Abstract
Autoimmune Type 1 Diabetes (T1D) in humans and NOD mice results from interactions between multiple susceptibility genes (termed Idd) located within and outside the MHC. Despite sharing ~88% of their genome with NOD, including the H2g7 MHC haplotype and other important Idd genes, the closely related NOR strain fails to develop T1D due to resistance alleles in residual genomic regions derived from C57BLKS mice mapping to Chromosomes (Chr.) 1, 2 and 4. We previously produced an NOD background strain developing a greatly decreased T1D incidence due to a NOR-derived 44.31 Mb congenic region on distal Chr. 4 containing disease resistance alleles decreasing the pathogenic activity of autoreactive B and CD4 T cells. In this study a series of subcongenic strains for the NOR-derived Chr. 4 region were utilized to significantly refine genetic loci regulating diabetogenic B and CD4 T cell activity. Analyses of these subcongenic strains revealed the presence of at least two NOR origin T1D resistance genes within this region. A 6.22Mb region between rs13477999 and D4Mit32, not previously known to contain a locus affecting T1D susceptibility and now designated Idd25, was found to contain the main NOR gene(s) dampening diabetogenic B cell activity, with Ephb2 and/or Padi2 being strong candidates as the causal variants. Penetrance of this Idd25 effect was influenced by genes in surrounding regions controlling B cell responsiveness and anergy induction. Conversely, the gene(s) controlling pathogenic CD4 T cell activity was mapped to a more proximal 24.26Mb region between the rs3674285 and D4Mit203 markers.
Keywords: Rodent, B cells, T cells, Diabetes, Gene Regulation
INTRODUCTION
Autoreactive T cells are responsible for destroying pancreatic β cells during the development of Type 1 Diabetes (T1D)3 in humans and the NOD mouse model (1, 2). A pathogenic role for B cells in this disease first became evident when NOD mice made deficient in this population through introduction of an inactivated Igµ heavy chain gene (NOD.Igµnull) or chronic treatment with anti-IgM antibodies were strongly protected from T1D (3, 4). More recent studies have shown that temporal B cell depletion in NOD mice at different stages of disease, using antibodies or chimeric proteins targeting CD20, CD22 or the B lymphocyte stimulator (Blys/BAFF), could be effective at delaying, inhibiting or reversing the onset of T1D (5, 6). These findings were mirrored in patients with recent-onset T1D treated with the B cell depleting CD20-specific Rituximab mAb, who on average, exhibited decreased loss of endogenous insulin production and reduced requirement for exogenous insulin over a one year follow-up period (7). This confirmed the role of B cells in the pathogenesis of human T1D, as well as showing that targeting this population could be a potentially effective therapeutic strategy.
Multiple lines of evidence indicate that the primary pathogenic function of B cells in T1D consists of acting as APC for CD4 T cells. Firstly, NOD.Igµnull mice were shown to produce poor CD4 T cell responses to T1D associated autoantigens (8–10). Secondly, autoreactive CD4 T cell responses and the development of insulitis and T1D could only be restored by reconstituting NOD.Igµnull mice with NOD B cells, but not autoantibodies (9). Finally, strong T1D protection was observed in NOD bone marrow (BM) chimera mice whose B cells were made specifically deficient in MHC class II molecules (11). The pathogenic APC function of B cells has been shown to be particularly dependent on their capacity to specifically capture antigens via surface Ig (12, 13). Thus, like T cells, B cells contributing to T1D must also arise from defects in self-tolerance mechanisms, leading to the generation and activation of autoreactive clones. Indeed, defects in various tolerance induction mechanisms, including receptor editing (14), anergy (15–17) and peripheral deletion of self-reactive clonotypes (15, 18) have been shown to contribute to the generation of autoreactive B cells in NOD mice. Similar defects in B cell tolerance have recently been reported in humans developing T1D (14, 19, 20). Given the important contribution of B cells to T1D, it was likely that their pathogenic capacity would be controlled by genes conferring susceptibility to this disease.
Predisposition to T1D in both NOD mice and humans is determined by multiple susceptibility genes located within and outside the MHC (21, 22). Presently, in NOD mice, these genes have been mapped to more than 40 loci (termed Idd) on 16 different chromosomes (21). While the MHC class II genes within the NOD H2g7 haplotype (Idd1) are the most influential determinants of T1D in this strain, contributions by non-MHC susceptibility genes are also necessary for disease development (21). This is highlighted by the closely related NOR mouse strain, which despite sharing approximately 88% of its genome with NOD mice, including the H2g7 MHC haplotype, exhibits strong protection from T1D (23). This protection is encoded within C57BLKS (BKS)-derived genetic regions (itself a combination of B6 and DBA/2 genomes) in NOR mice on Chromosomes (Chr.) 1, 2 and 4 (23–26). The NOR derived T1D resistance locus on Chr. 2, termed Idd13, had not been observed in outcrosses of NOD mice with other strains. Conversely, the NOR-derived T1D resistance genes on Chr. 1 and 4 were mapped to similar regions as the previously identified Idd5.2/5.3 and Idd9.1/9.2/11 loci, respectively, which were localized using NOD outcrosses with B10 or B6 disease-resistant strains (27–29). However, given their diverse origin from the BKS strain, it is also possible that the NOR-derived T1D resistance genes on Chr. 1 or 4 are not completely overlapping with those of B6 or B10 origin.
Our previous studies have shown that reconstitution of lethally irradiated NOD.Igµnull mice with syngeneic bone marrow (SBM) and purified NOD B cells, prevented rejection of grafted B cells by host CTL and restored their susceptibility to T1D (9, 30). On the other hand, those reconstituted with SBM only remained resistant to disease. Interestingly, reconstitution of NOD.Igµnull recipients with equivalent levels of B cells from NOR instead of NOD mice resulted in a significantly reduced incidence of disease (30). To identify the NOR-derived Idd resistance loci responsible for dampening the diabetogenic activity of B cells, NOD.Igµnull mice were reconstituted with B cells from NOD stocks containing NOR-derived congenic regions on either Chr. 1, 2 or 4 (30). Only those B cells containing a congenic region on Chr. 4 (NOD.NOR-Chr4, abbreviated to NR4), spanning from 104.99 to 149.30Mb (26), recapitulated the T1D resistance conferred by NOR B cells (30). Thus, genes within the Idd9.1/9.2/11 regions, or alternatively a previously unrecognized locus, are therefore important for inhibiting diabetogenic B cell activity in NOR mice.
NOD and NR4 B cells were subsequently compared for a number of characteristics to establish potential mechanisms by which polymorphic genes within the distal region of Chr. 4 controlled their diabetogenic activity. Whereas a previous study reported that genes within distal Chr. 4 contributed to exaggerated marginal zone (MZ) B cell development in NOD mice (31), splenic B cell subsets were comparable in NOD and NR4 mice (30). In addition, deletion of splenic transitional-1 (T1) B cells stimulated through the BCR was equivalent between strains. However, mature B cells from lymph nodes (mainly of the FO subset) or spleen (FO and MZ subsets) of NR4 congenic mice proliferated more vigorously to BCR stimulation than those from NOD mice, which was further accentuated by CD40 co-stimulation (30). The NR4 congenic region was also shown to contribute to the maintenance of anergy in self-reactive B cells (30). Thus, in contrast to hen-egg lysozyme (HEL) specific Ig (IgHEL) transgenic (tg) B cells on the NOD background, which showed equivalent levels of proliferation following stimulation through the BCR and CD40 irrespective of whether they developed in the presence or absence of their cognate soluble neo-self-antigen (sHEL), similar B cells from the NR4 stock anergized properly in response to self-antigen exposure. A subsequent study found that the NR4 congenic region also had the capacity to intrinsically dampen the pathogenic activity of CD4 T cells contributing to T1D (32). Whether the CD4 T cell effect is controlled by the same or a distinct NOR gene(s) to that controlling B cells remains to be elucidated.
The NOR-derived congenic interval in NR4 mice is large (44.31Mb, containing 889 known or predicted genes), making identification of genes regulating the pathogenic activity of B and CD4 T cells within this region difficult. In this study, we describe the generation of a series of novel subcongenic strains for the NOR-derived Chr. 4 region. B and CD4 T cells from subcongenic strains were utilized in various experimental strategies to significantly refine genetic regions within distal Chr. 4 that modulate development of T1D by controlling the pathogenic function of these lymphocyte populations.
MATERIALS AND METHODS
Mice
All mice were housed at the Garvan Institute or The Jackson Laboratory (TJL) animal facilities under standard specific pathogen free conditions with unrestricted access to food and acidified water. NOD/Lt mice were purchased from Australian BioResources (ABR; Moss Vale, NSW, Australia) or produced at TJL. Derivation of the NR4 strain (at N5) using a speed-congenic strategy was previously described (26). The proximal boundary of the congenic region in these mice lies between rs3674285 (104.99Mb) and D4Mit31 (106.78Mb). Screening of additional genetic markers has identified a more distal boundary for the congenic region than originally described, which lies between D4Mit129 (148.30Mb) and D4Mit127 (149.30Mb). Seven new subcongenic strains (NR4S1–S4 and NR4S6–7 at N7 and NR4S5 at N8) containing smaller NOR-derived congenic regions on Chr. 4 were developed by outcrossing the NR4 strain with NOD mice, and then backcrossing heterozygous progeny with NOD mice for an additional 2–3 generations. Progeny of the first backcross were screened for recombinations within the original congenic interval by determining the origin of microsatellite markers at the proximal (D4Mit31) and distal (D4Mit129) boundaries. Those that were NOD homozygous at one of these markers were screened with an additional 22 microsatellite (primer sequences obtained from the Mouse Genome Informatics website http://www.informatics.jax.org.au/ or (27)) or single nucleotide polymorphism (SNP) markers (carried out by KBioscience (Hoddesdon, UK)) to refine the location of recombinations. The NR4S5 strain, containing two recombinations within the original NR4 congenic interval, was generated by an additional backcross to NOD of a subcongenic mouse that was NOD/NOR heterozygous between microsatellites D4Mit203 and D4Mit129. Mice with informative recombinations underwent a final backcross to NOD to produce male and female offspring with the same recombination, which were intercrossed to fix the congenic regions to homozygosity. Generation of the NOD.Igµnull (3), NOD.CD4null, NOD.AI4αβ-tg (33), NOD.B6-Ptprcb (NOD.CD45.2) (34) as well as NOD and NR4 mice expressing IgHEL and sHEL transgenes (12, 15, 30) were previously described. Only female mice were used in experiments, which were approved by the Garvan Institute/St. Vincent's Hospital or TJL Animal Ethics Committees.
