Abstract
Serum albumin (SA) is the most abundant plasma protein in mammals. SA is a multifunctional protein with extraordinary ligand binding capacity, making it a transporter molecule for a diverse range of metabolites, drugs, nutrients, metals and other molecules. Due to its ligand binding properties, albumins have wide clinical, pharmaceutical, and biochemical applications. Albumins are also allergenic, and exhibit a high degree of cross-reactivity due to significant sequence and structure similarity of SAs from different organisms. Here we present crystal structures of albumins from cattle (BSA), horse (ESA) and rabbit (RSA) serums. The structural data are correlated with the results of immunological studies of SAs. We also analyze the conservation or divergence of structures and sequences of SAs in the context of their potential allergenicity and cross-reactivity. In addition, we identified a previously uncharacterized ligand binding site in the structure of RSA, and calcium binding sites in the structure of BSA, which is the first serum albumin structure to contain metal ions.
Keywords: Allergen, Serum Albumin (SA), Bovine Serum Albumin (BSA), Equine Serum Albumin (ESA), Rabbit Serum Albumin (RSA), cross-reactivity, calcium binding
1. Introduction
Serum albumin (SA) is the most abundant plasma protein in mammals. It is synthesized in the liver and exported as a non-glycosylated protein into the plasma, where it reaches a high concentration (ca. 0.6 mM) and greatly contributes to the colloid osmotic pressure. SA is a multifunctional protein with an extraordinary capacity for binding ligands. It is a reservoir of the signaling agent nitric oxide (Stamler et al., 1992) and serves as a transporter for a diverse range of metabolites, drugs, nutrients, and other molecules (de Wolf and Brett, 2000). Significantly, serum albumin is the major circulatory protein involved in the handling of Ca2+ (and Mg2+) in mammals, controlling the ionized or “biologically active” levels of these metals in the blood. These binding properties give SAs a wide range of clinical, pharmaceutical, and biochemical applications.
Many SAs are also allergenic, and antibodies that recognize SAs are typically cross-reactive (Boutin et al., 1988; Hilger et al., 1997; Spitzauer, 1999; Spitzauer et al., 1995; Spitzauer et al., 1994). Specifically, SAs of different mammals present in their milk, meat, or epithelia, have the capability to sensitize predisposed individuals which results in a high degree of cross-reactivity. For example, patients with an allergy to cow’s milk can develop allergy to dog or cat epithelia SAs without previous first direct contact with the respective animals (Vicente-Serrano et al., 2007). SAs are somewhat unusual allergens, as proteins with sequence identities to their human homolog above approximately 62% are rarely allergenic (Jenkins et al., 2007).
Serum albumins are relatively large (molecular weight of around 66kDa) and negatively charged proteins. A number of structures of human serum albumin (HSA) are present in the Protein Data Bank (PDB) (Berman et al., 2000), but there are no structures of other animal albumins. SAs are heart-shaped and comprise three helical domains, each comprising two subdomains. In order to compare the SAs from different mammalian source we determined the crystal structures of albumins from bovine (BSA), equine (ESA) and rabbit (RSA) serums. We conducted comparative analyses of their structures and sequences, correlating the structural data with results of different immunological studies of SAs. We also analyze the conservation of serum albumins as well as their divergence in the context of potential allergenicity and cross-reactivity.
2. Materials and Methods
2.1. Crystallization
BSA (Lot# 010M1506) and RSA (Lot#117K7565) were purchased from Sigma-Aldrich, whereas ESA was obtained from Rockland (Lot#4173). Tracking and analysis of the crystallization experiments were performed with the Xtaldb system (Zimmerman et al., 2005). Prior to crystallization, the proteins were dissolved in a buffer containing 10 mM Tris-HCl, 150 mM NaCl at pH 7.5, and were passed through a Superdex 200 column attached to the ÅKTA FPLC gel filtration system (GE Healthcare). After gel filtration, fractions containing monomeric proteins were pooled and concentrated to 27 mg/mL (ESA), 15 mg/mL (RSA) or 10 mg/mL (BSA). BSA and ESA crystals were grown using vapor diffusion and hanging drop setups, while RSA was crystallized using a sitting drop setup. In each case, the crystallization drops were composed of a 1:1 mixture of the protein solution and the precipitant solution from the wells. Crystallization and cryocooling conditions are summarized in Table 1.
Table 1.
Crystallization and cryo-cooling conditions.
| PDB code | 3V03 | 3V08 | 3V09 |
| Protein | BSA | ESA | RSA |
| Well solution | 0.2 M Ca acetate, 20% w/v PEG3350, 0.1 M Tris HCl pH 6.5 | 1.9 M NH4 sulfate, 2.5% w/v PEG8000, 0.1 M NaBr, 0.1 M Tris HCl pH 7.5 | 0.1 M MES pH 6.5, 30% v/v PEG400 |
| Crystallization temperature [°C] | 16 | 16 | 4 |
| Cryo-protectant | - | Mineral oil/paratone | - |
2.2. Data collection, structure determination and refinement
Data collection for ESA was performed at the 19-BM beamline of the Structural Biology Center (Rosenbaum et al., 2006) at the Advanced Photon Source (APS). Data collection for BSA and RSA was performed at the 21-ID-G beamline of the Life Sciences Collaborative Access Team at the APS. Data were collected at 100 K and processed with HKL-2000 (Otwinowski, 1997). All structures were solved using HKL-3000 (Minor et al., 2006) coupled with MOLREP (Vagin and Teplyakov, 1997). The structure of ESA was determined by molecular replacement (MR) using the published structure of HSA (PDB ID: 1ao6). Subsequently, the refined ESA structure was used as a search model to determine the structures of BSA and RSA by MR. Refinement was done using HKL-3000 coupled with REFMAC5 (Murshudov et al., 2011), COOT (Emsley et al., 2010), and selected programs from the CCP4 package (Collaborative Computional Project, 1994). The B-factors were refined in each case using Translation/Libration/Screw (TLS) groups assigned using the TLSMD server (Painter and Merritt, 2006). Non-crystallographic symmetry restraints were used during the refinement of BSA. Validation of the structures was performed using MOLPROBITY (Davis et al., 2007) and ADIT (Yang et al., 2004). The coordinates, structure factors, and intensities were deposited in the PDB (PDB IDs: 3v03 for BSA, 3v08 for ESA and 3v09 for RSA). Statistics describing crystallographic data collection and refinement are summarized in Table 2.
