Summary
Many neural lineages display a temporal pattern, but the mechanisms controlling the ordered production of neuronal subtypes remains unclear. Here, we show that Drosophila let-7 and miR-125, co-transcribed from the let-7-Complex (let-7-C) locus, regulate the transcription factor chinmo to control temporal cell fate in the mushroom body (MB) lineage. We find that let-7-C is activated in post-mitotic neurons born during the larval-to-pupal transition, when transitions between three MB subtypes occur. Loss or increase of let-7-C delays or accelerates these transitions, respectively, and lead to cell fate transformations. Consistent with our identification of let-7 and miR-125 sites in a recently identified ~6kb extension of the chinmo 3‘UTR, Chinmo is elevated in let-7-C mutant MBs. In addition, we show that let-7-C acts upstream of chinmo and let-7-C phenotypes are caused by elevated chinmo. Thus, these heterochronic miRNAs, originally identified in C. elegans, underlie progenitor cell multipotency during the development of diverse bilateria.
Keywords: let-7-C, let-7, miR-100, miR-125, chinmo, mushroom body, temporal identity, heterochronic
Introduction
Although the ordered production of neuronal subtypes is a pervasive feature of animal nervous system development, the underlying molecular mechanism of this phenomenon is not well understood (Jacob et al., 2008; Pearson and Doe, 2004). In Drosophila, for example, the post-transcriptional regulation of the BTB-zinc finger chronologically inappropriate morphogenesis (chinmo) appears to drive the sequential generation of mushroom body (MB) neurons (Zhu et al., 2006). A center for olfactory learning and memory, the MB is comprised of four subtypes that are born in an invariant order (γ → α’/β’ → pioneer α/β → α/β) by multipotent progenitor cells during development (Lee et al., 1999; Zhu et al., 2003; Zhu et al., 2006). These progenitors, or neuroblasts (Nbs), undergo self-renewing cell divisions to generate a ganglion mother cell (GMC), which divides once more to produce neurons that will differentiate into one of the MB subtypes. High Chinmo levels in post-mitotic neurons specify early born cell fates (γ, α’/β’) whereas low levels specify late born cell fates (pioneer α/β, α/β). The downregulation of Chinmo in progressively later-born neurons thereby determines the birth order of MB subtypes and occurs at the post-transcriptional level (Zhu et al., 2006). The mechanism responsible for generating this temporal gradient of Chinmo during development remains unknown, although it may involve the chinmo 5’ untranslated region (UTR) since this region controls Chinmo levels in the adult brain (Zhu et al., 2006).
Mechanisms controlling the ordered production of cells during C. elegans development have been identified by forward genetic analysis, and involve post-transcriptional regulation by microRNAs (miRNAs) (reviewed in Ambros, 2011). These miRNAs include members of the let-7 and miR-125/lin-4 families, which promote temporal cell fate transitions during C. elegans development by binding to sequences in the 3’UTRs of target mRNAs and repressing their expression (Lee et al., 1993; Reinhart et al., 2000). Deletion of these miRNAs leads to heterochronic phenotypes in which cell fates are inappropriately reiterated during development. The sequences and temporal expression patterns of heterochronic miRNAs are broadly conserved among diverse bilateria (Pasquinelli et al., 2000), suggesting a conserved function. However, there is no evidence in any system other than C. elegans that these miRNAs promote one stage-specific differentiation program over another. Thus, despite their conservation, it remains unclear whether heterochronic genes represent a fundamental pathway that controls temporal cell fate during animal development.
Here, we provide evidence that the switch-like function of let-7 and miR-125/lin-4 in C. elegans has been adapted to the formation of a temporal cell fate gradient of Chinmo in Drosophila. In Drosophila, let-7 and miR-125 are co-transcribed from a single locus, known as the let-7-Complex (let-7-C), along with a third ancient miRNA, miR-100 (Sokol et al., 2008). These let-7-C miRNAs are not detected early in development, but their upregulation during the larval-to-pupal transition coincides with the downregulation of Chinmo and is partially dependent on transcriptional activation by the Ecdysone Receptor (Chawla and Sokol, 2012). let-7-C mutants appear morphologically normal but display behavioral defects, indicating a role for these miRNAs during nervous system development (Caygill and Johnston, 2008; Sokol et al., 2008). The spatiotemporal expression profile of let-7-C miRNAs along with their known function in C. elegans suggest that these miRNAs may play a role in the temporal identity of MB neurons.