Assessment of T1D development
T1D development was assessed by measuring glycosuric values weekly with Ames Diastix (Bayer Corporation Diagnostics Division, Elkhart, IN). Values of ≥500 mg/dL were considered indicative of overt diabetes.
Mixed BM and B cell chimeras
CTL in otherwise unmanipulated NOD.Igµnull mice destroy passively transferred B cells (9). B cell reconstitution was therefore achieved by lethally irradiating NOD.Igµnull mice between 4–5 weeks of age with two 550-rad split doses from an X-Ray source followed by i.v. injection with 3×106 anti-CD4 (GK1.5) and anti-CD8 (53–6.7, eBioscience, San Diego, CA) coated SBM cells admixed with 7×106 purified splenic B cells from the indicated strains. Splenic B cells were purified using the previously described (9) MACS negative depletion system (Miltenyi Biotec, Bergisch Gladbach, Germany) which routinely achieved >93% purity as determined by flow cytometry. BM/B cell recipients were then monitored weekly between 8 and 23 weeks post-reconstitution for T1D development. Upon disease onset or at the end of incidence study (23 weeks post-reconstitution), spleens from recipient mice were assessed by flow cytometry to determine percentage reconstitution of B cells, and CD4 T cells using previously published methods and reagents (9). Flow cytometry was also utilized to establish reconstitution levels of lymphocytes expressing CD45.2 or CD45.1 allotypic markers in NOD.Igµnull recipients using the 104 and A20 antibody clones (BD Biosciences), respectively.
Mixed BM and CD4 T cell chimeras
NOD.CD4null mice were lethally irradiated (two 650-rad split doses from a 137Cs source) and reconstituted i.v. with 5×106 anti-CD8 (53–6.7, eBioscience) coated SBM cells admixed with 5×106 purified CD4 T cells from the indicated strains. Splenic CD4 T cells were purified using the previously described MACS negative depletion strategy (32), which routinely achieved >92% purity as determined by flow cytometry. BM/CD4 T cell recipients were monitored weekly between 8 and 20 weeks post-reconstitution for T1D development.
Adoptive transfer of AI4 T cells
The indicated recipient mice at 6–8 weeks of age were sublethally irradiated (600 rad from a 137Cs source) and injected i.v. with 5×106 NOD.Rag1null.AI4αβ-tg splenocytes (~1×106 AI4 T cells). T1D development was monitored daily for up to two weeks post-transfer.
B cell proliferation assays
B cells were purified from pooled spleens of three mice of the indicated strains (6–8 weeks of age) using the EasySep® Mouse B cell enrichment kit, followed by separation using an EasySep® magnet according to manufacturers instructions (Stemcell technologies inc., Vancouver, BC, Canada). This method routinely achieved >98% purity of B cells as determined by flow cytometry. Triplicate aliquots of 1×105 B cells were cultured in complete RPMI 1640 media with 10µg/ml of LPS (Sigma Aldrich, St Louis, MO) or 10µg/ml AffiniPure goat anti-mouse IgM F(ab’)2 fragments (Jackson ImmunoResearch Laboratories, West Grove, PA) and 5µg/ml of CD40-specific mAb HM40-3 (BD Biosciences, San Jose, CA) alone or in combination. Control wells contained no stimulatory agents. Proliferation of cells was assessed by addition of 1µCi/well [3H]-thymidine (Amersham Pharmacia Biotech, Uppsala, Sweden) in the final 24h of a 72h incubation period using previously described methods (12).
Microarrays and quantitative real-time PCR (q-rtPCR)
For microarrays, B cells from three independent lots of 7–8 pooled spleens from NOD or NR4 mice, respectively, were purified using a previously described MACS negative depletion strategy (9). B cells from each lot were resuspended at 1×107 cells/ml for 2h in complete RPMI 1640 media alone or with 10µg/ml AffiniPure goat anti-mouse IgM F(ab’)2 fragments (Jackson Immunoresearch Laboratories). Total RNA was then purified from B cells using TRIzol (Invitrogen, Carlsbad, CA) and subjected to one-cycle linear amplification, biotin-labeling and fragmentation according to the protocol provided by the GeneChip Expression 3’-Amplification One-cycle kit (Affymetrix, Santa Clara, CA). Ten micrograms of biotin-labeled and fragmented cRNA from two BCR-stimulated and three unstimulated NOD B cell samples plus three BCR-stimulated and three unstimulated NR4 B cell samples were then hybridized to Mouse Genome 430 2.0 Chips (Affymetrix) for 16 h at 45°C and scanned with a GeneChip Console Scanner (Affymetrix). All microarray data were submitted to the Gene Expression Omnibus public database (http://www.ncbi.nlm.nih.gov/geo/) under the submission number GSE37294. Analysis of expression data was performed using GeneSpring 12.0 software (Agilent Technologies, Santa Clara, CA). All samples were normalized by Robust Multiarray Analysis (RMA) on a per gene basis to the median of the total samples. Differential expression of genes was determined using one- or two-way ANOVA tests with Benjamini and Hochberg false detection rate (FDR) correction (q-value) to account for multiple testing. Gene symbols used in Figures and Tables are those prescribed by the Mouse Genomic Nomenclature Committee (MGNC). For q-rtPCR, B cells were purified from three pools of three spleens each from the indicated strains of mice (independent of microarray samples) utilizing the aforementioned EasySep® Mouse B cell enrichment kit. RNA was then immediately extracted from each sample using TRIzol (without culture) and transcribed into cDNA using M-MuL V reverse transcriptase (New England BioLabs, Ipswich, MA) via the first strand synthesis protocol provided by the manufacturer. Levels of gene transcripts were determined using a PRISM7900 HT machine (Applied Biosystems, Foster City, CA) utilizing triplicate q-rtPCR reactions containing 50–100ng cDNA and 0.9µM of each primer in LightCycler-RNA SYBR-Green-I Master Mix (Roche, Mannheim, Germany). Primers for each gene were designed such that one would bind across an exon-exon boundary (sequences in Supplementary Table I). The capacity of primers to quantify target genes by q-rtPCR was initially established using triplicate sets of four-fold serial dilutions (1/1, 1/4, 1/16, 1/64 and 1/256) of NR4 or NOD spleen cDNA. Amplification efficiency of all primers ranged between 95.7–99.6% (data not shown). Expression of target genes in B cell samples were normalized to the Hypoxanthine-guanine phosphoribosyl transferase (Hprt) housekeeping gene.
RESULTS
Determining the mode of inheritance of the NOR-derived Chr. 4 gene(s) that dampens the diabetogenic activity of B cells
In a previous study, we showed that in contrast to the low incidence of T1D exhibited by NR4 mice (25% by 30 weeks), mice that were NOD/NOR heterozygous at this congenic region developed a higher incidence of disease that was not significantly different from NOD homozygous littermates (71% vs. 92% by 30 weeks, respectively; NS, log-rank test)(26). This result indicated that NOR-derived allele(s) in the original full-length (FL) Chr. 4 congenic interval must be homozygous to confer significant resistance to T1D. To establish whether the gene(s) decreasing the diabetogenic capacity of B cells in the NR4 congenic region was consistent with this mode of inheritance, we compared the T1D incidence of lethally irradiated NOD.Igµnull mice reconstituted with SBM plus MACS-purified B cells from either, NOD, NR4 or (NOD×NR4)F1 donors (Fig. 1). The T1D incidence of NOD.Igµnull recipients reconstituted with NR4 B cells differed significantly from those reconstituted with NOD B cells after 23 weeks (31.3% (n=16) vs. 69.6% (n=23); p<0.05, log rank test). However, T1D development did not statistically differ in recipients reconstituted with (NOD×NR4)F1 or NOD B cells. This indicated that a NOR-derived allele(s) on distal Chr. 4 must be homozygous to significantly suppress diabetogenic B cell development. Differences in T1D susceptibility between cohorts was unlikely due to expansion of contaminating CD4 or CD8 T cells from the purified donor B cell preparations. This was made evident in two lethally irradiated NOD.Igµnull mice (expressing CD45.1) reconstituted with SBM plus B cells purified from NOD.CD45.2 congenic mice, whose spleens six weeks post-engraftment contained 1.2% and 6.0% CD45.2+ CD4 T cells, 2.4% and 6.4% CD45.2+ CD8+ T cells and 99.3% and 99.2% CD45.2+ B cells, respectively (data not shown).
FIGURE 1.
NOR-derived allele(s) on distal Chr. 4 must be homozygous to significantly suppress diabetogenic B cell development. T1D incidence was monitored weekly for up to 23 weeks in cohorts of female NOD.Igµnull mice that were lethally irradiated at 4–5 week of age and reconstituted with 3×106 syngeneic BM admixed with 7×106 splenic B cells from NOD (n=23), NR4 (n=16) and (NOD×NR4)F1 (n=15) mouse strains. P values for comparison of incidence curves by log-rank analysis are shown.
