Summary
The target range of a bacterial secretion system can be defined by effector substrate specificity or by the efficacy of effector delivery. Here, we report the crystal structure of Tse1, a type VI secretion (T6S) bacteriolytic amidase effector from Pseudomonas aeruginosa. Consistent with its role as a toxin, Tse1 has a more accessible active site than related housekeeping enzymes. The activity of Tse1 against isolated peptidoglycan shows its capacity to act broadly against Gram-negative bacteria, and even certain Gram-positive species. Studies with intact cells indicate that some Gram-positive bacteria remain vulnerable to Tse1 despite cell wall modifications. However, interbacterial competition studies demonstrate that Tse1-dependent lysis is restricted to Gram-negative targets. We propose that the previously observed specificity for T6S against Gram-negative bacteria is a consequence of high local effector concentration achieved by T6S-dependent targeting to its site of action, rather than inherent effector substrate specificity.
Introduction
Type VI secretions systems (T6SSs) provide bacteria a means of delivering protein effectors to both eukaryotic and prokaryotic cells (Jani and Cotter, 2010; Schwarz et al., 2010a). Like other complex bacterial export pathways, including type III and IV secretion, protein transfer via T6S requires likely involves direct translocation (Ma et al., 2009; Russell et al., 2011). Unlike these pathways, however, the T6SS displays likeness to bacteriophage (Cascales and Cambillau, 2012; Kanamaru, 2009). Structural, genetic and biochemical data support a model in which the T6SS punctures recipient cell membranes using a contractile phage tail-like apparatus (Basler et al., 2012; Russell et al., 2011).
The Pseudomonas aeruginosa Hcp Secretion Island I-encoded T6SS (H1-T6SS) targets at least three effector proteins to recipient bacterial cells (Hood et al., 2010). When delivered, these effector proteins, termed type VI secretion exported 1–3 (Tse1-3), act in the cytoplasm to induce stasis (Tse2) or in the periplasm to promote lysis by hydrolyzing peptidoglycan (Tse1 and Tse3) (Russell et al., 2011). P. aeruginosa employs cognate periplasmic immunity proteins, T6S immunity 1 and 3 (Tsi1 and Tsi3), to protect itself against intercellular self targeting of Tse1 and Tse3, respectively. The Tse proteins were shown to contribute significantly to the fitness of P. aeruginosa in competition against a close relative of the organism, P. putida, under cell contact-promoting conditions (Russell et al., 2011).
Tse1 is a peptidoglycan amidase with specificity toward the γ-D-glutamyl-meso-2,6-diaminopimelic acid (D-Glu–mDAP) bond and Tse3 is a muramidase that cleaves the β-1,4 linkage between N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc) (Russell et al., 2011). Tse1 belongs to a diverse superfamily of type VI amidase effector (Tae) proteins that consist of members broadly distributed amongst the proteobacteria (Russell et al., 2012). The Tae proteins segregate into four highly divergent families based on sequence homology; Tse1 is a member of Family 1. Reflective of their sequence divergence, representatives of the Tae families display varying cleavage specificities within the peptide stems of peptidoglycan (Russell et al., 2012).
Functionally, the T6SS appears to be quite plastic. Systems ostensibly devoted to interactions with bacterial or host cells have been defined, and one system involved in both has been reported (MacIntyre et al., 2010; Schwarz et al., 2010a; Schwarz et al., 2010b). Specificity within these organisms is largely unexplored, and, where specificity has been observed, its mechanistic basis is not known. For example, Burkholderia thailandensis T6SS-1 and the Vibrio cholerae Vas system appear to specifically target Gram-negative bacteria. Such specificity could arise in numerous ways, including regulatory mechanisms, the requirement for specific receptor(s) on recipient cells, the phylogenetic distribution of immunity proteins, susceptibility to effector activity, or a combination of these factors. Here, we determined the structure of Tse1 from P. aeruginosa. Our structural, genetic and biochemical analyses of Tse1 provide a molecular basis for the specificity of this effector against Gram-negative bacteria and establish a general framework for defining the target range of the T6SS.
Results
Crystal structure of Tse1
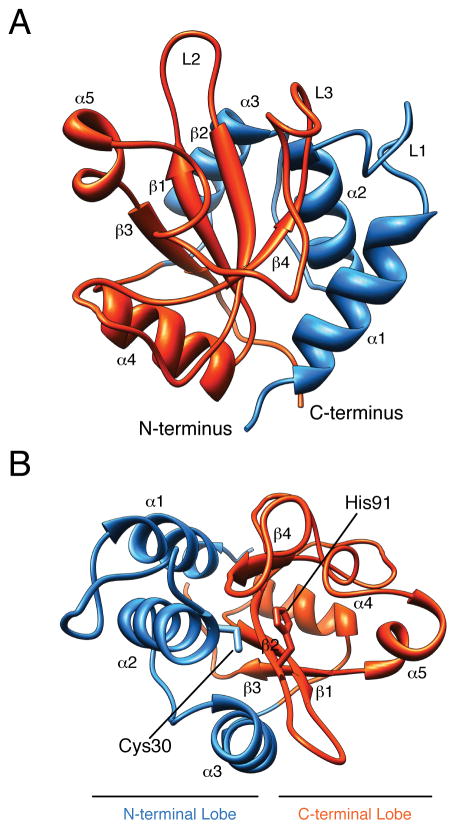
We recently discovered that Tse1 is an amidase-type T6S effector that degrades cell wall peptidoglycan of neighboring bacteria. To understand the structural basis for Tse1 activity and to define its relation to other amidases, we solved its X-ray crystal structure to a resolution of 2.6 Å (Figure 1A, Table S1, Figure S1A). Our final model contained 146 of the 154 residues present in the native protein. Although the asymmetric unit contained four Tse1 molecules, we observed a molecular mass in solution consistent with a monomeric species (Figure S1B and S1C). Given this finding and the high overall similarity between the Tse1 monomers (Cα r.m.s.d. = 0.21–0.36), we limit our discussion to one of the molecules (Chain A)
Figure 1. Overall structure of Tse1.