Results
let-7-C is expressed in MB neurons born during the L3-to-pupal transition
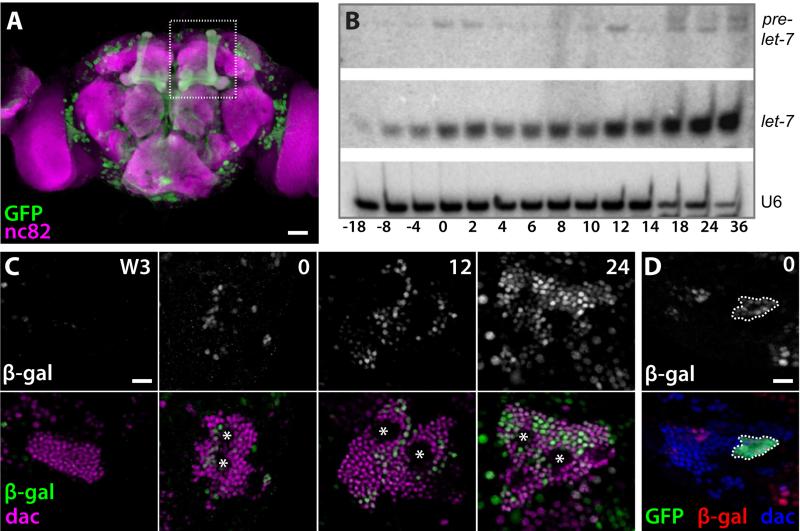
Expression of a let-7-C::Gal4 transcriptional reporter was detected in the adult MBs (Figure 1A), suggesting a potential role for let-7-C miRNAs during MB development. To correlate let-7-C expression with the sequential production of γ, α’/β’, pioneer α/β (p. α/β), and α/β neurons, we analyzed the temporal onset of let-7-C miRNAs during central nervous system (CNS) development. Levels of let-7 (Figure 1B) as well as miR-100 and miR-125 (Figure S1A, B) gradually increased in total CNS tissue during the larval-to-pupal transition, the time when α’/β’ → p. α/β and p. α/β → α/β transitions occur (Zhu et al., 2003; Zhu et al., 2006). To determine the timing of let-7-C onset specifically in MB neurons, we analyzed two additional transcriptional reporters, let-7-Cp12.5kb::lacZ and let-7-CΔ3miR::optGal4 (Chawla and Sokol, 2012, Figure S1C), within the CNS. These were used instead of let-7-C::Gal4, which displayed a ~21-hr developmental lag in CNS reporter expression relative to let-7-C miRNA northern blots (Y.W. and N.S., unpublished observations). let-7-Cp12.5kb::lacZ was detected in increasing numbers of MB neurons in 0-, 12-, and 24-h pupae (Figure 1C), correlating with the temporal gradient of let-7-C miRNAs detected by northern blot (Figure 1B, Figure S1A,B). In addition, let-7-Cp12.5kb::lacZ expression co-localized with the MB marker Dachshund (Dac), which labels post-mitotic MB neurons that are at least 8-10 hours old (Martini et al., 2000), but was conspicuously absent from regions containing MB Nbs and GMCs (Figure 1C, asterisks). Although let-7-Cp12.5kb::lacZ was not consistently detected in MB neurons from wandering third instar larvae (L3), it was expressed in almost all Dac-positive MB neurons in 24-h pupae (Figure 1C). Expression of the let-7-CΔ3miR::optGal4 reporter was similar though slightly delayed relative to let-7-Cp12.5kb::lacZ (Figure S1D-F), presumably reflecting a Gal4/UAS system lag. To identify the age of the neurons that first expressed let-7-Cp12.5kb::lacZ in white prepupae, we used the mosaic analysis with repressible marker (MARCM) technique (Lee and Luo, 1999) to analyze let-7-Cp12.5kb::lacZ expression in MB neurons born during the late L3 stage. let-7-Cp12.5kb::lacZ was detected in clones of recently born neurons in 0-h white prepupae (Figure 1D). Taken together, these data indicated that let-7-C is activated in a few MB neurons born in late larvae, and its expression expands during early metamorphosis to include almost all post-mitotic MB neurons.
Figure 1. Temporal onset of let-7-C in MB neurons.
(A) let-7-CGKI crossed to UAS-mCD8::GFP (green) is detected in the MBs (indicated by dotted box) of adult brains counterstained with neuropil marker nc82 (magenta). (B) An increasing gradient of let-7 miRNA is expressed in CNS tissue at the larval-to-pupal transition, along with miR-100 and miR-125 (see Figure S1). Total RNA was extracted from CNS tissues dissected from staged animals, and probed for U6 snRNA as loading control. (C) let-7-Cp12.5kb::lacZ (white in top panels, green in bottom panels) is not detected in wandering L3 MBs stained for Dac (magenta, bottom panels) but is detected in increasing numbers of MB neurons in white prepupae (0), 12 h pupae (12), and 24 h pupae (24). Asterisks mark the location of Nbs and GMCs. (D) let-7-Cp12.5kb::lacZ (white in top panel, red in bottom panel) is detected in a white prepupae MB clone expressing GFP (green, outlined) and Dac (blue) that was induced ~18 hours earlier. Scale bars: A, 25 μm; C-D, 10 μm. See also Figure S1.