Generation of novel subcongenic lines of the NR4 mouse strain
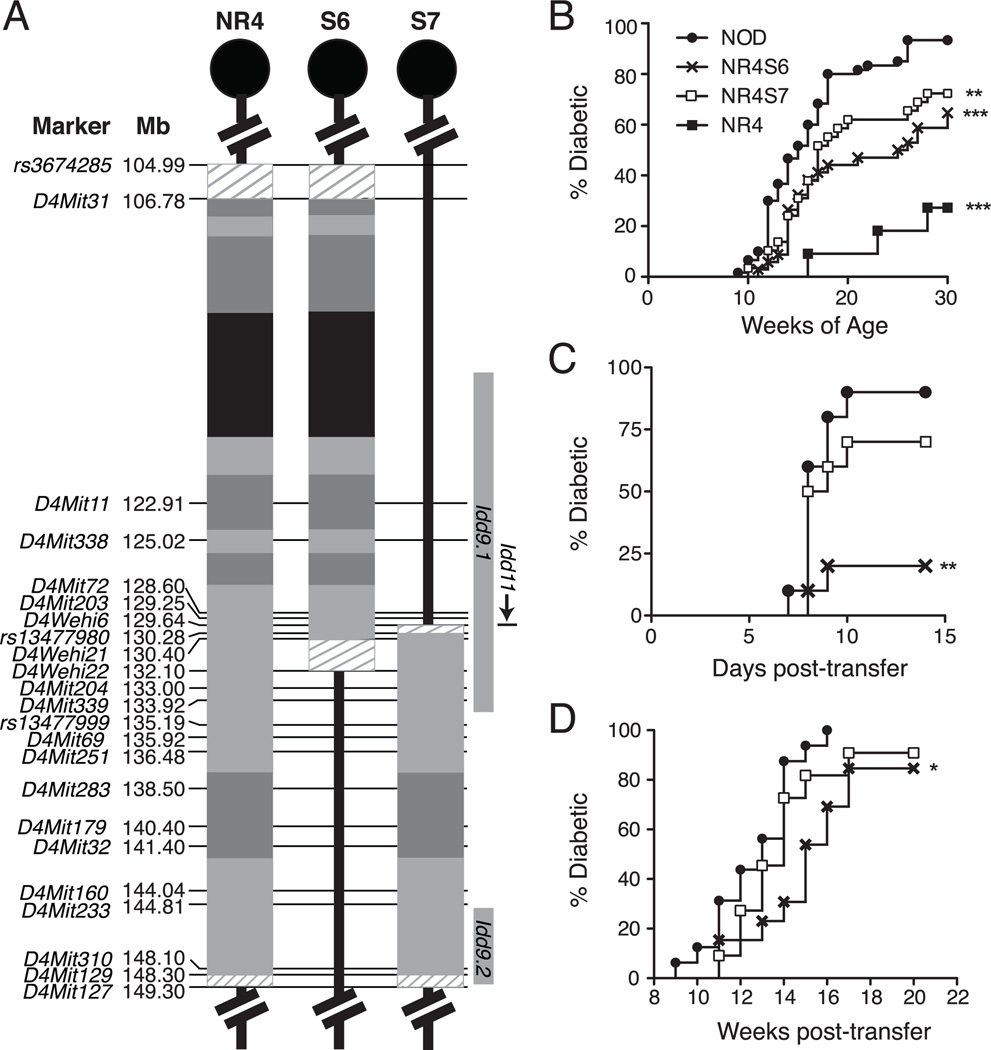
To facilitate identification of the NOR-derived distal Chr.4 region gene (or genes) controlling T1D resistance as well as the decreased pathogenic activity of B cells in the original FL NR4 congenic strain, subcongenic lines were generated as outlined in Materials and Methods. This resulted in the generation of five novel subcongenic NR4 mouse lines (referred to as NR4S1–S5) containing smaller homozygous NOR-derived regions spanning different sections of the original FL 44.31Mb congenic interval. The genetic boundaries of the NOR-derived regions in the NR4S1–S5 subcongenic strains relative to the original NR4 strain and previously identified Idd9 and Idd11 genes (27, 28) are illustrated on Fig. 2A.
FIGURE 2.
Generation of NR4 subcongenic strains reveals the presence of more that one T1D resistance gene in the distal Chr. 4 region of NOR mice. (A) Schematic illustration showing boundaries of NOR-derived congenic regions on Chr. 4 in the NR4 and NR4S1–S5 subcongenic mouse lines. Orientation of Chr. 4 is marked by a circle symbolising the centromere. Rectangular boxes within the broken lines represent NOR-derived congenic intervals. Boundaries of congenic intervals were determined by the polymorphic microsatellite and SNP markers shown. Markers are ordered according to their physical position in Mb according to Ensembl http://www.ensembl.org/; except WEHI markers, whose positions were obtained from (27). Colours within congenic regions mark NOR genome of NOD (black), B6 (light green) or DBA/2 (dark green) origin as previously defined by Reisnyder and colleagues (26). Regions of unknown origin between markers are represented by hatched lines. For comparison, the most current positions of the Idd9.1 (116.08–134.49Mb), Idd9.2 (144.97–149.10Mb), and Idd11 (129.63–129.64Mb) loci are shown (27, 28). (B) Cumulative incidence of T1D in female cohorts of NOD (n=33), NR4 (n=29), NR4S1 (n=33), NR4S2 (n=29), NR4S3 (n=36), NR4S4 (n=31), and NR4S5 (n=27) mice through to 40 weeks of age. (C) p-values from comparisons of T1D incidence curves using log-rank analysis (NS=non-significant).
T1D susceptibility in NR4 subcongenic mouse lines
To refine the NOR-derived region(s) on Chr. 4 responsible for conferring protection from T1D, the natural incidence of disease in female mice of the NR4S1–S5 subcongenic lines were compared to that of the NOD and the original FL NR4 congenic strain through forty weeks of age (Fig. 2B). Greater than 90% of female NOD mice developed T1D by forty weeks of age compared to only 23% in the NR4 stock (p<0.0001, log-rank analysis; Fig. 2C), consistent with the results of a previous study on the same strains performed in an independent animal facility (26). The NR4S1 (65%, n=33), NR4S2 (63%, n=29), NR4S3 (73%, n=36) and NR4S4 (59%, n=31) mouse strains all showed an intermediate T1D incidence (Fig. 2B) that differed significantly from both the NOD and FL NR4 strains, but not each other (Fig. 2C). In contrast, the NR4S5 mouse strain developed T1D at a similar incidence and rate (86%, n=27) as NOD mice (Fig. 2B), indicating that the central section of the NR4 congenic region between D4Mit72 and rs13477999 (Fig. 2A), which encompasses the Idd11 locus (27), does not contain an NOR origin gene conferring resistance to T1D development. Our observation that none of the subcongenic strains showed the same level of T1D protection as the original FL NR4 congenic strain provides evidence of more than one distal Chr. 4 region gene contributing to disease resistance in NOR mice. This was also confirmed by the fact that there was no overlap in the NOR-derived regions of two subcongenic strains showing T1D resistance, NR4S4 and NR4S2 (Fig. 2A–B). These two subcongenic strains therefore define at least two loci containing NOR T1D resistance genes on Chr. 4, which span the intervals between the markers rs3674285 (104.99Mb) to D4Mit203 (129.25Mb) and rs13477999 (135.18Mb) to D4Mit127 (149.30Mb). These regions are partially overlapping with the T1D protective Idd9.1 and Idd9.2 loci, respectively, which were found to be present in the B10 strain (28). However, given the T1D protective genes in the distal region of NOR Chr.4 originally derive from the BKS strain, the possibility cannot be excluded that their identity differs from those responsible for the Idd9.1 and/or Idd9.2 effects.
Diabetogenic capacity of B cells in NR4 subcongenic mouse lines
We next refined the location of the distal Chr. 4 region gene(s) suppressing the diabetogenic capacity of NOR B cells. T1D development was assessed in NOD.Igµnull mice that were lethally irradiated and reconstituted with SBM plus splenic B cells from the T1D-resistant subcongenic strains, NR4S1–S4 (Fig. 3A). Disease incidence in these groups up to 23 weeks were compared to control cohorts of NOD.Igµnull recipients reconstituted with SBM and splenic B cells from NOD or FL NR4 strains. Mirroring our previous study (30), NOD.Igµnull mice reconstituted with NOD B cells developed a high incidence of T1D (68%, n=33), which was reduced by approximately half (36%, n=30) in recipients of NR4 B cells (Fig. 3A) despite similar levels of B cell reconstitution between groups (Table I). Compared to those receiving NOD B cells, there was a significant reduction in the incidence of T1D shown by NOD.Igµnull mice reconstituted with either NR4S3 (20%, n=10) or NR4S4 (35%, n=17) B cells (Fig. 3A) In addition, these two groups of recipients developed a delayed onset of T1D (NR4S3 21.2±0.9, NR4S4 21.5±0.5 weeks) compared to the NR4 B cell reconstituted groups (18.5±0.6 weeks), suggestive of an enhanced level of protection (Fig. 3B). The difference in mean onset time of T1D was significant between NOD and NR4S4 B cell reconstituted recipients (p=0.006, Student’s t-test). Given that only two NOD.Igµnull mice containing NR4S3 B cells developed T1D in this study, the difference in mean time of disease onset compared to NR4 B cell recipients was not statistically significant, however, the trend for delay was still strong (p=0.08). In contrast, NOD.Igµnull mice reconstituted with B cells from NR4S1 (50%, n=16) and NR4S2 (54%, n=13) subcongenic strains showed no signficant decrease in T1D incidence compared to recipients of NOD B cells (Fig. 3A).
FIGURE 3.
B cells from NR4S3 and NR4S4 subcongenic mouse strains exhibit decreased diabetogenic capacity. (A) T1D incidence was monitored for up to 23 weeks in cohorts of female NOD.Igµnull mice that were lethally irradiated (1100 rad) at 4–5 weeks of age and reconstituted with 3×106 SBM admixed with 7×106 purified splenic B cells from NOD (n=33), NR4 (n=30), NR4S1 (n=16), NR4S2 (n=13), NR4S3 (n=10) or NR4S4 (n=17) strains. *p<0.05, **p<0.01, compared to NOD B cell reconstituted group (log-rank analysis). (B) Graph comparing mean time of disease onset in diabetic NOD.Igµnull mice reconstituted with NOD (n=23), NR4 (n=11), NR4S3 (n=2) or NR4S4 (n=6) B cells. The p-values between groups are shown (Student’s t-test).
Table I.
Levels of B220+ and CD4+ lymphocytes in SBM plus B cell reconstituted NOD.Igµnull recipients.
| B cell donor used to reconstitute NOD.Igµnull micea |
Diabetic (%±SEM) | Non-diabetic (%±SEM) | ||||
|---|---|---|---|---|---|---|
| n | B220+b | CD4+c | n | B220+b | CD4+c | |
| NOD | 22 | 41.8±6.9 | 44.9±6.0 | 11 | 37.8±3.4 | 40.8±2.4 |
| NR4 | 11 | 43.2±8.0 | 29.6±4.9 | 19 | 47.8±3.7 | 28.9±2.8 |
| NR4S1 | 8 | 40.6±3.6 | 39.4±3.0 | 8 | 37.3±3.9 | 31.8±3.2 |
| NR4S2 | 7 | 26.0±4.3 | 38.5±3.7 | 6 | 23.3±2.6 | 44.2±3.0 |
| NR4S3 | 2 | 31.3±7.4 | 34.9±7.2 | 8 | 22.6±1.3 | 38.9±3.4 |
| NR4S4 | 6 | 28.0±4.3 | 43.4±3.9 | 11 | 31.2±2.5 | 36.0±2.8 |
NOD.Igµnull mice were lethally irradiated at 4–5 weeks of age and reconstituted with 3×106 SBM and 7×106 splenic B cells from the indicated strain. Proportions of B220+ B cells and CD4+ T cells in spleens of each recipient mouse were determined by flow cytometry after T1D onset or at the end of study (23 weeks post-reconstitution).