Table 2.
Data collection and refinement statistics.
| PDB code | 3V03 | 3V08 | 3V09 |
| Protein | BSA | ESA | RSA |
| Data collection | |||
| Space group | C2 | P61 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | a=215.7, b=45.1, c=142.4 | a=b=94.6, c= 142.6 | a=75.5, b=81.9, c=104.9 |
| α, β, γ (°) | 90.0, 114.0, 90.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.00-2.70(2.70–2.75) | 50.00-2.45(2.49–2.55) | 50.00-2.27(2.31–2.27) |
| Rsym | 0.069(0.626) | 0.061(0.466) | 0.079(0.700) |
| I/σI | 29.1(2.2) | 21.0(2.7) | 22.1(2.3) |
| Completeness (%) | 99.6(100.0) | 97.7(98.9) | 99.9(100.0) |
| Redundancy | 4.0(4.0) | 3.2(3.1) | 4.3(4.3) |
| Refinement | |||
| Resolution (Å) | 2.7 | 2.45 | 2.27 |
| No. reflections | 33241 | 24701 | 29035 |
| Rwork/Rfree | 20.2/25.5 | 20.4/24.5 | 19.3/24.2 |
| RMSDs | |||
| Bond lengths (Å) | 0.012 | 0.012 | 0.014 |
| Bond angles (°) | 1.3 | 1.4 | 1.6 |
Numbers in parentheses refer to the highest resolution shell.
2.3. Sequence and structure analysis
The PDBeFold (Krissinel and Henrick, 2004) service was used to search for human serum albumin structures that adopt conformations most similar to those of BSA, ESA, or RSA, and calculate the root-mean-square deviations (RMSDs) between them. Structures with a Q-score equal or greater than 0.8 were chosen for further analysis. Solvent accessible surface areas were calculated using PyMOL (DeLano, 2002). PyMOL was also used for figure preparation. Multiple sequence alignments were calculated using ClustalW with the default parameter settings. PySHADE (Porebski et al. – manuscript in preparation) was used to plot sequence alignments and sequence variability plots.
3. Results and Discussion
3.1. Sequence analysis
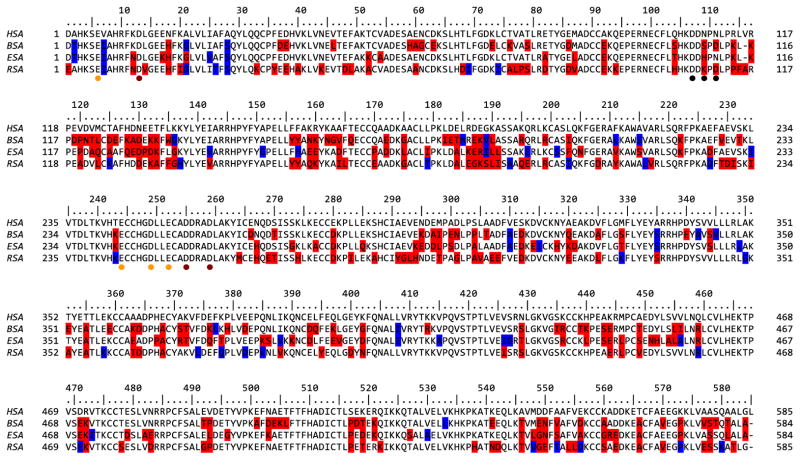
Albumins are initially expressed in vivo, as preproalbumins, including a signal sequence and a propeptide (Hawkins and Dugaiczyk, 1982). The bovine and horse preproalbumins are 607 amino acids long, whereas rabbit preproalbumin contains 608 amino acids, and the human protein, 609. The corresponding mature proteins are comprised of 583 (BSA, ESA), 584 (RSA), and 585 (HSA) amino acids. The overall sequence identities of mature BSA, ESA and RSA, compared to HSA has, are 75.6%, 76.1% and 74.2%, respectively and their sequence alignment is presented in Figure 1.
Figure 1. Sequence alignment of analyzed SA.
The residue numbering is shown according to HSA. Residues of BSA, ESA and RSA that differ from HSA are marked in either red or blue. Specifically, variable residues with a solvent accessible area >10 Å2 are red; those with a solvent accessible area ≤10Å are blue. Residues that are involved in calcium binding in BSA are marked with dots and colored with orange, red and black for first, second and third site respectively.