let-7-C has dosage-dependent effects on MB transitions
The spatial and temporal onset of let-7-Cp12.5kb::lacZ suggested that let-7-C miRNAs may promote the α’/β’ → p. α/β and p. α/β → α/β transitions in MB neuron identities. To test this, we used the MARCM technique to label individual neurons born in staged wildtype and let-7-C mutant animals (Figure 2A, Figure S2A-D). Consistent with previous reports (Zhu et al., 2003; Zhu et al., 2006), our fine scale temporal mapping indicated that the α’/β’ → p. α/β and p. α/β → α/β transitions began -18 to -8 hours and -4 to 0 hours before puparium formation in wildtype animals, respectively (Figure 2A, Figure S2E). In contrast, let-7-C mutants displayed delays in both of these transitions. For example, 100% of wildtype neurons born during the 6-h period after puparium formation displayed α/β characteristics whereas 29% of let-7-C mutant born during this same period displayed α’/β’ characteristics and another 59% displayed p. α/β characteristics (Figure 2A). Since these results suggested that removal of let-7-C miRNAs resulted in delayed transitions, we reasoned that an increase in let-7-C dosage might accelerate these transitions. To test this hypothesis, MARCM analysis was repeated in animals that carried two extra copies of the let-7-C locus and contained elevated levels of let-7 (Figure S2J). These 4Xlet-7-C animals displayed accelerated transitions: only 5% of neurons born ~4 hours before puparium formation of wildtype larvae displayed late-type α/β characteristics, whereas 38% of neurons born during the same timeframe did in 4Xlet-7-C larvae (Figure 2B). These data indicate that let-7-C has dosage sensitive effects on the temporal transitions between MB identities.
Figure 2. MB temporal identity is sensitive to let-7-C dosage.
(A) Percentages of different subtypes of MB neurons (y-axis) among single and two-cell clones that were induced in wildtype and let-7-C mutant animals at different times relative to puparium formation (x-axis). (B) Percentages of different subtypes of MB neurons (y-axis) among single and two-cell clones that were induced in wildtype and 4Xlet-7-C larvae at different times relative to puparium formation (x-axis). (C) Average number of c305a-Gal4 positive α’/β’ neurons in wildtype (2X) or let-7-CKO1 and let-7-CKO2 transheterozgyotes (0X). P = 0.0283. (D) Average number of c305a-Gal4 positive α’/β’ neurons in wildtype (2X) and 4Xlet-7-C (4X) adults. P < 0.0001. (E) Average number of c708a-Gal4 positive p. α/β neurons in wildtype (2X), let-7-CKO1 and let-7-CKO2 transheterozgyotes (0X), let-7-CKO2 heterozygotes harboring a let-7-C rescuing transgene (2X-R), and wildtype harboring a let-7-C rescuing transgene (3X). P < 0.0001 comparing either 2X and 0X or 2X and 3X. Figures in parentheses represent the total number of clones (A and B) or brains (C, D, E) analyzed for each genotype. Values are presented as mean ± SEM. (F, G) OK107-Gal4, UAS-mCD8::GFP labeled wildtype (F) and let-7-C KO2 (G) adult MB clones generated in newly hatched larvae and stained with anti-FasII antibodies. Scale bar in F is 50 μm, refers to G. See also Figure S2.
Such changes in the transitions between MB subtypes during neurogenesis might be reflected as alterations in the ratios of α’/β’, p. α/β, and α/β neurons in the adult MBs. To test this, we used previously described MB subtype-specific Gal4 reporters (Aso et al., 2009; Lin et al., 2007; Zhu et al., 2006) to quantify the number of each class of MB neuron in animals with varying doses of let-7-C. Consistent with the MARCM analysis, we found that the number of earlier born α’/β’ neurons as detected by the c305a-Gal4 driver was increased in let-7-C adults (393.8 ± 9.3, n=6) relative to wildtype adults (363.4 ± 10.9, n=5) (Figure 2C) and decreased in 4Xlet-7-C adults (303 ± 15.3, n=9) relative to wildtype adults (354 ± 5.5, n=8) (Figure 2D). Conversely, the number of later born p. α/β neurons as detected by the c708a-Gal4 driver was decreased in let-7-C adults (23.9 ± 7.4, n=31) and increased in 3Xlet-7-C adults (74.6 ± 8.4, n=24) relative to wildtype adult (57.5 ± 3.3, n=16)(Figure 2E). No evidence for changes in the size of either the γ or α/β populations were identified using the H24- and c739-Gal4 reporters, respectively (Figure S2G, H). Importantly, the total number of MB neurons generated by a single MB Nb was also not affected by removal of let-7-C (Figure S2I). Despite these penetrant temporal identity defects, the morphology of let-7-C mushroom bodies was grossly normal although we noted some p. α/β and α/β axons that inappropriately crossed the midline (Figure 2G and Figure S2N-Q). Taken together, these data indicated that manipulation of let-7-C dosage leads to cell fate transformations.