Two-way ANOVA test of B220+ reconstitution dataset did not find significant interaction between diabetes status and B cell donor. Furthermore, diabetes status did not contribute significantly to variation in B220+ reconstitution. In contrast, the strain of B cell donor was found to significantly contribute to variation in B220+ reconstitution (p=0.03). However, Bonferroni post-tests did not identify a significant difference between any two specific B cell donors in diabetic or non-diabetic groups.
Two-way ANOVA test of the CD4+ T cell reconstitution dataset did not find significant interaction between disease status and B cell donor. In addition, neither diabetes status nor B cell donor contributed significantly to variation in CD4 T cell reconstitution.
Two-way ANOVA tests were performed to examine differences in B cell or CD4 T cell reconstitution levels in spleens of diabetic and non-diabetic mice of the various recipient groups (Table I). The strain of B cell donor did contribute to variance in B cell (p = 0.03), but not CD4 T cell (NS), reconstitution levels in the different recipient groups. However, this affect was not strong given that Bonferroni post-tests did not identify a significant difference in percentages of B cells between recipients of any two specific B cell donors in either the diabetic or non-diabetic groups. Added to this, it is unlikely that differences in T1D susceptibility between recipient groups were a consequence of variance in B cell reconstitution levels, as in all transfers, there was no significant difference in B cell proportions in spleens of diabetic vs. non-diabetic mice (Table I; NS, two-way ANOVA, Bonferroni post-tests). Furthermore, recipient groups with a higher incidence of T1D (i.e. NOD, NR4S1 and NR4S2 donors) did not necessarily have higher mean levels of B cell reconstitution than those with a lower incidence (i.e. NR4, NR4S3 and NR4S4 donors) (NS, Student t-test).
The results of these incidence studies indicate that the main NOR-derived gene(s) responsible for dampening the diabetogenic activity of B cells lies within the 6.22Mb overlap of the NR4S3 and NR4S4 congenic regions (i.e. between rs13477999 and D4Mit32; Fig. 2A). This region of Chr. 4 has not been previously identified to contain an Idd locus. Thus, we now designate this newly identified locus on distal Chr.4 differentially controlling the development of diabetogenic B cells in NOD and NOR mice as Idd25. However, it is interesting that the protective B cell phenotype mediated by the Idd25 resistance variant seems to be abrogated by a combined interaction between at least two other NOR genes within the intervals bound by D4Wehi6 to D4Mit69 and D4Mit179 to D4Mit127; which are both present in the NR4S1, but not coexisting in NR4S3 and NR4S4 strains (Fig. 2A). The original FL NR4 congenic region also dampens diabetogenic B cell activity despite containing all of the above mentioned subregions. Thus, it is likely that another NOR origin gene(s) in the most proximal region, bound by the markers rs3674285 and D4Mit203, also contributes to the dampened diabetogenic activity of B cells. However, this NOR gene(s) does not mediate a reduction in pathogenic B cell activity when inherited alone, given that B cells from the NR4S2 congenic stock are as diabetogenic as those from standard NOD mice (Fig. 3A).
Hyperresponsiveness of B cells in NR4 subcongenic mouse lines
B cells from FL NR4 mice undergo significantly higher levels of proliferation upon BCR or BCR plus CD40 stimulation than those from NOD mice (30). This difference in responsiveness to stimulation was evident in B cells from spleen (FO and MZ subsets) or lymph nodes (FO subset only). To identify the genetic region(s) within the original FL NR4 congenic region that are responsible for this B cell hyperproliferative phenotype, we subjected purified splenic B cells from the NR4S1–5 congenic strains to in-vitro stimulation with anti-IgM-F(ab’)2 fragments or anti-CD40 antibodies alone or in combination, or LPS. Consistent with our previous study, NR4 B cells proliferated significantly more than those from NOD mice when stimulated with anti-IgM-F(ab’)2 or anti-IgM-F(ab’)2 and anti-CD40 (1.3±0.07 and 1.5±0.1 fold higher, respectively; Fig. 4A–B). This difference was probably the direct consequence of variability in signalling downstream of the BCR given the observed equivalent responses between B cells stimulated through CD40 alone or through the TLR4 receptor with LPS (Fig. 4C–D). Of the subcongenic B cells, only those isolated from the NR4S1 stock exhibited the hyperproliferative phenotype of the FL NR4 strain in response to stimulation with BCR, with or without CD40 co-stimulation (1.5±0.1 and 1.4±0.2 fold higher proliferation than NOD, respectively; Fig. 4A–B). NR4S4 and NR4S3 B cells did not exhibit a hyperproliferative response to BCR stimulation. This suggested that the hyperproliferation phenotype of NR4S1 B cells is the result of an interaction between NOR genes located in two regions bound by D4Wehi6 and D4Mit69 and D4Mit179 and D4Mit127 that are not co-existant in the NR4S4 and NR4S3 lines. Interestingly, these were also the two regions that combined to lessen the ability of the NOR gene(s) within the rs13477999-D4Mit32 region to suppress diabetogenic B cell activity (Figs. 2A and 3A). These results dissociate the induction of a stronger BCR signaling response from the ability of a NOR-derived Chr. 4 gene(s) to dampen diabetogenic B cell activity.
FIGURE 4.
Hyperresponsive and anergy phenotypes are not necessary for decreased diabetogenic activity of NR4 B cells. (A–D) To test hyperresponsiveness, B cells were purified from pooled spleens of three 6–8 week-old female NOD, NR4, NR4S1, NR4S2, NR4S3, NR4S4 and NR4S5 mice. Triplicate aliquots of 1×105 B cells were stimulated in culture with (A) 10µg/mL anti-IgM-F(ab’)2; (B) 10µg/mL anti-IgM-F(ab’)2 and 5µg/mL anti-CD40; (C) 5µg/mL anti-CD40 or; (D) 10µg/mL LPS. Proliferation over the final 24h of a 72h incubation period was measured by [3H]thymidine incorporation and is shown as a mean fold-change compared to NOD B cells (±SEM) in 2 to 4 independent experiments for each congenic strain. *p<0.05, **p<0.01, ***p<0.001 compared to NOD B cells (one-way ANOVA, Tukey post-test). (E–H) To test anergy induction, B cells were purified from pooled spleens of three 6–8 week-old female IgHEL single-tg or IgHEL/sHEL double-tg mice with NOD, NR4, (NOD×NR4)F1, (NR4S2×NR4)F1, (NR4S3× NR4)F1, (NR4S4×NR4)F1 or (NR4S5×NR4)F1 genetic backgrounds. Triplicate aliquots of 1×105 B cells were stimulated in culture with: (E) 10µg/mL anti-IgM-F(ab’)2; (F) 10µg/mL anti-IgM-F(ab’)2 and 5µg/mL anti-CD40; (G) 5µg/mL anti-CD40; or (H) 10µg/mL LPS. Proliferation over the final 24h of a 72h incubation period was measured by [3H]thymidine incorporation. Bars represent the mean fold-change in proliferation of B cells from IgHEL/sHEL double-tg mice compared to single IgHEL counterparts (±SEM) in 2 to 4 independent experiments for each subcongenic background, while the NOD, NR4 and (NOD × NR4)F1 background controls were performed 5, 4 and 8 times, respectively. *p<0.05 compared to NOD and (NOD × NR4)F1 background group (one-way ANOVA, Tukey post-test).
