Abstract
The genetic code specifies 20 common amino acids and is largely preserved in both single and multicellular organisms. Unnatural amino acids (Uaas) have been genetically incorporated into proteins by using engineered orthogonal tRNA/aminoacyltRNA synthetase (RS) pairs, enabling new research capabilities and precision inaccessible with common amino acids. We show here that Escherichia coli tyrosyl and leucyl amber suppressor tRNA/RS pairs can be evolved to incorporate different Uaas in response to the amber stop codon UAG into various proteins in Caenorhabditis elegans. To accurately report Uaa incorporation in worms, we found that it is crucial to integrate the UAG-containing reporter gene into the genome rather than to express it on an extrachromosomal array from which variable expression can lead to reporter activation independent of the amber-suppressing tRNA/RS. Synthesizing a Uaa in a dipeptide drives Uaa uptake and bioavailability. Uaa incorporation has dosage, temporal, tRNA copy, and temperature dependencies similar to endogenous amber suppression. Uaa incorporation efficiency was improved by impairing the nonsense-mediated mRNA decay pathway through knockdown of smg-1. We have generated stable transgenic worms capable of genetically encoding Uaas, enabling Uaa exploitation to address complex biological problems within a metazoan. We anticipate our strategies will be generally extendable to other multicellular organisms.
Introduction
The canonical genetic code specifies 20 common amino acids and is used by almost all life forms. This code has been artificially expanded to include unnatural amino acids (Uaas) in single cells including bacteria, yeast, mammalian somatic cells and stem cells (1–7). Genetically encoded Uaas allow new physical and chemical properties to be selectively introduced into proteins in vivo, providing novel means to investigate and manipulate biological processes in native cellular environments (8, 9). Although single cells are valuable models, research into development, intercellular communication, differentiation, cancerous transformation, and a variety of other processes necessitates multicellular organisms. The ability to genetically incorporate Uaas in multicellular organisms will provide tools to address complex questions with chemical precision previously unattainable.
A Uaa can be genetically encoded in single cells by using an orthogonal tRNA/aminoacyl-tRNA synthetase (RS) pair that is specific for the Uaa, does not crosstalk with endogenous tRNA/RS pairs, and functionally couples with the endogenous translation machinery. The amber stop codon TAG is often introduced into the target gene to encode the Uaa. The orthogonal tRNA, after being charged with the Uaa by the orthogonal RS, recognizes the UAG codon and incorporates the Uaa into the target protein during translation. Expanding this methodology from single cells to a multicellular organism poses additional challenges (Figure 1). An orthogonal tRNA/RS pair must be identified that is functional in animal cells, and the tRNA, the RS, and the target protein need to be simultaneously expressed in the same cell. Appropriate and consistent expression of UAG-containing genes in C. elegans depends on both the gene delivery method and the UAG position within the gene. Uaas must also be efficiently delivered to the target cells without being sequestered or metabolized for expression of Uaa-containing proteins. Nonsense-mediated mRNA decay (NMD), a eukaryotic quality-control mechanism that degrades mRNAs containing premature stop codons, is also a potential concern for efficient Uaa incorporation.
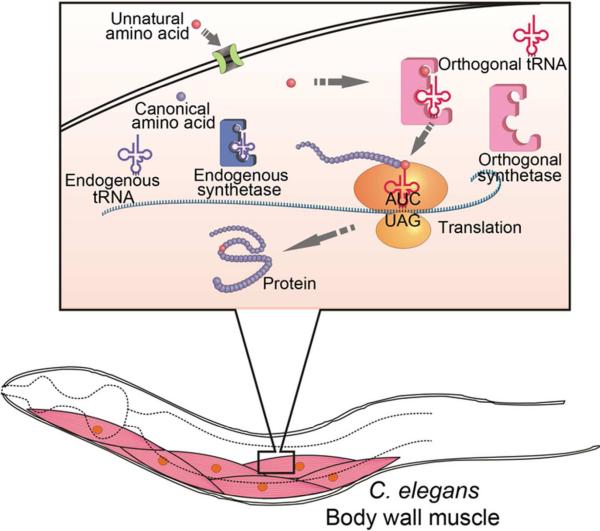
Figure 1. Scheme for incorporating unnatural amino acids into proteins in C. elegans.
The Uaa must be able to enter the worm and be transported into cells of the target tissue. An orthogonal tRNA/RS pair evolved to be specific for the Uaa should be expressed in the target cell together with the gene of interest. An amber stop codon UAG is introduced at the desired site for Uaa insertion in target gene. The orthogonal RS charges the Uaa onto the orthogonal tRNA, which recognizes the UAG codon and incorporates the Uaa during translation.
We choose the nematode Caenorhabditis elegans for genetic code expansion because it is a valuable model organism that has been extensively used for researching development, neurobiology, aging, meiosis, and epigenetics (10, 11). Besides a wealth of knowledge on the C. elegans genome (12, 13), epigenetic regulation (14), and cell lineage (15), certain characteristics of this animal may particularly facilitate Uaa incorporation and application. It is the only metazoan in which endogenous amber suppressor tRNAs have been identified (16, 17), demonstrating the worms' tolerance for amber suppression. In addition, the transparent body of C. elegans enables the use of light for experimentation. Photo-responsive Uaas can be encoded for fluorescent imaging, photocrosslinking and modulation via photolysis, and should greatly expand research in C. elegans with broader biophotonic technologies.
Here we report strategies addressing many aspects of Uaa incorporation in C. elegans and the successful expansion of its genetic code with amber suppressing tRNA/RS pairs derived from E. coli tRNATyr/TyrRS and tRNALeu/LeuRS. While our work was reviewed elsewhere, Greiss et al. reported the use of the Methanosarcina mazei pyrrolysyl tRNA/RS pair in worms to incorporate Uaas in a UAG site introduced into a linker region between two genes expressed from extrachromosomal arrays (18). We found that for accurate reporting of Uaa incorporation in C. elegans, the UAG-containing reporter gene must be chromosomally integrated. We selectively incorporated different Uaas into various reporter proteins in C. elegans with tissue specificity. Uaa-containing reporter proteins remained functional in C. elegans, demonstrating the feasibility of genetically encoding Uaas to study protein biology in vivo. The considerations and solutions devised for C. elegans should be valuable for expanding the code of other multicellular organisms.
Results and Discussion
Accurately reporting amber suppression in C. elegans requires chromosomal integration of the reporter gene
We used the amber stop codon UAG to encode Uaas in C. elegans because amber suppression has been successfully employed to incorporate Uaas in single cells, and natural amber suppressors have been isolated in C. elegans. To identify orthogonal tRNA/RS pairs that decode UAG in C. elegans, we constructed a fluorescent reporter for amber suppression by introducing the UAG codon at a permissive site in the fluorescent protein mCherry for body wall muscle expression. Initially, the reporter (unc-54p∷mCherryTAG156∷unc-54 3'UTR) was injected alone to generate extrachromosomal arrays. These injected worms, however, showed diffuse red fluorescence in body wall muscles, with some muscles showing fairly bright fluorescence (Figure 2A). Of worms expressing the co-injection marker, approximately half exhibited detectable fluorescence in the absence of any amber suppressor tRNA/RS. The UAG codon may have been mutated or deleted during array formation (19). Moreover, nonspecific stop codon readthrough has been reported in worms (16). To avoid these problems, we integrated a single copy of the fluorescent reporter gene within Chromosome II using the MosSCI method (20) and confirmed the integrity of the UAG codon with DNA sequencing. The resultant reporter strain, LWA1560 (wlSi151[unc54∷mCherryTAG156 cb-unc-119(+)]II) (Table 1), showed zero detectable fluorescence (other than known intestinal autofluorescence) in the absence of a suppressor tRNA/RS but consistent red body wall muscle fluorescence when crossed to the C. elegans endogenous amber suppressor sup-7(st5) (Figure 2B,C) (21), indicating that this reporter reliably reflected amber suppression. We therefore used a single integrated copy of reporter genes for all experiments.
