Abstract
A series of compounds that target reactive transition metal chelates to somatic Angiotensin Converting Enzyme (sACE-1) have been synthesized. Half maximal inhibitory concentrations (IC50) and rate constants for both inactivation and cleavage of full length sACE-1 have been determined and evaluated in terms of metal-chelate size, charge, reduction potential, coordination unsaturation, and coreactant selectivity. Ethylenediamine-tetraacetic acid (EDTA), nitrilotriacetic acid (NTA), 1,4,7,10-tetraazacyclo-dodecane-1,4,7,10-tetraacetic acid (DOTA), and tripeptide GGH were linked to the lysine sidechain of lisinopril by EDC/NHS coupling. The resulting amide-linked chelate-lisinopril (EDTA-lisinopril, NTA-lisinopril, DOTA-lisinopril, and GGH-lisinopril) conjugates were used to form coordination complexes with iron, cobalt, nickel and copper, such that lisinopril could mediate localization of the reactive metal chelates to sACE-1. ACE activity was assayed by monitoring cleavage of the fluorogenic substrate Mca-RPPGFSAFK(Dnp)-OH, a derivative of bradykinin, following pre-incubation with metal-chelate-lisinopril compounds. Concentration-dependent inhibition of sACE-1 by metal-chelate-lisinopril complexes revealed IC50 values ranging from 44 nM to 4,500 nM for Ni-NTA-lisinopril and Ni-DOTA-lisinopril, respectively, versus 1.9 nM for lisinopril. Stronger inhibition was correlated with smaller size and lower negative charge of the attached metal chelates. Time-dependent inactivation of sACE-1 by metal-chelate-lisinopril complexes revealed a remarkable range of catalytic activities, with second order rate constants as high as 150,000 M−1min−1 (Cu-GGH-lisinopril), while catalyst-mediated cleavage of sACE-1 typically occurred at much lower rates, indicating that inactivation arose primary from sidechain modification. Optimal inactivation of sACE-1 was observed when the reduction potential for the metal center was poised near 1000 mV, reflecting the difficulty of protein oxidation. This class of metal-chelate-lisinopril complexes possesses a range of high-affinity binding to ACE, introduces the advantage of irreversible catalytic turnover, and marks an important step toward the development of multiple-turnover drugs for selective inactivation of sACE-1.
Keywords: metallodrug, lisinopril, angiotensin converting enzyme, multiple-turnover
INTRODUCTION
Angiotensin converting enzyme (ACE) is an important therapeutic target for treatment of hypertension and heart failure because of its critical physiological role in cardiovascular function, primarily the hydrolysis of Angiotensin I to form Angiotensin II, which results in vasoconstriction, and hydrolysis of the vasodilator bradykinin.1–4 Human ACE-1 exists in multiple active forms: somatic ACE (sACE-1), a full length two-domain (N and C) membrane-anchored enzyme, testicular ACE (tACE), a membrane-anchored enzyme with identical sequence to the C-terminal domain of sACE-1 (except for an additional N-terminal 36 amino acid segment),5 and N-domain form, a diffusible N-terminal cleavage product of sACE-1.6,7 The N- and C-terminal domains share ~ 60% sequence homology, similar active site structures,5,6 similar substrate binding selectivity,2 and the requirement for an active site zinc cation for activity. Several inhibitors of sACE-1, such as lisinopril (currently marketed as Zestril and Prinivil), successfully alleviate hypertension and congestive heart failure, resulting in decreased mortality among patients,1,8 by directly competing with the natural substrates for binding to sACE-1 active sites, resulting in overall vasodilation.
One significant limitation of lisinopril, and reversible inhibitors in general, is the requirement for stoichiometric saturation of the target; a typical active dose of lisinopril ranges from 5 to 40 mg for humans.1 There is significant interest in the development of multiple-turnover drugs that irreversibly modify therapeutic targets, allowing substoichiometric concentrations of drug to be used with the potential for therapeutic effect at lower dosage with reduced side-effects.9–14 To this end, several research groups have recently developed various metallodrugs and/or artificial proteases that use target-selective metal catalysts, including hydrolytic Co- and Cu-cyclen, oxidative Ni- and Cu-ATCUN, and photo-activated Ru-complexes to effect protein modification.9,10,13,15–25 The goal of this study was to develop a class of multiple-turnover metallodrugs that selectively and irreversibly inactivate sACE-1, using a variety of oxidative metal catalysts, with lisinopril as the targeting molecule.
A series of metallodrugs was developed in which transition metal chelates (M-chelates) with known ability to generate reactive oxygen species with multiple-turnover26 were attached to the lysine sidechain of lisinopril, such that the inhibitor lisinopril targeted each reactive M-chelate to the active sites of sACE-1. The M-chelates used were based on combinations of the transition metals Fe3+, Co2+, Ni2+, and Cu2+ and the chelators DOTA, EDTA, NTA, and tripeptide GGH. These complexes provided variability in reduction potential, coordination unsaturation, size, and charge.12 The ability of each M-chelate-lisinopril complex to both reversibly inhibit and catalytically inactivate sACE-1 was assessed, and varying binding affinities and rates of targeted irreversible inactivation (and cleavage) of full length sACE-1 by M-chelate-lisinopril complexes were observed. This class of compounds illustrates the potential for catalytic irreversible inactivation of sACE-1 as an efficient alternative to reversible inhibition and demonstrates that catalyst parameters such as reduction potential, coordination unsaturation, geometric alignment, size, and charge may be tuned to optimize catalytic efficiency. The results of this study are likely to prove valuable in the design and development of catalytic metallodrugs directed toward other therapeutic targets.
MATERIALS AND METHODS
Chemicals and Reagents
Lisinopril was purchased from Cayman Chemical Company and stored at −20 °C in powder form, and ESI-TOF-MS analysis confirmed the expected mass of 404 amu. Recombinant human somatic ACE (sACE-1: Leu30-Leu1261, with C-terminal His tag, >95 % purity by SDS-PAGE under reducing conditions), originally isolated from an NS0-derived murine myeloma cell line, was purchased from R&D Systems as a stock solution containing 12.5 mM Tris, 75 mM NaCl, 0.5 µM ZnCl2, and 40% (v/v) glycerol, pH 7.5 with [sACE-1] = 0.434 mg/mL, and divided into single use aliquots prior to storing at −20 °C. Fluorogenic substrate Mca-RPPGFSAFK(Dnp)-OH was purchased from R&D Systems, dissolved in DMSO, divided into single use aliquots, and stored at −20 °C. The bifunctional compound NHS-DOTA was purchased from Macrocyclics and stored at −20 °C in powder form. N-hydroxysuccinimide (NHS) was purchased from GenScript, and 1-ethyl-3-[3-dimethyl aminopropyl] carbodiimide hydrochloride (EDC) was purchased from Pierce and stored at −20 °C. Ethylenediamine-tetraacetic acid (EDTA) was purchased from Aldrich. Nitrilotriacetic acid (NTA) was purchased from Sigma. The tripeptides GGH-OH (GGH) and Z-GGH-OH (Z-GGH; Z = carboxybenzyl) were obtained from Bachem, and 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) was purchased from Macrocyclics. The Fe(II) sulfate heptahydrate, Co(II) chloride hexahydrate, Ni(II) acetate tetrahydrate, Cu(II) chloride dihydrate, and Zn(II) chloride salts were purchased from ACROS, J.T.Baker, Aldrich, J. T. Baker, and MCB Reagents, respectively. Sodium chloride, sodium hydroxide, and ammonium persulfate were purchased from Fisher. HEPES was purchased from Sigma. Acetonitrile, SDS, and Na2HPO4 were purchased from Sigma-Aldrich, NaHCO3 was purchased from Mallinckrodt, 40% acrylamide/Bis solution (19:1) was purchased from Bio-Rad, and TFA was purchased from ACROS. The silver stain kit was purchased from Pierce. The C18 preparatory and analytical columns used for RP-HPLC were purchased from Vydac, and the PolyWAX LP column used for anion exchange HPLC was purchased from PolyLC. D2O (99.96%) for 1H-NMR was purchased from Cambridge Isotopes Laboratory.