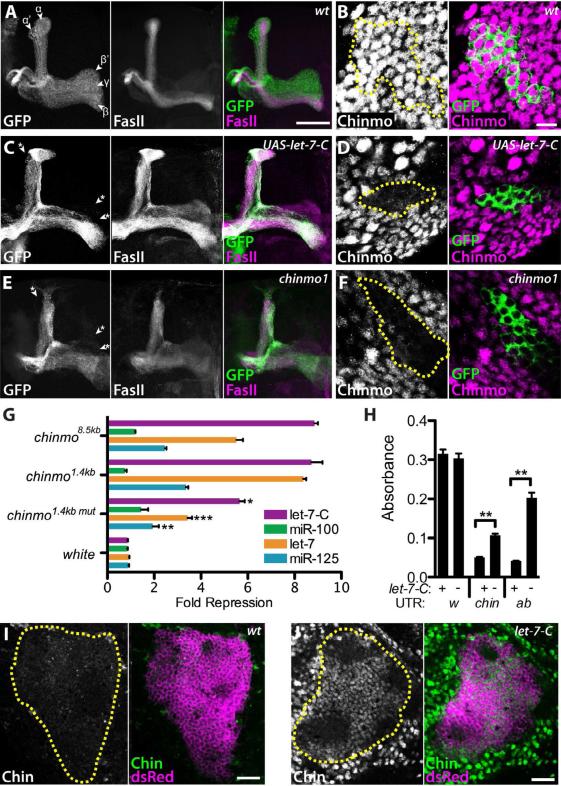
In contrast to the relatively mild morphological defects associated with let-7-C loss, forced expression of let-7-C beginning in L1 MB lineages resulted in dramatic effects on adult MB morphology (Figure 3A, C). These phenotypes included the apparent elimination of γ as well as α’/β’ lobes, indicating that let-7-C miRNAs promote later born α/β cell fates at the expense of earlier born γ and α’/β’ cell fates. This transformation appeared identical to the phenotypes resulting from loss of the temporal identity factor chinmo (compare Figures 3C and 3E; Zhu et al., 2006). To test whether Chinmo expression levels were affected by precocious expression of let-7-C, larval MB clones ectopically expressing let-7-C were stained for Chinmo protein. Whereas abundant Chinmo was detected in wildtype MB neurons at this stage (Figure 3B), Chinmo levels were depleted in MB neurons either expressing a UAS-let-7-C transgene (Figure 3D) or homozygous for the protein null chinmo1 allele (Figure 3F). These data indicated that the let-7-C temporal identity phenotypes are likely due to changes in Chinmo levels, and also suggested that chinmo might be a direct target of let-7-C miRNAs. Prior analysis found that post-transcriptional control of chinmo was mediated via its 5’UTR but not its ~2.5kb 3’UTR (Zhu et al., 2006), which is where miRNA binding sites are usually found. These results are consistent with the absence of predicted let-7-C miRNA binding sites in the annotated chinmo 3‘UTR (Brennecke et al., 2005; Grun et al., 2005; Lewis et al., 2005). Recent microarray and deep sequencing analyses, however, indicate that the full-length chinmo 3’ UTR is ~8.5kb, considerably longer than previous gene annotations suggested (Cherbas et al., 2011; Graveley et al., 2011; Smibert et al, 2012; Figure S3A). Thus, let-7-C miRNAs might regulate Chinmo expression via binding sites in the ~6kb portion of the chinmo 3‘UTR that was previously unannotated.
Figure 3. let-7-C microRNAs regulate chinmo expression.