3.2. Overview of the structures
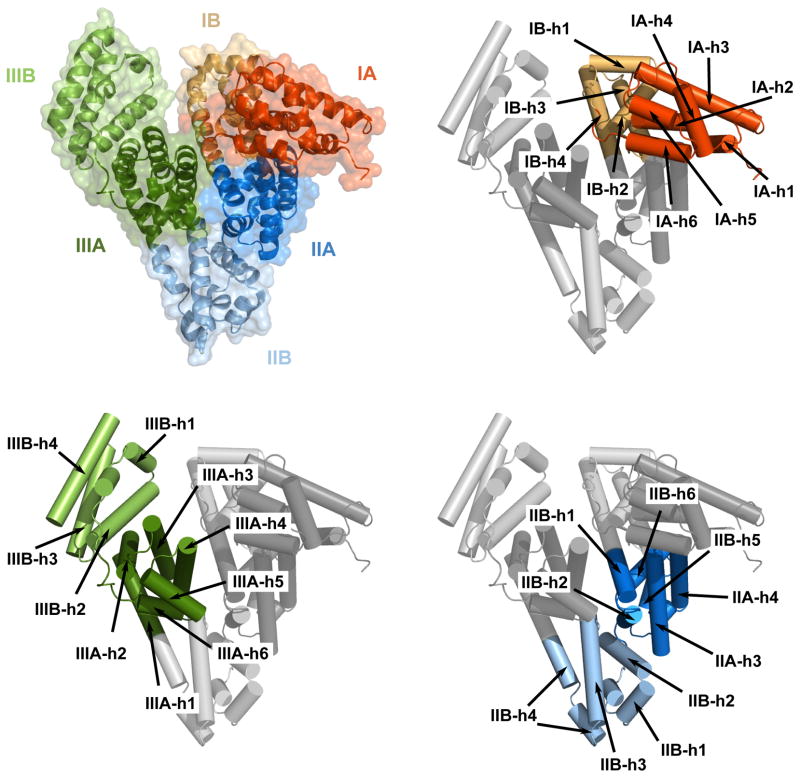
Bovine serum albumin crystallized in a monoclinic crystal form, in the C2 space group, serum albumin crystallized in a hexagonal crystal form in the P61 space group, as described previously (Ho et al., 1993). However, despite the fact that crystal structure of ESA was determined (Ho et al., 1993), the coordinates were never deposited in the PDB. Rabbit albumin crystallized in an orthorhombic crystal form, in the P212121 space group. Both ESA and RSA crystallized with one molecule in the asymmetric units. In all three structures, the albumins are composed of three structurally similar helical domains (I, II, and III) arranged in a heart-shaped molecule. Each domain can be divided into two subdomains (A and B) (Fig. 2). The helical content of the three albumins is 74% (BSA), 75% (ESA) or 72% (RSA), and each has 17 conserved disulfide bonds and a free thiol group associated with Cys34. The structures of all three albumins show strong similarity to the structure of the HSA. The averaged RMSDs between a particular albumin structure and HSA structures (where the number of structures for which RMSDs were calculated is indicated in parentheses) adopting the most similar conformations is 1.1 Å (N=24) for BSA, 1.2 Å for ESA (N=8) and 1.2 Å for RSA (N=21).
Figure 2. Domain and secondary structure element assignment for serum albumins, as represented by the BSA molecular structure.
Each domain is marked with a different color, and each subdomain is marked with a different shade. The subdomains and secondary structure elements are assigned based on (Sugio et al., 1999).
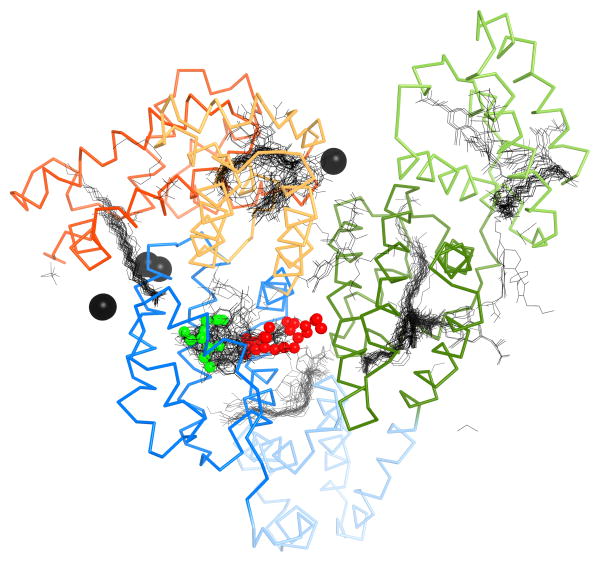
Unidentified endogenous ligands were observed in the crystal structures of equine and rabbit albumin (Fig. 3). However, the shape of the electron density does not allow for unambiguous identification of the ligands. In ESA, the ligand is positioned in domain IIA, in the pocket that corresponds to one of the two primary drug binding sites in HSA, known as Sudlow’s site I (Sudlow et al., 1975). That site has been characterized in HSA as a conformationally adaptable region with up to three subcompartments (Petitpas et al., 2001). The ligand in the equine protein is a relatively small molecule and its position is approximately the same as the position of an outer ring of thyroxine bound in the HSA structure with the PDB id 1hk1, or warfarin in the structure with the PDB id 2bxd. Thyroxine and warfarin are two examples out of many ligands that have been observed to bind to this pocket in HSA. The unidentified ligand in RSA is a longer molecule— possibly a fatty acid—positioned partly in site I, but mostly in the central part of the protein, between the IIA, IIB and IIIB subdomains. This site is not characterized as a binding site in any published structures of HSA.
Figure 3. Ligand binding sites of RSA and ESA.
The structure of RSA is shown in ribbon representation, colored according to the domain assignment presented in Figure 2. All HSA structures with ligands were superimposed. Black lines represent small molecules present in the structures of HSA. The two unknown ligands in the RSA and ESA structures are shown as spheres in red and green, respectively. Calcium ions that are bound to BSA are shown as black spheres.