Synthesis and Characterization
Compound EDTA-lisinopril was prepared by making a solution containing 500 mM EDTA, 500 mM NHS, 500 mM EDC in DMSO and reacting for 20 min at ambient temperature. After 20 min, 48 µL of this reaction mixture was mixed with 552 µL of a solution that contained 22 mM lisinopril in 100 mM NaHCO3, pH 8.0. The reaction proceeded overnight at RT in the dark, followed directly by anion exchange HPLC purification. Anion exchange elution conditions used a gradient method, running from 0 to 100% B from 0 to 50 min, and 100% B from 50 to 55 min, where mobile phase A = 10 mM Na2HPO4, pH 5.7; and B = 1 M NaCl, 10 mM Na2HPO4, pH 5.7. The anion exchange HPLC fraction for product EDTA-lisinopril was collected and further separated by RP-HPLC. RP-HPLC elution conditions used a gradient method, running from 15 to 65% B from 0 to 45 min, 65 to 95% B from 45 to 50 min, and 95% B from 50 to 55 min where mobile phase A = H2O, 0.1% TFA; B = acetonitrile, 0.1% TFA. The RP-HPLC fraction for product EDTA-lisinopril was collected, lyophilized, resuspended in water, and ESI-TOF MS analysis provided the expected mass of 678 amu (negative mode), as well as the +Na+-1H+ adduct at 700 amu, with no evidence of uncoupled lisinopril reactant (404 amu). EDTA-lisinopril concentration was quantified via UV/Vis (270 nm) titration with a solution of known concentration of copper(II) chloride.
Compound NTA-lisinopril was prepared by making a solution containing 500 mM NTA, 500 mM NHS, and 500 mM EDC in DMSO and reacting for 20 min, after which time 48 µL of the reaction volume was mixed with 552 µL of a solution that contained 22 mM lisinopril in 100 mM NaHCO3, pH 8.0. The reaction proceeded overnight at RT in the dark, followed by consecutive HPLC purification (first anion exchange, then RP-HPLC) using the same elution conditions as used for EDTA-lisinopril. The RP-HPLC fraction for product NTA-lisinopril was collected, lyophilized, resuspended in water, and ESI-LCQ MS analysis (in the laboratory of Dr. Michael Freitas) provided the expected mass of 578 amu (negative mode), with no evidence of uncoupled lisinopril reactant. NTA-lisinopril concentration was quantified via UV/Vis (250 nm) titration with a solution of known concentration of iron(II) sulfate.
Compound DOTA-lisinopril was prepared by making a solution containing 40 mM NHS-DOTA (10× stock made in DMSO) and 20 mM lisinopril in 100 mM NaHCO3, pH 8.0. The reaction proceeded overnight at RT in the dark, followed by consecutive HPLC purification (first anion exchange, then RP-HPLC) using the same elution conditions as used for EDTA-lisinopril. The RP-HPLC fraction for product DOTA-lisinopril was collected, lyophilized, resuspended in water, and ESI-TOF MS analysis provided the expected mass of 791 amu (negative mode), with no evidence of uncoupled lisinopril reactant. DOTA-lisinopril concentration was quantified via UV/Vis (240 nm) titration with a solution of known concentration of nickel(II) acetate.
Compound GGH-lisinopril was prepared by making a solution containing 200 mM Z-GGH (Z = carboxybenzyl), 200 mM NHS, 200 mM EDC in DMSO and reacting for 20 min, after which time 54 µL of this reaction volume was mixed with 30 µL of 200 mM lisinopril in DMSO. The reaction proceeded overnight at RT in the dark, followed by consecutive HPLC purification (first anion exchange, then RP-HPLC) using the same elution conditions as used for EDTA-lisinopril. The RP-HPLC fraction for product Z-GGH-lisinopril was collected and lyophilized. ESI-TOF MS analysis provided the expected mass for Z-GGH-lisinopril of 789 amu with no evidence of uncoupled lisinopril reactant. The lyophilized Z-GGH-lisinopril was deprotected by dissolving in 1 mL TFA, and 5 mg 20 % Pd(OH)2 on charcoal was added and stirred to form a slurry. The mixture was Ar-purged and the anaerobic solution was reacted under a positive pressure of H2 for ~ 6 hrs. The mixture was dried under vacuum, redissolved in water, and centrifuged to remove solid catalyst, and the resulting deprotected GGH-lisinopril was purified by RP-HPLC using the same RP-HPLC conditions as used for EDTA-lisinopril. The RP-HPLC fraction for product GGH-lisinopril was collected, lyophilized, resuspended in water, and ESI-TOF MS analysis provided the expected mass of 655 amu (negative mode), as well as the +Na+-1H+ adduct at 677 amu, with no evidence of uncoupled lisinopril reactant. The concentration of product GGH-lisinopril was quantified via UV/Vis (245 nm) titration with a solution of known concentration of nickel(II) acetate.
The identities and isomeric purity of the synthesized chelate-lisinopril compounds were validated by 1H-NMR analysis, analytical RP-HPLC, and ESI-MS (Figures SM1–SM19). Divalent iron, cobalt, nickel, and copper complexes of DOTA-lisinopril, EDTA-lisinopril, NTA-lisinopril, and GGH-lisinopril were prepared by mixing the respective metal salts with the chelate-lisinopril species in a buffer containing 20 mM HEPES, 100 mM NaCl, pH 7.4 and mixing at RT for 30 min prior to each measurement. M-chelate complex formation was verified by metal ion titration monitored by UV/vis (Figures SM20–SM23). Metal:chelate ratios of 1:1 and 1:1.1 (to ensure that essentially all metal was chelated) were used for concentration-dependent and time-dependent inactivation experiments, respectively. The Fe2+ complex with GGH-lisinopril was not used in later experiments due to weak complex formation. Experimentally determined extinction coefficients for all M-chelate-lisinopril and M-chelate species, as well as stability constants for M-chelates, are summarized in the supporting information (Tables SM1 and SM2).