(A, C, E) OK107-Gal4, UAS-mCD8::GFP labeled wildtype (A), UAS-let-7-C, (C), and chinmo1 (E) adult MB clones generated in newly hatched larvae and stained with anti-FasII antibodies. In wildtype, γ lobes are weakly stained with FasII whereas α’ and β’ are devoid of FasII staining. Note the absence of γ, α’ and β’ lobes in UAS-let-7-C and chinmo1 MBs (asterisks). Scale bar in A is 50 μm; refers to C, E. (B, D, F) OK107-Gal4 labeled wildtype (B), UAS-let-7-C (D), and chinmo1 mutant (F) L3 MB clones labeled with GFP generated in newly hatched larvae and stained with anti-Chinmo antibodies. Clones are outlined in yellow in the Chinmo channel. Scale bar in B is 7.5 μm; refers to D, F. (G) Fold repression of luciferase reporters containing the 8.5kb full-length chinmo 3‘UTR, a 1.4kb chinmo 3‘UTR fragment, a mutated 1.4kb 3‘UTR fragment, or the white 3‘UTR in cell cultures in which let-7-C miRNAs were ectopically expressed either together or individually. P = 0.0135, 0.0003, and 0.0049 comparing repression of chinmo1.4kb vs. chinmo1.4kbmut by let-7-C miRNAs, let-7 alone, or miR-125 alone, respectively. (H) Quantification of pigment levels from flies harboring white reporter transgenes containing the white (w), chinmo (chin) or abrupt (ab) 3‘UTR and containing either wildtype (+) or let-7-CKO1 mutant (-) eye cells. P = 0.0028 or 0.0060 comparing chinmo or abrupt 3’UTR in wildtype and let-7-CKO1 cells. Values are presented as mean ± SEM. (I, J) Wildtype (I) and let-7-C (J) adult brains harboring OK107-Gal4 and UAS-mCD8::ChRFP insertions and stained with anti-Chinmo antibodies. MBs are outlined. Scale bars: I, J, 25 μm. See also Figures S3 and S4.
Chinmo is regulated by let-7 and miR-125 via its 3‘UTR
To investigate whether chinmo was directly regulated by let-7-C miRNAs, we used a cell culture assay to test whether ectopic let-7-C miRNAs repressed expression of a luciferase reporter containing the full-length 8.5 kb chinmo 3‘UTR (Figure 3G). Ectopic expression of all three miRNAs resulted in an 8.8 fold repression of luciferase. This was predominantly due to let-7 and miR-125: ectopic let-7 alone repressed luciferase 5.5 fold and ectopic miR-125 alone repressed luciferase 2.5 fold, whereas ectopic miR-100 alone repressed luciferase only 1.1 fold. Control assays indicated that all three miRNAs were expressed in cell culture and could repress control reporters (Figure S3B-G). Consistent with this cell culture experiment, we found that precocious expression of let-7 and miR-125 but not miR-100 in L1 resulted in reduced Chinmo levels in L3 MBs and morphological defects in adult MBs (Figure S4). We therefore searched the 8.5kb chinmo 3‘UTR for putative let-7 and miR-125 binding sites using the RNAhybrid program (Rehmsmeier et al., 2004), and identified six let-7 sites and four miR-125 binding sites that are conserved between Drosophila species and have a free energy of at most -18 kcal/mol (Figure S3A). Since most of these sites clustered in a 1.4kb region of the chinmo 3‘UTR, we prepared two new luciferase reporters containing either a wildtype or mutated version of this fragment in which seed sequences of the let-7 and miR-125 sites were deleted (Figure S3A). Ectopic expression of all three let-7-C miRNAs together or either let-7 alone or miR-125 alone repressed a luciferase reporter containing the wildtype fragment 8.7-, 8.4-, and 3.4-fold, respectively (Figure 3G). In contrast, repression of the mutated version was significantly reduced, indicating that let-7 and miR-125 binding sites mediated reporter repression.
To determine whether the chinmo 3‘UTR was regulated by endogenous let-7-C miRNAs, we analyzed the effect let-7-C dosage on the expression of a reporter transgene. In this assay, flies containing the eye pigmentation gene white fused to its own 3'UTR displayed dark eye color with an absorbance of 0.315 absorbance units (Figure 3H, Figure S3H). In contrast, flies carrying the white open reading frame fused to either the chinmo 3'UTR or the known let-7 regulated abrupt 3'UTR displayed a significantly lighter eye color (Figure 3H, Figure S3J, L). Flies whose eyes were comprised exclusively of let-7-CKO1 mutant cells and that carried the same reporter transgenes displayed darker eye color (Figure 3G, Figure S3K, M). Therefore, endogenous let-7-C miRNAs are required for chinmo 3'UTR mediated regulation of white reporter expression. Having established that endogenous let-7-C miRNAs can regulate protein expression through the chinmo 3’UTR, we looked for aberrant Chinmo protein expression in let-7-C mutant tissue. Chinmo is ordinarily downregulated during metamorphosis and is virtually absent in adults (Figure 3I; Zhu et al., 2006). In contrast, Chinmo is abundantly expressed in adult let-7-C brains, including throughout the MB (Figure 3J). Elevated levels of Chinmo are first detected in 48-h let-7-C pupae (Y. W., data not shown). Collectively, these data indicate that chinmo is a direct target of let-7-C miRNAs.