In the crystal structure of bovine albumin we observe binding of three calcium ions. The interaction of calcium with serum albumins is of significant physiological and biomedical importance and have been intensively studied (Besarab and Caro, 1981; Besarab et al., 1981; Martin and Perkins, 1950a; Martin and Perkins, 1950b; Martin and Perkins, 1953). Although the interaction with Ca2+ is relatively weak (Ka = 1.5×103 M−1 for human albumin (Kragh-Hansen and Vorum, 1993), approximately 45% of the 2.4 mM of circulating Ca2+ in the serum is albumin-bound (Peters, 1995). However, the locations of specific Ca2+ sites on albumin are unknown. In addition, it is thought that the binding of Mg2+ to serum albumins will parallel Ca2+ in terms of binding sites (Peters, 1995). Mg2+ is also transported by albumin but binds with slightly lower affinity than Ca2+ (Ka = 1×102 M−1; (Pedersen, 1972)). The concentration of Mg2+ in blood plasma is around 1.2 mM and like Ca2+, approximately 45% is complexed with albumin (Peters, 1995). Hence, given that 1.1 mM Ca2+ and 0.5 mM Mg2+ are bound to 0.6 mM circulating albumin, there should be at least three sites for ‘hard’ metal ions, which is in agreement with our findings. It has been suggested that Ca2+ associates with albumin non-specifically (Peters, 1995). However, further evidence (in addition to the presented bovine albumin structure) for the existence of distinct sites comes from the observation that unsaturated and short-chain fatty acids have been shown to influence the binding of Ca2+ to HSA (Aguanno and Ladenson, 1982). Fatty acid-binding to mammalian albumins is known to allosterically disrupt the major Zn2+ site, which is located in close proximity to the three Ca2+ sites (Lu et al., 2012; Stewart et al., 2003). In fact, all three Ca2+ binding sites identified in BSA are localized in close proximity to fatty acid binding sites identified in the structure of HSA (Fig. 3), thus Ca2+ binding could potentially be disrupted in the presence of fatty acids.
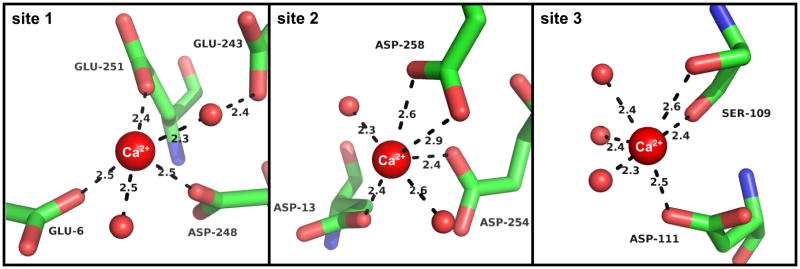
All Ca2+ binding sites are located in domain I (Fig. 3). The relatively low resolution and high temperature factors of the BSA structure make detailed interpretation of the exact metal binding geometry ambiguous to some extent. Therefore during the refinement we restrained the ligand distances based on the data in Zheng et al. (Zheng et al., 2008). For the first Ca2+ binding site that we identifed in the BSA structure (Fig. 4) we observed some flexibility in the binding of Ca2+. The site is composed of residues E6, E243, D248, and E251, with the electron density for the Ca2+ in chain A more disordered than in chain B. In chain B, Ca2+ is coordinated by residues D248, E251, and E243. In chain A, D248 and E251 also coordinate the Ca2+, but the ion is shifted closer to residue E6 and is coordinated by it directly, while the distance to E243 is increased enough to require water-mediated interactions with the ion. As NCS was used during refinement, there is no difference in the position of the side chains between A and B chains. The first Ca2+ binding site is located in close proximity to the previously determined Zn2+ binding site (Stewart et al., 2003), with residue D248 being shared between the two sites. This suggests that crosstalk may exist between Zn2+ and Ca2+/Mg2+ transport in the blood. Serum albumins bind Zn2+ with a much higher affinity (1.9×107 M−1 for bovine albumin; (Masuoka et al., 1993)) hence Zn2+ binding at this region would be preferential. The second Ca2+ site (Fig. 4) is close in space to the first site and is composed of residues D13, D254, and D258. All of the contacts with the first two Ca2+ ions are mediated by side chain interactions, and all residues in the two sites are conserved in all analyzed species. The third Ca2+ binding site (Fig. 4) utilizes side chain of D111, the side chain and main chain oxygen of S109, and a water mediated interactions with D107. In this site D107 is conserved, D11 is replaced by N in HSA and ESA and S109 is not conserved at all (Fig. 1). In each case the coordination is completed by the water molecules, however due to resolution of the data the exact position of water molecules is uncertain.
Figure 4. Binding sites of the calcium ions.
The three calcium binding sites from chain A of BSA are shown. Calcium ions and waters are shown as spheres.
3.3. Analysis of known and potential epitopes
3.3.1. Bovine serum albumin
The antigenic activity of serum albumins of different animals has been extensively studied (Spitzauer, 1999). Albumins can act as allergens either through ingestion (meat, milk, eggs) or inhalation (animal dander) (Liccardi et al., 2011; Vicente-Serrano et al., 2007). BSA, ESA, and RSA have been classified as allergens (Bos d 6, Equ c 3 and Ory c RSA, respectively). BSA has been identified as one of the most important allergens in bovine meat, whereas equine and rabbit albumins are mostly considered inhaled allergens. In addition, BSA was identified as a trigger of life-threatening anaphylaxis during some medical procedures (Matheu et al., 2002; Wuthrich et al., 1995). Several regions in BSA have been identified as epitopes, but in most cases the fragments identified as containing epitopes are relatively long (Alting et al., 1997; Beretta et al., 2001; Karjalainen et al., 1992; Restani et al., 1998; Tanabe et al., 2002; Ueno et al., 1994). To narrow the size of the possible epitope region we compared differences between HSA and BSA and considered the surface exposure of these residues (Fig. 5). Although several residues differ between the human and bovine albumin, only some of them are sufficiently exposed on the surface to allow recognition and binding to the protein in its native folded form. Other regions are only exposed after protein digestion or denaturation. We also analyzed the conservation of serum albumins in the available sequences of Artiodactyla (bovine, goat, sheep, and deer) and rodents/lagomorphs (rat, mouse, rabbit, hamster and guinea pig) in context of the possible epitopes and their possible cross-reactivity (Spitzauer, 1999; Spitzauer et al., 1995). For the odd-toed ungulate there are only two serum albumin sequences known, for horse (Equus ferus caballus) and donkey (Equus africanus asinus), which are very similar.
