CD8+ T cell epitope and MHC-I-restricting allele allowed us to fold MHC-I tetramers and stain these cells directly (Fig. 5D). Cumulatively, these data indicate that both CD8+ and CD4+ T cell against self-antigen can be generated via vaccination in primates and that they are able to perform multiple effector functions.
Figure 5. Characteristics of ERE-specific T cells.
A) Intracellular cytokine stain (ICS) of the largest responses against each ERE antigen directly ex vivo at two weeks post second boost using single 15-mer peptides identified by ELISPOT. Graphs were produced by gating on either CD4+CD3+ or CD8+ CD3+ cells as indicated. B) Representative data of hL1O2 response showing that ERE-specific T cells are able to mediate multiple effector functions. C) Summary of the characteristics of the four T cell responses shown in figure 5A. n.d. = not determined. D) Mamu-A*02 ERV-K Env LM9 tetramer staining of PBMC directly ex vivo in a Mamu-A*02 negative and positive animal two weeks following the final DNA boost. Also shown is a two week old in vitro expanded cell line as a positive control.
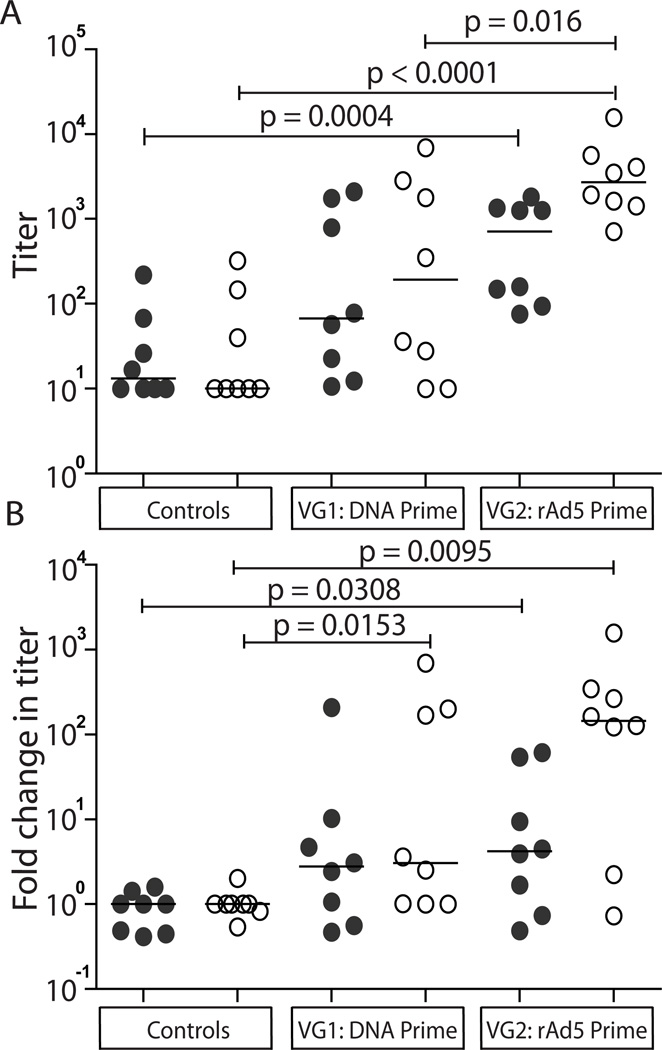
We next investigated the IgG responses to ERE vaccination. Our studies were constrained by the significant technical challenge of deriving protein of sufficient quality (>90% purity, soluble, low proportion of aggregates and degraded product). This was achieved for the CA subprotein of SERV-K Gag, and for the SU and TM subproteins of HERV-K Env. We determined that HERV-K Env is 84% identical to SERV-K Env, favouring sero-crossreactivity, although potentially underestimating the true response. Sera taken at baseline and two weeks following the final vaccination were used in our ELISA studies. In control animals, spontaneous weak IgG responses to both SERV-K Gag CA and HERV-K Env were detected in up to half of the animals, but their levels remained constant throughout the study. Conversely, amongst the vaccinated animals 14/16 macaques made IgG responses crossreacting to HERV-K Env ranging in titer from 1:28 – 1:15,700 (Fig. 6A). The IgG responses detected by both HERV-K SU and TM ELISAs were significantly greater in vaccine group 2 than in the controls (Fig. 6A, [ANOVA P < 0.0001]). Vaccine group 2 also attained greater titers of IgG against HERV-K TM protein than vaccine group 1 (P = 0.0160). When the fold ELISA titer-change from baseline was examined vaccine group 2 achieved statistically significant 145-fold and 4.2-fold increases in HERV-K Env TM and SU-specific IgG titers, whereas vaccine group 1 demonstrated a median 3-fold increase in HERV-K Env TM titer only (Fig. 6B, [Kruskal-Wallis P = 0.0041]). Only two macaques made modest vaccine-induced IgG responses to SERV-K Gag CA (data not shown). Although we were not successful at purifying recombinant protein for hL1O2, we were able to screen the baseline and final sera by western blot on lysates of hL1O2 Fr2-transfected HEK293 cells. We detected a clear vaccine-induced response in a single animal (r99080) from vaccine group 2 (data not shown). The findings for SERV-K Gag CA and hL1O2 suggest that these antigens are not potent stimulators of antibody responses, although this may in part be explained by the lack of a secretory leader sequence in both of these vaccine constructs, which was included in the SERV-K Env construct.