let-7-C MB phenotypes are due to elevated chinmo
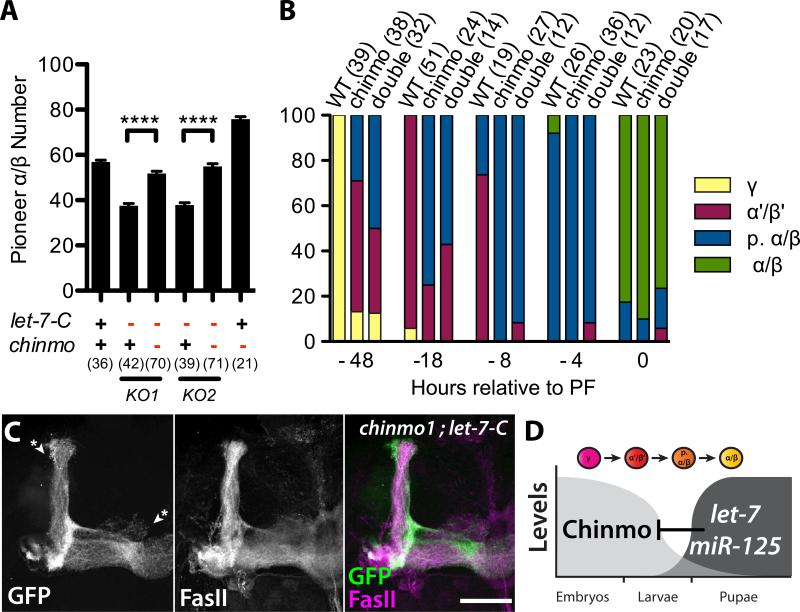
Given that the downregulation of Chinmo is required for the ordered production of MB neurons, we reasoned that let-7-C mutant MB phenotypes were due to elevated Chinmo. Consistently, phenotypes resulting from either reduced let-7-C or increased chinmo dosage were similar: adults with one copy of let-7-C or three copies of chinmo had a decrease in p. α/β number from ~58 to between 36 and 45 (Figure 4A; Zhu et al., 2006). We therefore tested whether reduction in chinmo dosage could suppress the heterozygous let-7-C MB phenotypes, using p. α/β number as our assay to analyze genetic interaction. Removal of one copy of chinmo in flies heterozygous for two different let-7-C null alleles restored p. α/β number to wildtype (Figure 4A). We also tested whether chinmo was epistatic to let-7-C by analyzing chinmo1; let-7-CKO double mutant phenotypes. MARCM of single/two-cell clones in staged animals indicated that chinmo1; let-7-CKO double mutant neurons aberrantly acquired late-type neurite trajectories (Figure 4B), a phenotype identical to that displayed by chinmo1 single mutant neurons (Figure 4B; Zhu et al., 2006), and opposite to that displayed by let-7-C single mutant neurons (compare Figure 4B with Figure 2A). Similarly, MBs generated from chinmo1; let-7-CKO double mutant Nbs appeared identical to MBs generated from chinmo1 single mutant Nbs (Figure 4C). This analysis indicated that chinmo was epistatic to let-7-C, and that the let-7-C MB temporal identity phenotypes were a consequence of chinmo misregulation.
Figure 4. chinmo suppresses let-7-C mushroom body phenotypes and is epistatic to let-7-C.
(A) Average number of c708a Gal4 positive p. α/β neurons in adults that are wildtype (+) or heterozygous (-) for chinmo1 (columns 3, 5, 6), let-7-CKO1 (columns 2, 3) or let-7-CKO2 (columns 4, 5). P < 0.0001 comparing either column 2 and 3 or column 4 and 5. Values are presented as mean ± SEM. (B) Percentages of different subtypes among single and two-cell wildtype, chinmo1 or chinmo1, let-7-CKO2 mutant clones that were induced at different times relative to puparium formation (x-axis).Figures in parentheses represent the total number of brains (A) or clones (B) analyzed for each genotype. (C) OK107-Gal4, UAS-mCD8::GFP labeled chinmo1 , let-7-CKO2 double mutant adult MB clone generated in newly hatched larvae and stained with anti-FasII antibodies. Note the absence of γ, α’ and β’ lobes (asterisks). Scale bar: C, 50 μm. (D) let-7 and miR-125 are upregulated at the larval-to-pupal transition and promote transitions in MB temporal identity by repressing chinmo.