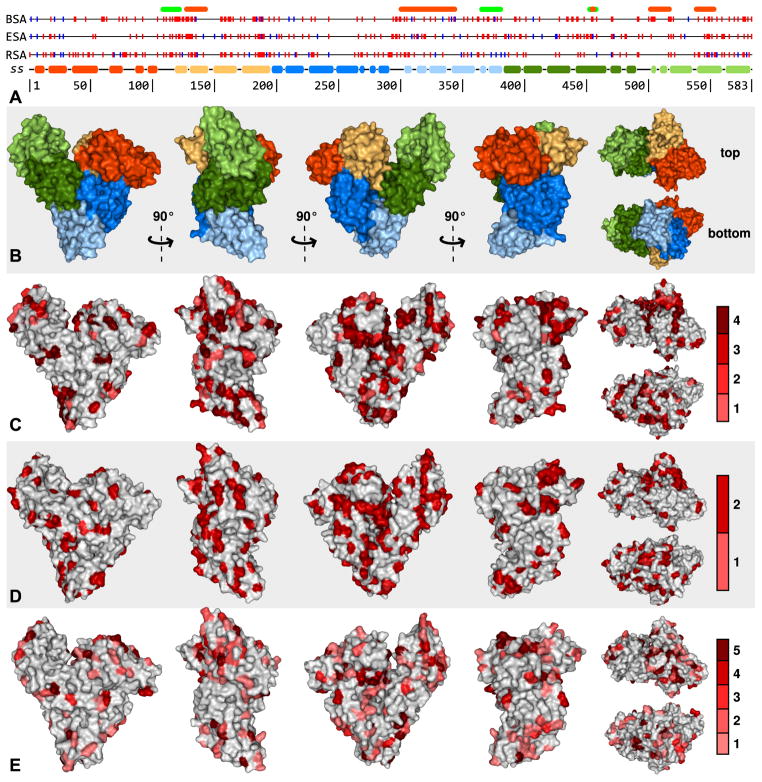
Figure 5. Variability of SA surface amino acids.
A – Sequence variability of BSA, ESA and RSA as compared to HSA. Residues in the specified SA that differ from the HSA sequence are marked with vertical lines. Amino acids with solvent accessible area >10 Å2 are marked in red and those with ≤10Å2 in blue. Secondary structure (SS) is colored according to the domain assignment presented in Figure 2. Above the alignment, regions of BSA that are known B-cell epitopes described in the text are marked in orange and regions that are known T-cell epitopes are marked in green. B – BSA molecular surface colored according to domain assignment. C, D, E – Molecular surfaces of BSA, ESA and RSA (respectively) with residues that are not conserved between the particular SA and HSA colored according to the conservation of that residue in other SAs of known sequence in the same or similar order (BSA – Artiodactyla (bovine, goat, sheep, and deer), ESA – Perissodactyla (horse and donkey), RSA – Rodentia/Lagomorpha (rat, mouse, rabbit, hamster and guinea pig)). The scales on the right shows the colors used to mark residues, which indicate the number of other SAs in which the residue is conserved.
There are two regions showing the significant differences between HSA and BSA, both localized on the subdomain IB. The first region is localized on the first tow α-helices of the IB subdomain, comprising residues 114 to 135 (numbered with respect to mature BSA), with most of the different residues exposed at the protein surface. BSA residues S109, D111, D37 and E38 also differ from the HSA sequence and lie in close proximity, possibly extending the recognition region. Residues 126–144 (collectively known as the ABBOS peptide) have been described as a reactive epitope (Alting et al., 1997; Karjalainen et al., 1992), though residues 136–144 are identical between BSA and HSA. Residues 107–123 have been found to induce T cell proliferation (Tanabe et al., 2002).
The second region with an accumulation of surface-exposed residues that differ between BSA and HSA is found on the third and fourth α-helices of the IB domain, comprising residues 155–189. Specifically, the BSA residues in this region that both (1) differ from those of HSA and (2) are highly solvent-exposed are Y155, N158, K159, N161, G162, V163, Q165 (in the third α-helix) and E182, T183, E186, K187 (in the fourth α-helix). This region is also relatively conserved in the albumins of Artiodactyla, suggesting this could be a cross-reactive epitope in these species. BSA residues K294, A296, I297, E299, and N300, also differ from those of HSA and constitute a continuous patch on the protein’s surface. The residues are located on the long loop connecting subdomains IIA and IIB and could potentially be recognized as either a conformational or linear epitope.
It has been shown that anti-BSA AB6 mAb probably recognizes an epitope of the undigested form of BSA present in a peptide corresponding to residues 299–338 (Ueno et al., 1994). Region 364–382 has been found to induce T cell proliferation (Tanabe et al., 2002). Our observations indicate more precisely the recognizable residues corresponding to this region. The BSA residues that differ from those of HSA make two parallel patches on the third and fourth α-helix of subdomain IIB. Specifically residues E351, A354, E358, K362, and D363 form the first patch and A367, S370, T371, K375, H378, and D381 from the second. Some of these amino acids are conserved in Artiodactyla, especially on the patch on the fourth helix (A367, T371, K375, and D381), making them a potentially cross-reactive epitope. Residues P303 and T305 lie close to the second patch, and may also be recognized. Of residues 336–345, which have been shown to be a B-cell epitope (Tanabe et al., 2002), the only residue exposed to the surface that differs between human and bovine albumin is E338. The remaining residues of this region that differ from those of HSA are not exposed to the protein surface, and likely only recognized as epitopes after protein digestion, denaturation, or significant conformational changes. Peptides corresponding to residues 451–459 induce T cell proliferation, and a subset of those residues (453–457) act as a B-cell (IgE-binding) epitope (Tanabe et al., 2002). This includes BSA residues L453, I454 and R457 located in subdomain IIIA, that are different from HSA, but are not exposed in this conformation of BSA.