Determination of IC50 Values
sACE-1 (1 nM) and variable concentrations of each M-chelate-lisinopril complex (prepared in the absence of Zn2+) were incubated for 20 min at 37 °C in a buffer containing 50 mM HEPES, 300 mM NaCl, 10 µM ZnCl2, 0.05% Brij35, pH 7.4. After 20 min, 68.6 µL of each preincubated mixture of sACE-1 and inhibitor were mixed with 1.4 µL of 0.5 mM fluorogenic substrate in a fluorescence cuvette, and substrate cleavage by sACE-1 was immediately monitored by real-time fluorimetry at 37 °C, with excitation at 320 nm and emission at 405 nm. Initial rates of fluorescence increase were determined for each concentration of M-chelate-lisinopril, and these initial rates were expressed as a percentage (% maximal activity) of the average of several initial rates of uninhibited substrate cleavage by sACE-1 in the absence of M-chelate-lisinopril complexes. Plots of % maximal activity vs. M-chelate-lisinopril concentration were fit to equation (1), where A, [I], n, and [IC50] are the % maximal activity, inhibitor concentration, fitted cooperativity, and fitted IC50 respectively. IC50 values were determined for each M-chelate-lisinopril, chelate-lisinopril, M-chelate, and chelator species.
| (1) |
Time-Dependent Inactivation of sACE-1
sACE-1 and each M-chelate-lisinopril complex (prepared in the absence of Zn2+) were preincubated for 20 min at 37 °C in a buffer containing 50 mM HEPES, 300 mM NaCl, 10 µM ZnCl2, 0.05% Brij35, pH 7.4, and after 20 min, coreactants ascorbate and/or H2O2 (or no coreactants) were added to initiate each reaction. Reaction concentrations were 1 nM sACE-1, a concentration of M-chelate-lisinopril that gave approximately 80% activity (calculated by use of equation (1), where A = 80%; concentrations listed in Table 1), and 1 mM ascorbate and/or H2O2 (or no coreactants). Each time-dependent ACE-inactivation reaction proceeded at 37 °C for a period of 2 h, and at each specific intervening time point, a 68.6 µL aliquot of the reaction mixture containing sACE-1 was mixed with 1.4 µL of 0.5 mM fluorogenic substrate in a fluorescence cuvette. Substrate cleavage by sACE-1 was immediately monitored by real-time fluorimetry at 37 °C, with excitation at 320 nm and emission at 405 nm. Initial rates were determined for each time point for the time-dependent inactivation of sACE-1, and these initial rates were expressed as a percentage (% maximal activity) of the average of several initial rates for uninhibited substrate cleavage by sACE-1, determined in the absence of both M-chelate-lisinopril complexes and coreactants. Plots of % maximal activity vs. time were fit to a first-order exponential decay model, and initial rates of inactivation of sACE-1 by M-chelate-lisinopril complexes were determined. Second-order rate constants for inactivation of sACE-1 were obtained using equation (2), where k2 is the second-order rate constant (M−1min−1), R is the initial rate of enzyme inactivation (M/min) after subtraction of the corresponding background rate in the absence of M-chelate-lisinopril complex, but with the same coreactants, [I] is the concentration of M-chelate-lisinopril complex used (M), and [E] is the concentration of sACE-1 used (1×10−9 M). Control experiments with M-chelate complexes lacking attached lisinopril were performed in the same manner and conditions used for the respective M-chelate-lisinopril complexes.
| (2) |
Table 1.
IC50’s for sACE-1 inhibition by metal-chelate-lisinopril complexes, metal chelates, chelator-lisinopril compounds, and chelators.
| complex | IC50 (nM) | TD conc. (nM)a |
|---|---|---|
| [NTA]2-–lisinopril | 210 ± 40 | --b |
| [Fe-NTA]1+–lisinopril | 54 ± 9 | 7.8 |
| [Co-NTA]0–lisinopril | 45 ± 8 | 4.8 |
| [Ni-NTA]0–lisinopril | 44 ± 4 | 4.8 |
| [Cu-NTA]0–lisinopril | 90 ± 10 | 15 |
| [GGH]1+–lisinopril | 50 ± 10 | --b |
| [Co-GGH]0–lisinopril | 90 ± 10 | 20 |
| [Ni-GGH]0–lisinopril | 150 ± 30 | 25 |
| [Cu-GGH]0–lisinopril | 1,470 ± 20 | 300 |
| [EDTA]3-–lisinopril | 1,100 ± 100 | --b |
| [Fe-EDTA]0–lisinopril | 88 ± 5 | 14 |
| [Co-EDTA]1-–lisinopril | 1,200 ± 40 | 210 |
| [Ni-EDTA]1-–lisinopril | 1,420 ± 50 | 250 |
| [Cu-EDTA]1-–lisinopril | 960 ± 60 | 230 |
| [DOTA]3-–lisinopril | 6,800 ± 900 | --b |
| [Fe-DOTA]0–lisinopril | 3,800 ± 400 | 870 |
| [Co-DOTA]1-–lisinopril | 4,100 ± 200 | 790 |
| [Ni-DOTA]1-–lisinopril | 4,500 ± 200 | 980 |
| [Cu-DOTA]1-–lisinopril | 4,400 ± 400 | 1,000 |
| lisinopril | 1.9 ± 0.3 | --b |
| Fluorescein-lisinopril | 290 ± 20 | --b |
| DOTA | 7,000 ± 1,000 | --b |
| [Cu-DOTA]2- | 30,000 ± 2,000 | --c |
| EDTA | 7,200 ± 600 | --b |
| [Cu-EDTA]2- | 26,000 ± 5,000 | --c |
| NTA | 32,000 ± 3,000 | --b |
| [Cu-NTA]1- | 36,000 ± 4,000 | --b |
| GGH | > 100,000 | --b |
| [Cu-GGH]1- | 17,000 ± 2,000 | --c |
The concentration of M-chelate-lisinopril that inhibited sACE-1 by 20% (TD conc.) was used in time-dependent sACE-1 inactivation experiments so that the remaining 80% activity could be used to monitor the kinetics of inactivation. The concentration required for 20% saturation was higher for cleavage assays, due to the higher concentration of sACE-1 used (20 nM).
A time-dependent inactivation experiment was not performed.
The concentration used for time-dependent inactivation was the same as that used for the corresponding M-chelate-lisinopril complex.