Figure 6. Pre-existing and elicited IgG responses to ERV-K Env SU and TM proteins in Indian rhesus macaques.
Baseline and week 35 sera from each group were tested in ELISAs using HERV-K Env SU and TM proteins (84% identical to SERV-K sequences). A) ELISA titers: defined as the dilution at which OD600nm = 1.0, a value falling consistently within the linear range at 40% of the typical plateau value. B) Fold-change in ELISA titer compared to the baseline titer. Closed circles indicate the response to HERV-K Env SU, whereas open circles indicate the response to HERV-K Env TM. Medians are indicated. Statistical significance was determined for: titer data by log-transformation followed by ANOVA then Student’s t-tests; fold-change data by Kruskal-Wallis analysis then Mann-Whitney tests.
ERE vaccination is safe in Indian rhesus macaques
Blood samples were taken for clinical chemistry and hematological analysis twice before vaccination commenced (baseline values) and following each vaccination to provide correlation between any findings and the increasing immune response magnitude or duration. No vaccine-related changes from baseline or standard reference ranges for rhesus macaques were evident (data not shown). Emerging immunohistochemistry data showing HERV-K Env expression in the kidneys prompted us to include urinanalysis during the experiment with urine samples taken at week 20 and 35. Although we lacked a true urinanalysis baseline, no significant changes were detected over the 15 week period, which included the rAd5-boost in vaccine group 1 and two PMED DNA boosts in vaccine group 2. These in-life safety findings encouraged us to transfer the animals to an SIV-challenge protocol to determine the safety and efficacy of immune response to these antigens at preventing, controlling and eradicating SIV infection. Animals were repeatedly challenged until they became infected. After 10–12 weeks of SIV infection the animals were culled for detailed safety readouts before chronic SIV pathology could become a confounding factor. Extensive analysis of the challenge outcome is under way and will be reported shortly (Sheppard et al, manuscript in preparation).
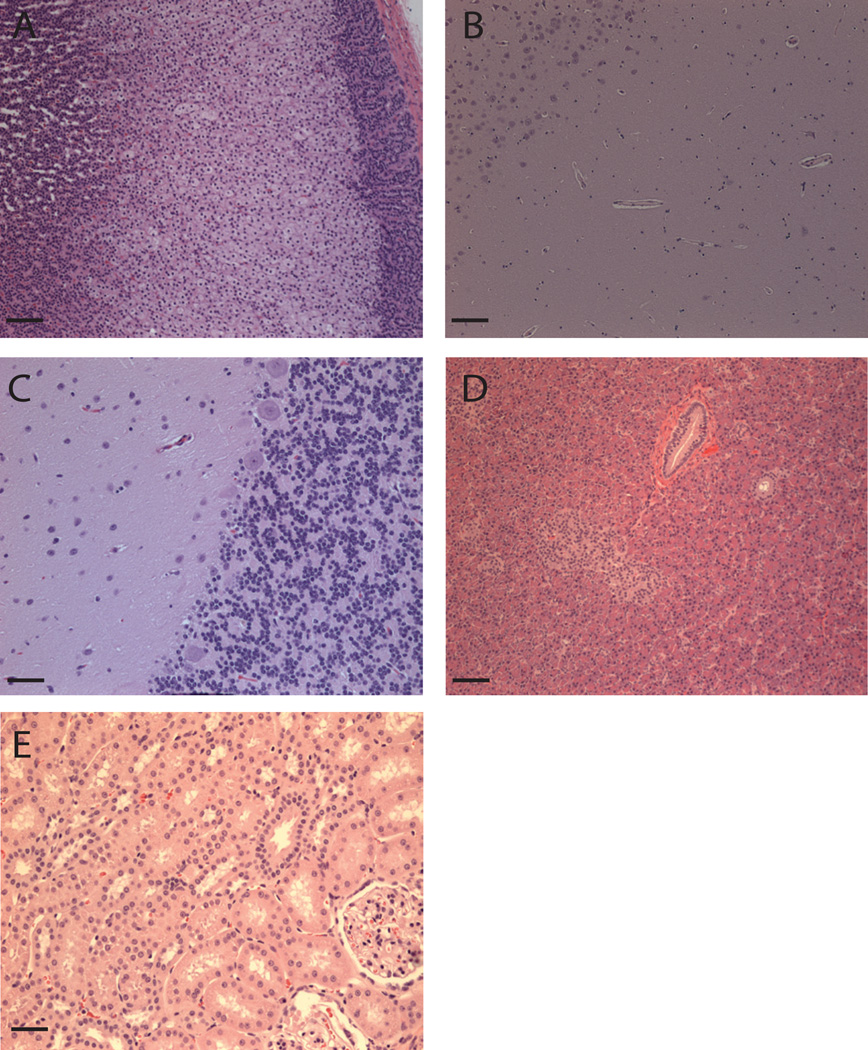
Following necropsy, the animals underwent extensive histopathological analysis focusing on tissues that had been flagged by our immunoprobe (Fig. 1 & Table I) and by prior reports as potential target tissues (37–39, 42). Early-stage SIV infection could be eliminated as a confounding factor since we had unvaccinated controls for comparison and SIV-related pathology has been extensively characterized (61). Even in animals with the strongest immune responses the tissues were