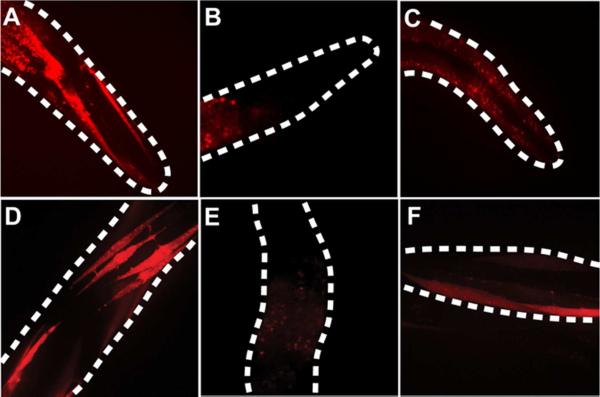
Figure 2. Screening for functional amber suppression with mCherry.
(A) The extrachromosomal array unc-54p∷mCherryTAG156∷unc-54 3'UTR showed fluorescence in muscle cells in the absence of any amber suppressor tRNA/RS. (B) A single integrated copy of the same mCherry amber reporter in strain LWA1560 (wlSi151[unc54∷mCherryTAG156 cb-unc-119(+)]) had no detectable fluorescence in muscle cells. The observed autofluorescence was from intestinal granules, which are distinct from muscle cells in morphology and location. (C) LWA1560 crossed with an endogenous amber suppressor (sup-7(st5); wlSi151) showed red fluorescence in muscle cells. Because the sup-7 suppressor tRNA is expressed in all muscle cells, all of the cells uniformly displayed red fluorescence. (D) LWA1561 (LWA1560 with wlEx1[unc-54∷LeuRS_rpr-1∷ + pRF4(rol-6)]) showed strong fluorescence in some body wall muscles. Only a subset of muscles were fluorescent due to the mosaic nature of the extrachromosomal array expressing the E. coli . (E) LWA1560 with rpr-1p: + pRF4(rol-6) had no muscle fluorescence. The observed autofluorescence was from intestinal granules. (F) LWA1562 (LWA1560 with wlEx22[unc-54∷TyrRS_rpr-1∷ + pRF4(rol-6)]) showed red fluorescence in some body wall muscles. Because the was expressed in an extrachromosomal array, the red fluorescence was mosaic. Dashed lines indicate worm outline. All images are confocal Z stacks and representative of each genotype at young adulthood.
Table 1.
Genotypes and strain names created for this work.
| Strain | Genotype |
|---|---|
| LWA1560 | wlSi151[unc54∷mCherryTAG156 cb-unc-119(+)] II. |
| LWA1561 | wlSi151 II, wlEx1[unc-54∷LeuRS_rpr-1∷tRNACUALeu + pRF4(rol-6)]. |
| LWA1562 | wlSi151 II, wlEx22[unc-54∷TyrRS_rpr-1∷tRNACUATyr + pRF4(rol-6)]. |
| LWA1563 | wlSi151 II, wlEx13[unc-54∷OmeRS_rpr-1∷tRNACUATyr + pRF4(rol-6)]. |
| LWA1564 | wlSi151 II, wlEx35[unc-54∷DanRS_rpr-1∷tRNACUALeu + pRF4(rol-6)]. |
| LWA1565 | wlSi151 II, wlls1565[unc-54∷DanRS_rpr-1∷tRNACUALeu + myo-2∷GFP]X. |
| LWA1850 | wlSi1852[unc-54∷luciferaseTAG185 cb-unc-119 (+)]I. |
| LWA1851 | wlSi1852 I, wlls1851[unc-54∷TyrRS_rpr-1∷tRNACUATyr + myo-2∷GFP]X. |
| LWA1852 | wlSi1852 I, wlls1853[unc-54∷OmeRS_rpr-1∷tRNACUATyr + myo-2∷GFP]X. |
| LWA1031 | wlSi1852 I wlls1565[unc-54∷DanRS_rpr-1∷tRNACUALeu + myo-2∷GFP]X. |
| LWA1008 | wlSi1852 I, wlSi13[rpr-1∷tRNACUALeu]II, wlSi550[unc-54∷LeuRS]IV. |
| LWA1009 | wlSi1852 I wlSi17[(rpr-1∷tRNACUALeu)3x]II, wlSi550 IV. |
| LWA1855 | wlSi1852 I, wlSi1082[(rpr-1∷tRNACUATyr)3x]IV, wlls1853 X. |
| LWA1856 | wlSi1852 I, wlSi17 II, wlls1565 X. |
| LWA1857 | wlSi1852 I, wlSi1082 IV, wlls1851 X. |
| LWA1580 | wlSi1580[unc-54∷JFFluciferase cb-unc-119 (+)]I |
| LWA1582 | wlSi1582[unc-54∷JFFluciferaseTAG158 cb-unc-119 (+)]I. |
| LWA717 | wlSi1582 I, wlSi1082 IV, wlls1853 X. |
| LWA718 | wlsi1582 I, wlSi17 II, wlls1565 X. |
Identification and expression of orthogonal tRNA/RS pairs
Effective Uaa incorporation requires that a tRNA/RS does not interact with any endogenous tRNA/RS pairs. A tRNA/RS pair taken from a different kingdom of life is likely to be orthogonal (22), directing our focus to bacterial pairs. One obstacle to incorporating Uaas in eukaryotic organisms is the functional expression of orthogonal bacterial tRNAs (3, 5). Prokaryotes and eukaryotes differ in tRNA transcription and processing. Prokaryotic tRNAs have external 5' promoters; eukaryotic tRNAs are transcribed by RNA polymerase III (Pol III) using internal promoters within the tRNA itself. These sequences are conserved among eukaryotic tRNAs but are often missing from bacterial tRNAs. Therefore, transplanting bacterial tRNA genes that don't have matching A-box and B-box sequences into eukaryotic tRNA expression cassettes often results in tRNAs with no or weak function (3–6, 23). We previously circumvented this problem in yeast (3), mammalian (5), and stem cells (7) using external Pol III promoters, which contain all necessary promoter elements within.
We tested type-3 Pol III C. elegans promoters to drive the expression of E. coli tRNA in worms. The promoters cen-4, cen-38, and rpr-1 were identified from the C. elegans noncoding transcriptome (24) as type-3 Pol III candidates. These promoters were placed at the 5' end of the E. coli amber suppressor tRNA (without the last trinucleotide CCA) and followed by the 3' flanking sequence of the C. elegans tRNALys. The endogenous 3' flanking sequences of tRNAs encourage proper processing of prokaryotic tRNAs in eukaryotic systems, as does deleting the last trinucleotide from the introduced tRNA (3, 5). Plasmids containing the candidate promoter driving the E. coli leucyl amber suppressor tRNA () and the E. coli leucyl-tRNA synthetase (LeuRS) driven by the unc-54 promoter (unc-54p∷LeuRS∷unc-54 3'UTR_promoter∷ ) were microinjected into the mCherry reporter line and maintained as extrachromosomal arrays to create LWA1561(wlSi151 II, ). Only if the tRNA is correctly expressed and processed will it be charged with Leu to suppress the amber codon within mCherry, generating red fluorescence. After screening we found that the rpr-1 promoter showed strong reporter activation (Figure 2D), indicating that it can drive the functional expression of E. coli in C. elegans. Moreover, when a plasmid containing rpr-1 driven was injected in the absence of LeuRS, no red fluorescence was detected (Figure 2E), indicating that the E. coli is orthogonal to C. elegans.