(A) Ribbon diagram showing the overall structure of Tse1. Secondary elements referred to in the text are labeled. (B) Top-down view of the Tse1 active site. The active site cleft is formed by the N- and C-terminal lobes, with catalytic residues Cys30 and His91 positioned at the N-terminal ends of α2 and β2. See also Figure S1.
Tse1 adopts the classical α + β papain-like amidase fold, comprised of an antiparallel β-sheet surrounded by α-helices (Figure 1A–B, Figure S1D). The N-terminal lobe, consisting of α1–3, folds together with the C-terminal lobe, comprising β1–4 and α4–5, to form a V-shaped active site cleft (Dubey et al., 2007). The active site is composed of conserved secondary structure elements α2 and β2. Similar to other amidases, the reactive cysteine of Tse1, Cys30, is positioned at the N-terminal end of α2, and the catalytic histidine, His91, resides at the N-terminal end of β2 (Figure 1B).
The active site of Tse1 is highly accessible
To prevent the toxic consequences of inappropriate cell wall degradation, peptidoglycan hydrolase activity in the cell is tightly regulated. Accordingly, these enzymes are often members of larger complexes or contain structural features implicated in controlling their activity (Typas et al., 2012). For instance, structural studies of peptidoglycan papain amidases have invariably revealed either a closed active site architecture or occlusion of the active site by non-catalytic regulatory domains (Figure 2A). In contrast, Tse1 has an exposed active site with highly accessible catalytic residues. Superposition of Tse1 with its closest structural homologs identified using the DALI server, the NlpC/P60-family amidases B. subtilis YkfC (Z score = 9.7, Cα r.m.s.d. = 3.9) and E. coli Spr (Z score = 9.4, Cα r.m.s.d. = 2.8), reveals that differences in the loops surrounding the conserved catalytic center account for its open active site architecture (Figure 2B) (Anantharaman and Aravind, 2003; Aramini et al., 2008; Russell et al., 2011; Xu et al., 2010). On the N-terminal lobe, Loop 1 extends outward from the catalytic cysteine by approximately 10.0 Å relative to its position in YkfC and Spr (Figure 2B). On the C-terminal lobe, Loop 2 extends parallel to β1 and 2, thereby lengthening the substrate-binding cleft. In Spr and YkfC, the curvature of this loop causes it to abut α5, forming a surface that truncates the cleft. Our observation that Tse1 lacks repressive structural features typically associated with these enzymes is consistent with its role as a cell wall-degrading toxin.
Figure 2. Tse1 has an open active site relative to housekeeping amidase enzymes.
(A) Comparison of bacterial amidase structures. Surface representation of B. subtilis YkfC (PDB ID 3H41), M. tuberculosis RipB (PDB ID 3PBI), E. coli Spr (PDB ID 2K1G) in the same orientation as Tse1 highlights regulatory structural element types not found in the toxin. Regulatory structural elements and catalytic cysteine residues are shown in red and yellow, respectively. YkfC has an additional domain adjacent to its active site and RipB contains an N-terminal extension that occludes its substrate binding cleft (Both et al., 2011; Xu et al., 2010). In Spr, residues proximal to the active site limit accessibility to catalytic residues (Aramini et al., 2008). (B) Superposition of the catalytic center of Tse1 with YkfC and Spr. Homologs were identified using the DALI server (Cα r.m.s.d. YkfC, 3.9 Å,127 residues; Spr, 2.8 Å, 129 residues).