morphologically normal (Fig. 7). Lesions in all animals other than r99002 (see below) were mild and consistent with the common background pathology of rhesus macaques maintained in colony at the Wisconsin National Primate Center, combined with their early-stage SIV infection status. Such background lesions include: mild focal capsular fibrosis of the liver, minimal focal lymphocytic myocarditis, mild prostatitis, mild cholecystitis of the gall bladder, minimal to mild esophagitis, mild inguinal hernia, and very mild gastritis. We observed no histologic evidence of pathology induced by an immune response (vasculitis, glomeulonephritis, or retinitis), or immune complex formation and/or deposition (vacular, glomerular and occular inflammation). In one case (macaque r99002) an adenocarcinoma involving the ileum and ileocecocolic junction was diagnosed. However, intestinal neoplasia accounts for approximately 48% of all neoplasms diagnosed in this colony thus this is not thought to be vaccine-related (62). In summary, we observed no adverse safety affects from vaccination with ERE.
Figure 7. No evidence of autoimmune pathology in potential target tissues of macaques with the strongest immune responses to hL1O2 and SERV-K Gag & Env.
Potential target tissues (Table I) of rhesus macaques that made the strongest immune responses to the vaccine antigens were examined at post-mortem (10–12 weeks following SIV infection). No auto-immune or SIV-related pathology is evident. A) Adrenal gland of r01061. B) Hippocampus of r01061. C) Cerebellum of r99080. D) Pancreas of r99080 and E) Kidney of r01359. Magnification is ×10 in A–D and ×20 in E with scale bars of 100 and 50µm respectively.
Discussion
Mounting evidence suggests that HERV-K and L1 may encode tumor- or viral-infection-associated antigens (1–7, 10–12, 14, 16, 17, 20–25). As such the ORFs of these EREs are novel candidates for targeting with vaccines or immunotherapeutics. Their utility will depend on the consistency of their association with the targeted pathological state and a lack of expression in healthy somatic cells accessible to the immune system. While evidence for the former has emerged from various laboratories in recent years, evidence of the latter is particularly sparse when one considers only proof of expression at the protein level or direct evidence of increases in genomic copy number within a given tissue. To our knowledge, ours is the first report of somatic tissue array analysis by immunohistochemistry in mice, macaques and humans for our three candidate antigens: L1O2 and HERV/SERV-K Gag and Env, using antibody reagents carefully selected and validated to enable cross-species investigation.