A relatively small patch composed of residues that are not linear in terms of sequence is located between subdomains IIIA and IIIB of BSA, including R412, T491, P492 and E540. The C-terminal subdomain of BSA (IIIB) has also been shown to contain important epitopes. Region 500–574 has been suggested as an epitope-containing region for human species, with 500–518 being the most critical (Beretta et al., 2001). Residues 537–554 have been shown to be recognized by AB3 mAb (Ueno et al., 1994), and residues 520–542 have been suggested to be important (Restani et al., 1998). Within this region of the BSA structure, four patches of residues are exposed to the protein surface and differ from HSA. Each of these four patches is located on a different helix of subdomain IIIB. Residues A500, D502, E503, K504 and L505 constitute the first patch on one side of the subdomain, whereas residues P516, D517, T518 and K520 constitute the second patch on the opposite site of the subdomain. The last two patches are located on the last two helices of BSA and are long and parallel; specifically T545, E548, N549, V551, D555, and A559 (third patch) and A565, V569, P572, V576, S577, T580, and A583 (fourth patch). Residues V551, D555, and A559 are separated from A565, V569, and P572, by residues that are identical between BSA and HSA. Residues T545, E548, and N549 from one helix and V576, S577, and T580 from the other helix are spatially close, and thus could be recognized as a single epitope.
3.3.2. Equine serum albumin
ESA has not been well characterized in terms of epitopes (Fjeldsgaard and Paulsen, 1993; Ponterius et al., 1973). Three tryptic peptides of ESA (corresponding to residues 21–113, 188–275, and 503–560) have been isolated and characterized (Goubran Botros et al., 1996). These peptides were able to inhibit the binding of allergic patients’ IgE and IgG antibodies to horse albumin as well as to dog and cat serum albumins, indicating that they are involved in cross-reactions. These peptides do not correspond to regions with a high number of residues different between human and equine albumin, which are, to some extent, similar to the differing regions in BSA. Here we analyze ESA based on the differences versus HSA and correlate them with surface accessibility, suggesting the location of regions that could potentially function as epitopes (Fig. 5).
ESA amino acids A121, Q122, A124 and 127–132 (QEDPDK) differ from those in HSA and constitute a patch that is corresponding to the BSA patch (part of the ABBOS peptide). There is also a patch on the third and fourth α-helices of the IB domain, including residues E158, E159, A183, 185–190 (KERILL), with residues K17, H18 and Q284 also contributing to that patch. As in BSA, some ESA residues located on the long loop connecting subdomains IIA and IIB (including residues K293, E294, D296, L297 and S299) differ from their cognates, so could be recognized either as conformational or linear epitope. ESA residues D311, E313, H317, K319 and D320 on subdomain IIB constitute a patch that is not present in BSA. There is also a potentially recognizable patch on the fourth α-helix of subdomain IIB, including residues P366, A367, R370, T371, Q375, and T377, with residue A309 contributing: and a second patch including residues 384–401 (KSLVKKVCDLFEEVGETD; where ESA residues different from those in HSA are depicted in bold).
Several ESA residues in the range of 428–458 differ from the human albumin sequence, constituting a continuous patch that is exposed to solvent. However, most of these residues are located in a central part of ESA, at the bottom of deep clefts which would occlude the binding of large antibody proteins (without significant conformation changes). Only the residues of the loop and at the ends of the two helices are more accessible (R435, L439, S442, and E443). Multiple residues in the range of 470–482 of ESA also differ from the HSA sequence.
The surface patches of non-conserved residues located on the last two helices of ESA correspond to some extent to the analogous patches on BSA. The first one, comprising residues 545–561 (TVLGNFSAFVAKCCGRE) is elongated and continuous, and may extended by residues D392, E396, V397 and D400, which are also exposed and differ from the HSA sequence. The sequence of the last helix of ESA differs less from HSA than BSA, with only 5 residues (A565, P572, S577, L580 and A583) that are not conserved in the HSA sequence.
3.3.3. Rabbit serum albumin
Examples of possible epitopes on the RSA (Fig. 5) include the third helix of the IA subdomain, residues 33–52 (KCPYEEHAKLVKEVTDLAKA). Residue E38 is proximal to the second patch made by residues 76–82 (ALPSLRD) of the loop between fourth and fifth helix of subdomain IA, which is relatively well-conserved in rodents, making it a potentially cross-reactive epitope. The patch formed by residues 33–52 contacts a second patch formed by residues 130–137 (DEKAFFGH) of the second helix of subdomain IB. In the central part of the heart-shaped RSA structure there are three connected patches. The first is located on the third helix of the subdomain IB comprising residues 156–164 (YYAQKYKAI). The second comprises residues 179–199 (TPKLDALEGKSLISAAQERLR) from the fourth helix of the subdomain IB, which span into domain IIA. The third includes residues from domain IIA, specifically H274 and 291–294 (YGLH). The patches mentioned above include some residues that differ from the HSA sequence but are conserved in rodents, making them good candidates for possible cross-reactivity. There is also a patch on the helices of subdomain IIA including residues D227, T229, D230, I231, H267, E269, and T270, which is much bigger in RSA than in BSA or ESA. The patch located on subdomain IIB is composed of residues A304, V305, E307, E308, D312, L325, and K329. One of the patches most conserved in rodents, but different from HSA is located on the helix spanning the subdomains IIB and IIIA (L374, Q378, D382, K385, and V388). The patches located on the last two helices of the RSA are composed of residues 538–562 (HATNDQLKTVVGEFTALLDKCCSAE) of the penultimate helix and 566–583 (ACFAVEGPKLVESSKATLG) of the last helix.
4. Conclusions
Until now, crystallographic coordinates of only human serum albumin have been deposited in the PDB. Here we report crystal structures of albumins from bovine, horse and rabbit sera. Since mammalian serum albumins are highly cross-reactive allergens, we correlate the structural data with results of different immunological studies of SAs (Spitzauer, 1999). We also analyze structures and conservation as well as the divergence of serum albumins in context of potential allergenicity and cross-reactivity. These results provide a tool for better understanding the potential epitopes in these albumins, and can be applied in planning experiments on allergenic properties of specific fragments of these albumins. Moreover, the structure of RSA reveals the presence of a new, previously uncharacterized ligand binding site. Finally our structure of BSA is the first serum albumin structure to contain ordered metal ions. We observe three Ca2+ ions bound, which is in agreement with previous studies that have suggested the existence of 2–3 calcium binding sites on albumin.