Discussion
Drosophila let-7-C is activated throughout the developing nervous system at the larval-to-pupal transition, where it triggers cell fate transitions in MB cell lineages and likely other lineages as well. Here, we provide evidence that let-7-C miRNAs participate in the formation of a temporal gradient of Chinmo in post-mitotic MB neurons via sequences in a recently annotated extension of the chinmo 3’UTR. Post-transcriptional regulation of chinmo is complex, and involves both its 5’ as well as its 3’ UTRs. The 5’UTR regulates Chinmo expression in adult MB cells (Zhu et al., 2006), although the identity of the responsible trans-acting factors and their relationship with miRNA-mediated silencing pathways remain to be identified. Chinmo expression is also post-transcriptionally regulated in MB Nbs (Zhu et al., 2006) and may involve either the 5’ and/or 3’UTR although likely not the let-7-C miRNAs since they appear to be expressed exclusively in post-mitotic cells. The work presented here can be reconciled with a previous observation that the general disruption of miRNA processing does not affect MB temporal cell fate specification (Yu and Lee, 2007) if let-7-C miRNAs are either counteracted by additional MB miRNAs or are processed by non-canonical mechanisms in the MBs (Yang and Lai, 2011).
Chinmo is closely related to another known let-7 target, Abrupt (Caygill and Johnston, 2008). Like chinmo, abrupt appears to repress morphologies associated with adult neuronal cell fates (Li et al., 2004; Sugimura et al., 2004), suggesting that this family of BTB-ZFs may generally function as temporal cell fate determinants during nervous system formation. Whether let-7-C miRNAs regulate abrupt and/or chinmo to control temporal identity in other adult lineages in the developing fly nervous system remains to be determined.
Our finding that let-7-C miRNAs promote one neural differentiation program over another suggests that these conserved miRNAs may underlie the multipotency of vertebrate neural stem cells as well. Available evidence indicates that these miRNA display temporal patterns of onset during nervous system formation in a variety of vertebrates. During zebrafish development, for example, let-7 is first detected 72 hours after fertilization (Pasquinelli et al., 2000) and is found specifically in the spinal cord and brain, along with miR-100 and miR-125 (Wienholds et al., 2005). In addition, miR-125 and let-7 are enriched in human and mouse brain samples (Sempere et al., 2004), and are also sharply upregulated in pluripotent P19 mouse cells as they differentiate into neurons (Wulczyn et al., 2007). The ordered production of neurons with distinct morphologies may therefore generally involve let-7-C miRNAs, and their post-developmental modulation may regulate the neuronal remodeling that contributes to adult function as well.
Experimental Procedures
Drosophila strains
Mutants included: let-7-CGKI (Sokol et al., 2008), let-7-CKO1 (Sokol et al., 2008), let-7-CKO2 (this study) and chinmo1 (Zhu et al., 2006). Transgenic insertions included: let-7-Cp12.5kb::lacZ (Chawla and Sokol, 2012), let-7-CΔ3miR::optGal4 (this study), P(W8, let-7-C) (Sokol et al., 2008), c708a-Gal4 (Zhu et al., 2006), c305a-Gal4 (Lin et al., 2007), H24-Gal4 (Lin et al., 2007), c739-Gal4 (Lin et al., 2007), OK107-Gal4 (BL854 ), UAS-let-7-C (Sokol et al., 2008), P(wORF::w3’UTR) (this study), P(wORF::ab3’UTR) (this study), P(wORF::chin3’UTR) (this study), UAS-GFP::nls (BL4775 and 4776), UAS-mCD8::GFP (BL5130) and UAS-mCD8::ChRFP (BL27391 and 27392). Strains for MARCM analysis included: (1) UAS-mCD8::GFP, hsFLP; FRT3L2A ; OK107, (2) FRT3L2A, tubP-Gal80, (3) UAS-mCD8::GFP, hsFLP; let-7-CKO1/CyO ; FRT3L2A; OK107, (4) let-7-CKO2; FRT3L2A, tubP-GAL80 /CyO-TM6b, (5) UAS-mCD8::GFP, hsFLP ; FRT40A, tubGal80 ; OK107, (6) FRT40A, (7) P(W8, let-7-C); FRT40A; P(W8, let-7-C), (8) FRT40A; UAS-let-7-C, (9) FRT40A, chinmo1, UAS-mCD8::GFP / CyO, (10) UAS-mCD8::GFP, hsFLP; chinmo1, let-7-CKO2, FRT40A/CyO; OK107, and (11) let-7-CKO1, FRT40 tubPGal80/CyO. New transgenics were generated by Rainbow Transgenic Services (California USA). Stocks with multiple genetic elements were obtained by meiotic recombination and/or genetic crosses. The let-7-CKO2 allele was generated in parallel with the let-7-CGKI allele (Sokol et al., 2008), and is identical except that the endogenous let-7-C locus was replaced with white rather than white and gal4.