Characterization of Residual Activity for Inactivated sACE-1
sACE-1 (1 nM), H2O2 (1 mM), ascorbate (1 mM), and each M-chelate-lisinopril (IC20 concentration) were preincubated for 15 h at 37 °C. Following preincubation, 68.6 µL aliquots of the reaction mixture containing inactivated sACE-1 were separately mixed with 1.4 µL of variable concentration of fluorogenic substrate in a fluorescence cuvette at 37 °C. Substrate cleavage by sACE-1 was immediately monitored by real-time fluorimetry, and initial rates (fluorescence intensity per min) were recorded. The inner-filter effect for the fluorogenic substrate, which became significant at higher substrate concentrations, was corrected for by use of equation (3),27 where c is the concentration of substrate, Fobs and Fcorr are the uncorrected and corrected fluorescence intensities, respectively, ℓex and ℓem are the fluorescence cuvette pathlengths for excitation and emission, respectively, and εex and εem are the fluorogenic substrate extinction coefficients at 320 nm and 405 nm, respectively. Initial rate units of corrected fluorescence intensity per min were converted to units of µM substrate per min through use of a linear standard curve, which related the total change in fluorescence intensity resulting from complete substrate cleavage to the substrate concentration used. Plots of initial rate (µM substrate / min) vs substrate concentration (µM) were constructed and were fit to the Michaelis-Menten equation to obtain kcat, KM, and kcat/KM values for inactivated sACE-1. Metal complex preparation, complex concentration, and preincubation conditions were the same as used in the time-dependent inactivation assay.
| (3) |
SDS-PAGE Analysis
sACE-1 (20 nM), H2O2 (1 mM), ascorbate (1 mM), and each M-chelate-lisinopril complex (concentration that gave 20% saturation of 20 nM sACE-1, ~ IC50/4) were preincubated for 15 h at 37 °C. The resulting mixtures were separated by 7.5% SDS-PAGE and silver staining used to visualize sACE-1 and cleavage products. To quantify the sACE-1 remaining after each time-dependent incubation reaction, the intensity of each starting material band was quantified and converted to % full length sACE-1 remaining. Initial rates of cleavage of sACE-1 were determined by incubating M-chelate-lisinopril complexes, coreactants, and sACE-1 for varied time intervals (0, 10, 20, 50, 80 min) and fitting of the change in % full length sACE-1 remaining over time to a first order equation; these initial rates were expressed as % full length sACE-1 per min. In the case of the 15 h incubation reactions, the mole % of full length sACE-1 and each cleavage product were determined by use of the intensities of each band and apparent molecular weights. Control reactions were also performed for reactions with M-chelates lacking attached lisinopril, as well as for reactions lacking catalyst, coreactants, or both (Figures SM34, SM37, and SM38; Table SM5).
Molecular Modeling
Interactions between both the N- and C-domains of sACE-1 and the M-chelate-lisinopril complexes were modeled using a combination of Spartan and Gaussian software. From the X-ray crystal structure of the N-domain of sACE-1 in complex with the inhibitor lisinopril (PDB ID: 2C6N), a sphere with 15 Å radius, centered on the N atom of the lisinopril lysine sidechain, was generated in Gaussian and used to model the active site. This process was repeated using the X-Ray crystal structure of the C-domain of ACE (tACE) in complex with lisinopril (PDB ID: 1O86). Molecular mechanics energy minimization was performed in Spartan using the Merck molecular force field MMFFaq, in which structural energy minimization occurred without solvent considerations. Model geometries were constrained to the X-Ray crystal structures, while the lisinopril lysine sidechain and lysine-chelate linker geometries were allowed to vary during energy minimization. Structures were accepted once 10 successive rounds of energy minimization lowered the energy by no more than 0.1 kcal/mol. Modeling parameters: equilibrium geometry at ground state, subject to frozen atoms and symmetry, multiplicity = singlet, total charge = 0 (partial charges handled by MMFFaq28). Gaussian software was used to generate figures.
Complex Kinetic Stability
To observe whether Zn2+ was able to displace each transition metal from each chelate, a solution containing 100 µM M-chelate complex and 100 µM ZnCl2 was incubated at 37 °C (in cuvette) in a buffer containing 20 mM HEPES, 100 mM NaCl, pH 7.4, and a UV/Vis scan (800 nm to 200 nm) was taken every 1.5 min. This process was repeated for each M-chelate used, and the rate of the transition (if present) from {M-chelate + Zn2+} to {Zn2+-chelate + metal} was determined by following the change in absorbance at the specified wavelengths for each M-chelate over time (Figure SM40, Table SM7).
RESULTS
Synthesis and Characterization
Chelate-lisinopril conjugates (DOTA-lisinopril, EDTA-lisinopril, GGH-lisinopril, and NTA-lisinopril) were synthesized by coupling the respective chelators with the lysine sidechain of lisinopril to form amide linkages. Synthesis of DOTA-lisinopril was achieved by direct reaction of lisinopril with the bifunctional chelator NHS-DOTA. Synthesis of both EDTA-lisinopril and NTA-lisinopril were each performed by activation of EDTA and NTA with EDC/NHS in DMSO, followed by addition of a limiting concentration of lisinopril. Synthesis of GGH-lisinopril was achieved in three steps. First, Z-GGH was reacted with EDC/NHS in DMSO to form the activated NHS-ester of Z-GGH. Second, a limiting concentration of lisinopril was added (in DMSO) to form the product Z-GGH-lisinopril. Catalytic hydrogenation over Pd(OH)2/C in TFA yielded the final deprotected product GGH-lisinopril.
Separation of chelate-lisinopril species from reactants was generally achieved by two consecutive HPLC steps. First, chelate-lisinopril products were isolated by anion exchange HPLC, and second, anion exchange fractions containing product were purified by RP-HPLC. RP-HPLC alone was generally not sufficient to effect adequate separation, because the RP-HPLC chelate-lisinopril elution times were too similar to that of lisinopril (with the exception of Z-GGH-lisinopril, which differed in elution time from lisinopril by 7 min), whereas anion exchange allowed separation of NTA-lisinopril, EDTA-lisinopril, and DOTA-lisinopril from the lisinopril reactant by at least 12 min in elution time. Subsequent purification by RP-HPLC allowed separation from any remaining free chelator or other reactants (by at least 10 min), and simultaneously served to desalt the chelate-lisinopril products. Following synthesis and HPLC purification, mass spectrometric analysis revealed the expected masses for EDTA-lisinopril, NTA-lisinopril, DOTA-lisinopril, and GGH-lisinopril (678 amu, 578 amu, 791 amu, and 655 amu, respectively) with no observed mass for remaining uncoupled lisinopril (404 amu) in any of the products. 1H-NMR analysis confirmed the identity and isomeric purity of the synthesized products.