Ergün and colleagues had earlier reported on L1O2 expression in human Leydig, Sertoli and vascular endothelial cells of the testes using chicken IgY pAbs to the L1O2 endonuclease domain (39). We did not observe the same pattern of staining with our affinity purified anti-L1O2 RT pAb. The discrepancy with the results of Ergün and colleagues may be due to differential antibody specificity (endonuclease peptide versus RT peptide), avidity, host species (chicken IgY versus rabbit IgG) or procedural differences. The principal procedural difference was that we conducted peptide competition by premixing antibody with peptide and adding the mixture to the slide, whereas Ergün and colleagues first depleted their anti-L1O2 endonuclease IgY preparation with a ~1:270 molar excess of the endonuclease peptide coupled to Sepharose beads before using cleared supernatant to repeat the stain. Such a procedure may deplete specific antibody entirely from the staining reaction, and in our opinion this does not prove the specificity of their staining.
The use of HERV-K Env mAb HERM-1811 in western blots, immunofluorecence and immunohistochemistry has been reported previously (44, 63) (and in Jones et al, submitted). We conducted our own validation for this mAb and proved that it could recognize SERV-K Env. We did not observe any specific staining of human tissues, however we did observe staining in macaques.
We developed and validated four of our own pAbs against HERV-K Gag that cross-reacted with SERV-K Gag. The most convincing staining was that shown by antibodies to both the p15 and CA components of HERV-K Gag, which occurred in the Purkinje cell body and axons of the brain, and in the β-islets of the pancreas of both human and non-human primates. We also observed differential staining whereby only the p15 or the CA antibodies stained a particular tissue. Although the Gag protein of retroviruses is produced as a single full-length pre-cursor protein, differential stability might lead to unequal half-lives of p15 and CA in tissues and differential pAb affinities might also contribute to this observation. Moreover, expression of small fragments of degraded HERV ORFs has been reported (19), therefore it is conceivable that p15 and CA might be present in different quantities, preventing us ruling out our immunohistochemistry findings.
The results of our immunohistochemistry studies together with literature reports of L1O2 expression in the testes and brain (39, 42, 45) guided our preclinical safety strategy for in-life and post-mortem investigation. Our consistent L1O2 findings in the adrenal medulla are potentially predictive of a cardiovascular risk as the resident chromaffin cells are responsible for the production of adrenaline and noradrenaline. The potential targeting of HERV-K Gag in the Purkinje cells of the brain could conceivably cause autoimmune cerebellar ataxia. If the β-islets of the pancreas were to be targeted due to expression of HERV-K Gag, glucose homeostasis could be affected. The potential expression of HERV-K Gag and/or Env in the bone marrow and/or liver also predicts certain safety risks. Throughout the in-life phase of mouse and non-human primate experiments no behavioural changes (ataxia) or clinical signs other than those typically associated with vaccination were observed. At post-mortem no vaccine-antigen related findings were observed in mice. Infiltrates at the injection site of the rAd5 vector were seen independently of the immune response against mL1O2, which was equally strong in mice immunized by DNA in the skin via PMED. For the rhesus macaque study, the clinical chemistry, hematology and urinanalysis did not reveal any significant changes from baseline values or standard reference ranges. Moreover, at post-mortem there was no evidence autoimmune reaction to the adrenal glands, Purkinje cell layers, pancreas, kidneys, bone marrow or any other tissue that was not consistent with findings typical for members of this rhesus macaque colony early in the course of SIV infection.
There are several potential explanations for the apparent safety observed in these studies. Firstly; it is possible that all the observed staining is caused by cross-reactive proteins. However, we were unable to find any close matches for our immunogen peptides by Blast searches, making this possibility less likely. Secondly; the immune responses induced might not have been potent enough or of the correct CD4/CD8 balance to drive autoimmunity, especially if antigen-specific CD4 T cell responses are required to provide the cue for migration of CD8 CTLs into tissues expressing the self-antigens, as shown for virus-specific CTLs (64). This is potentially the case for SERV-K Gag to which only modest CD8 T cell responses were observed in 6/16 macaques in the absence of detectable CD4 response, and for L1O2, which elicited strong CD8 responses without detectable CD4 responses in both mice and macaques. However, the lack of CD4 responses cannot explain the safety of SERV-K Env immunization, as we observed both CD4 and CD8 T cell responses in addition to IgG responses in the majority of immunized animals. Thirdly; regulatory T cells (Treg) may suppress autoimmunity in healthy tissues in which there are no inflammatory events. Fourthly; for L1O2, the use of the human form in macaques might also explain the safety, however the antigens are highly similar (92% identity, 96% similarity) thus antigen mismatch seems an unlikely explanation for the safety of L1O2 immunization, especially given the immunogenicity and safety of mL1O2 in mice. Finally; for the staining related to the brain, the existence of the antigen within an immune-privileged site might be sufficient to prevent autoimmune pathology as the responding T cells cannot access the tissue.