To test if the rpr-1 promoter can be generally used to express other E. coli tRNAs, we replaced the E. coli with E. coli . When the E. coli was expressed with its cognate E. coli TyrRS, the strain LWA1562 (wlSi151 II, ) showed red fluorescence in body wall muscles (Figure 2F). Additionally, in the absence of the E. coli TyrRS, no red fluorescence was detected, indicating that the E. coli is orthogonal to C. elegans.
Amber suppression in C. elegans using our orthogonal tRNA/RS system showed robust temperature dependence: at 15 °C there was strong fluorescence in body wall muscles, but at 20 °C fluorescence was weak. This result is consistent with the previously reported temperature dependence of endogenous C. elegans amber suppressors (17), further suggesting our system is functional for amber suppression.
Delivery and incorporation of Uaas
Delivery of Uaas into the cells of interest is another potential hurdle to Uaa incorporation in multicellular animals. For tissue culture cells and unicellular organisms, addition of the Uaa to the growth medium is sufficient for the uptake of many Uaas (25). However, C. elegans has a protective cuticle that excludes many compounds from internalization and an active digestion system that could potentially degrade Uaas before assimilation by the intestine. We reasoned that Uaas might be taken in as food via the intestinal route and trafficked to multiple tissues, though intestinal transporters may not recognize Uaas with drastically different structures. By incorporating Uaas into proteins specific for a tissue that is not exposed to the external environment, we can monitor whether specific Uaas can be transported to that tissue. We chose two Uaas to study their uptake in C. elegans: o-methyl-L-tyrosine (OmeY), structurally similar to tyrosine, and 2-amino-3-(5-(dimethylamino) napththalene-1-sulfonamido) propanoic acid (DanAla), a fluorescent amino acid that shares no structural similarity to any natural amino acid (Figure 3A).
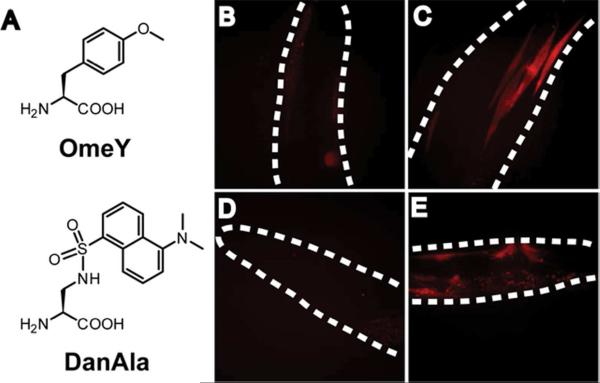
Figure 3. Incorporating Uaas into mCherry.
(A) Structure of Uaas. (B) LWA1563 (LWA1560 with wlEx13[unc-54∷OmeRS_rpr-1∷ + pRF4(rol-6)]) showed very little fluorescence in the absence of OmeY. (C) LWA1563 grown with 5 mM OmeY showed strong mosaic activation of the mCherry reporter in muscle cells. (D) LWA1564 (wlEx35[unc-54∷DanRS_rpr-1∷ + pRF4(rol-6)]) showed no fluorescence in the absence of Uaa. (E) LWA1564 grown with 1 mM Ala-DanAla dipeptide showed strong mosaic activation of the mCherry reporter in body wall and vulval muscles. All images are confocal Z stacks and representative of each genotype. Dashed lines indicate worm outline. Animals were grown in liquid culture for 1 generation and imaged at young adulthood.
Orthogonal synthetases specific for Uaas have been evolved in E. coli and later in yeast from large mutant synthetase libraries consisting of >109 members (1, 2). Similar evolution strategies cannot be used in C. elegans due to inefficient transgene delivery and low throughput of phenotypic selection. We and others have shown that transferring a synthetase evolved in bacteria or yeast to tissue culture cells is a useful strategy to generate Uaa-specific synthetases for systems in which directed evolution is not possible (5, 6, 26). Mutant synthetases specific for OmeY (OmeRS, derived from E. coli TyrRS) (27) and DanAla (DanRS, derived from E. coli LeuRS) (28) have been generated in S. cerevisiae, suggesting that they could be transferred into C. elegans for incorporation of the respective Uaa.
To incorporate OmeY into mCherry, we exposed gravid mCherry reporter animals harboring an array encoding unc-54p∷OmeRS∷unc-54 3'UTR_rpr-1p∷ (LWA1563) to OmeY in liquid culture. Five millimolar OmeY led to reporter activation in more than half of L3-L4 progeny containing the array (Figure 3B, C). Only animals reared on OmeY from the embryo stage exhibited reporter activation, whereas animals placed in OmeY culture after hatching lacked fluorescence. In addition, most negative controls grown in the absence of OmeY lacked fluorescence; only 10% showed faint fluorescence in 1 or 2 muscle cells. These results indicate that the E. coli and the evolved OmeRS specifically insert OmeY into mCherry in C. elegans.
For the incorporation of DanAla, feeding free Uaa at 1 or 2.5 mM did not activate the reporter in animals with unc-54p∷DanRS∷unc-54 3'UTR_rpr-1p∷ (LWA1564). OmeY might enter C. elegans and reach muscle cells through pathways used by Tyr or Phe due to their structural similarity, but DanAla is too divergent to follow natural uptake routes. Initial attempts to localize DanAla were hindered by the overlap of gut granule fluorescence (Figure 4A). Using a glo-4 (ok623) (29) mutant that lacks fluorescent gut granules (Figure 4B), we found that DanAla was sequestered in the intestinal cells, which became fluorescent only after feeding DanAla (Figure 4C).
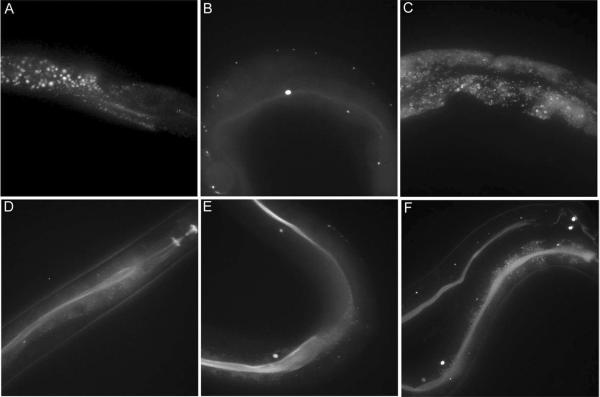
Figure 4. Fluorescence imaging shows that DanAla is sequestered in intestinal cells but Ala-DanAla dipeptide can be transported to other tissues inside C. elegans.
(A) N2 worms has autofluorescent intestinal granules. (B) glo-4 (ok623) does not have autofluorescent intestinal granules. (C) After exposure to 1 mM DanAla, intestinal granules of sequestered DanAla appeared in glo-4 (ok623). (D) glo-4 (ok623) after exposure to 1 mM Ala-DanAla dipeptide. There was some diffuse fluorescence in the lumen and intestinal cells, but few punctate granules. (E) glo-4 (ok623) after exposure to 2.5 mM Ala-DanAla dipeptide. (F) glo-4 (ok623) after exposure to 5 mM Ala-DanAla dipeptide. At this high concentration of dipeptide punctate fluorescent granules began to appear. All animals were exposed for 1 generation on solid plates and assayed as young adults. Images are confocal Z stacks.
PEPT-1 and PEPT-2 are dipeptide transporters present on the surface of many C. elegans cells (30). We reasoned that an Ala-Uaa dipeptide should enter cells through dipeptide transporters. The dipeptide would then be hydrolyzed by cellular peptidases to generate the free Uaa inside cells. We devised a strategy for intestinal uptake of DanAla by using an Ala-DanAla dipeptide. Unlike free DanAla, the Ala-DanAla dipeptide was not trapped within the intestine unless high concentrations were used (Figure 4D–F). When gravid worms were fed 1 mM Ala-DanAla dipeptide, mCherry reporter activation was observed in body wall and vulval muscle cells of L3-L4 stage progeny (Figure 3D, E). In the absence of the dipeptide, no mCherry expression was detected. As the DanRS is specific for DanAla, these results suggest that DanAla entered muscle cells and was incorporated into mCherry.