B cell anergy in NR4 subcongenic mouse lines
Our previous study demonstrated that homozygous expression of the FL NOR-derived Chr. 4 congenic region on the NOD background resulted in the correction of faulty B cell anergy induction in the IgHEL/sHEL tg model (30). Based on the observation that the NOR-derived FL NR4 congenic interval must be inherited in a homozygous manner to significantly suppress diabetogenic B cell development (Fig. 1), an array of F1 hybrids were made to fix the various NR4 subcongenic intervals to homozygosity to assess their effects on B cell anergy induction. The strategy consisted of crossing the previously developed FL NR4 congenic strain expressing both IgHEL and sHEL transgenes (30) with the NR4S2, NR4S3, NR4S4 or NR4S5 subcongenic mouse lines. The resulting hybrid progeny were homozygous at the NOR-derived region shared between the NR4 and subcongenic strain, but heterozygous throughout the remainder of the FL congenic region; whilst also expressing a combination of hemizygous IgHEL and sHEL transgenes. Anergy was assessed by measuring the proliferative response of B cells from IgHEL/sHEL double-tg mice versus IgHEL single-tg mice from each hybrid cross following in vitro stimulation with LPS or anti-IgM-F(ab’)2 fragments and anti-CD40 antibodies alone or in combination (Fig. 4E–H). The occupancy of HEL on the majority of BCR of B cells of double-tg, but not single-tg, mice precluded us from using specific antigen stimulation (17). B cells from NOD, (NOD×NR4)F1 and NR4 background mice expressing the same transgenes were utilized as controls. Consistent with our previous study (30), HEL-specific B cells in NOD mice were not effectively anergized when maturing in the presence of sHEL, and thus proliferated at similar levels as naïve B cells in culture, irrespective of the stimuli (Fig. 4E–H). Conversely, a significant reduction in proliferation was observed in NR4 B cells from IgHEL/sHEL double-tg compared to IgHEL single-tg mice in response to anti-IgM-F(ab’)2 with or without anti-CD40 (Fig. 4E–F). Such a reduction was not evident when B cells were stimulated with LPS or anti-CD40 alone (Fig. 4G–H). B cells from (NOD×NR4)F1 mice expressing IgHEL and sHEL transgenes showed a similar defect in B cell anergy as NOD background mice (Fig. 4E–F), highlighting the requirement for homozygous expression of NOR alleles in the FL-NR4 congenic region to correct this immunoregulatory process. Of the hybrid subcongenic progeny, only B cells homozygous for the NR4S2 region exhibited a significant decrease in proliferation following stimulation with anti-IgM-F(ab’)2 alone or in combination with anti-CD40 after having developed in the presence as opposed to the absence of their cognate antigen (Fig. 4E–F). This decrease (~30%) was not as distinct as that observed for IgHEL/sHEL double-tg B cells from the FL NR4 strain (~50%). This indicated the NR4S2 subcongenic region contributes to a partial decrease in responsiveness of self-reactive B cells, but is not the sole region affecting this phenotype in the original FL NR4 congenic mouse strain. In this regard, it is worth highlighting the result of tg B cells derived from hybrid mice homozygous for the NR4S3 region. Despite also being homozygous throughout the NR4S2 region, NR4S3 B cells showed equivalent levels of proliferation in response to anti-IgM-F(ab’)2 and anti-IgM-F(ab’)2 plus anti-CD40 antibodies whether they matured in the presence or absence of their cognate self-antigen, similar to NOD background mice (Fig. 4E–F). This result would imply that the NOR region differentiating the NR4S2 and NR4S3 strains (between D4Mit203 and D4Mit32; Fig. 2A) contains a gene(s) that contributes to the aberrant induction of anergy in self-reactive B cells. Moreover, the clear difference in responsiveness of self-reactive B cells in mice homozygous for the NR4S3 subcongenic region compared to those carrying the FL NR4 congenic segment would indicate that a NOR-derived region between D4Mit179 and D4Mit127 (Fig. 2A) must also contribute to the correction of B cell anergy induction. The gene within this region, can only mediate its effect on B cell anergy when inherited in combination with a homozygous NOR-derived gene(s) in the NR4S2 congenic interval. This conclusion is supported by the finding that strains homozygous for NOR genes in the distal region alone (i.e. NR4S1 and NR4S4 hybrid strains) displayed defects in B cell anergy induction that were indistinguishable from that in standard NOD mice (Fig. 4E–F). B cells showing partially corrected tolerance induction in the NR4S2 line were as diabetogenic as those from standard NOD mice, while NR4S3 and NR4S4 B cells with dampened pathogenic activity exhibited anergy defects equivalent to NOD (Figs. 3A and 4E–F). Thus, it would appear that homozygous expression of NOR genes in both the rs3674285 to D4Mit203 and D4Mit179 to D4Mit127 intervals, which combinatorially restore anergy induction processes, do not independently suppress diabetogenic B cell activity, but may augment this process.
Differentially expressed genes from microarray analysis of NOD vs. NR4 B cells
Microarrays were conducted on B cells purified from spleens of NOD and NR4 mice to highlight differentially expressed genes within the distal Chr. 4 locus. B cells were either cultured in media alone (unstimulated) or with BCR cross-linking anti-IgM-F(ab’)2 fragments (stimulated) for 2h before RNA was extracted for transcript analysis. Several significant expression differences were noted between NOD and NR4 B cell samples using a one-way ANOVA test, and as expected, most of these genes (24/32 mapped probes) were situated within the distal Chr. 4 region encompassed by the FL NR4 congenic interval (Fig. 5A, Supplementary Fig. 1A–B and Supplementary Table II). These 24 differentially expressed genes represented 1.8% of the total probesets (n=1299) that detect transcripts within the Chr. 4 congenic region. Interestingly, none of the genes that were differentially expressed between NOD and NR4 B cells changed significantly upon 2h of BCR cross-linking (Supplementary Fig. 1A–B). Nevertheless, polymorphic genes within the distal Chr. 4 region had the capacity to differentially alter stimulation responsive genes located at other sites, given more than approximately half of the genes that changed significantly after BCR cross-linking (≥2-fold change and q<0.1, one-way ANOVA), were not shared by the NOD and NR4 strains (Supplementary Fig. 1A and Supplementary Table III).
FIGURE 5.
Differentially expressed genes in NOD vs. NR4 B cells within the distal Chr. 4 locus. (A) Differentially expressed genes between NOD vs. NR4 splenic B cells (q<0.05 FDR, one-way ANOVA) that map within the region of distal Chr. 4 distinguishing the two strains were determined using Affymetrix Mouse 430 2.0 microarray chips after 2h culture in media alone (n=3 for NOD, n=3 for NR4) or with 10µg/ml of anti-IgM-F(ab’)2 fragments (n=2 for NOD, n=3 for NR4). None of the Chr. 4 gene probes showed significant changes in expression with stimulation (see Supplementary Fig. 1A–B). Therefore, each line represents the mean fold-change in expression between combined NOD (n=5) vs. NR4 (n=6) B cell groups for each differentially expressed gene probe plotted against its physical position in Mb according to the Ensembl database. The location of the congenic region in NR4 mice is demarcated by the thick grey line. All differentially expressed genes reside in genomic areas that are B6-derived in NR4 mice, except Padi2 (underlined), which is in a DBA/2-derived region. (B) Differentially expressed genes in microarray analyses were confirmed by performing q-rtPCR of selected genes using cDNA from three independent lots of purified NOD and NR4 splenic B cells that were not subjected to culture. Rt-qPCR was conducted on triplicate aliquots of cDNA and normalized to expression of the house-keeping gene Hprt. Data are presented as mean fold-change differences in expression in three NR4 samples relative to the mean of the NOD B cell samples. *p<0.05, **p<0.01, ***p<0.001 (Student’s t-test). (C) q-rtPCR was performed for the Tceb3, Ephb2 and Padi2 genes using cDNA obtained from three independent lots of splenic B cells from 6–8 week-old female NOD, NR4, NR4S3 and NR4S4 mice. Gene expression was normalized to Hprt. Data for each strain are presented as the mean fold-change difference in expression relative to mean of the NOD B cell samples. *p<0.05, **p<0.01 and ***p<0.001 compared to NOD (one-way ANOVA, Tukey post-test).
Of the 24 genes within distal Chr. 4 that were deemed to be differentially expressed between NOD and NR4 B cells (Fig. 5A), eleven were chosen for further examination by q-rtPCR due to their potential involvement in tolerance mechanisms (e.g. cell death), BCR signaling, lymphocyte function or as a consequence of large differences in expression levels between mouse strains. Differential expression of these genes were analyzed in an independent set of cDNA derived from purified NOD and NR4 splenic B cells that were not subjected to culture. Consistent with the results of the microarrays, the expression of the selected genes: Low density lipoprotein receptor-related protein 8 (Lrp8), Coiled-coil domain containing (Ccdc)28b, Ccdc21, Ephrin Receptor B2 (Ephb2) and Taxilin α (Txlna) were higher in B cells from NR4 than NOD mice; while Zinc finger protein 69 (Zfp69), Transcription elongation factor B (SIII), polypeptide 3 (Tceb3), Peptidyl arginine deiminase, type II (Padi2), Riken cDNA 2610305D13 (2610305D13Rik) and Mammalian target of rapamycin (mTOR) were more highly expressed by B cells from NOD compared to NR4 mice (Fig. 5B). The only gene that was not consistent with the results of the microarrays was Protein tyrosine phosphatase, receptor type, F (Ptprf), which did not show a significant difference in expression between NOD and NR4 B cells by q-rtPCR.
B cells from the subcongenic strain NR4S3 and NR4S4 strains showed a decrease in diabetogenic activity (Fig. 3A). Of the genes confirmed to be differentially expressed between NOD and NR4 B cells by microarray, only Tceb3, Ephb2 and Padi2 are located within the overlap of the NR4S3 and NR4S4 congenic regions (between rs13477999 and D4Mit32; Fig. 2A). We therefore assessed the differential expression of these genes by rt-qPCR in NR4S3 and NR4S4 B cells compared to those from NOD and NR4 mice (Fig. 5C). Consistent with their expression in NR4 B cells, we observed a significant decrease in expression of Padi2 and a significant increase in expression of Ephb2 in NR4S3 and NR4S4 B cells compared to those from NOD mice. Whilst a significant decrease in Tceb3 expression was also observed in NR4S3 relative to NOD B cells, this difference was not evident in the comparison with NR4S4 B cells. Confirmation of differential Padi2 and Ephb expression within the NR4S3 and NR4S4 mouse strains implicates these genes as potential Idd25 candidates for the control of diabetogenic B cell activity in NOD mice.
Subcongenic analysis of the NR4 region at the level of CD4 T cells
We previously showed that the FL NR4 congenic interval also conferred T1D resistance at the level of CD4 T cells (32). The complex interaction of the NR4 region genes described above raised the possibility that some of the B cell phenotypes may be influenced to some extent by the quality of CD4 T cell help. Conversely, B cell phenotypes controlled by polymorphic NR4 region genes could also differentially effect activation of diabetogenic CD4 T cells. In an independent effort carried out at TJL, two additional NR4 subcongenic lines (NR4S6 and NR4S7) were generated to further define the critical interval that regulates the diabetogenic activity of CD4 T cells as depicted in Fig. 6A. T1D incidence studies performed at TJL demonstrated that both subcongenic lines were less susceptible to disease than standard NOD mice (Fig. 6B). However, neither subregion alone was sufficient to confer the same level of T1D protection provided by the FL NR4 congenic interval. These results are consistent with the observations at the Garvan Institute and confirm that at least two genes contribute to T1D resistance in NR4 mice.
FIGURE 6.
Genes within the NR4S6 subcongenic region regulate T1D development by modulating the pathogenic activity of CD4 T cells. (A) Polymorphic microsatellite and SNP markers were genotyped to define the boundaries of the NR4S6 and NR4S7 subcongenic lines developed at TJL. Colours within congenic regions mark NOR genome of NOD (black), B6 (light grey) or DBA/2 (dark grey) origin. For comparison, the most current positions of the Idd9.1 (116.08–134.49Mb), Idd9.2 (144.97–149.10Mb), and Idd11 (129.63–129.64Mb) loci are shown (27, 28). (B) Cumulative incidence of T1D in female cohorts of NOD (n=60), NR4 (n=11), NR4S6 (n=34), and NR4S7 (n=29) mice was monitored through to 30 weeks of age at TJL. **p<0.01, ***p<0.001 compared to NOD group (log-rank analysis). (C) Five million NOD.Rag1null.AI4-tg splenocytes were transferred into the indicated sublethally (600 rad) irradiated female recipients (n=10/group). T1D development was monitored daily for up to two weeks post-transfer. **p<0.01 compared to NOD recipients (log-rank analysis). (D) T1D development in NOD.CD4null females lethally irradiated (1300 rad) at 4–6 weeks of age and reconstituted with equal numbers (5×106) of SBM and purified CD4 T cells from NR4S6 (n=13), NR4S7 (n=11) or NOD mice (n=16) was followed for 20 weeks. *p<0.05 compared to NOD CD4 T cell recipients (log-rank analysis).