Our use of two different vaccination schedules in rhesus macaques revealed that the DNA prime, rAd5 boost regimen showed greater immunodominance of hL1O2 than the reverse regimen. Since all antigen-encoding plasmids were co-formulated onto gold particles and delivered to the same anatomical site, whereas the rAd5 vectors each encoded a single antigen were delivered at separate anatomical sites, this pattern might reflect competition at the level of the antigen presenting cells in the local draining lymph nodes. Alternatively, the SERV-K antigens might require more potent danger signals in order to prime responses, and these would be better provided by our rAd5 vectors than our DNA vaccines. We conclude that future multi antigen studies consider regimens that overcome immunodominance such as rAd5 prime, DNA boost.
In summary, we have expanded the knowledge regarding protein level expression of HERV-K Gag and Env and L1O2 in healthy tissues. Further we have shown that it is possible to induce both T and B cell responses to self and near-self ERE antigens in preclinical species and the responses we induced did not cause autoimmune pathology. These findings enable the investigation of ERE antigens in relevant efficacy models as tumor- or HIV infection-association targets and form a foundation for pre-clinical safety studies of lead candidates emerging from such efficacy studies.
Supplementary Material
Acknowledgements
The authors would like to thank Professor Deborah Fuller of Albany Medical College; Fred Immermann of Pfizer for assistance with statistics; and Professor David Watkins and Dr Nancy Wilson-Schlei of the University of Wisconsin for helpful discussions and guidance. We also thank the WNPRC Immunology Services and Virology Services for experimental assistance and members of the WNPRC Animal Care, SPI and Pathology units for NHP care and experimental manipulation.
This study was funded by Pfizer Inc, with additional support to JBS by NIH R21 AI087474.
Abbreviations
- ERE
Endogenous retrotransposable element
- ERV
Endogeous retrovirus
- Fr
Fragement
- FL
Full-length
- HML
Human mouse mammary tumor virus-like
- HERV
Human ERV
- LINE-1 or L1
Long interspersed nuclear element 1
- L1O2
LINE-1 open reading frame 2
- LTR
Long terminal repeat
- ORF
Open reading frame
- PMED
Particle mediated epidermal delivery
- pAb
Polyclonal antibody
- rAd5
Recombinant adenovirus serotype 5
- RT
Reverse transcriptase
- SERV
Simian ERV
References
- 1.Asch HL, Eliacin E, Fanning TG, Connolly JL, Bratthauer G, Asch BB. Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncology research. 1996;8:239–247. [PubMed] [Google Scholar]
- 2.Callinan PA, Batzer MA. Retrotransposable elements and human disease. Genome dynamics. 2006;1:104–115. doi: 10.1159/000092503. [DOI] [PubMed] [Google Scholar]
- 3.Saito K, Kawakami K, Matsumoto I, Oda M, Watanabe G, Minamoto T. Long interspersed nuclear element 1 hypomethylation is a marker of poor prognosis in stage IA non-small cell lung cancer. Clin Cancer Res. 16:2418–2426. doi: 10.1158/1078-0432.CCR-09-2819. [DOI] [PubMed] [Google Scholar]
- 4.Sunami E, de Maat M, Vu A, Turner RR, Hoon DS. LINE-1 Hypomethylation During Primary Colon Cancer Progression. PloS one. 6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, Rivera MN, Bardeesy N, Maheswaran S, Haber DA. Science. Vol. 331. New York, N.Y: Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers; pp. 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buscher K, Hahn S, Hofmann M, Trefzer U, Ozel M, Sterry W, Lower J, Lower R, Kurth R, Denner J. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma research. 2006;16:223–234. doi: 10.1097/01.cmr.0000215031.07941.ca. [DOI] [PubMed] [Google Scholar]
- 7.Contreras-Galindo R, Kaplan MH, Leissner P, Verjat T, Ferlenghi I, Bagnoli F, Giusti F, Dosik MH, Hayes DF, Gitlin SD, Markovitz DM. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. Journal of virology. 2008;82:9329–9336. doi: 10.1128/JVI.00646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank O, Verbeke C, Schwarz N, Mayer J, Fabarius A, Hehlmann R, Leib-Mosch C, Seifarth W. Variable transcriptional activity of endogenous retroviruses in human breast cancer. Journal of virology. 2008;82:1808–1818. doi: 10.1128/JVI.02115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galli UM, Sauter M, Lecher B, Maurer S, Herbst H, Roemer K, Mueller-Lantzsch N. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene. 2005;24:3223–3228. doi: 10.1038/sj.onc.1208543. [DOI] [PubMed] [Google Scholar]