Integration of an orthogonal tRNA/RS array and Uaa feeding on solid medium improve Uaa incorporation
To improve incorporation efficiency and generate animals with 100% transmission of the Uaa incorporation machinery, the extrachromosomal arrays encoding the orthogonal tRNA/RS genes were integrated using γ-irradiation. In contrast to the mosaic expression of the extrachromosomal array caused by random mitotic segregation, the integrated strain LWA1565 () displayed uniform non-mosaic expression of the fluorescent reporter in all body wall muscles (Figure S1A–C). After integration, expression of the reporter gene was less variable, ensuring all animals within a population had similar levels of Uaa incorporation and target gene expression.
Further optimization indicated that the stably integrated animals exhibited more robust reporter activation on solid medium compared to liquid (Figure S2A–F). We also noted that LWA1565 animals grown in the presence of Ala-DanAla dipeptide developed slower with fewer progeny than those grown without Uaa, but these effects were more pronounced in liquid than solid medium. Animals grown on plates appeared healthier for most conditions with dipeptide concentrations up to 3 mM.
Quantification of Uaa incorporation
A quantifiable reporter would enable us to optimize system components for amber suppression and protein expression. A different target protein for Uaa incorporation would also test whether our strategy can be generally applied to other proteins. We decided to use a luciferase because the enzymatic oxidation of luciferin offers sensitive detection of Uaa incorporation by measuring the resultant luminescence.
A permissive site for mutagenesis (Ser185) in the Photinus pyralis luciferase was identified based on its structure and verified using amber suppression in mammalian cells. A single copy of unc-54p∷luciferaseTAG185∷unc-54 3'UTR was integrated within Chromosome I using MosSCI to generate the stable luciferase reporter strain LWA1850 (wlSi1852[unc-54∷luciferaseTAG185 cb-unc-119 (+)]I). Wild-type luciferase was similarly integrated as a positive control, from which robust luciferase activity was measured after worm lysis. When the (Ser185TAG) luciferase reporter line was crossed to a line expressing an integrated array to generate LWA1851 (wlSi1852; ), luciferase activity was readily detected in LW1851 (Figure S3). In the absence of the suppression machinery, no luciferase activity was detected from the reporter line, confirming that single integration of the amber-containing luciferase gene had no nonspecific UAG readthrough. These results demonstrate the suitability of the luciferase reporter strain for quantification.
Uaa incorporation in luciferase has Uaa dose dependency
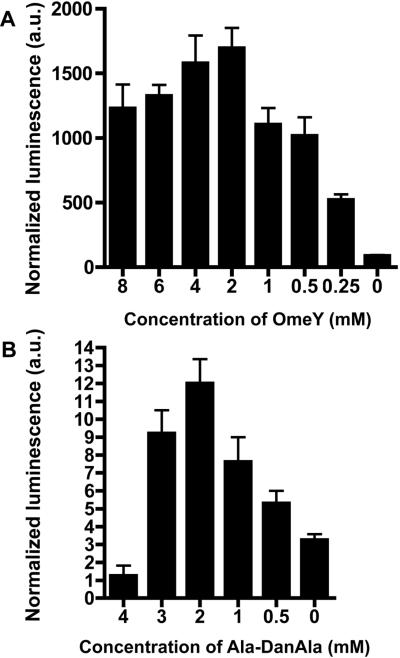
To test for Uaa incorporation efficiency and the optimal Uaa dosage, we crossed the luciferase reporter line to integrated tRNA/RS array lines, creating strain LWA1852 (wlSi1852; ) and LWA1031 (wlSi1852 I; ). Animals were grown on solid media with varying concentrations of Uaas. A dose dependent increase in luciferase activity was observed for both OmeY and Ala-DanAla dipeptide, up to concentrations where growth and fertility became limiting (Figure 5A, B). For instance, at concentrations above 3 mM the Ala-DanAla dipeptide made the medium acidic, inhibiting bacterial lawn growth and leading to few surviving animals. Additionally, similarly increasing the dosage of Phe or Tyr had little to no effect on luciferase activity. Uaa dosage dependence and the established specificity of the evolved RS for the Uaa suggest that the Uaa was not metabolized but incorporated unaltered into the luciferase protein in C. elegans.
Figure 5. Quantification of Uaa incorporation in luciferase shows dependence on Uaa concentration.
(A) Luciferase activities measured from strain LWA1852 (wlSi1852 [unc-54∷luciferaseTAG185 cb-unc-119 (+)]I; wlls1851[unc-54∷OmeRS_rpr-1∷ + myo-2∷GFP]X) grown at different OmeY concentrations. (B) Luciferase activities measured from strain LWA1031 (wlSi1852 I; wlls1565[unc-54∷DanRS_rpr-1∷ + myo-2∷GFP]X) grown at different Ala-DanAla concentrations. Both strains were grown on solid plates at 15 °C for 1 generation in duplicate. Luminescence was normalized to total protein concentration. Error bars represent s.e.m.; n ≥ 2.
We also analyzed incorporation efficiency in relation to Uaa placement within the dipeptide. Surprisingly, we found a dramatic increase of DanAla incorporation using DanAla-Ala instead of Ala-DanAla (Figure S4), which may be caused by enhanced accessibility of DanAla-Ala to cellular peptidases or dipeptide transporters.
Uaa incorporation efficiency increases with tRNA copy number
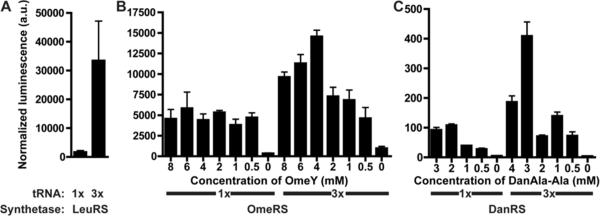
Previously, we found that a higher ratio of tRNA to RS improves Uaa incorporation efficiency in mammalian cells (31). To test if this extends to C. elegans, the ratio of tRNA to RS expression cassettes in the animal was altered. A single expression cassette (rpr-1p∷ 3' flanking sequence), as well as the wild type LeuRS and the luciferase amber reporter, both driven by the unc-54 promoter, were integrated using MosSCI into different chromosomes to create LWA1008 (wlSi1852 I; wlsi13[(rpr-1∷ )]II; wlSi550[unc-54∷LeuRS]IV). Another strain, LWA1009 (wlSi1852 I; wlsi17[rpr-1∷ 3x]II; wlSi550), was similarly generated except that the single expression cassette was replaced with 3 tandem copies. A comparison of these 2 lines showed that an increase in tRNA copy number leads to 19 times higher luciferase activity (Figure 6A).
Figure 6. Uaa incorporation shows dependence on the tRNA to RS ratio.
(A) Comparison of LWA1008 (wlSi1852 I; wlSi13[rpr-1∷ ]II; wlSi550[unc-54∷LeuRS]IV) with LWA1009 (wlSi1852 I; wlSi17[(rpr-1∷ )3x]II; wlSi550 IV) indicates that 3x increased Leu incorporation into luciferase. (B) Comparison of LWA1852 (1x ) with LWA1855 (with wlSi1082[(rpr-1∷ )3x]IV) indicates that additional increased OmeY incorporation into luciferase. (C) Comparison of LWA1031 with LWA1856 (with wlSi13 II) indicates that additional increased DanAla incorporation into luciferase. All animals were grown for 3 generations on solid plates with indicated concentration of Uaa when applicable at 15 °C in duplicate. All error bars represent s.e.m., n ≥ 2.