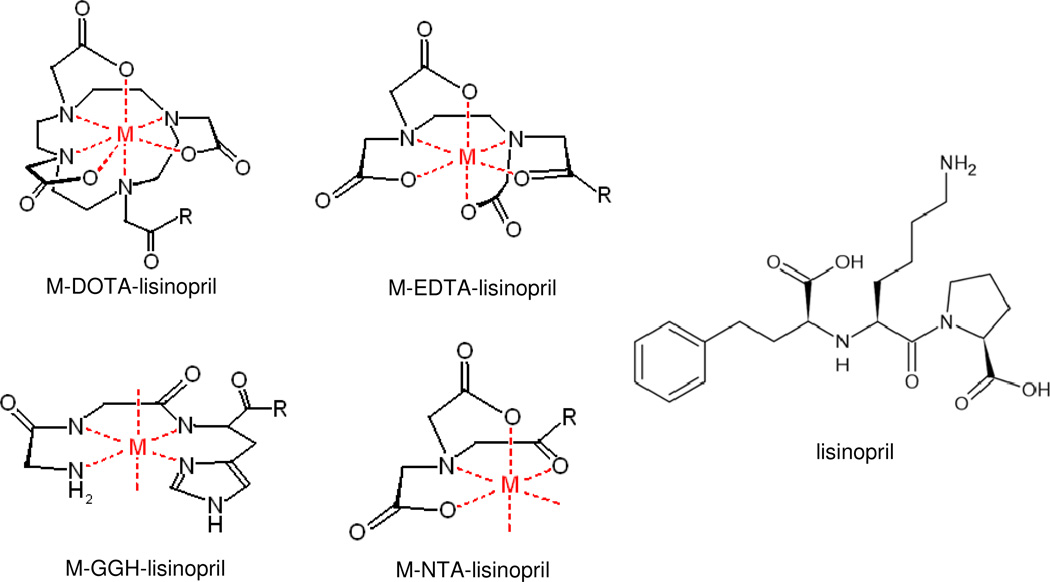
Complex formation between each metal ion (Fe2+, Co2+, Ni2+, and Cu2+) and each chelator-lisinopril species, as well as each chelator lacking lisinopril, was monitored by UV/Vis spectroscopy, and metal complex extinction coefficients were determined. As expected, complex formation between Fe2+ and GGH was not observed under the experimental conditions used, and so the combination of Fe2+ with GGH-lisinopril was not used in later experiments. Fe2+ is known to convert to Fe3+ following chelation under the aerobic conditions used in this study; therefore, the 3+ oxidation state of iron complexes of NTA-lisinopril, EDTA-lisinopril, and DOTA-lisinopril differed from the 2+ oxidation state of the other metal complexes. Absorption spectra for each of the M-chelate-lisinopril species closely matched the characteristic absorption spectra for each of the respective M-chelates obtained without the attached lisinopril, although the extinction coefficients for the charge transfer bands of Fe-DOTA and Cu-NTA were observed to decrease and increase, respectively, following coupling to lisinopril. The M-chelate-lisinopril species used in this study are summarized in Figure 1.
Figure 1.
Summary of the metal chelate complexes and their attachment to the lysine sidechain of lisinopril. M = Fe3+, Co2+, Ni2+, Cu2+; R = N(H)-lisinopril.
Concentration-Dependent Inhibition of sACE-1
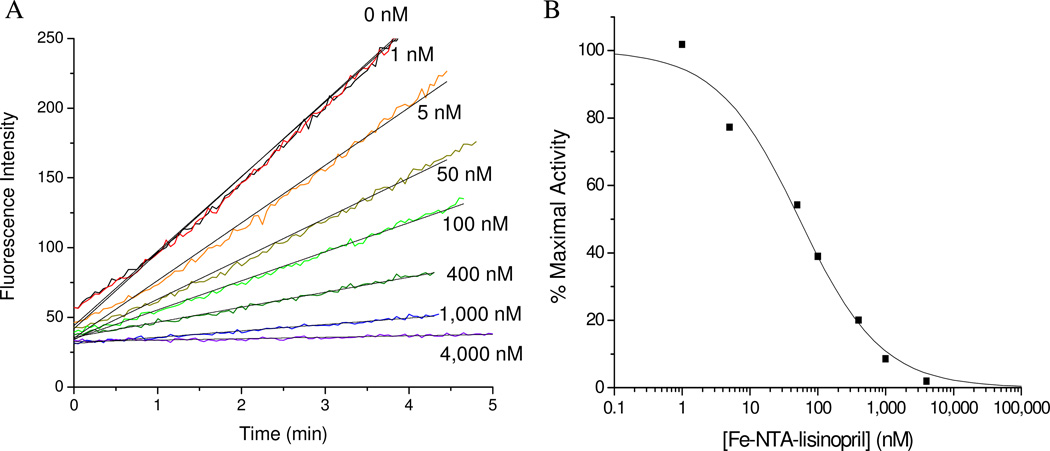
The relative binding affinity of each M-chelate-lisinopril species to sACE-1, as well as other species lacking either metal, lisinopril, or both, were determined by incubating the enzyme with variable concentrations of each inhibitor and following the change in rate for sACE-1-mediated cleavage of the fluorogenic substrate Mca-RPPGFSAFK(Dnp)-OH (Figure 2). IC50 values for sACE-1 inhibition were determined for each complex and are summarized in Table 1. Lisinopril was found to have an IC50 of 1.9 nM, consistent with previous reports.29 Following attachment of M-chelates to lisinopril, IC50 values were found in the range 44 – 4,500 nM, with IC50 values increasing (affinity decreasing) in the order M-NTA-lisinopril < M-GGH-lisinopril ~ M-EDTA-lisinopril < M-DOTA-lisinopril. These results confirm the expected inverse correlation between target affinity and steric bulk, as well as negative charge, of the attached M-chelate.29 Among metals, it was generally found that Fe complexes had the lowest IC50 values, most likely reflecting the trivalent oxidation state of iron and the higher positive charge associated with the resulting complex, consistent with the inverse correlation observed between the overall negative charge on the chelate-lisinopril complex and affinity for sACE-1. Experiments in which both inhibitor concentration and substrate concentration were varied were performed for lisinopril and Cu-GGH-lisinopril, and these experiments confirm a competitive mode of inhibition of sACE-1, with active site binding (Dixon plots shown in Figure SM27).
Figure 2.
Concentration-dependent inactivation of sACE-1 by Fe-NTA-lisinopril. (A) sACE-1 (1 nM) was preincubated with variable concentrations of Fe-NTA-lisinopril, and subsequently sACE-1 activity was measured from the initial cleavage rates of fluorogenic substrate. (B) Dependence of sACE-1 activity on Fe-NTA-lisinopril concentration. IC50 values were determined for all synthesized Mchelate-lisinopril complexes in the same manner.
Comparison of IC50 values obtained for M-chelate-lisinopril complexes with those obtained for M-chelates lacking lisinopril confirms the requirement for lisinopril to direct the attached M-chelates to the sACE-1 active sites. Weak inhibition of sACE-1 by M-chelates alone presumably reflected weaker interactions with either sACE-1 or the substrate through metal coordination, hydrogen bonding, or electrostatic interactions, while strong inhibition of sACE-1 by M-chelate-lisinopril complexes resulted from specific binding to the active site. Weak inhibition was also observed for metal-free chelators, with IC50 values ranging from 7,000 nM to over 100,000 nM, and IC50 values increased in the order DOTA < EDTA < NTA < GGH, although sACE-1 inhibition by metal-free chelators lacking lisinopril appeared to depend solely upon the ability of each chelator to bind Zn2+ (present at 10,000 nM), a cofactor necessary for sACE-1 activity, rather than active site binding, consistent with prior investigations.30,31 Chelation of Zn2+ was expected to be much less prominent for metal-free GGH than for the metal-free chelators DOTA, EDTA, and NTA, as a result of the distinct mode of chelation for GGH,32–34 and this was indeed reflected by the relatively high IC50 for sACE-1 inhibition by metal-free GGH. In principle, metal-bound chelators also possess the potential to undergo slow exchange with active site Zn2+, although such exchange was found to be insignificant under the conditions used, as discussed quantitatively in a later section. Following attachment of metal-free chelators to lisinopril, sACE-1 inhibition was enhanced by as much as 2,000-fold (for GGH). In summary, attachment to lisinopril was a requirement for high-affinity targeting of M-chelates to the active sites of sACE-1, yielding a variety of ACE-affinities that depended on the chemical nature of the M-chelate.