We next asked whether any of the NOR-derived Chr. 4 subregions could suppress the diabetogenic activity of CD4 T cells. We previously showed that the ability of adoptively transferred β cell-autoreactive AI4-αβTCR-tg CD8+ effectors to induce rapid T1D onset in sublethally irradiated NOD recipients was enhanced by help provided by pathogenic CD4 T cells (32). Therefore, the susceptibility of NR4S6 and NR4S7 mice to AI4-induced T1D development was examined to initially evaluate the pathogenic helper activity of their CD4 T cells. As shown in Fig. 6C, the ability of AI4 T cells to induce rapid onset of T1D was significantly reduced in NR4S6, but not NR4S7 mice. These results suggested that CD4 T cells from the NR4S6 stock were less diabetogenic than those from the standard NOD mice. This was indeed the case as demonstrated in a combined BM/CD4 T cell reconstitution system. NR4S6 CD4 T cells had a reduced ability compared to those from standard NOD mice to promote T1D development in NOD.CD4null recipients (Fig. 6D). In contrast, NR4S7 and NOD CD4 T cells had comparable T1D promoting capacity in NOD.CD4null recipients. The NR4S6 congenic region partially overlaps that in NR4S5 mice (Figs. 2A and 6A). As the congenic interval in the NR4S5 stock did not confer T1D resistance (Fig. 2B), it appears that the NOR-derived region regulating diabetogenic CD4 T cells can be defined in a 24.26Mb region between the markers rs3674285 and D4Mit203. This region partially overlaps with the previously defined Idd9.1 locus (Fig. 6A and (28)). In the B cell studies, the same region was shown to promote anergy induction. Collectively, these results suggest that genes within this region may regulate T1D through modulating the interplay between B and CD4 T cells.
DISCUSSION
Congenic NOD mouse strains have facilitated discovery of various T1D susceptibility genes in this model (e.g. β2 microglobulin, Interleukin 2, and Ctla4) (24, 35, 36), which have provided important insights into the molecular and cellular mechanisms influencing the human disease (21, 22). We previously described the generation of the NR4 strain (26), which contained a 44.31Mb NOR-derived congenic region on distal Chr. 4 that decreased susceptibility to T1D in NOD background mice by dampening the pathogenic activity of B and CD4 T cells (30, 32). To significantly refine the location of genes contributing to these phenotypes, we generated seven novel subcongenic NOD mouse lines (termed NR4S1–S7) with truncated NOR-derived regions spanning different parts of the original FL NR4 congenic interval (Figs. 2A and 6A). T1D incidence studies revealed that six of the seven subcongenic strains displayed some protection from disease, although none exhibited the same level of resistance seen in the original NR4 congenic mouse strain (Figs. 2B and 6B). This implied that the NOR-derived region contained more than one gene controlling T1D resistance; which was confirmed by the fact that the NR4S2 and NR4S4 lines containing non-overlapping subcongenic regions both affording similar levels of disease protection. The result of these studies mapped the NOR genes conferring T1D resistance within the Chr. 4 locus to two regions: (i) rs3674285 (104.99Mb) to D4Mit203 (129.25Mb) and; (ii) rs13477999 (135.18Mb) to D4Mit127 (149.30Mb) (Fig. 7). These partly overlap with the previously described Idd9.1 (128.37–131.18Mb) and Idd9.2 (144.97–149.10Mb) susceptibility loci, respectively, mapped using B10 congenic regions on the NOD background (28). Whilst the NOR and B10 strains may share T1D resistance alleles at these loci, the distinct origin of the NOR genome, which is primarily derived from NOD, B6 and DBA/2 strains, appears to also endow it with other novel genes regulating disease (23–26).
FIGURE 7.
Model delineating the distinct segments controlling pathogenic B and CD4 T cell function in the distal Chr. 4 locus of NOR mice. Results of our experiments in the NR4S1–S7 subcongenic strains have identified four regions (R1–R4) on distal Chr. 4 that contain genes controlling pathogenic B and CD4 T cell function. Markers demarcating the boundaries of each region are shown. T1D is independently controlled by NOR-derived genes in the R1 region and the R3–R4 region. The hyperresponsive phenotype of NR4 B cells to BCR stimulation is the result of an interaction of genes within R2 and R4. The maintenance of anergy in NR4 B cells is controlled by genes within R1 and R4, with counter-balancing negative regulation by R2. R3 contains the gene(s) responsible for dampening the diabetogenic activity of NR4 B cells. The penetrance of this gene is inhibited by genes within R2 and R4, potentially by making B cells hyperresponsive to BCR stimulation. Genes within R1 and R4 could also contribute to the dampening of B cell diabetogenic activity in the presence of the R3 region, potentially through their effects on B cell anergy. Finally, the R1 region contains a gene(s) responsible for dampening the diabetogenic function of NR4 CD4 T cells. For comparison, the most current Mb positions of the Idd9.1, Idd9.2, and Idd11 resistance loci from B10 or B6 mice are shown (27, 28).
Of the seven subcongenic strains described in this study, only the NR4S5 strain, with a NOR-derived congenic interval spanning from 128.60 to 135.19Mb (Fig. 2A), did not contribute any protection from the development of T1D (Fig. 2B). This result was surprising given that refinement of the locus housing the Idd11 gene using NOD mice with B6-derived congenic regions showed that significant resistance to T1D was conferred within a 6.9kb interval between 129.63–129.64Mb that lies within a predicted gene of unknown function named Ak005651 (27). Given that the NOR strain was found to have an identical sequence to B6 mice over this 6.9kb interval (27), we would have predicted the NR4S5 strain to show significant protection from disease. This result may be suggestive of genetic drift in the B6-derived region surrounding the Idd11 locus in NOR mice, which may possess polymorphisms that decrease the penetrance of the protective Ak005651 allele.
The other incentive for generating NR4 subcongenic mouse lines was to map the NOR gene(s) within the distal end of Chr. 4 that reduced the diabetogenic activity of B cells; as well as determining whether B cell hyperresponsiveness and the restoration of B cell anergy remained linked to this gene. Our experiments with the subcongenic strains demonstrated that the original NR4 congenic interval could be broken up into four subregions (termed R1–R4) that control different aspects of B cell function (Fig. 7). B cells from the NR4S3 and NR4S4 subcongenic strains had a reduced capacity to induce T1D in NOD.Igµnull mice compared to those from standard NOD donors (Fig. 3A). This led us to hypothesize that the principal gene(s) controlling the diabetogenic capacity of B cells is encompassed within the overlap of the NR4S3 and NR4S4 congenic regions. This subregion, termed R3, consists of a 6.22Mb interval, located between 135.18–141.40Mb (Fig. 7). This region has not previously been associated with the development of T1D and thus is likely to represent a distinct susceptibility gene from those mapped to Idd9.1, Idd9.2 and Idd11 loci, that we now designate as Idd25. Interestingly, the NOD.Igµnull recipients reconstituted with NR4S3 and NR4S4 B cells also exhibited a delayed onset of disease compared to those reconstituted with NR4 B cells (Fig. 3B). Hence, the gene(s) dampening diabetogenic B cell activity in the NR4S3 and NR4S4 strains may have been separated from NOR genes with opposing effects that exist within the FL congenic region.
Importantly, neither NR4S3 nor NR4S4 B cells exhibited the hyperresponsive (Fig. 4A–B) or the restoration of anergy phenotype (Fig. 4E–F) that characterize B cells from the NR4 strain (30). This confirmed that these phenotypes are not directly associated with the NOR gene(s) dampening diabetogenic B cell activity. Nevertheless, our experiments suggest that genes controlling these associated phenotypes may have the capacity to modulate the diabetogenic activity of B cells. In this regard, it is worth noting the B cells from the NR4S1 strain, whose congenic interval also encompasses the R3 region (Fig. 2A), but showed a similar diabetogenic capacity as those from standard NOD mice (Fig. 3A). The potential reason for this is that in addition to R3, the congenic interval in NR4S1 mice contains two additional regions located between 129.64–135.18Mb (termed R2) and 141.40–149.30Mb (termed R4; Figs. 2A and 7). Our experiments showed that the combination of NOR genes within R2 and R4 regions were necessary for B cells to display hyperresponsiveness to BCR stimulation (Fig. 4A–B). NR4S3 and NR4S4 B cells were not hyperresponsive since they only separately contained the R2 or R4 region, respectively (Figs. 2A and 7). From these experiments, we predict that the hyperresponsive phenotype encoded by NOR genes within R2 and R4 leads to increased diabetogenic activity of B cells, overriding the protective B cell phenotype encoded by R3.
Despite containing the R2, R3 and R4 regions (and the hyperresponsive phenotype), B cells from the original NR4 congenic mouse were less diabetogenic than those from NR4S1 donors (Fig. 3A). This difference indicated that an additional region on NOR Chr. 4, between 104.99–129.25Mb (termed R1; Fig. 7), was capable of decreasing the diabetogenic activity of B cells. Nonetheless, it is important to note that genes within the R1 region are incapable of independently decreasing diabetogenic B cell activity, as demonstrated by the high incidence of disease in NOD.Igµnull mice reconstituted with NR4S2 B cells (Fig. 3A). NOR genes within R1, as well as the R4 region, were shown to contribute to the decreased responsiveness (i.e. anergy) of self-reactive B cells to BCR stimulation (Fig. 4E–F). Such a finding may explain the capacity of a R1 region gene(s) to reduce the diabetogenic capacity of B cells in an augmentative, but not independent fashion.