We tested if this effect could be recapitulated in animals incorporating Uaas. Three tandem copies of the E. coli expression cassette were integrated into Chromosome IV for use with the OmeRS. We crossed the integrated tRNA/RS array lines to those with the triple tRNA expression cassettes, generating LWA1855 (wlSi1852 I; wlSi1082[(rpr-1∷ )3x]IV; wlls1853 X) to incorporate OmeY and LWA1856 (wlSi1852I; wlSi17 II; wlls1565 X) to incorporate DanAla. When incorporating either OmeY or DanAla (using either dipeptide orientation), there was indeed an increase in luciferase activity with a higher tRNA copy number (Figure 6B, C, Figure S5).
Dependence of Uaa incorporation on multi-generational exposure
Unlike single cell systems, a multicellular organism will alter its gene expression and small molecule uptake throughout the course of development and aging. To perform precisely timed studies, it is necessary to discern the timing of Uaa uptake and clearance. Understanding Uaa uptake can improve incorporation efficiency and permit pulse-chase experiments using Uaas.
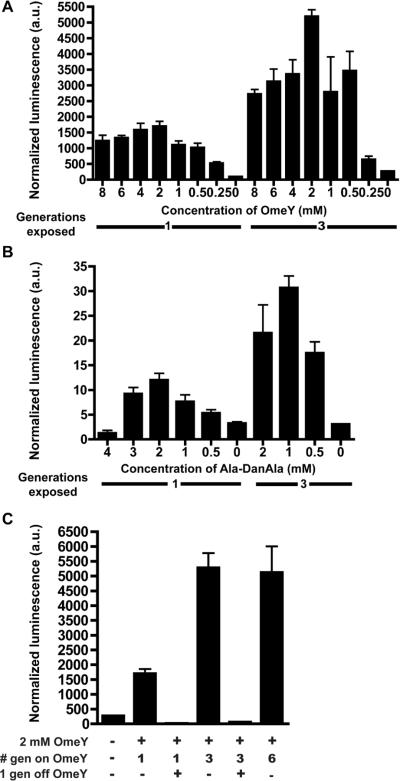
We previously observed that only animals raised in the presence of Uaa from embryonic stages were responsive to Uaa incorporation, whereas those exposed to Uaa during later stages showed no reporter activation. Aliquoting fewer animals initially allowed worms to grow for 3 generations on Uaa before assaying for luciferase activity. Whereas negative control worms showed no change, worms exposed to the Uaa over 3 generations showed an increase in luciferase activity compared to those exposed for only 1 generation (Figure 7A, B). This increase suggests enrichment or transmission of Uaas from hermaphrodite to progeny occurred. The effect was observed for both Uaas examined and may be general for other Uaas. However, there was no further increase in luciferase activity with growth for 6 generations on Uaa containing media (Figure 7C).
Figure 7. Uaa incorporation increases over multiple generations and is reversible.
(A) LWA1852 animals show an increase in OmeY-dependent luciferase activity over 3 generations compared to 1. (B) LWA1031 animals show an increase in Ala-DanAla-dependent luciferase activity over 3 generations compared to 1. For (A) and (B), animals were grown in duplicate on solid plates of indicated Uaa concentrations at 15 °C, and assayed after 1 or 3 generations' exposure to Uaa. (C) Comparison of multi-generation data and reversibility of Uaa incorporation. LWA1852 animals were grown in duplicate at 15 °C with 2 mM OmeY on solid plates for indicated generations. Removal of worms from the Uaa for 1 generation dropped the luciferase activity to baseline levels. All error bars represent s.e.m.; n ≥ 2.
To determine if Uaa incorporation persists after worms have been removed from Uaa plates, animals were placed on unsupplemented media for 1 generation after exposure to 2 mM OmeY. For animals grown on Uaa plates for 1 or 3 generations, the luciferase activity dropped to background levels after a single generation on normal plates, with no lingering protein expression (Figure 7C). This effect was consistent for all concentrations of Uaa examined (Figure S6A, B). The ability to turn reporter activity on and off with Uaa exposure confirms both the necessity of the Uaa for reporter activity and Uaa incorporation into the reporter.
Detection of Uaa incorporation by Western blot and Uaa fluorescence
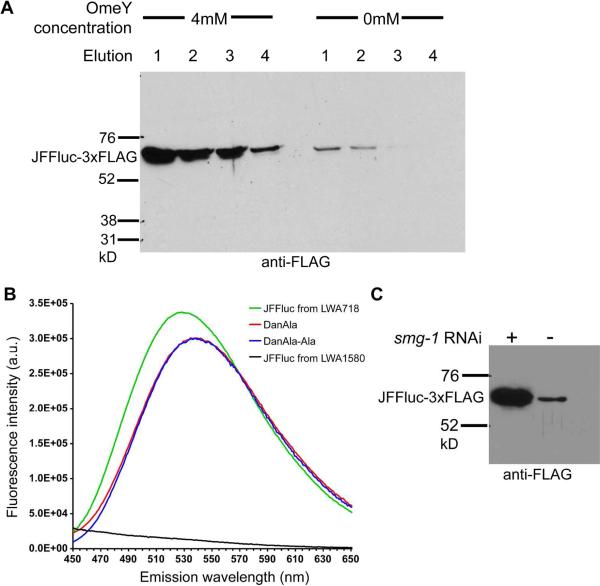
To biochemically demonstrate that robust expression of target proteins is dependent on Uaa supplementation, we integrated a third reporter, a luciferase protein from Luciola cruciata, a Japanese firefly. This protein (JFFluc) tolerates the addition of a 3xFLAG tag at its C-terminus to facilitate Western detection, and is well expressed in C. elegans body wall muscle. We identified a permissive site (Val 158) in JFFluc in tissue culture and cloned the Val158TAG mutant to generate the plasmid unc-54∷JFFluciferaseTAG158_ICRH2B:GFP. This plasmid was used for MosSCI integration into chromosome I to generate the JFFluc amber reporter line LWA1582 (wlSi1582[unc-54∷JFFluciferaseTAG158 cb-unc-119 (+)]I), which was crossed to LWA1855 (a line containing the integrated 3x and OmeRS) to generate LWA717 (wlSi1582 I, wlSi1082 IV, wlls1853 X). These animals were incubated at 15 °C on 4 mM or 0 mM OmeY plates. After three generations, immunoprecipitation with FLAG beads was performed on lysates from these animals.
Incubating the animals with 4 mM OmeY enabled a large amount of JFF luciferase to be purified, while in the absence of OmeY, only small amounts of JFFluc were detected in the first two elutions by Western blotting (Figure 8A). The OmeRS used here was evolved in yeast, and may charge a natural amino acid on the with low efficiency in the absence of the cognate Uaa OmeY. Natural synthetases have also been known to mischarge amino acid analogues when the concentration of the cognate amino acid is low (32). In the presence of OmeY, a clear response and marked increase in protein expression was observed: band densitometry indicated that the amount of JFF luciferase was 27-fold more than that purified from animals grown without OmeY. These results further support the incorporation of OmeY into the luciferase protein in C. elegans.
Figure 8. Biochemical analyses of Uaa incorporation and NMD effect.