Structural Modeling
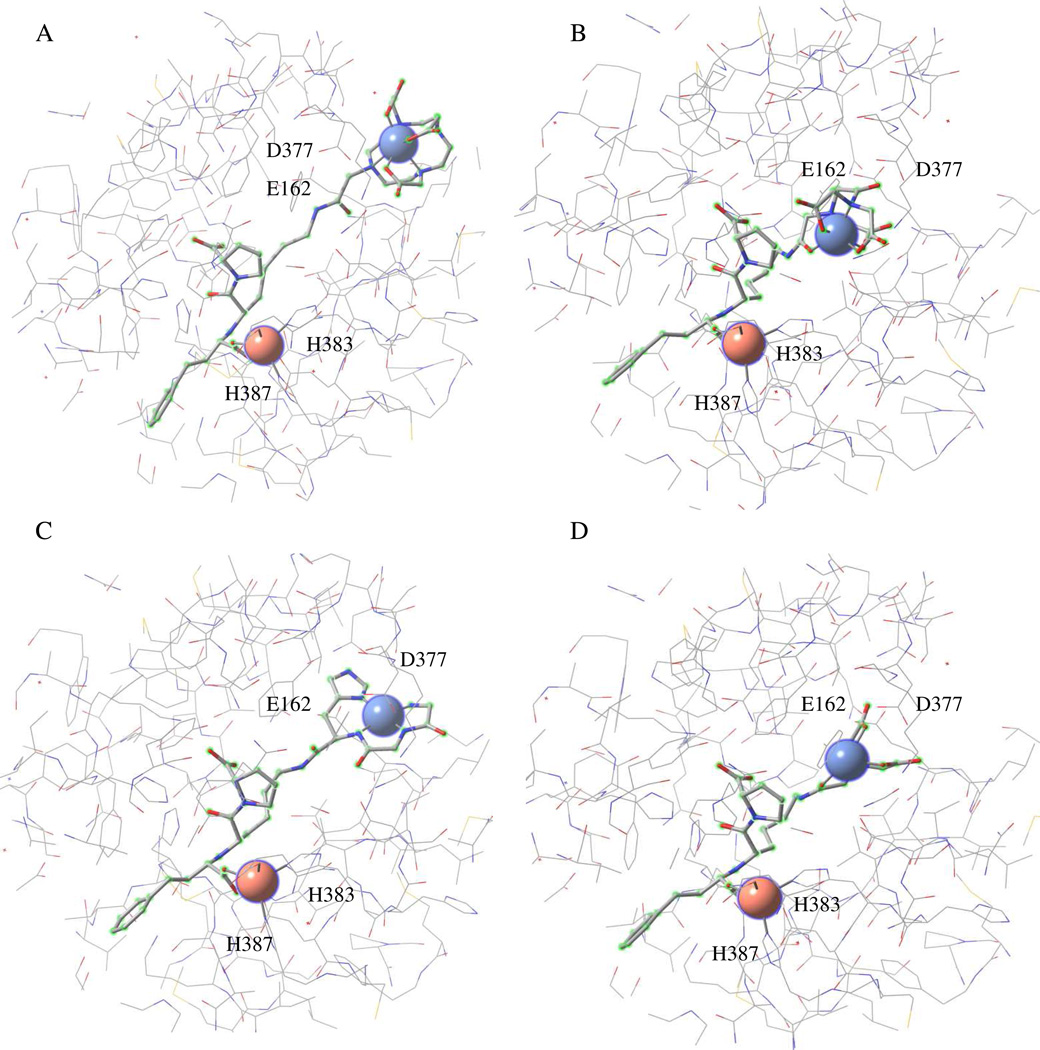
Molecular models were constructed to visually assess the alignment of M-chelate-lisinopril complexes within the active sites of ACE (Figure 3, Figure SM39). Model geometries were constrained to that of the X-ray crystal structure of ACE (N- and C-domains, separately) in complex with lisinopril, while geometries for the lysine sidechain of lisinopril and attached M-chelates were allowed to vary, and molecular mechanics energy minimization was performed. Structural models revealed that the M-DOTA portion of the M-DOTA-lisinopril complex preferred not to settle into the active sites of either the N-domain or C-domain of ACE, consistent with the relatively large size/charge of M-DOTA relative to M-EDTA, M-NTA and M-GGH, as well as the relatively high IC50 values observed for the M-DOTA-lisinopril complexes, while M-EDTA-lisinopril, M-NTA-lisinopril, and M-GGH-lisinopril all preferred complete burial of the respective M-chelates within the active sites of both the N- and C-domains of ACE. Several negatively-charged residues of ACE (D140 for N-domain ACE; D377 and E162 for tACE) were positioned in close proximity to the M-chelate portion of the M-chelate-lisinopril complexes, and these residues most likely contribute to electrostatic repulsion of negatively-charged M-chelates, while attracting positive functional groups, such as the lysine sidechain of unmodified lisinopril. Additionally, many residues with known susceptibility to sidechain oxidation (His, Pro, Arg, Lys, Thr, Tyr, Cys, Trp),35,36 are positioned in this binding pocket. Several of these have known roles in binding and/or hydrolysis of ACE substrates, including binding of active site Zn2+ (H361, H365 in N-domain ACE; H383, H387 in tACE) and Cl− ions (R186, W485, R489 for chloride I and Y224, R522 for chloride II in tACE; Y202, Y501 for chloride II in N-domain ACE).5,6 Oxidation of such residues is the most likely cause of the oxidative inactivation of ACE.
Figure 3.
Energy-minimized structural models of tACE active site (equivalent to the active site of C-domain of sACE except for 36 additional residues at the N-terminus of tACE). The models show binding by (A) Fe-DOTA-lisinopril, (B) Fe-EDTA-lisinopril, (C) Cu-GGH-lisinopril, and (D) Fe-NTAlisinopril. The redox-active metal for each metal-chelate-lisinopril complex is shown as a blue sphere (upper right within each model), and the active site Zn2+ is shown as a pink sphere (bottom left within each model). Residues E162 and D377 can interact with the lysine sidechain of unmodified lisinopril, while H383 and H387 form part of the conserved HEXXH Zn2+-binding motif.