Conserved surfaces coordinate substrate binding by Tse1
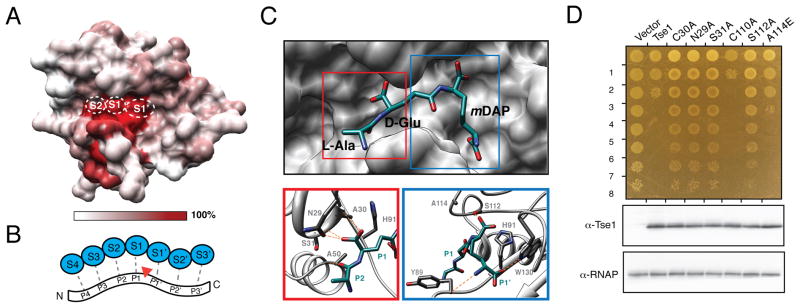
In an effort to understand the molecular basis for Tse1 function, we investigated the residues important for peptidoglycan recognition using a combination of conservation mapping and ligand docking studies. To overcome the high degree of sequence diversity within Tae effector Family 1, we threaded family members onto the Tse1 structure to generate an accurate alignment (Figure S2A) (Kelley and Sternberg, 2009). This analysis revealed considerable surface variation within Family 1 Tae effectors, with the notable exception of a conserved patch encompassing residues within the active site cleft and a wide, adjacent perpendicular groove (Figure 3A). To explore the significance of sequence conservation within these regions, we aligned the Tse1 structure to other papain-like amidases with finely mapped substrate interaction sites (Yao et al., 2009). While we recognize the peptide stems of peptidoglycan differ in many respects from typical peptide protease substrates, for consistency we will use the subsite (S) and substrate position (P) nomenclature of Schechter and Berger in our description of the Tse1 structure (Figure 3B) (Schechter and Berger, 1967). Conservation of active site architecture between Tse1 and papain proteases allowed us to confidently map S2, S1 and S1′ sites onto the Tse1 structure (Figure 3A). Interestingly, our structural alignment shows that the residues defining each of these sites constituting the Tse1 catalytic center are perfectly conserved within Family 1 effectors (Figure S2A).
Figure 3. Conserved Family 1 active site residues mediate substrate recognition in Tse1.
(A) Surface representation of Tse1 with color corresponding to amino acid conservation with Tae Family 1 effectors. The position of subsites S2, S1, and S1′ within the Tse1 structure are shown.
(B) Schematic representation of canonical subsites that mediate papain protease–substrate interactions. Enzyme subsites (blue, S4–S3′) interact with positions along the peptide ligand (P4–P3′). P-sites are assigned according to their positions relative to the site of cleavage (red arrow).
(C) Molecular docking of a peptidoglycan fragment into the Tse1 active site identifies putative substrate recognition sites. Docked L-Ala-D-Glu-mDAP (cyan) is shown with Tse1 (gray, upper panel). Tse1 residues mediating L-Ala-D-Glu-mDAP contacts are labeled (lower panels), and lines representing hydrogen bonds (orange) are shown. See also Figure S2.
(D) Growth of E. coli expressing periplasmic tse1 (peri-Tse1) harboring indicated mutations (top panel). Numbers at left represent ten-fold serial dilutions. Western blot analysis of peri-Tse1 expression in corresponding cells is shown below.
To expand on our identification of evolutionarily conserved substrate interaction sites, we attempted to determine the structure of a Tse1 catalytic mutant (C30A) in the presence of its minimal peptide ligand, L-alanyl-γ-D-glutamyl-meso-diaminopimelic acid (L-Ala-D-Glu-mDAP), which includes the preferred cleavage site of Tse1 (Table S1). Although we observed additional electron density near the active site S1′ position not present in the apo structure, this density was insufficient for determining the overall placement of the ligand. We therefore turned to molecular modeling and employed Glide to dock the L-Ala-D-Glu-mDAP molecule into the refined Tse1 catalytic mutant structure. In the top scoring pose (XPscore = −11.46 kcal/mol), the scissile bond of L-Ala-D-GlumDAP lies between the catalytic residues Cys30 and His91 (Figure 3C).
The docked ligand fragment is engaged in an extensive hydrogen bond network with the Tse1 active site cleft. N-terminal to the scissile bond of the substrate, Ala30 and Ser31 each have backbone hydrogen bond contacts to the peptide at P1 (Figure 3C). Asn29 contacts the peptide via a side-chain hydrogen bond at P2 (D-Glu), while Ala50 forms a backbone hydrogen bond with L-Ala. On the C-terminal side of the scissile bond, the carboxylate of the D-stereocenter of mDAP (P1′) engages in hydrogen bonding contacts to the side chain of Ser112 (Figure 3C). Also, Tyr89 accepts a hydrogen bond from the P1′ amide and Trp130 donates a hydrogen bond to the P1′ hydroxyl at the C-terminal end of L-Ala-D-Glu-mDAP (Figure 3C). Several other residues within the active site, including Leu84, Ala114, and Val129, also mediate hydrophobic contacts with the peptide (Figure S2B).
To test the importance of these modeled peptidoglycan interactions for Tse1 activity, we mutated a subset of the contacting residues and measured effects on periplasmic Tse1 toxicity in E. coli (Figure 3D). In line with our predictions, conservative substitutions within residues engaged in hydrogen bonds with the ligand (N29A, S31A, S112A) completely abolished Tse1 activity. A substitution disrupting a predicted hydrophobic interaction (A114E) had a weaker effect on toxicity. In contrast, a substitution in a residue near the active site – at a position not predicted to make substrate contacts – did not inhibit Tse1 toxicity (C110A). Importantly, none of the mutations influenced overall Tse1 levels. Together with our conservation analyses, these substrate modeling and mutational studies of the Tse1 active site define the residues that likely mediate key interactions with peptidoglycan.