(A) Western blot analysis of JFFluc protein expressed in LWA717 animals grown on solid plates containing 4 mM or 0 mM OmeY. Animals were lysed, and the JFFluc protein was immunoprecipitated. Four elutions for each sample were collected and equal volumes were loaded into each lane. After transferring, the blot was probed with an anti-FLAG antibody to detect the purified JFFluc-3xFLAG. At the molecular weight corresponding to the JFFluc-3xFLAG protein (64 kDa) a strong band was visible for each elution of purified protein from animals grown on 4 mM OmeY. Only a weak band in the first 2 elutions was seen for animals grown in the absence of OmeY. Band densitometry revealed that in the absence of OmeY, the total protein detected was only 3.5% of that purified in the presence of 4 mM OmeY. (B) Fluorometric analysis of JFFluc proteins. The JFFluc protein purified from strain LWA718, which has JFFluc gene with a TAG codon at position 158, showed a fluorescence emission peak at 526 nm. The JFFluc protein purified from the control stain LWA1580 expressing the wild-type JFFluc gene (without the TAG158 codon) showed no detectable fluorescence. The same amount of protein from each strain was used for measurement. For comparison, the fluorescence emission spectra of the free Uaa DanAla and DanAla-Ala dipeptide were also measured, both of which had an emission peak at 538 nm. The dansyl fluorophore is sensitive to the environment, and the emission maximum changes after being incorporated into a protein. (C) Inactivation of NMD increased Uaa incorporation efficiency. LWA717 animals grown with 4 mM OmeY expressed more JFFluc protein when smg-1 was knocked down by RNAi (left lane), in comparison to the control RNAi against GFP (right lane). JFFluc protein was immunoprecipitated and detected by an anti-FLAG antibody in the Western. The mean fold change determined by band density when comparing smg-1 treated animals to the GFP control was 5.6, n=2.
To incorporate DanAla into JFFluc, the JFFluc amber reporter line LWA1582 was crossed to LWA1856 (a line expressing the integrated 3x and DanRS) to generate LWA718 (wlsi1581 I, wlSi17 II, wlls1565 X). The LWA718 worms were grown on NG media supplemented with the DanAla-Ala dipeptide. The JFFluc protein was purified with anti-FLAG beads and subjected to fluorometry. Fluorescence emission with a peak at 526 nm, characteristic of DanAla, was observed (Figure 8B). For comparison, the free Uaa DanAla and DanAla-Ala dipeptide showed an emission peak at 538 nm. Because the fluorescence of DanAla is sensitive to the environment (28), the emission maximum shift of the JFFluc sample suggests that DanAla was incorporated into the JFFluc protein. In control worms, in which no TAG codon was introduced into the JFFluc gene and DanAla and DanAla-Ala were additionally spiked to the worm lysate, no fluorescence was detected from the JFFluc protein. The detection of DanAla fluorescence in the JFFluc protein purified from LWA718 worms support that DanAla was specifically incorporated into this protein in response to the amber codon.
Inactivation of nonsense-mediated mRNA decay increases Uaa incorporation efficiency
Nonsense-mediated mRNA decay (NMD) is a eukaryotic surveillance mechanism that degrades mRNAs with premature stop codons (33). Because an amber stop codon is used to encode Uaas, NMD can result in a decreased lifetime for the target mRNA and thus a lower yield of the Uaa-incorporating protein. Previously, we demonstrated that inactivation of NMD in yeast increases Uaa incorporation efficiency (3). To test if NMD has a similar effect on Uaa incorporation in C. elegans, we knocked down the NMD component smg-1 in C. elegans using RNA interference (RNAi) and measured Uaa incorporation into the JJFluc reporter using Western blotting after immunoprecipitation. More protein was purified from samples in which smg-1 had been knocked down, compared to control samples in which RNAi is targeting GFP (Figure 8C). Two independent experiments showed similar results, with a mean fold increase of 5.6 fold measured by band densitometry. Greiss et al. reported that an NMD deficient mutant smg-2(e2008) increased GFP fluorescence than wild type worms (18). However, the TAG codon was introduced outside of the GFP gene, so GFP fluorescence does not require the TAG codon to be suppressed and the TAG codon was actually not suppressed in the experiment. Therefore, the observed GFP fluorescence increase does not reflect NMD effect on amber suppression and Uaa incorporation, but rather increased stability of UAG-containing mRNA as reported before (33). In contrast, by placing the TAG codon within the gene and suppressing it with the orthogonal tRNA/RS pair, our Western results here directly showed that disabling NMD increased Uaa incorporation in worms.
Conclusions
We demonstrate the successful expansion of the genetic code in a metazoan with components derived from two different E. coli tRNA/RS pairs. Uaa incorporation is nonlethal to the transgenic worms and shows dosage, temporal, and temperature dependence. The mCherry fluorescence and luciferase activity we observed in C. elegans were Uaa-dependent with dosage and on-and-off switch effects, indicating that reporter protein expression is dependent on the incorporation of the Uaa. The different tRNA/RS pairs, Uaas and target proteins we employed suggest that our approach can be generally applied to various Uaas and proteins. The tyrosyl and leucyl tRNA/RS pairs have been evolved to incorporate a large number of Uaas and widely used in E. coli, yeast and mammalian cells (9, 22); our demonstration of their compatibility in C. elegans should enable the genetic encoding of these reported Uaas in worms. Strikingly, important concerns when expressing Uaa-containing proteins in C. elegans have been uncovered in this research, including the transgenesis techniques used to express the target protein and the methods used to deliver the Uaa to the cells of interest.
A common challenge for incorporating Uaas in eukaryotes is the expression of a functional orthogonal tRNA, which often lacks the intragenic promoter elements conserved in eukaryotic tRNAs necessary for transcription by Pol III. We previously developed a general method to express orthogonal tRNAs by using a post-transcriptionally cleavable Pol III promoter in yeast (3) and type-3 Pol III promoters in mammalian and stem cells (5, 7). Here we demonstrate that this strategy can be extended to C. elegans. The rpr-1 promoter efficiently drives the expression of E. coli and for functional translation in worms. The rpr-1 promoter is a homolog of the H1 promoter in human cells that we used to express orthogonal tRNAs in mammalian cells (5). Both promoters are type-3 Pol III promoters for expressing the RNA component of RNaseP, which is involved in tRNA processing and required for all cells at various developmental stages. Therefore, use of the rpr-1 promoter for orthogonal tRNA expression should be generally compatible with Uaa incorporation in different tissues and stages in C. elegans. Recently Greiss et al. demonstrated the use of another Pol III promoter, the cen74-1 promoter, to express the M. mazei tRNAPyl in worms (18).
There are several methods used to introduce transgenes in C. elegans. We have discovered that a target gene containing a UAG codon in C. elegans is most reliably expressed from a chromosomally integrated low copy number construct. Both microinjection and microparticle bombardment can result in the formation of extrachromosomal arrays containing multiple copies of the transgene for high-level expression. When a UAG-containing gene is expressed in an extrachromosomal array, background UAG readthrough is detectable in the absence of an amber suppressor tRNA. Low levels of stop codon readthrough have been reported in C. elegans (16) and are confirmed by our observation of red fluorescence in worms injected with only the mCherry amber reporter. Nonspecific UAG readthrough is possibly due to insertions or deletions near the UAG and/or context codons, which can occur during the recombination that forms the extrachromosomal array (19, 34). In contrast, single-copy integration of the same mCherry reporter gene into a chromosome showed absolutely no background red fluorescence. Consistently, single integration of the luciferase amber reporter into a chromosome showed virtually no luciferase activity. Therefore, a chromosomally integrated, low copy, sequence confirmed UAG-containing reporter gene is important to maintain high fidelity when reporting amber suppression and Uaa incorporation.
A critical component of effective and experimentally relevant Uaa incorporation is the generation of animals with a high level of expression and a minimum of genetic mosaicism. The rate of generating transgenic worms in Greiss's report is very low (1–5 per hundreds of worms), and even within an established line maintained with antibiotics, only 5% of the animals containing the transgenes express the reporter. The low success rate in generating animals harboring introduced genes in combination with the high level of mosaicism due to array transmission would make it nearly impossible to identify a strain expressing an exogenous gene that does not have a sensitive and straightforward readout such as fluorescence. In contrast, our integrated single copy reporter had zero false-positive signals. When an orthogonal tRNA/RS was injected into the reporter line, we obtained strains that incorporate Uaas with a success rate of 25–50%. To achieve the genetic uniformity of our animals, we integrated all of the genetic components into chromosomes and bred the worms to homozygosity, enabling the transmission rate of our transgenes to be 100%. Each of these transgenic worms showed Uaa-dependent responses at reproducible levels in the tissue targeted. Our high transmission and incorporation rate should facilitate the generation of transgenic worms to incorporate Uaa into different target genes.