Time-Dependent Inactivation of sACE-1
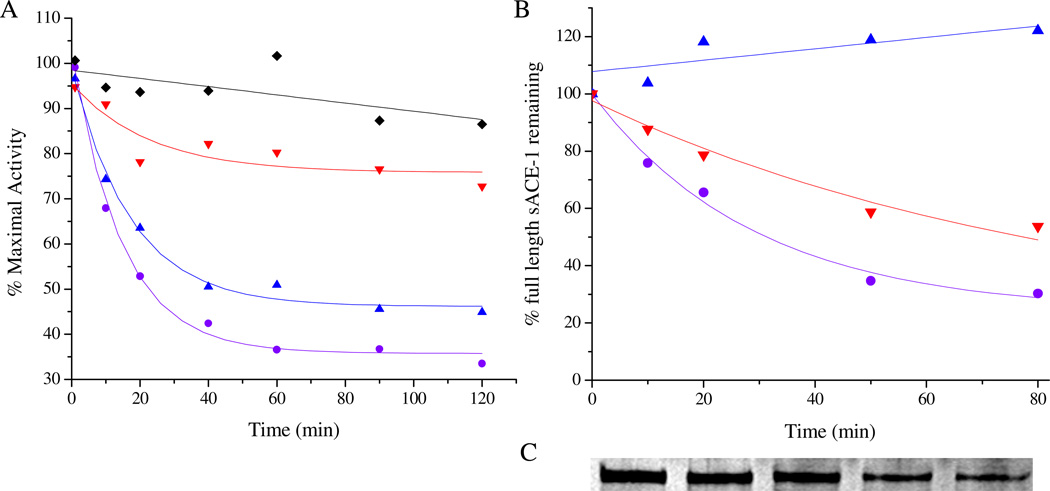
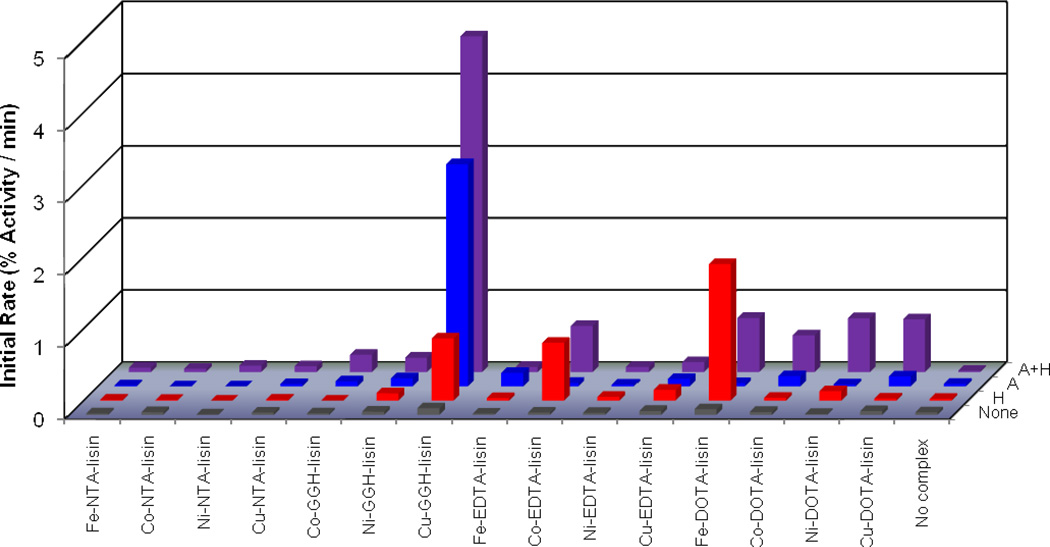
The time-dependent catalytic inactivation of sACE-1 by each M-chelate-lisinopril complex was determined by incubating each complex with sACE-1 in the presence of coreactants and assaying the substrate cleavage activity of sACE-1 at various time intervals during the incubation (Figure 4A). Catalytic inactivation of sACE-1 promoted by each M-chelate-lisinopril complex was tested aerobically under a variety of coreactant conditions that included 1 mM ascorbate + 1 mM H2O2, 1 mM ascorbate, 1 mM H2O2, and with no added coreactants. For catalysts and conditions promoting above-background initial rates of inactivation of sACE-1, the initial rates of cleavage of full length sACE-1 were determined by SDS-PAGE (Figures 4B/C). Comparisons between initial rates of inactivation and initial rates of protein cleavage (Table 2) allowed an assessment of whether the observed inactivation arose from either side chain modification or backbone cleavage. Initial rates of cleavage were significantly lower than the overall rates of inactivation, suggesting that the observed inactivation arose primarily from side chain oxidation, rather than backbone cleavage, except for several reactions with Fe- and Co-DOTA-lisinopril and Cu-GGH-lisinopril, for which the initial rate of cleavage was both > 50% of the initial rate of inactivation and greater than the background rate of cleavage. The observed rates for both inactivation and cleavage of full length sACE-1 by the redox active M-chelate-lisinopril catalysts depended on the presence of a coreactant.
Figure 4.
Time-dependent inactivation (A) and cleavage (B) of full length sACE-1 by Cu-GGH-lisinopril with the coreactants 1 mM ascorbate and 1 mM H2O2 (●), 1 mM ascorbate (▲), 1 mM H2O2 (▼), or no coreactant (♦). Time-dependent cleavage of sACE-1 was quantified by SDS-PAGE (C), shown here for the reaction of Cu-GGH-lisinopril with ascorbate and H2O2.
Table 2.
Initial rates for both inactivation and cleavage of full length sACE-1 by M-chelate-lisinopril complexes and reduction potentials of the attached M-chelates
| complexa | initial rates for inactivation {and cleavage} of full length sACE-1 (% sACE-1 / min) by M-chelate-lisinopril complexes and coreactantsb |
red. potential vs. NHE of M-chelate (mV)c |
|||
|---|---|---|---|---|---|
| ascorbate + H2O2 |
ascorbate | H2O2 | none | ||
| Fe-NTA-lisin | 0.06 ± 0.01 | 0.01 ± 0.02 | 0.01 ± 0.03 | 0.02 ± 0.02 | 464 |
| Co-NTA-lisin | 0.04 ± 0.04 | < 0.03 | 0.01 ± 0.02 | 0.04 ± 0.04 | 274 |
| Ni-NTA-lisin | 0.09 ± 0.01 | < 0.04 | < 0.02 | < 0.03 | 176 |
| Cu-NTA-lisin | 0.08 ± 0.02 | 0.03 ± 0.03 | 0.01 ± 0.03 | 0.03 ± 0.04 | 215 |
| Co-GGH-lisin | 0.24 ± 0.02 {< 0.2} | 0.07 ± 0.03 | < 0.06 | 0.02 ± 0.03 | −119 |
| Ni-GGH-lisin | 0.20 ± 0.04 {< 0.05} | 0.11 ± 0.03 {0.12 ± 0.08} | 0.10 ± 0.05 | 0.04 ± 0.07 | 1000 |
| Cu-GGH-lisin | 4.7 ± 0.2 {2.6 ± 0.6} | 3.1 ± 0.2 {< 0.09} | 0.9 ± 0.2 {0.6 ± 0.1} | 0.09 ± 0.04 | 1038 |
| Fe-EDTA-lisin | 0.07 ± 0.02 | 0.19 ± 0.03 {< 0.1} | 0.03 ± 0.03 | 0.02 ± 0.04 | 391 |
| Co-EDTA-lisin | 0.6 ± 0.2 {< 0.08} | 0.05 ± 0.01 | 0.8 ± 0.2 {0.29 ± 0.09} | 0.03 ± 0.03 | 146 |
| Ni-EDTA-lisin | 0.07 ± 0.03 | 0.02 ± 0.02 | 0.05 ± 0.03 | 0.03 ± 0.02 | 172 |
| Cu-EDTA-lisin | 0.14 ± 0.02 | 0.10 ± 0.03 {0.28 ± 0.07} | 0.15 ± 0.06 {< 0.09} | 0.06 ± 0.04 | 47 |
| Fe-DOTA-lisin | 0.75 ± 0.09 {0.43 ± 0.08} | 0.04 ± 0.02 | 1.9 ± 0.4 {< 0.04} | 0.08 ± 0.03 | 396 |
| Co-DOTA-lisin | 0.51 ± 0.04 {0.4 ± 0.2} | 0.14 ± 0.05 {0.1 ± 0.1} | 0.04 ± 0.03 | 0.04 ± 0.03 | 142 |
| Ni-DOTA-lisin | 0.75 ± 0.08 {< 0.01} | 0.03 ± 0.02 | 0.13 ± 0.01 {< 0.1} | 0.01 ± 0.04 | −35 |
| Cu-DOTA-lisin | 0.73 ± 0.03 {< 0.1} | 0.14 ± 0.03 {0.2 ± 0.1} | 0.03 ± 0.04 | 0.05 ± 0.05 | 180 |
| None | 0.02 ± 0.05 {0.17 ± 0.07} | 0.03 ± 0.03 {0.2 ± 0.2} | < 0.03 {< 0.2} | 0.04 ± 0.02 | |
lisin = lisinopril.