Tse1 requires a P3′ residue for cleavage
Our earlier work demonstrated that Tse1 preferentially cleaves the donor peptide stem of tetrapeptide-tetrapeptide crosslinks in peptidoglycan (Russell et al., 2011). Although both tetrapeptide donor and acceptor stems contain the preferred cleavage site of Tse1, the configuration of the P1′ and P2′ residues, mDAP and D-Ala, respectively, differ between the two. In the acceptor, but not the donor stem peptide, mDAP participates at its D-stereocenter in the crosslink and thus is at the vertex of a branched structure to two D-Ala residues. Likewise, at the P2′ position, the asymmetric nature of the crosslink results in either a D-Ala bonded to mDAP (donor) or a free C-terminal D-Ala (acceptor).
To gain further insights into the specificity of Tse1 and to determine how the enzyme distinguishes between donor and acceptor stems, we studied its activity in vitro against mutant E. coli sacculi enriched in pentapeptide stems (Meberg et al., 2001). Interestingly, Tse1 readily cleaved pentapeptide-tetrapeptide crosslinked peptidoglycan in both the donor and acceptor stems (Figure 4A and S3A–B). The enzyme also processed uncrosslinked pentapeptides, and consistent with our earlier findings, it did not efficiently degrade free uncrosslinked tetrapeptides (Russell et al., 2011). Tse1 retained specificity for the D-Glu–mDAP bond in each of these contexts. These data indicate that peptidoglycan cleavage by Tse1 is insensitive to P1′ (mDAP) participation in a branched structure. Rather, the data strongly suggest that the critical determinant for Tse1 cleavage at the D-Glu–mDAP bond is the presence of a P3′ residue. This residue can either be mDAP in the donor stem of tetrapeptide-tetrapeptide and pentapeptide-tetrapeptide crosslinks or D-Ala (position 5) in uncrosslinked pentapeptide stems and the acceptor stem of pentapeptide-tetrapeptide crosslinks. The cleft in the S1′ region of the Tse1 active site is wide and bifurcates into two channels, thus helping to explain how Tse1 could accommodate a branch in peptidoglycan at this position. However, given the conformational flexibility of the substrate, it is not currently possible to determine which of these channels would hold the crosslink versus the terminal D-Ala residues.
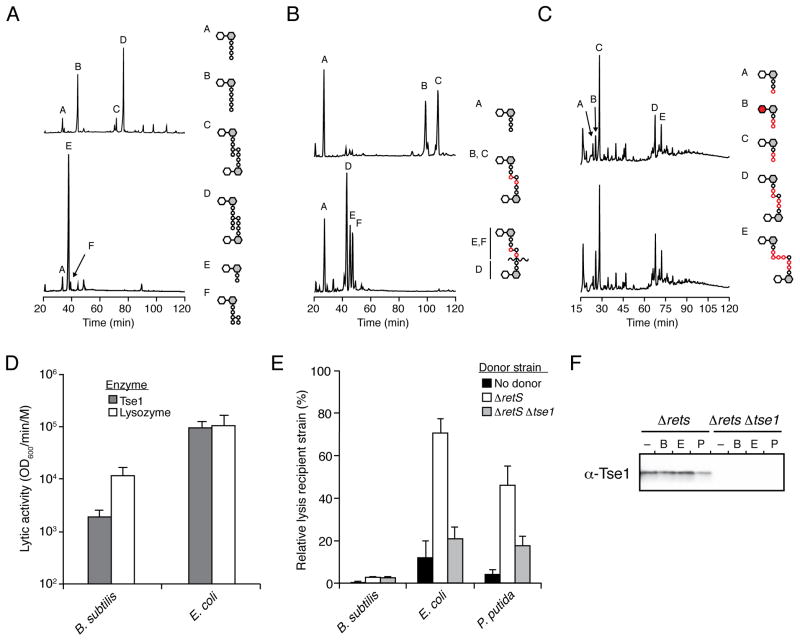
Figure 4. Tse1 is specific for Glu-mDAP-type peptidoglycan.
(A–C) Tse1 degrades only Glu-mDAP-type peptidoglycan of E. coli and B. subtilis and not the Lys-type peptidoglycan of S. pneumoniae. Partial HPLC chromatograms of sodium borohydride-reduced soluble E. coli (A), B. subtilis (B) or S. pneumoniae(C) peptidoglycan sacculi products resulting from digestion with cellosyl (top) or with Tse1 followed by cellosyl (bottom). Assignments of Tse1 cleavage products were made based on MS analysis (Panel A, Peaks E and F; Figure S3A) or prior work (Bui et al., 2012; Russell et al., 2011). Structures corresponding to the major peaks are shown schematically with hexagons and circles representing sugars and amino acid residues, respectively. Reduced sugar moieties are shown with grey fill. Modifications or substitutions relative to typical Gram-negative peptidoglycan are indicated with red fill.
(D) Tse1 can lyse B. subtilis cells in vitro. Activity of purified Tse1 and lysozyme against B. subtilis and EDTA-permeabilized E. coli. Error bars +/− s.d. n=3. See also Figure S3C.