Low-copy integration of amber-containing target genes is desirable for the generation of Uaa-containing target proteins to investigate biological processes in C. elegans. Nonspecific amber readthrough in extrachromosomal arrays can result in natural amino acids at the amber site in the target protein, and this lack of fidelity when incorporating Uaas will impair rigorous study of the target protein. In addition, array expression results in inconsistency both between animals of the same genotype and within a single animal, which can make results difficult to interpret. By integrating the extrachromosomal array expressing the orthogonal tRNA/RS pair, we generated a population of genetically identical animals, and abrogated the need to constantly select for transgenic animals. A stable transgenic worm capable of Uaa incorporation should facilitate analysis wherein mosaic transgenic worms can impose complications. Moreover, the ability to detectably suppress the amber codon in a single-copy gene suggests a fairly high efficiency for our Uaa incorporation machinery. Therefore, we may be able to recapitulate natural levels of gene expression, rather than overexpression, when generating Uaa-containing proteins. This close-to-native condition will help obtain physiologically relevant results.
A multicellular organism such as C. elegans imposes more challenges on the bioavailability of Uaa than single cells. DanAla, a Uaa deviating from natural amino acids in structure, was sequestered in the intestine when unmodified. Supplying DanAla in dipeptide form enables DanAla to traverse the intestine and reach muscle cells, suggesting that dipeptide transporters tolerate structural deviations of substrates more than amino acid transporters. Because DanAla is so drastically different from natural amino acids, successful uptake in dipeptide form suggests that other Uaas can be delivered into C. elegans using this strategy. As dipeptide transporters are present on most of C. elegans cells, this method has the potential to deliver Uaas to different tissues.
The development of a multicellular animal involves many complex interactions between proteins. Genetically encoded Uaas will afford new chemistries to enable finer investigation and manipulation of protein functions directly in living C. elegans. Our strategies should be generally applicable for the incorporation of various Uaas in worms, and should be valuable in guiding the genetic code expansion of other multicellular organisms.
Methods
C. elegans Strains
EG4322, EG5003, and EG6247 for MosSCI were provided by E. Jorgensen. RB811(glo-4 (ok623)) was provided by Y. Jin. N2 (Bristol) was obtained from the Caenorhabditis Genetics Center.
Unnatural Amino Acids and Dipeptides
O-methyl-L-tyrosine was purchased from Chem-Impex International. DanAla was synthesized as described previously (28).
Dipeptide Ala-DanAla was synthesized with two methods. In method 1 as shown in Scheme S1A, compound 4 was synthesized from Boc-Dap-OH and dansylchloride via 2 steps according to the known procedure. After deprotection of the Boc group, compound 5 was coupled with Boc-Ala-OH to give compound 6. Debenzylation and deprotection of the Boc group furnished the Ala-DanAla dipeptide. In method 2 (Scheme S1B), Ala-DanAla was prepared by condensation of Boc-Ala-OSu and DanAla in aqueous bicarbonate (dioxane:water 2:1, pH 8.5). The dipeptide was purified via flash chromatography and deprotected with TFA:CH2Cl2 1:1. Solvents were evaporated and the final product re-precipitated from MeOH/Et2O. The overall yield was about 40%. Ala-DanAla synthesized by both methods behaved the same in C. elegans experiments.
Dipeptide DanAla-Ala was synthesized using standard peptide synthesis methods in solution (Scheme S1C). The precursor Boc-Dap(Fmoc)-Ala-OtBu was prepared by condensation of Boc-Dap(Fmoc)-OH and H-Ala-OH using EDC/HOBt/NMM in DCM, and purified via flash chromatography. The removal of the Fmoc group and the Dansylation were performed in one pot by treatment with 1 equiv. of DBU in DCM followed by the addition of an excess (2.5 equiv.) of dansyl chloride (35). After flash chromatographic purification, the protecting groups were removed in a TFA solution (50% in CH2Cl2), the volatiles evaporated and the final product re-precipitated from MeOH/Et2O. The overall yield was about 70%.
Microinjection, Extrachromosomal Array Formation and Integration
Microinjection of plasmid DNA was performed as previously described (36). Construction of strains LWA1560, LWA1850, LWA1008, LWA1009 and LWA1855, LWA1582 was performed as described (20). Candidate lines were bred to homozygosity, tested by PCR, and sequenced to confirm insertions.
The unc-54p∷RS constructs were injected as follows: unc-54p∷LeuRS 15 ng/μL, unc-54p∷DanRS 20 ng/μL, unc-54p∷TyrRS 20 ng/μL, unc-54p∷OmeRS 15 ng/μL. The injection mix also contained 5 ng/μL myo-2:GFP (pPD118.33 from Fire Lab Vector Kit) and 75 ng/μL pBSK2+ to facilitate extrachromosomal array formation. To integrate extrachromosomal arrays containing orthogonal tRNA/RS genes into chromosome, animals harboring each array from injection above were collected. Using γ-irradiation from a Co-60 source, 50 young adult hermaphrodites were exposed to 4100 rads on an unseeded plate. After a recovery period, the Po generation animals were picked to single plates. Isolation of F1 and F2 animals with high myo-2∷GFP expression levels led to the identification of animals propagating progeny with 100% co-injection marker expression. The integrated arrays were outcrossed four times to remove any unwanted mutations and compared to the original extrachromosomal arrays in terms of expression pattern and suppression efficiency.
Culture and Uaa Feeding
General culture and manipulation were performed as previously described (37). Unless indicated, strains were grown at 15 °C. Animals were grown in liquid culture in the absence of antibiotics (38) to facilitate Uaa uptake. Gravid worms harboring the array were placed in sterile-filtered S basal with cholesterol and incubated in the presence of Uaa or dipeptide with agitation for five days; progeny containing the array were then scored. Small samples were performed in a 24-well tissue culture dish in 0.85 mL total volume, which can support 10–15 gravid adults for one generation. Negative controls used the same procedure but equivalent amounts of NaOH or HCl as in the Uaa were added to the medium to ensure identical pH. For solid culture, NG medium was supplemented with Uaa immediately before pouring. Plates were seeded with OP50 according to standard techniques (39).
Luciferase Assays
Animals were grown on Uaa-containing plates in duplicate for 1 or 3 generations. Plates were incubated at 15 °C until food is exhausted. Animals were washed off of plates with M9 buffer, spun down, and washed twice with M9 and twice with chilled water. After pelleting the worms, 450 μL 1× CCLR lysis buffer (Promega) was added. Animals were lysed in a Precellys24 with ceramic and glass beads for 3 cycles, 10 sec at 6,500 rpm with a 45 sec pause. Samples were spun at 10,000 rpm for 5 min at 4°C. Lysate (80 μL) was placed in a 96-well Nunclon Surface dish. After 100 μL of luciferase assay reagent (Promega) was added to each well, samples were read on a Tecan plate reader using 20 sec integration time. All samples were assayed in duplicate. Luminescence reading was normalized to protein concentration as determined by absorption at 280 nm.
Immunoprecipitation and Western Blotting
LWA717 animals were grown on solid media until food was exhausted and were collected with M9 buffer. Thirty-five plates for each condition were used. Animals were flash frozen, then ground into a powder with a mortar and pestle on dry ice, and resuspended in 400 μL cell lysis buffer (Cell Signaling) supplemented with protease inhibitors (Roche, Sigma). The lysate was clarified by centrifugation at 16,000 g for 30 minutes at 4 °C and pre-incubated with protein A/G PLUS agarose beads (Santa Cruz). FLAG tagged protein was immunoprecipitated with EZview Red anti-FLAG M2 affinity gel (Sigma) for 2 h at 4 °C, and then beads were washed extensively. Samples were eluted with 1 mg/mL 3xFLAG peptide (Sigma) and run on an 8% SDS-PAGE gel. After transfer to a PVDF membrane, the sample was blocked overnight in 5% milk in TBS-T, then incubated with anti-FLAG M2-peroxidase (Sigma), washed for 30 minutes and visualized with SuperSignal West Chemiluminescent Substrate (Thermo Scientific). Quantification of Western blot bands was performed using the gel analyzer tool in the Image J software (NIH).