In addition to their effects on B cells, NOR genes in the R1 region were shown to contribute to T1D resistance by dampening the pathogenic activity of CD4 T cells. This raised the possibility that modulation of CD4 T cells by R1 genes may indirectly regulate B cell diabetogenic activity and anergy. We do not believe this to be the case, given that: (i) our previous experiments have shown that the B cell anergy defect in NOD mice is an intrinsic trait controlled by genes within Chr. 1 and 4 loci (17) and; (ii) as previously mentioned, the R1 region on its own had little effect on the diabetogenic activity of B cells (Fig. 3A). Alternately, while the congenic region in NR4S4 mice was shown to have a strong dampening effect on the diabetogenic activity of B cells (Fig. 3A), a virtually identical congenic interval in the NR4S7 strain (Fig. 6A) had no effect on CD4 T cell pathogenesis (Fig. 6C–D). From these studies, we could conclude that the NOR distal Chr.4 region contains distinct sets of resistance alleles that respectively mediate independent effects on B and CD4 T cells contributing to T1D. Inheritance of both sets of NOR genes is likely to significantly affect the interplay between these cell types, leading to greater protection from T1D.
Through our analyses of subcongenic mouse strains, we have managed to reduce the region containing a gene(s) controlling the diabetogenic activity of B cells from 44.31Mb (containing 889 genes) to a 6.22Mb region (containing 152 genes) encompassing the newly identified Idd25 locus contributing to this phenotype. The smaller size of this region now makes it much more amenable to sequencing, which has the capacity to reveal all SNP between NOD and NOR origin genes that may be responsible for the Idd25 effect regulating B cell diabetogenic function. In the meantime, our microarray (Fig. 5A) and rt-qPCR studies (Fig. 5B–C) have identified two promising Idd25 candidate genes that were differentially expressed between NOD and NR4 B cells in this 6.22Mb region: Ephb2 and Padi2. EphB2 encodes a protein tyrosine kinase receptor with the capacity to bind various Ephrin B ligands on neighboring cells (37). While the function of Ephb2 has been studied most extensively in the nervous system (38), a role for this receptor has more recently been revealed in T cells, where it has been shown to potentially act as a co-stimulatory molecule that alters TCR mediated signaling (39). As a result, mice containing a deletion of EphB2 exhibit defects in T cell development (40) and peripheral T cell responses (41, 42). As we also found in this study, expression of EphB2 has been reported for B cells, though its function in this population remains to be elucidated (37). Padi2 encodes the Ca2+-dependent enzyme peptidylarginine deiminase-2 (Pad2) that catalyzes the conversion of arginine residues to citrulline (43). This post-translational conversion results in an ablation of the positive charge in proteins, potentially altering protein structure and function, as well as their recognition and presentation by the immune system (44–46). In humans, PADI2 and its closely related family member, PADI4, have been identified as susceptibility genes for multiple sclerosis and rheumatoid arthritis, respectively (47, 48). In both diseases, excess citrullination of proteins caused by overexpression of PADI2 or PADI4 leads to a breakdown in B and T cell tolerance (43, 47, 49, 50). Excessive expression of Padi2 by NOD B cells could play a similar role in the breakdown of tolerance to pancreatic β cell antigens.
In conclusion, the refinement of the large congenic region in NR4 mice into smaller subcongenic regions has revealed a complexity of interactions between multiple genes on Chr. 4 that regulate susceptibility to T1D by controlling the pathogenic activity of B and CD4 T cells. Identifying these gene(s) can provide significant insights into the mechanisms underlying the development of T1D as well as having the potential to highlight new molecular pathways that can be targeted as a disease intervention approach with greater specificity than current pan-B or T cell depleting drugs (6, 51). The distinct set of polymorphisms in the NOR background also make our subcongenic mice useful for mapping allelic variants affecting several other immune defects in NOD mice which have been linked to the distal Chr. 4 region (21).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Christopher Brownlee for technical assistance with flow cytometry and Jenny Kingham, Eric Schmeid, John Fisher and other staff of the Garvan Institute Biological Testing Facility and Australian BioResouces for their help with animal husbandry. We also thank Tom Brodnicki for providing microsatellite primers.
Footnotes
This work was supported by grants from the National Health and Medical Research Council of Australia (402727 and 427620), National Institute of Health (DK46266 and DK077443), the American Diabetes Association and the JDRF. J.S. and S.L.C. were recipients of Postgraduate Research Awards from the University of New South Wales.
Abbreviations: B6, C57BL/6; B10, C57BL/10; BKS, C57BKLS; BM, bone marrow; Chr., Chromosome; Ephb2, Ephrin receptor B2; FL, full-length; FDR, false detection rate; FO, follicular; HEL, hen-egg lysozyme; IgHEL, hen-egg lysozyme specific Ig; Idd, insulin dependent diabetes susceptibility gene; MZ, marginal zone; non-obese resistant, NOR; NR4, NOD.NOR-Chr4; Padi, Peptidyl arginine deiminase; sHEL, soluble HEL; SBM, syngeneic bone marrow; SNP, single nucleotide polymorphism; T1D, Type 1 Diabetes; tg, transgenic; TJL, The Jackson Laboratory.
REFERENCES
- 1.Anderson MS, Bluestone JA. The NOD mouse: a model of immune dysregulation. Annu Rev Immunol. 2005;23:447–485. doi: 10.1146/annurev.immunol.23.021704.115643. [DOI] [PubMed] [Google Scholar]
- 2.Kelemen K, Liu E, Davidson HW. The role of T cells in beta cell damage in NOD mice and humans. In: Eisenbarth GS, editor. Type I Diabetes: Molecular, Cellular and Clinical Immunology. Online Edition Version 2.5 ed. Springer; 2005. [Google Scholar]
- 3.Serreze DV, Chapman HD, Varnum DS, Hanson MS, Reifsnyder PC, Richard SD, Fleming SA, Leiter EH, Shultz LD. B lymphocytes are essential for the initiation of T cell-mediated autoimmune diabetes: analysis of a new "speed congenic" stock of NOD.Ig mu null mice. J Exp Med. 1996;184:2049–2053. doi: 10.1084/jem.184.5.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noorchashm H, Noorchashm N, Kern J, Rostami SY, Barker CF, Naji A. B-cells are required for the initiation of insulitis and sialitis in nonobese diabetic mice. Diabetes. 1997;46:941–946. doi: 10.2337/diab.46.6.941. [DOI] [PubMed] [Google Scholar]
- 5.Serreze DV, Chapman HD, Niens M, Dunn R, Kehry MR, Driver JP, Haller M, Wasserfall C, Atkinson MA. Loss of intra-islet CD20 expression may complicate efficacy of B-cell-directed type 1 diabetes therapies. Diabetes. 2011;60:2914–2921. doi: 10.2337/db11-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marino E, Silveira PA, Stolp J, Grey ST. B cell-directed therapies in type 1 diabetes. Trends Immunol. 2011;32:287–294. doi: 10.1016/j.it.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, Becker DJ, Gitelman SE, Goland R, Gottlieb PA, Marks JB, McGee PF, Moran AM, Raskin P, Rodriguez H, Schatz DA, Wherrett D, Wilson DM, Lachin JM, Skyler JS. Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N Engl J Med. 2009;361:2143–2152. doi: 10.1056/NEJMoa0904452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falcone M, Lee J, Patstone G, Yeung B, Sarvetnick N. B lymphocytes are crucial antigen-presenting cells in the pathogenic autoimmune response to GAD65 antigen in nonobese diabetic mice. J Immunol. 1998;161:1163–1168. [PubMed] [Google Scholar]
- 9.Serreze DV, Fleming SA, Chapman HD, Richard SD, Leiter EH, Tisch RM. B lymphocytes are critical antigen-presenting cells for the initiation of T cell-mediated autoimmune diabetes in nonobese diabetic mice. J Immunol. 1998;161:3912–3918. [PubMed] [Google Scholar]
- 10.Wheat W, Kupfer R, Gutches DG, Rayat GR, Beilke J, Scheinman RI, Wegmann DR. Increased NF-kappa B activity in B cells and bone marrow-derived dendritic cells from NOD mice. Eur J Immunol. 2004;34:1395–1404. doi: 10.1002/eji.200324490. [DOI] [PubMed] [Google Scholar]
- 11.Noorchashm H, Lieu YK, Noorchashm N, Rostami SY, Greeley SA, Schlachterman A, Song HK, Noto LE, Jevnikar AM, Barker CF, Naji A. I-Ag7-mediated antigen presentation by B lymphocytes is critical in overcoming a checkpoint in T cell tolerance to islet beta cells of nonobese diabetic mice. J Immunol. 1999;163:743–750. [PubMed] [Google Scholar]
- 12.Silveira PA, Johnson E, Chapman HD, Bui T, Tisch RM, Serreze DV. The preferential ability of B lymphocytes to act as diabetogenic APC in NOD mice depends on expression of self-antigen-specific immunoglobulin receptors. Eur J Immunol. 2002;32:3657–3666. doi: 10.1002/1521-4141(200212)32:12<3657::AID-IMMU3657>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 13.Hulbert C, Riseili B, Rojas M, Thomas JW. B cell specificity contributes to the outcome of diabetes in nonobese diabetic mice. J Immunol. 2001;167:5535–5538. doi: 10.4049/jimmunol.167.10.5535. [DOI] [PubMed] [Google Scholar]
- 14.Panigrahi AK, Goodman NG, Eisenberg RA, Rickels MR, Naji A, Luning Prak ET. RS rearrangement frequency as a marker of receptor editing in lupus and type 1 diabetes. J Exp Med. 2008;205:2985–2994. doi: 10.1084/jem.20082053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silveira PA, Dombrowsky J, Johnson E, Chapman HD, Nemazee D, Serreze DV. B cell selection defects underlie the development of diabetogenic APCs in nonobese diabetic mice. J Immunol. 2004;172:5086–5094. doi: 10.4049/jimmunol.172.8.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acevedo-Suarez CA, Hulbert C, Woodward EJ, Thomas JW. Uncoupling of anergy from developmental arrest in anti-insulin B cells supports the development of autoimmune diabetes. J Immunol. 2005;174:827–833. doi: 10.4049/jimmunol.174.2.827. [DOI] [PubMed] [Google Scholar]