(E) Tse1 delivered by the H1-T6SS does not contribute to lysis of B. subtilis. Relative lysis as measured by supernatant LacZ activity of the indicated recipient organisms in growth competition under T6SS-promoting conditions with the indicated P. aeruginosa strains. Error bars +/− s.d. n=3.
(F) Tse1 is secreted by Δrets P. aeruginosa. Western blot analysis of total extracellular Tse1 levels in competition assays against B. subtilis, E. coli, and P. putida.
Tse1 can cleave Gram-positive peptidoglycan
The surprising finding that Tse1 is promiscuous with regard to the involvement of the P1′ residue (mDAP) in a crosslink led us to hypothesize that the enzyme might possess activity against other types of peptidoglycan. To probe more broadly the activity of Tse1 against peptidoglycan, we incubated the purified enzyme with sacculi prepared from two Gram-positive organisms, Streptococcus pneumoniae and Bacillus subtilis. B. subtilis peptidoglycan is similar to that of Gram-negative bacteria, however notable differences exist. The mDAP residues of B. subtilis bear an amide modification and its uncrosslinked and acceptor peptide stems are most often tripeptides generated through the action of a LD-carboxypeptidase (Vollmer, 2012; Vollmer et al., 2008). Peptidoglycan of S. pneumoniae is more typical of Gram-positive bacteria; it contains an amidated D-Glu residue at position two and L-Lys instead of mDap at position 3. Both direct crosslinks and crosslinks with a Ser-Ala or Ala-Ala interpeptide bridge occur simultaneously in this organism (Bui et al., 2012; Severin and Tomasz, 1996).
Analysis of B. subtilis peptidoglycan cleavage products using HPLC and MS showed that Tse1cleaves both of the major crosslinked species found in this organism – mono- and di-amidated tripeptide–tetrapeptide – at the expected position between D-Glu and mDAP (Figure 4B). The enzyme did not efficiently cleave uncrosslinked tripeptide stems present in the sample. Under similar conditions, Tse1 displayed no detectable activity against S. pneumoniae peptidoglycan (Figure 4C). These data suggest that Tse1 exhibits a strong preference for mDAP-type, directly crosslinked peptidoglycan.
T6S-exported Tse1 is not effective against B. subtilis
Studies of the activity of T6SSs against different bacteria have yielded evidence that the secretion pathway is specifically active against Gram-negative bacteria (MacIntyre et al., 2010; Murdoch et al., 2011; Schwarz et al., 2010b). Our in vitro evidence that Tse1 is active against the mDAP-type peptidoglycan of Gram-positive bacteria provided an opportunity to define the basis of this apparent cell targeting selectivity. Since peptidoglycan modifications, including wall techoic acids, are lost during preparation of sacculi, we first probed whether Tse1 could act on intact B. subtilis cells (Bui et al., 2012). For reference, we compared the activity of Tse1 against B. subtilis and permeabilized E. coli with that of the indiscriminant muramidase lysozyme (Nakimbugwe et al., 2006). Quantitative studies showed that lysozyme and Tse1 display similar lytic activity against E. coli, while a Tse1 catalytic mutant (C30A) showed no appreciable activity(Figure 4D and Figure S3C). The rate of lysis of B. subtilis by Tse1 was lower than that of lysozyme, however the enzyme retained significant activity against these cells.
The capacity of Tse1 to lyse B. subtilis cells left open the possibility that the enzyme – exported via the T6SS – could contribute to the fitness of P. aeruginosa in competition against this organism. A common feature of T6SSs is that they are tightly repressed under in vitro culturing conditions (Bernard et al., 2010; Hood et al., 2010; Silverman et al., 2011). The systems can become activated in the presence of competing bacteria, by physical cues, or by activation through natural or engineered mutations. For example, a deletion of retS in P. aeruginosa renders the H1-T6SS highly expressed and constitutively active (Goodman et al., 2004; Laskowski et al., 2004). Therefore, to eliminate repression as a confounding factor in our effort to understand the fundamental determinant of cell targeting specificity by the T6SS, we employed the ΔretS genetic background of P. aeruginosa in these studies (Goodman et al., 2004; Laskowski et al., 2004). Key properties of this strain are that it indiscriminately releases Tse effectors into the milieu at levels greater than are observed in natural isolates (data not shown), yet it retains that capacity to translocate effector proteins when in contact with Gram-negative cells (Hood et al., 2010; Russell et al., 2011). As a sensitive measure of the impact of Tse1 activity, we quantified lysis of recipient cells by release of the cytoplasmic LacZ enzyme during interbacterial competition assays. Our results showed that P. aeruginosa ΔretS causes significant lysis of E. coli and that deletion of tse1 restores levels to nearly those of the controls, suggesting that the majority of this effect is attributable to the Tse1 enzyme (Figure 4E). A similar effect was observed with P. putida, an organism previously described as a target of the H1-T6SS (Russell et al., 2011). However, we found that Tse1 does not contribute to lysis of B. subtilis by P. aeruginosa.