RNAi feeding
RNAi feeding was performed as described (40) on solid plates supplemented with IPTG, carbenicillin, and 4 mM OmeY, allowing animals to grow as in Uaa feeding experiments until food was exhausted at 15 °C. Approximately equal numbers of animals were used for each experiment. smg-1 RNAi plasmid is from the Ahringer library (41, 42).
Microscopy
All fluorescent images are maximum Z-stack projections. Imaging of live worms was performed on 2% agarose pads and 10 mM NaN3 as an anesthetic using an Olympus IX81 equipped with a spinning disk and a Hamamatsu EM-CCD (Model C9100-02) camera. The light source was a xenon lamp from Sutter Instruments. For DanAla, excitation filter was 360/60 nm and emission filter was 535/40 nm; For mCherry, excitation filter was 580/20 nm and emission filter was 675/130 nm.
Fluorometry
Worms were raised on solid NG media containing 3 mM DanAla-Ala dipeptide. After the worms were grounded into fine powder, an additional 3 mM of DanAla-Ala and 3 mM DanAla were spiked into the control sample LWA1580. The FLAG-tagged JFFluc proteins were then immunoprecipitated as described above and eluted with the 3X FLAG peptide. The elutions were pooled together and the total protein was concentrated by acetone precipitation. The protein samples were re-suspended in dimethyl sulfoxide, and the fluorescence emission spectrum was measured on a Fluorolog 3 (Horiba Jobin Yvon) using the same amount of protein for each sample. The excitation wavelength was 350 nm, the slits were 5 nm, and the emission was recorded from 450 nm to 650 nm at 1 nm increments. For comparison, the emission spectra were also recorded for DanAla and DanAla-Ala dipeptide dissolved in dimethyl sulfoxide.
Details for plasmid construction and mass spectrometric analyses are included in Supporting Methods.
Supplementary Material
Acknowledgments
We thank C. Frokjaer-Jensen, E. Jorgensen, Y. Jin for strains and reagents and V. Lacey for editing. This work was supported by the Ray Thomas Edwards Foundation, March of Dimes Foundation (#5-FY08-110) and National Institutes of Health (1DP2OD004744) to L.W.
References
- 1.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 2.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J. Am. Chem. Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- 4.Sakamoto K, Hayashi A, Sakamoto A, Kiga D, Nakayama H, Soma A, Kobayashi T, Kitabatake M, Takio K, Saito K, Shirouzu M, Hirao I, Yokoyama S. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee KF, Slesinger PA, Wang L. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat. Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 7.Shen B, Xiang Z, Miller B, Louie G, Wang W, Noel JP, Gage FH, Wang L. Genetically encoding unnatural amino acids in neural stem cells and optically reporting voltage-sensitive domain changes in differentiated neurons. Stem Cells. 2011;29:1231–1240. doi: 10.1002/stem.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Parrish AR, Wang L. Expanding the genetic code for biological studies. Chem. Biol. 2009;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 10.Jin Y. Synaptogenesis. In: The C. elegans Research Community, editor. WormBook. 2005. doi/10.1895/wormbook.1891.1844.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Vol. 33. Cold Spring Harbor Laboratory Press; New York: 1997. [PubMed] [Google Scholar]
- 12.C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 13.Antoshechkin I, Sternberg PW. The versatile worm: genetic and genomic resources for Caenorhabditis elegans research. Nat. Rev. Genet. 2007;8:518–532. doi: 10.1038/nrg2105. [DOI] [PubMed] [Google Scholar]
- 14.Meyer BJ. Targeting X chromosomes for repression. Curr. Opin. Genet. Dev. 2010;20:179–189. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 16.Hodgkin J. Novel Nematode Amber Suppressors. Genetics. 1985;111:287–310. doi: 10.1093/genetics/111.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waterston R. A second informational suppressor, SUP-7 X, in Caenorhabditis elegans. Genetics. 1981;97:307–325. doi: 10.1093/genetics/97.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greiss S, Chin JW. Expanding the Genetic Code of an Animal. J. Am. Chem. Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolten SL, Powell-Abel P, Fischhoff DA, Waterston RH. The sup-7(st5) X gene of Caenorhabditis elegans encodes a tRNATrpUAG amber suppressor. Proc. Natl. Acad. Sci. U.S.A. 1984;81:6784–6788. doi: 10.1073/pnas.81.21.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Schultz PG. Expanding the genetic code. Angew. Chem. Int. Ed. Engl. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Schultz PG, Brock A. An improved system for the generation and analysis of mutant proteins containing unnatural amino acids in Saccharomyces cerevisiae. J. Mol. Biol. 2007;371:112–122. doi: 10.1016/j.jmb.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Deng W, Zhu X, Skogerbø G, Zhao Y, Fu Z, Wang Y, He H, Cai L, Sun H, Liu C, Li B, Bai B, Wang J, Jia D, Sun S, He H, Cui Y, Wang Y, Bu D, Chen R. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome Res. 2005;16:20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takimoto JK, Xiang Z, Kang JY, Wang L. Esterification of an unnatural amino acid structurally deviating from canonical amino acids promotes its uptake and incorporation into proteins in mammalian cells. Chembiochem. 11:2268–2272. doi: 10.1002/cbic.201000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takimoto JK, Dellas N, Noel JP, Wang L. Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem. Biol. 2011;6:733–743. doi: 10.1021/cb200057a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takimoto JK, Adams KL, Xiang Z, Wang L. Improving orthogonal tRNA-synthetase recognition for efficient unnatural amino acid incorporation and application in mammalian cells. Mol. Biosyst. 2009;5:931–934. doi: 10.1039/b904228h. [DOI] [PubMed] [Google Scholar]
- 28.Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. A genetically encoded fluorescent amino acid. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, Grant BD, Priess JR. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fei YJ, Fujita T, Lapp DF, Ganapathy V, Leibach FH. Two oligopeptide transporters from Caenorhabditis elegans: molecular cloning and functional expression. Biochem J. 1998;332(Pt 2):565–572. doi: 10.1042/bj3320565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coin I, Perrin MH, Vale WW, Wang L. Photo-cross-linkers incorporated into G-protein-coupled receptors in mammalian cells: a ligand comparison. Angew. Chem. Int. Ed. Engl. 2011;50:8077–8081. doi: 10.1002/anie.201102646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richmond MH. The Effect of Amino Acid Analogues on Growth and Protein Synthesis in Microorganisms. Bacteriol. Rev. 1962;26:398–420. doi: 10.1128/br.26.4.398-420.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 34.Stinchcomb DT, Shaw JE, Carr SH, Hirsh D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol Cell Biol. 1985;5:3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coin I, Beerbaum M, Schmieder P, Bienert M, Beyermann M. Solid-phase synthesis of a cyclodepsipeptide: cotransin. Org Lett. 2008;10:3857–3860. doi: 10.1021/ol800855p. [DOI] [PubMed] [Google Scholar]
- 36.Kimble J, Hodgkin J, Smith T, Smith J. Suppression of an amber mutation by microinjection of suppressor tRNA in C. elegans. Nature. 1982;299:456–458. doi: 10.1038/299456a0. [DOI] [PubMed] [Google Scholar]
- 37.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 39.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahringer J. Reverse genetics. In: The C. elegans Research Community, editor. WormBook. 2006. doi/10.1895/wormbook.1891.1847.1891. [Google Scholar]
- 41.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 42.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.