- 17.Cox SL, Stolp J, Hallahan NL, Counotte J, Zhang W, Serreze DV, Basten A, Silveira PA. Enhanced responsiveness to T-cell help causes loss of B-lymphocyte tolerance to a beta-cell neo-self-antigen in type 1 diabetes prone NOD mice. Eur J Immunol. 2010;40:3413–3425. doi: 10.1002/eji.201040817. [DOI] [PubMed] [Google Scholar]
- 18.Quinn WJ, 3rd, Noorchashm N, Crowley JE, Reed AJ, Noorchashm H, Naji A, Cancro MP. Cutting edge: impaired transitional B cell production and selection in the nonobese diabetic mouse. J Immunol. 2006;176:7159–7164. doi: 10.4049/jimmunol.176.12.7159. [DOI] [PubMed] [Google Scholar]
- 19.Menard L, Saadoun D, Isnardi I, Ng YS, Meyers G, Massad C, Price C, Abraham C, Motaghedi R, Buckner JH, Gregersen PK, Meffre E. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J Clin Invest. 2011;121:3635–3644. doi: 10.1172/JCI45790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habib T, Funk A, Rieck M, Brahmandam A, Dai X, Panigrahi AK, Luning Prak ET, Meyer-Bahlburg A, Sanda S, Greenbaum C, Rawlings DJ, Buckner JH. Altered B Cell Homeostasis Is Associated with Type I Diabetes and Carriers of the PTPN22 Allelic Variant. J Immunol. 2011 doi: 10.4049/jimmunol.1102176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Driver JP, Serreze DV, Chen YG. Mouse models for the study of autoimmune type 1 diabetes: a NOD to similarities and differences to human disease. Semin Immunopathol. 2010;33:67–87. doi: 10.1007/s00281-010-0204-1. [DOI] [PubMed] [Google Scholar]
- 22.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Serreze DV, Prochazka M, Reifsnyder PC, Bridgett MM, Leiter EH. Use of recombinant congenic and congenic strains of NOD mice to identify a new insulin-dependent diabetes resistance gene. J Exp Med. 1994;180:1553–1558. doi: 10.1084/jem.180.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton-Williams EE, Serreze DV, Charlton B, Johnson EA, Marron MP, Mullbacher A, Slattery RM. Transgenic rescue implicates beta2-microglobulin as a diabetes susceptibility gene in nonobese diabetic (NOD) mice. Proc Natl Acad Sci U S A. 2001;98:11533–11538. doi: 10.1073/pnas.191383798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serreze DV, Bridgett M, Chapman HD, Chen E, Richard SD, Leiter EH. Subcongenic analysis of the Idd13 locus in NOD/Lt mice: evidence for several susceptibility genes including a possible diabetogenic role for beta 2-microglobulin. J Immunol. 1998;160:1472–1478. [PubMed] [Google Scholar]
- 26.Reifsnyder PC, Li R, Silveira PA, Churchill G, Serreze DV, Leiter EH. Conditioning the genome identifies additional diabetes resistance loci in Type I diabetes resistant NOR/Lt mice. Genes Immun. 2005;6:528–538. doi: 10.1038/sj.gene.6364241. [DOI] [PubMed] [Google Scholar]
- 27.Tan IK, Mackin L, Wang N, Papenfuss AT, Elso CM, Ashton MP, Quirk F, Phipson B, Bahlo M, Speed TP, Smyth GK, Morahan G, Brodnicki TC. A recombination hotspot leads to sequence variability within a novel gene (AK005651) and contributes to type 1 diabetes susceptibility. Genome Res. 2010;20:1629–1638. doi: 10.1101/gr.101881.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamanouchi J, Puertas MC, Verdaguer J, Lyons PA, Rainbow DB, Chamberlain G, Hunter KM, Peterson LB, Wicker LS, Santamaria P. Idd9.1 locus controls the suppressive activity of FoxP3+CD4+CD25+ regulatory T-cells. Diabetes. 2010;59:272–281. doi: 10.2337/db09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter K, Rainbow D, Plagnol V, Todd JA, Peterson LB, Wicker LS. Interactions between Idd5.1/Ctla4 and other type 1 diabetes genes. J Immunol. 2007;179:8341–8349. doi: 10.4049/jimmunol.179.12.8341. [DOI] [PubMed] [Google Scholar]
- 30.Silveira PA, Chapman HD, Stolp J, Johnson E, Cox SL, Hunter K, Wicker LS, Serreze DV. Genes within the Idd5 and Idd9/11 diabetes susceptibility loci affect the pathogenic activity of B cells in nonobese diabetic mice. J Immunol. 2006;177:7033–7041. doi: 10.4049/jimmunol.177.10.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rolf J, Motta V, Duarte N, Lundholm M, Berntman E, Bergman ML, Sorokin L, Cardell SL, Holmberg D. The enlarged population of marginal zone/CD1d(high) B lymphocytes in nonobese diabetic mice maps to diabetes susceptibility region Idd11. J Immunol. 2005;174:4821–4827. doi: 10.4049/jimmunol.174.8.4821. [DOI] [PubMed] [Google Scholar]
- 32.Chen YG, Scheuplein F, Osborne MA, Tsaih SW, Chapman HD, Serreze DV. Idd9/11 genetic locus regulates diabetogenic activity of CD4 T-cells in nonobese diabetic (NOD) mice. Diabetes. 2008;57:3273–3280. doi: 10.2337/db08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graser RT, DiLorenzo TP, Wang F, Christianson GJ, Chapman HD, Roopenian DC, Nathenson SG, Serreze DV. Identification of a CD8 T cell that can independently mediate autoimmune diabetes development in the complete absence of CD4 T cell helper functions. J Immunol. 2000;164:3913–3918. doi: 10.4049/jimmunol.164.7.3913. [DOI] [PubMed] [Google Scholar]
- 34.Hamilton-Williams EE, Martinez X, Clark J, Howlett S, Hunter KM, Rainbow DB, Wen L, Shlomchik MJ, Katz JD, Beilhack GF, Wicker LS, Sherman LA. Expression of diabetes-associated genes by dendritic cells and CD4 T cells drives the loss of tolerance in nonobese diabetic mice. J Immunol. 2009;183:1533–1541. doi: 10.4049/jimmunol.0900428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Araki M, Chung D, Liu S, Rainbow DB, Chamberlain G, Garner V, Hunter KM, Vijayakrishnan L, Peterson LB, Oukka M, Sharpe AH, Sobel R, Kuchroo VK, Wicker LS. Genetic evidence that the differential expression of the ligand-independent isoform of CTLA-4 is the molecular basis of the Idd5.1 type 1 diabetes region in nonobese diabetic mice. J Immunol. 2009;183:5146–5157. doi: 10.4049/jimmunol.0802610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, Gonzalez-Munoz A, Clark J, Veijola R, Cubbon R, Chen SL, Rosa R, Cumiskey AM, Serreze DV, Gregory S, Rogers J, Lyons PA, Healy B, Smink LJ, Todd JA, Peterson LB, Wicker LS, Santamaria P. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet. 2007;39:329–337. doi: 10.1038/ng1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Palmer A, Klein R. Multiple roles of ephrins in morphogenesis, neuronal networking, and brain function. Genes Dev. 2003;17:1429–1450. doi: 10.1101/gad.1093703. [DOI] [PubMed] [Google Scholar]
- 39.Wu J, Luo H. Recent advances on T-cell regulation by receptor tyrosine kinases. Curr Opin Hematol. 2005;12:292–297. doi: 10.1097/01.moh.0000166497.26397.9f. [DOI] [PubMed] [Google Scholar]
- 40.Stimamiglio MA, Jimenez E, Silva-Barbosa SD, Alfaro D, Garcia-Ceca JJ, Munoz JJ, Cejalvo T, Savino W, Zapata A. EphB2-mediated interactions are essential for proper migration of T cell progenitors during fetal thymus colonization. J Leukoc Biol. 2010;88:483–494. doi: 10.1189/jlb.0210079. [DOI] [PubMed] [Google Scholar]
- 41.Yu G, Mao J, Wu Y, Luo H, Wu J. Ephrin-B1 is critical in T-cell development. J Biol Chem. 2006;281:10222–10229. doi: 10.1074/jbc.M510320200. [DOI] [PubMed] [Google Scholar]
- 42.Alfaro D, Garcia-Ceca JJ, Cejalvo T, Jimenez E, Jenkinson EJ, Anderson G, Munoz JJ, Zapata A. EphrinB1-EphB signaling regulates thymocyte-epithelium interactions involved in functional T cell development. Eur J Immunol. 2007;37:2596–2605. doi: 10.1002/eji.200737097. [DOI] [PubMed] [Google Scholar]
- 43.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 44.Tarcsa E, Marekov LN, Mei G, Melino G, Lee SC, Steinert PM. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J Biol Chem. 1996;271:30709–30716. doi: 10.1074/jbc.271.48.30709. [DOI] [PubMed] [Google Scholar]
- 45.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 46.Arita K, Shimizu T, Hashimoto H, Hidaka Y, Yamada M, Sato M. Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4. Proc Natl Acad Sci U S A. 2006;103:5291–5296. doi: 10.1073/pnas.0509639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono M, Ohtsuki M, Furukawa H, Yoshino S, Yukioka M, Tohma S, Matsubara T, Wakitani S, Teshima R, Nishioka Y, Sekine A, Iida A, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 48.Kenealy SJ, Babron MC, Bradford Y, Schnetz-Boutaud N, Haines JL, Rimmler JB, Schmidt S, Pericak-Vance MA, Barcellos LF, Lincoln RR, Oksenberg JR, Hauser SL, Clanet M, Brassat D, Edan G, Yaouanq J, Semana G, Cournu-Rebeix I, Lyon-Caen O, Fontaine B. A second-generation genomic screen for multiple sclerosis. Am J Hum Genet. 2004;75:1070–1078. doi: 10.1086/426459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim JK, Mastronardi FG, Wood DD, Lubman DM, Zand R, Moscarello MA. Multiple sclerosis: an important role for post-translational modifications of myelin basic protein in pathogenesis. Mol Cell Proteomics. 2003;2:453–462. doi: 10.1074/mcp.M200050-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Lundberg K, Nijenhuis S, Vossenaar ER, Palmblad K, van Venrooij WJ, Klareskog L, Zendman AJ, Harris HE. Citrullinated proteins have increased immunogenicity and arthritogenicity and their presence in arthritic joints correlates with disease severity. Arthritis Res Ther. 2005;7:R458–R467. doi: 10.1186/ar1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaufman A, Herold KC. Anti-CD3 mAbs for treatment of type 1 diabetes. Diabetes Metab Res Rev. 2009;25:302–306. doi: 10.1002/dmrr.933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.