- 10.Herbst H, Sauter M, Mueller-Lantzsch N. Expression of human endogenous retrovirus K elements in germ cell and trophoblastic tumors. The American journal of pathology. 1996;149:1727–1735. [PMC free article] [PubMed] [Google Scholar]
- 11.Iramaneerat K, Rattanatunyong P, Khemapech N, Triratanachat S, Mutirangura A. HERV-K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance. Int J Gynecol Cancer. 21:51–57. doi: 10.1097/IGC.0b013e3182021c1a. [DOI] [PubMed] [Google Scholar]
- 12.Ishida T, Obata Y, Ohara N, Matsushita H, Sato S, Uenaka A, Saika T, Miyamura T, Chayama K, Nakamura Y, Wada H, Yamashita T, Morishima T, Old LJ, Nakayama E. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun. 2008;8:15. [PMC free article] [PubMed] [Google Scholar]
- 13.Ruprecht K, Ferreira H, Flockerzi A, Wahl S, Sauter M, Mayer J, Mueller-Lantzsch N. Human endogenous retrovirus family HERV-K(HML-2) RNA transcripts are selectively packaged into retroviral particles produced by the human germ cell tumor line Tera-1 and originate mainly from a provirus on chromosome 22q11.21. Journal of virology. 2008;82:10008–10016. doi: 10.1128/JVI.01016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauter M, Schommer S, Kremmer E, Remberger K, Dolken G, Lemm I, Buck M, Best B, Neumann-Haefelin D, Mueller-Lantzsch N. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. Journal of virology. 1995;69:414–421. doi: 10.1128/jvi.69.1.414-421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. International journal of cancer. 2007;120:81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 16.Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, Piyathilake CJ, Hunt KK, Johanning GL. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68:5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goering W, Ribarska T, Schulz WA. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis. 32:1484–1492. doi: 10.1093/carcin/bgr181. [DOI] [PubMed] [Google Scholar]
- 18.Harris CR, Normart R, Yang Q, Stevenson E, Haffty BG, Ganesan S, Cordon-Cardo C, Levine AJ, Tang LH. Association of Nuclear Localization of a Long Interspersed Nuclear Element-1 Protein in Breast Tumors with Poor Prognostic Outcomes. Genes & cancer. 1:115–124. doi: 10.1177/1947601909360812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy JP, Hanada K, Mena O, Kurlander R, Tawab A, Srinivasan R, Lundqvist A, Malinzak E, Geller N, Lerman MI, Childs RW. Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. The Journal of clinical investigation. 2008;118:1099–1109. doi: 10.1172/JCI34409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutkowski N, Conrad B, Thorley-Lawson DA, Huber BT. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity. 2001;15:579–589. doi: 10.1016/s1074-7613(01)00210-2. [DOI] [PubMed] [Google Scholar]
- 21.Contreras-Galindo R, Kaplan MH, Markovitz DM, Lorenzo E, Yamamura Y. Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS research and human retroviruses. 2006;22:979–984. doi: 10.1089/aid.2006.22.979. [DOI] [PubMed] [Google Scholar]
- 22.Contreras-Galindo R, Lopez P, Velez R, Yamamura Y. HIV-1 infection increases the expression of human endogenous retroviruses type K (HERV-K) in vitro. AIDS research and human retroviruses. 2007;23:116–122. doi: 10.1089/aid.2006.0117. [DOI] [PubMed] [Google Scholar]
- 23.Laderoute MP, Giulivi A, Larocque L, Bellfoy D, Hou Y, Wu HX, Fowke K, Wu J, Diaz-Mitoma F. The replicative activity of human endogenous retrovirus K102 (HERV-K102) with HIV viremia. AIDS (London, England) 2007;21:2417–2424. doi: 10.1097/QAD.0b013e3282f14d64. [DOI] [PubMed] [Google Scholar]