To further probe the differential activity of Tse1 against B. subtilis and the Gram-negative organisms, we quantified extracellular Tse1 levels during interbacterial competitions. Using band densitometry relative to purified recombinant Tse1, we calculated an average of 3 ng of extracellular Tse1 in our competition assays (Figure 4F). While it is difficult to approximate the extracellular volume available to Tse1 in a colony of bacteria, the effective Tse1 concentration likely falls below the 0.01 μM required to observe significant B. subtilis lysis in our in vitro assays. Taken together with our in vitro and cell-based assays, which show that Tse1 can act enzymatically against B. subtilis cell wall peptidoglycan, the most parsimonious explanation for the lack of lysis of B. subtilis cells in bacterial competition assays is that physiological levels of Tse1 do not reach effective concentrations in this setting. We posit that a fundamental barrier to T6S targeting of bacteriolytic effectors – one that supersedes enzymatic specificity – is the failure of the toxins to reach an effective concentration unless targeted appropriately by the T6S apparatus.
Discussion
Our structural, biochemical, and functional dissection of the Tse1 effector protein from P. aeruginosa offers key insights into several facets of T6S. Tse1 belongs to Family 1 of the T6S amidase effector (Tae) superfamily, a superfamily with representatives distributed widely throughout the γ and β proteobacteria (Russell et al., 2012). Like other known enzyme superfamilies, the Tae group comprises dozens of enzymes that share a common catalytic motif and strategy, although with extensive divergence in sequence and substrate specificity (Khersonsky and Tawfik, 2010; Russell et al., 2012). Despite the sequence divergence of Tae effectors, our structure shows that Tse1 shares a conserved amidase fold, albeit with considerable modifications. In contrast to its housekeeping counterparts, Tse1 is a naked enzyme with a highly accessible active site. This observation suggests that it may be constitutively active and thus highly potent in vivo. Consistent with this, we observed that E. coli undergoes rapid lysis upon induction of periplasmic-targeted Tse1, and in our current study we found Tse1 activity accounts for the majority of the bacteriolytic effects of the H1-T6SS (Russell et al., 2011).
Sequence divergence within the Tae superfamily provides insights into the molecular basis of overlapping and unique substrate preferences between its constituent families. Mapping of Tae conservation on the Tse1 structure revealed a highly conserved core with considerable surface variability outside of the catalytic center. This finding highlights the evolvability of T6S effectors, even under the constraints of enzymatic function and T6SS export requirements. Given our finding that the corresponding T6S amidase immunity superfamily displays less sequence conservation than their cognate effector proteins, the extent of Tae surface diversity could allow for variable immunity interactions (Russell et al., 2012).
Delineating all substrates exported by a secretion system is essential to understanding the full spectrum of effects it can mediate. However, determinants for export through the T6SS remain obscure. Previous studies have focused on identifying substrates either by proteomics or through informatics-based searches that rely on sequence or functional homology (Hood et al., 2010; Russell et al., 2012). While these approaches have been successful, defining the precise determinants of T6S export is required to generate an unbiased approach to substrate identification. The three-dimensional structure of a T6S effector provides a first step towards this end. Using the structure of Tse1, we were able to generate a confident alignment of Family 1 effectors. Interestingly, we did not observe highly conserved features outside of the active sites of these enzymes, suggesting that a simplistic sequence or structure-based export signal is unlikely. Structural comparison of different Tse effectors (Tse1-3) within the same organism in combination with a more precise understanding of the secretion mechanism will likely be important for the dissection of T6S export determinants.
The path that T6S effector proteins transit en route to recipient cells has not been delineated. Although there is no direct evidence, effectors have been postulated to exit the T6SS through a pore formed most proximally by an abundant ring-shaped hexameric component of the apparatus termed hemolysin co-regulated protein (Hcp) (Basler et al., 2012; Cascales, 2008; Mougous et al., 2006). If effectors do pass through the pore of Hcp, the structures of Hcp1 and Tse1 from P. aeruginosa offer glimpses into how this process might proceed. These structures show that the inner diameter of the Hcp pore is approximately 40 Å, while Tse1 adopts a globular shape with a diameter of approximately 32 Å at its narrowest point and 47 Å at its widest. Of note, extended loops of Tse1, and not core structural regions, define its widest dimension. Conformational flexibility within these regions or partial unfolding may therefore facilitate accommodation of the native protein within the Hcp1 pore. Tse1 is considerably smaller than Tse3 (44.4 kDa), another substrate of the H1-T6SS that is translocated to recipient cell periplasm. The region of this protein responsible for its muramidase activity is strongly predicted to adopt a G-type lysozyme fold – a domain slightly larger than Tse1 (>20 kDa) (Russell et al., 2011). Therefore, if transit through Hcp is a general property of T6S effectors, Tse3 would also likely require significant rearrangements to its native structure.