Highlights.
First crystal structures of animal serum albumins from cattle, horse and rabbit
Identification of calcium transport sites on bovine albumin
Identification of potential epitopes; sequence divergence in structural context
Effect of albumin conservation on potential cross-reactivity
Acknowledgments
The authors would like thank Alexander Wlodawer and Heimo Breiteneder for critical reading of the manuscript. We would like to thank Steve Almo and members of New York Structural Genomics Research Consortium for their contribution to this work. The work described here was supported by the NIH Protein Structure Initiative grant GM094662. The structural results shown in this report are derived from work performed at the Argonne National Laboratory, at the Structural Biology Center and Life Sciences Collaborative Access Team of the Advanced Photon Source. Argonne is operated by University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
Abbreviations
- SA
Serum Albumin
- BSA
Bovine Serum Albumin
- ESA
Equine Serum Albumin
- RSA
Rabbit Serum Albumin
- RMSD
Root Mean Standard Deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguanno JJ, Ladenson JH. Influence of fatty acids on the binding of calcium to human albumin. Correlation of binding and conformation studies and evidence for distinct differences between unsaturated fatty acids and saturated fatty acids. J Biol Chem. 1982;257:8745–8. [PubMed] [Google Scholar]
- Alting AC, Meijer RJ, van Beresteijn EC. Incomplete elimination of the ABBOS epitope of bovine serum albumin under simulated gastrointestinal conditions of infants. Diabetes Care. 1997;20:875–80. doi: 10.2337/diacare.20.5.875. [DOI] [PubMed] [Google Scholar]
- Beretta B, Conti A, Fiocchi A, Gaiaschi A, Galli CL, Giuffrida MG, Ballabio C, Restani P. Antigenic determinants of bovine serum albumin. Int Arch Allergy Immunol. 2001;126:188–95. doi: 10.1159/000049513. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besarab A, Caro JF. Increased absolute calcium binding to albumin in hypoalbuminaemia. J Clin Pathol. 1981;34:1368–74. doi: 10.1136/jcp.34.12.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besarab A, DeGuzman A, Swanson JW. Effect of albumin and free calcium concentrations on calcium binding in vitro. J Clin Pathol. 1981;34:1361–7. doi: 10.1136/jcp.34.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin Y, Hebert H, Vrancken ER, Mourad W. Allergenicity and cross-reactivity of cat and dog allergenic extracts. Clin Allergy. 1988;18:287–93. doi: 10.1111/j.1365-2222.1988.tb02871.x. [DOI] [PubMed] [Google Scholar]
- Collaborative Computional Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr, Sect D: Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wolf FA, Brett GM. Ligand-binding proteins: their potential for application in systems for controlled delivery and uptake of ligands. Pharmacol Rev. 2000;52:207–36. [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002. [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjeldsgaard BE, Paulsen BS. Comparison of IgE-binding antigens in horse dander and a mixture of horse hair and skin scrapings. Allergy. 1993;48:535–41. doi: 10.1111/j.1398-9995.1993.tb01111.x. [DOI] [PubMed] [Google Scholar]
- Goubran Botros H, Gregoire C, Rabillon J, David B, Dandeu JP. Cross-antigenicity of horse serum albumin with dog and cat albumins: study of three short peptides with significant inhibitory activity towards specific human IgE and IgG antibodies. Immunology. 1996;88:340–7. doi: 10.1046/j.1365-2567.1996.d01-669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JW, Dugaiczyk A. The human serum albumin gene: structure of a unique locus. Gene. 1982;19:55–8. doi: 10.1016/0378-1119(82)90188-3. [DOI] [PubMed] [Google Scholar]
- Hilger C, Kohnen M, Grigioni F, Lehners C, Hentges F. Allergic cross-reactions between cat and pig serum albumin. Study at the protein and DNA levels. Allergy. 1997;52:179–87. doi: 10.1111/j.1398-9995.1997.tb00972.x. [DOI] [PubMed] [Google Scholar]
- Ho JX, Holowachuk EW, Norton EJ, Twigg PD, Carter DC. X-ray and primary structure of horse serum albumin (Equus caballus) at 0.27-nm resolution. Eur J Biochem. 1993;215:205–12. doi: 10.1111/j.1432-1033.1993.tb18024.x. [DOI] [PubMed] [Google Scholar]
- Jenkins JA, Breiteneder H, Mills EN. Evolutionary distance from human homologs reflects allergenicity of animal food proteins. J Allergy Clin Immunol. 2007;120:1399–405. doi: 10.1016/j.jaci.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Karjalainen J, Martin JM, Knip M, Ilonen J, Robinson BH, Savilahti E, Akerblom HK, Dosch HM. A Bovine Albumin Peptide as a Possible Trigger of Insulin-Dependent Diabetes Mellitus. The New England Journal of Medicine. 1992:302–307. doi: 10.1056/NEJM199207303270502. [DOI] [PubMed] [Google Scholar]
- Kragh-Hansen U, Vorum H. Quantitative analyses of the interaction between calcium ions and human serum albumin. Clin Chem. 1993;39:202–8. [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–68. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Liccardi G, Asero R, D’Amato M, D’Amato G. Role of sensitization to mammalian serum albumin in allergic disease. Curr Allergy Asthma Rep. 2011;11:421–6. doi: 10.1007/s11882-011-0214-7. [DOI] [PubMed] [Google Scholar]
- Lu J, Stewart AJ, Sleep D, Sadler PJ, Pinheiro TJ, Blindauer CA. A molecular mechanism for modulating plasma Zn speciation by fatty acids. J Am Chem Soc. 2012;134:1454–7. doi: 10.1021/ja210496n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NH, Perkins DJ. The interaction of serum albumins with calcium. Biochem J. 1950a;47:323–6. doi: 10.1042/bj0470323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NH, Perkins DJ. The interactions of purified serum albumins with calcium. Biochem J. 1950b;2:16–7. [PubMed] [Google Scholar]