- 24.Garrison KE, Jones RB, Meiklejohn DA, Anwar N, Ndhlovu LC, Chapman JM, Erickson AL, Agrawal A, Spotts G, Hecht FM, Rakoff-Nahoum S, Lenz J, Ostrowski MA, Nixon DF. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS pathogens. 2007;3:e165. doi: 10.1371/journal.ppat.0030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sengupta D, Tandon R, Vieira RG, Ndhlovu LC, Lown-Hecht R, Ormsby CE, Loh L, Jones RB, Garrison KE, Martin JN, York VA, Spotts G, Reyes-Teran G, Ostrowski MA, Hecht FM, Deeks SG, Nixon DF. Strong human endogenous retrovirus (HERV)-specific T cell responses are associated with control of HIV-1 in chronic infection. Journal of virology. doi: 10.1128/JVI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tandon R, Sengupta D, Ndhlovu LC, Vieira RG, Jones RB, York VA, Vieira VA, Sharp ER, Wiznia AA, Ostrowski MA, Rosenberg MG, Nixon DF. Identification of Human Endogenous Retrovirus-Specific T Cell Responses in Vertically HIV-1-Infected Subjects. Journal of virology. 85:11526–11531. doi: 10.1128/JVI.05418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nature reviews. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 29.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome research. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 30.Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Human genetics. 2005;117:411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 31.Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 32.Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. Journal of virology. 2005;79:12507–12514. doi: 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Barbulescu M, Turner G, Seaman MI, Deinard AS, Kidd KK, Lenz J. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol. 1999;9:861–868. doi: 10.1016/s0960-9822(99)80390-x. [DOI] [PubMed] [Google Scholar]
- 35.Hughes JF, Coffin JM. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1668–1672. doi: 10.1073/pnas.0307885100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- 37.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Molecular and cellular biology. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, Chalajour F, Kilic N, Stratling WH, Schumann GG. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. The Journal of biological chemistry. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 40.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 41.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 42.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kammerer U, Germeyer A, Stengel S, Kapp M, Denner J. Human endogenous retrovirus K (HERV-K) is expressed in villous and extravillous cytotrophoblast cells of the human placenta. Journal of reproductive immunology. doi: 10.1016/j.jri.2011.06.102. [DOI] [PubMed] [Google Scholar]
- 45.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic acids research. 38:3909–3922. doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dombroski BA, Scott AF, Kazazian HH., Jr Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medstrand P, Mager DL. Human-specific integrations of the HERV-K endogenous retrovirus family. Journal of virology. 1998;72:9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loudon PT, Yager EJ, Lynch DT, Narendran A, Stagnar C, Franchini AM, Fuller JT, White PA, Nyuandi J, Wiley CA, Murphey-Corb M, Fuller DH. GM-CSF increases mucosal and systemic immunogenicity of an H1N1 influenza DNA vaccine administered into the epidermis of non-human primates. PloS one. 5:e11021. doi: 10.1371/journal.pone.0011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy MJ, Wu MS, Barr LJ, Fuller JT, Tussey LG, Speller S, Culp J, Burkholder JK, Swain WF, Dixon RM, Widera G, Vessey R, King A, Ogg G, Gallimore A, Haynes JR, Heydenburg Fuller D. Induction of antigen-specific CD8+ T cells, T helper cells, protective levels of antibody in humans by particle-mediated administration of a hepatitis B virus DNA vaccine. Vaccine. 2000;19:764–778. doi: 10.1016/s0264-410x(00)00302-9. [DOI] [PubMed] [Google Scholar]
- 51.Kaizu M, Borchardt GJ, Glidden CE, Fisk DL, Loffredo JT, Watkins DI, Rehrauer WM. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics. 2007;59:693–703. doi: 10.1007/s00251-007-0233-7. [DOI] [PubMed] [Google Scholar]