Our structure and in vitro analyses of Tse1 have also lent insight into the previous observation that T6S targets Gram-negative and not Gram-positive organisms (MacIntyre et al., 2010; Schwarz et al., 2010b). Our data show that not only is the enzyme more broadly adapted to Gram-negative than Gram-positive peptidoglycan, but that even in an instance wherein the effector protein is biochemically competent for an attack on a Gram-positive cell, it fails to elicit significant effects. It remains possible that Gram-positive adapted effectors of the T6SS await discovery. However, our data suggest that the fundamental barrier to targeting of known T6S bacteriolytic effectors to Gram-positive cells is their inability to reach an effective concentration without directed localization to their peptidoglycan substrate by the secretory apparatus. Together these observations strongly support the model that selectivity of the T6SS is not encoded in enzyme activity per se, but rather through proper delivery of effectors to their appropriate targets.
Materials and methods
Bacterial strains and growth conditions
P. aeruginosa, B. subtilis, and P. putida strains used in this study were derived from the strains PAO1, JH642, and KT2440, respectively (Nelson et al., 2002; Srivatsan et al., 2008; Stover et al., 2000). Details are included in Extended Experimental Procedures.
Tse1 structure determination and analyses
Tse1 crystals were obtained by sitting drop vapor diffusion at 25° C from a 1:1 mixture of 10 mg/mL protein with 0.2 M sodium thiocyanate, 0.1 M HEPES pH 7.5, 20% PEG3350 for 3 days. Crystals were cryopreserved in 20% glycerol and diffraction data were collected at the Lawrence Berkeley National Laboratory Advanced Light Source Beamline 8.3.1 (MacDowell et al., 2004). Phases were obtained experimentally with data from a selenomethionine-substituted crystal using the PHENIX software suite. Coot (Emsley and Cowtan, 2004) and maximum likelihood refinement with PHENIX (Adams et al., 2010) were used for iterative building and refinement. Structure was validated by MOLProbity (Davis et al., 2004). Coordinates and structure factors were deposited in the Protein Data Bank (PDB ID 4EOB and 4F4M). Structural homologs identified using the DALI server (Holm and Rosenstrom, 2010) were visualized with Chimera (Pettersen et al., 2004). Structural models of Family 1 homologs were built using the PHYRE One to One threading algorithm (Kelley and Sternberg, 2009) and aligned with Chimera.
Ligand docking
The refined structure of Tse1 was optimized in Maestro (Schrödinger) by the Protein Preparation Wizard (Schrodinger, 2010). Ligand conformers were flexibly docked by Glide 5.6 (Friesner et al., 2004; Friesner et al., 2006; Halgren et al., 2004) with the OPLS2005 force field. Details are included in Extended Experimental Procedures.
In vitro peptidoglycan assays
Purified peptidoglycan sacculi from E. coli MC1061 or CS703-1 (300 μg) (Meberg et al., 2001), B. subtilis 168 (300 μg) or from S. pneumoniae R6 (120 μg) were incubated with Tse1 (1 μM)in 300 μl of 20 mM Tris/HCl, pH 8.0 for 4 h at 37 °C. Muropeptides were prepared as detailed in Extended Experimental Procedures and analyzed by HPLC and MS as described previously (Bui et al., 2009; Glauner, 1988).
Whole cell lysis assays
Overnight cultures of were grown to late-logarithmic phase. E. coli K12 and B. subtilis JH642 cells grown to an OD600 of 1.0 in Luria-Bertani (LB) media at 27°C were harvested by centrifugation, resuspended in permeabilization buffer (4μM EDTA, 0.5M sucrose, 0.2% v/v Tween-20, and 40mM Tris-HCl pH 8.0), and three concentrations of either Tse1 or lysozyme were added. Lysis rate was defined as the buffer subtracted-average OD600 decrease of cells. Details are included in Extended Experimental Procedures.
Competitive lysis assays
Overnight cultures of β-galactosidase recipient strains and donor P. aeruginosa strains were washed, mixed 1:1, and spotted on a nitrocellulose membrane placed on 3% agar LB low-salt media for 4 hr at 37°C (B. subtilis and E. coli) or 30°C (P. putida). Cells were suspended in phosphate buffered saline. Cellular and extracellular LacZ fractions were harvested as described previously without TCA precipitation (Mougous et al., 2006). Cellular LacZ was released by permeablization with 16.8% v/v chloroform. LacZ activity in fractions was determined using the Tropix Glacto-Light kit (Applied Biosystems).
Supplementary Material
Highlights.