- Martin NH, Perkins DJ. The calcium binding of human serum albumin in health and disease. Biochem J. 1953;54:642–5. doi: 10.1042/bj0540642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuoka J, Hegenauer J, Van Dyke BR, Saltman P. Intrinsic stoichiometric equilibrium constants for the binding of zinc(II) and copper(II) to the high affinity site of serum albumin. J Biol Chem. 1993;268:21533–7. [PubMed] [Google Scholar]
- Matheu V, Caloto M, de Barrio M, Baeza ML, Rubio M. Life-threatening anaphylaxis after artificial insemination. Lancet. 2002;359:1779. doi: 10.1016/s0140-6736(02)08627-0. [DOI] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–66. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Methods in enzymology: Macromolecular crystallography, part A. Vol. 276. Academic Press; New York: 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- Pedersen KO. Binding of calcium to serum albumin. III. Influence of ionic strength and ionic medium. Scand J Clin Lab Invest. 1972;29:427–32. doi: 10.3109/00365517209080262. [DOI] [PubMed] [Google Scholar]
- Peters T. All about albumin: Biochemistry, genetics, and medical applications. Academic Press; 1995. [Google Scholar]
- Petitpas I, Bhattacharya AA, Twine S, East M, Curry S. Crystal structure analysis of warfarin binding to human serum albumin: anatomy of drug site I. J Biol Chem. 2001;276:22804–9. doi: 10.1074/jbc.M100575200. [DOI] [PubMed] [Google Scholar]
- Ponterius G, Brandt R, Hulten E, Yman L. Comparative studies on the allergens of horse dandruff and horse serum. Int Arch Allergy Appl Immunol. 1973;44:679–91. doi: 10.1159/000230970. [DOI] [PubMed] [Google Scholar]
- Restani P, Fiocchi A, Beretta B, Velona T, Giovannini M, Galli CL. Effects of structure modifications on IgE binding properties of serum albumins. Int Arch Allergy Immunol. 1998;117:113–9. doi: 10.1159/000023997. [DOI] [PubMed] [Google Scholar]
- Rosenbaum G, Alkire RW, Evans G, Rotella FJ, Lazarski K, Zhang RG, Ginell SL, Duke N, Naday I, Lazarz J, Molitsky MJ, Keefe L, Gonczy J, Rock L, Sanishvili R, Walsh MA, Westbrook E, Joachimiak A. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J Synchrotron Radiat. 2006;13:30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzauer S. Allergy to mammalian proteins: at the borderline between foreign and self? Int Arch Allergy Immunol. 1999;120:259–69. doi: 10.1159/000024278. [DOI] [PubMed] [Google Scholar]
- Spitzauer S, Pandjaitan B, Soregi G, Muhl S, Ebner C, Kraft D, Valenta R, Rumpold H. IgE cross-reactivities against albumins in patients allergic to animals. J Allergy Clin Immunol. 1995;96:951–9. doi: 10.1016/s0091-6749(95)70233-4. [DOI] [PubMed] [Google Scholar]
- Spitzauer S, Schweiger C, Sperr WR, Pandjaitan B, Valent P, Muhl S, Ebner C, Scheiner O, Kraft D, Rumpold H, et al. Molecular characterization of dog albumin as a cross-reactive allergen. J Allergy Clin Immunol. 1994;93:614–27. doi: 10.1016/s0091-6749(94)70073-7. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel D, Valeri CR, Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci U S A. 1992;89:7674–7. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AJ, Blindauer CA, Berezenko S, Sleep D, Sadler PJ. Interdomain zinc site on human albumin. Proc Natl Acad Sci U S A. 2003;100:3701–6. doi: 10.1073/pnas.0436576100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow G, Birkett DJ, Wade DN. The characterization of two specific drug binding sites on human serum albumin. Mol Pharmacol. 1975;11:824–32. [PubMed] [Google Scholar]
- Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 1999;12:439–46. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- Tanabe S, Kobayashi Y, Takahata Y, Morimatsu F, Shibata R, Nishimura T. Some human B and T cell epitopes of bovine serum albumin, the major beef allergen. Biochem Biophys Res Commun. 2002;293:1348–53. doi: 10.1016/S0006-291X(02)00381-9. [DOI] [PubMed] [Google Scholar]
- Ueno H, Masuko T, Wang J, Hashimoto Y. Epitope mapping of bovine serum albumin using monoclonal antibodies coupled with a photoreactive crosslinker. J Biochem. 1994;115:1119–27. doi: 10.1093/oxfordjournals.jbchem.a124467. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. Journal of Applied Crystallography. 1997;30:1022–1025. [Google Scholar]
- Vicente-Serrano J, Caballero ML, Rodriguez-Perez R, Carretero P, Perez R, Blanco JG, Juste S, Moneo I. Sensitization to serum albumins in children allergic to cow’s milk and epithelia. Pediatr Allergy Immunol. 2007;18:503–7. doi: 10.1111/j.1399-3038.2007.00548.x. [DOI] [PubMed] [Google Scholar]
- Wuthrich B, Stern A, Johansson SG. Severe anaphylactic reaction to bovine serum albumin at the first attempt of artificial insemination. Allergy. 1995;50:179–83. doi: 10.1111/j.1398-9995.1995.tb05077.x. [DOI] [PubMed] [Google Scholar]
- Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallographica Section D. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- Zheng H, Chruszcz M, Lasota P, Lebioda L, Minor W. Data mining of metal ion environments present in protein structures. J Inorg Biochem. 2008;102:1765–76. doi: 10.1016/j.jinorgbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman MD, Chruszcz M, Koclega KD, Otwinowski Z, Minor W. The Xtaldb system for project salvaging in high-throughput crystallization. Acta Crystallogr Sect A. 2005;61:c178–c179. [Google Scholar]