- 52.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. Journal of virology. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones S, Evans K, McElwaine-Johnn H, Sharpe M, Oxford J, Lambkin-Williams R, Mant T, Nolan A, Zambon M, Ellis J, Beadle J, Loudon PT. DNA vaccination protects against an influenza challenge in a double-blind randomised placebo-controlled phase 1b clinical trial. Vaccine. 2009;27:2506–2512. doi: 10.1016/j.vaccine.2009.02.061. [DOI] [PubMed] [Google Scholar]
- 54.Reynolds MR, Sacha JB, Weiler AM, Borchardt GJ, Glidden CE, Sheppard NC, Norante FA, Castrovinci PA, Harris JJ, Robertson HT, Friedrich TC, McDermott AB, Wilson NA, Allison DB, Koff WC, Johnson WE, Watkins DI. The TRIM5{alpha} genotype of rhesus macaques affects acquisition of simian immunodeficiency virus SIVsmE660 infection after repeated limiting-dose intrarectal challenge. Journal of virology. 85:9637–9640. doi: 10.1128/JVI.05074-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynolds MR, Weiler AM, Piaskowski SM, Kolar HL, Hessell AJ, Weiker M, Weisgrau KL, Leon EJ, Rogers WE, Makowsky R, McDermott AB, Boyle R, Wilson NA, Allison DB, Burton DR, Koff WC, Watkins DI. Macaques vaccinated with simian immunodeficiency virus SIVmac239Delta nef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. Journal of virology. 84:9190–9199. doi: 10.1128/JVI.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Most RG, Sette A, Oseroff C, Alexander J, Murali-Krishna K, Lau LL, Southwood S, Sidney J, Chesnut RW, Matloubian M, Ahmed R. Analysis of cytotoxic T cell responses to dominant and subdominant epitopes during acute and chronic lymphocytic choriomeningitis virus infection. J Immunol. 1996;157:5543–5554. [PubMed] [Google Scholar]
- 57.Taguchi T, McGhee JR, Coffman RL, Beagley KW, Eldridge JH, Takatsu K, Kiyono H. Detection of individual mouse splenic T cells producing IFN-gamma and IL-5 using the enzyme-linked immunospot (ELISPOT) assay. Journal of immunological methods. 1990;128:65–73. doi: 10.1016/0022-1759(90)90464-7. [DOI] [PubMed] [Google Scholar]
- 58.Reynolds MR, Weiler AM, Weisgrau KL, Piaskowski SM, Furlott JR, Weinfurter JT, Kaizu M, Soma T, Leon EJ, MacNair C, Leaman DP, Zwick MB, Gostick E, Musani SK, Price DA, Friedrich TC, Rakasz EG, Wilson NA, McDermott AB, Boyle R, Allison DB, Burton DR, Koff WC, Watkins DI. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. The Journal of experimental medicine. 2008;205:2537–2550. doi: 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loo CP, Long BR, Hecht FM, Nixon DF, Michaelsson J. HIV-1-specific T Cell-dependent natural killer (NK) cell activation: major contribution by NK cells to interferon-gamma production in response to HIV-1 antigens. AIDS research and human retroviruses. 2009;25:603–605. doi: 10.1089/aid.2008.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dincer Z, Jones S, Haworth R. Preclinical safety assessment of a DNA vaccine using particle-mediated epidermal delivery in domestic pig, minipig and mouse. Exp Toxicol Pathol. 2006;57:351–357. doi: 10.1016/j.etp.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 61.Letvin NL, King NW. Immunologic and pathologic manifestations of the infection of rhesus monkeys with simian immunodeficiency virus of macaques. Journal of acquired immune deficiency syndromes. 1990;3:1023–1040. [PubMed] [Google Scholar]
- 62.Simmons HA, Mattison JA. The incidence of spontaneous neoplasia in two populations of captive rhesus macaques (Macaca mulatta) Antioxidants & redox signaling. 14:221–227. doi: 10.1089/ars.2010.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanke K, Kramer P, Seeher S, Beimforde N, Kurth R, Bannert N. Reconstitution of the ancestral glycoprotein of human endogenous retrovirus k and modulation of its functional activity by truncation of the cytoplasmic domain. Journal of virology. 2009;83:12790–12800. doi: 10.1128/JVI.01368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakanishi Y, Lu B, Gerard C, Iwasaki A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature. 2009;462:510–513. doi: 10.1038/nature08511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.