Crystal structure of Tse1 reveals an open active site
Conserved type VI Family 1 amidase effector residues define Tse1 specificity
Tse1 displays activity against Gram (−) and (+) mDAP-type peptidolgycan
The type VI secretion system does not effectively target Tse1 to Gram (+) cells
Acknowledgments
We wish to thank Joe Gray of the Pinnacle Laboratory of Newcastle University for MS analysis, Dr. Houra Merrikh for providing B. subtilis, members of the Mougous laboratory and Dr. Christoph Grundner for helpful discussions, Dr. Corie Ralston at the Advanced Light Source and Dr. Tom Alber for assistance with X-ray data collection, and Dr. Randy Read and Dr. Nathaniel Echols for help with data processing. Research was supported by grants to J.D.M. from the National Institutes of Health (AI080609) and to W.V. from the European Commission within the DIVINOCELL programme. A.B.R. was supported by a grant from the National Science Foundation (DGE-0718124), S.S. was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) and J.D.M. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome biology. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramini JM, Rossi P, Huang YJ, Zhao L, Jiang M, Maglaqui M, Xiao R, Locke J, Nair R, Rost B, et al. Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli: structural evidence for a novel cysteine peptidase catalytic triad. Biochemistry. 2008;47:9715–9717. doi: 10.1021/bi8010779. [DOI] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard CS, Brunet YR, Gueguen E, Cascales E. Nooks and Crannies in type VI secretion regulation. J Bacteriol. 2010;192:3850–3860. doi: 10.1128/JB.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both D, Schneider G, Schnell R. Peptidoglycan remodeling in Mycobacterium tuberculosis: comparison of structures and catalytic activities of RipA and RipB. J Mol Biol. 2011;413:247–260. doi: 10.1016/j.jmb.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Bui NK, Eberhardt A, Vollmer D, Kern T, Bougault C, Tomasz A, Simorre JP, Vollmer W. Isolation and analysis of cell wall components from Streptococcus pneumoniae. Anal Biochem. 2012;421:657–666. doi: 10.1016/j.ab.2011.11.026. [DOI] [PubMed] [Google Scholar]
- Bui NK, Gray J, Schwarz H, Schumann P, Blanot D, Vollmer W. The peptidoglycan sacculus of Myxococcus xanthus has unusual structural features and is degraded during glycerol-induced myxospore development. J Bacteriol. 2009;191:494–505. doi: 10.1128/JB.00608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E. The type VI secretion toolkit. EMBO reports. 2008;9:735–741. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Cambillau C. Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci. 2012 doi: 10.1098/rstb.2011.0209. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic acids research. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey VK, Pande M, Singh BK, Jagannadham MV. Papain-like proteases: Applications of their inhibitors. Afr J Biotechnol. 2007;6:1077–1086. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of medicinal chemistry. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of medicinal chemistry. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. Journal of medicinal chemistry. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic acids research. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell host & microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell host & microbe. 2010;8:2–6. doi: 10.1016/j.chom.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru S. Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc Natl Acad Sci U S A. 2009;106:4067–4068. doi: 10.1073/pnas.0901205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annual review of biochemistry. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- Laskowski MA, Osborn E, Kazmierczak BI. A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol Microbiol. 2004;54:1090–1103. doi: 10.1111/j.1365-2958.2004.04331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell host & microbe. 2009;5:234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDowell AA, Celestre RS, Howells M, McKinney W, Krupnick J, Cambie D, Domning EE, Duarte RM, Kelez N, Plate DW, et al. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J Synchrotron Radiat. 2004;11:447–455. doi: 10.1107/S0909049504024835. [DOI] [PubMed] [Google Scholar]
- MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg BM, Sailer FC, Nelson DE, Young KD. Reconstruction of Escherichia coli mrcA (PBP 1a) mutants lacking multiple combinations of penicillin binding proteins. J Bacteriol. 2001;183:6148–6149. doi: 10.1128/JB.183.20.6148-6149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. The opportunistic pathogen Serratia marcescens utilises Type VI Secretion to target bacterial competitors. J Bacteriol. 2011 doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakimbugwe D, Masschalck B, Atanassova M, Zewdie-Bosuner A, Michiels CW. Comparison of bactericidal activity of six lysozymes at atmospheric pressure and under high hydrostatic pressure. Int J Food Microbiol. 2006;108:355–363. doi: 10.1016/j.ijfoodmicro.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VA, Fouts DE, Gill SR, Pop M, Holmes M, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environmental microbiology. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DA, Schwarz S, Goodlett DR, Vollmer W, et al. A widespread type VI secretion effector superfamily identified using a heuristic approach. Cell host & microbe. 2012 doi: 10.1016/j.chom.2012.04.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1 2010 [Google Scholar]
- Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010a;18:531–537. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010b;6 doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin A, Tomasz A. Naturally occurring peptidoglycan variants of Streptococcus pneumoniae. J Bacteriol. 1996;178:168–174. doi: 10.1128/jb.178.1.168-174.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol. 2011;82:1277–1290. doi: 10.1111/j.1365-2958.2011.07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Han Y, Peng J, Tehranchi AK, Gibbs R, Wang JD, Chen R. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS genetics. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W. Bacterial outer membrane evolution via sporulation? Nat Chem Biol. 2012;8:14–18. doi: 10.1038/nchembio.748. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Xu Q, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Cai X, Carlton D, Chen C, Chiu HJ, Chiu M, et al. Structure of the gamma-D-glutamyl-L-diamino acid endopeptidase YkfC from Bacillus cereus in complex with L-Ala-gamma-D-Glu: insights into substrate recognition by NlpC/P60 cysteine peptidases. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1354–1364. doi: 10.1107/S1744309110021214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Cui J, Zhu Y, Wang G, Hu L, Long C, Cao R, Liu X, Huang N, Chen S, et al. A bacterial type III effector family uses the papain-like hydrolytic activity to arrest the host cell cycle. Proc Natl Acad Sci U S A. 2009;106:3716–3721. doi: 10.1073/pnas.0900212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.