Immunostaining and microscopy
Antibodies used for immunostaining included: rat-anti-Chinmo (1:500; this study), mouse anti-FasII and anti-Dac (1:5 and 1:100; Developmental Studies Hybridoma Bank), rabbit anti-dsRed (1:5000; Clontech), rabbit anti-GFP (1:1000; Invitrogen), rabbit anti-β-gal (1:500; MP Biomedicals), chicken anti-β-gal (1:500; Abcam). Immunostaining was performed as previously described (Chawla and Sokol 2012). Antibodies were generated in rats (Cocalico Biologicals) against a His-tagged version of full-length Chinmo-PA protein that was expressed and purified according to standard methods (Cultivation and Bioprocessing Facility, Indiana University)
Images were collected on a Leica SP5 (Light Microscopy Imaging Center, Indiana University) or a TCS confocal microscope. Confocal stacks were merged using Leica LSM software or Adobe Photoshop. Samples whose staining was directly compared were prepared and imaged in parallel and under identical conditions. For Nb clone, H24, c305a, and c739 neuron counts, MBs were optically sectioned in 0.5 μm increments and the total number of neurons was determined by manually counting the number of β-galor GFP-positive cells section-by-section, ensuring that cells present on consecutive sections were counted only once. Statistical analysis was performed and histograms generated using GraphPad Prism software. P values were calculated using a two-tailed pair t-test. Values are presented as mean ± SEM.
Cell Culture Procedures
Drosophila KC-167 cells were cultured in CCM3 at 23C. Cells were transfected in 48-well plates with 25 ng of tub-Gal4 plasmid DNA, 100 ng UAS-miRNA plasmid DNA, and 25 ng of 3‘UTR-containing pSiCheck2 (Promega) plasmid DNA using Effectene (Qiagen). Luciferase assays were performed using the Dual-Luciferase reporter system (Promega). Transfections were performed in quadruplicate, and resulting luciferase levels were averaged. Fold repression was calculated relative to control samples transfected with UAS-let-7-CΔ3miR. instead of miRNA-encoding plasmids. Additional molecular biology techniques as well as schemes used for plasmid construction are described in detail in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
let-7-C is transcribed in MB neurons born during α’/β’ → p α/β → α/β transitions
Changes in let-7-C dosage lead to temporal cell fate transformations
let-7 and miR-125 directly regulate temporal identity factor chinmo
let-7-C temporal identity phenotypes are suppressed by chinmo reduction
Acknowledgements
We thank Erika Bach, Ann-Shyn Chiang, Pam Geyer, Yuh-Nung Jan, Tzumin Lee, Liz Perkins, Barret Pfeiffer, Hiromu Tanimoto, Peizhang Xu, the Bloomington Drosophila Stock Center, and the Developmental Studies Hybridoma Bank for reagents; Peter and Lucy Cherbas, Thomas Kaufman and the Light Microscopy Center for access to equipment; Victor Ambros, Chris Hammell and Arthur Luhur for comments on the manuscript; and the National Institute of Mental Health (Award R01MH087511) for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Grubel K, Busch S, Friedrich AB, Siwanowicz I, Tanimoto H. The mushroom body of adult Drosophila characterized by GAL4 drivers. J Neurogenet. 2009;23:156–172. doi: 10.1080/01677060802471718. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G, Sokol NS. Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development. 2012;139:1788–97. doi: 10.1242/dev.077743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Maurange C, Gould AP. Temporal control of neuronal diversity: common regulatory principles in insects and vertebrates? Development. 2008;135:3481–3489. doi: 10.1242/dev.016931. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li W, Wang F, Menut L, Gao FB. BTB/POZ-sinc finger protein abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron. 2004;43(6):823–34. doi: 10.1016/j.neuron.2004.08.040. [DOI] [PubMed] [Google Scholar]
- Lin HH, Lai JS, Chin AL, Chen YC, Chiang AS. A map of olfactory representation in the Drosophila mushroom body. Cell. 2007;128:1205–1217. doi: 10.1016/j.cell.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Martini SR, Roman G, Meuser S, Mardon G, Davis RL. The retinal determination gene, dachshund, is required for mushroom body cell differentiation. Development. 2000;127:2663–2672. doi: 10.1242/dev.127.12.2663. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm J, Shenker S, May G, Duff M, Zhang D, Eads B, Carlsom J, Brown J, et al. Global patterns of Tissue-specific alternative polyadenylation in Drosophila. Cell Reports. 2012;1:277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K, Satoh D, Estes P, Crews S, Uemura T. Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein Abrupt. Neuron. 2004;43(6):809–22. doi: 10.1016/j.neuron.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB journal. 2007;21(2):415–26. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Lee T. Neuronal temporal identity in post-embryonic Drosophila brain. Trends in neurosciences. 2007;30(10):520–6. doi: 10.1016/j.tins.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Zhu S, Chiang AS, Lee T. Development of the Drosophila mushroom bodies: elaboration, remodeling and spatial organization of dendrites in the calyx. Development. 2003;130:2603–2610. doi: 10.1242/dev.00466. [DOI] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.