For reactions with initial rates of inactivation (monitored by fluorogenic substrate cleavage) that were above background (shown in bold), initial rates of cleavage of full length sACE-1 (monitored by SDS-PAGE) are listed for comparison (shown in brackets).
Reduction potentials of the M-chelate domains were determined previously;12 redox couples are 3+/2+ for Fe, Co, Ni-ATCUN, and Cu-ATCUN complexes and 2+/1+ for all other Ni and Cu complexes.
Catalytic inactivation of sACE-1 was observed for several M-chelate-lisinopril complexes, among which Cu-GGH-lisinopril was found to be the fastest. In the presence of both ascorbate and H2O2, rates of sACE-1 inactivation decreased in the order Cu-GGH-lisinopril > Fe-DOTA-lisinopril ~ Ni-DOTA-lisinopril ~ Cu-DOTA-lisinopril > Co-EDTA-lisinopril > Co-DOTA-lisinopril > Co-GGH-lisinopril ~ Ni-GGH-lisinopril. In the presence of ascorbate (without H2O2), rates of sACE-1 inactivation decreased in the order Cu-GGH-lisinopril > Fe-EDTA-lisinopril > Co-DOTA-lisinopril ~ Cu-DOTA-lisinopril ~ Ni-GGH-lisinopril ~ Cu-EDTA-lisinopril. In the presence of H2O2 (without ascorbate), rates of sACE-1 inactivation decreased in the order Fe-DOTA-lisinopril > Cu-GGH-lisinopril > Co-EDTA-lisinopril > Cu-EDTA-lisinopril ~ Ni-DOTA-lisinopril. Control experiments were performed for M-chelates lacking attached lisinopril, and under these conditions the rates of sACE-1 inactivation were found to be dramatically reduced (generally at least 10-fold reduction), reflecting the requirement for lisinopril to promote targeted inactivation of sACE-1 (Table 4 and Figure SM32). Catalytic inactivation of sACE-1 by M-chelate-lisinopril complexes was generally dependent on the presence of a redox coreagent (ascorbate, H2O2, or O2), each of which is physiologically available, and in most cases the combination of a reductant (ascorbate) with an oxidant (O2 or H2O2) yielded the fastest rates of inactivation. The rate of sACE-1 inactivation in the absence of either M-chelate-lisinopril complex or coreactant was negligible, and no substrate cleavage was observed with catalyst and coreactants in the absence of enzyme (Figure SM33). The initial rates of catalytic inactivation and cleavage of full length sACE-1 by each M-chelate-lisinopril complex, under each set of coreactant conditions, as well as previously determined reduction potentials for each attached M-chelate,12 are summarized in Table 2. All initial rates of inactivation are plotted in Figure 5 for visual comparison.
Table 4.
Second order rate constants for both inactivation and cleavage of full length sACE-1 by several M-chelate-lisinopril complexes and control experiment second order rate constants for the corresponding M-chelates lacking lisinopril.
| Complex a | k2 for inactivation {and cleavage} of full length sACE-1 (M−1min−1)b | Conditionsc | |
|---|---|---|---|
| M-chelate-lisinopril | without lisinopril | ||
| Cu-GGH-lisin | 152,000 ± 7,000 {70,000 ± 20,000} | 10,000 ± 5,000 {< 3,000} | Ascorbate + H2O2 |
| Fe-EDTA-lisin | 110,000 ± 30,000 {< 100,000} | 10,000 ± 50,000 {< 100,000} | Ascorbate |
| Cu-GGH-lisin | 102,000 ± 6,000 {< 6,000} | < 1,000 {--d} | Ascorbate |
| Co-GGH-lisin | 70,000 ± 40,000 {< 70,000} | < 20,000 {--d} | Ascorbate + H2O2 |
| Co-EDTA-lisin | 40,000 ± 10,000 {10,000 ± 7,000} | < 4,000 {--d} | H2O2 |
| Co-EDTA-lisin | 30,000 ± 10,000 {< 3,000} | < 3,000 {--d} | Ascorbate + H2O2 |
| Ni-GGH-lisin | 30,000 ± 20,000 {< 60,000} | < 5,000 {--d} | Ascorbate |
| Cu-GGH-lisin | 28,000 ± 9,000 {15,000 ± 6,000} | < 2,000 {--d} | H2O2 |
| Fe-DOTA-lisin | 22,000 ± 5,000 {< 2,000} | < 600 {--d} | H2O2 |
| Fe-DOTA-lisin | 8,000 ± 1,000 {3,000 ± 1,000} | < 400 {--d} | Ascorbate + H2O2 |
| Ni-DOTA-lisin | 7,000 ± 1,000 {< 600} | < 700 {--d} | Ascorbate + H2O2 |
| Cu-DOTA-lisin | 6,200 ± 900 {< 1,000} | < 500 {--d} | Ascorbate + H2O2 |
| Cu-EDTA-lisin | 5,000 ± 4,000 {< 9,000} | < 3,000 {--d} | H2O2 |
| Co-DOTA-lisin | 5,000 ± 1,000 {2,000 ± 2,000} | < 700 {--d} | Ascorbate + H2O2 |
| Cu-EDTA-lisin | 3,000 ± 2,000 {2,000 ± 9,000} | 2,000 ± 3,000 {--d} | Ascorbate |
| Cu-DOTA-lisin | 1,000 ± 400 {< 2,000} | < 1,000 {--d} | Ascorbate |
lisin = lisinopril.
Second order rate constants for inactivation of sACE-1 (measured by substrate cleavage) and for cleavage of full length sACE-1 (monitored by SDS-PAGE) are listed for comparison (shown in brackets).
All experiments throughout this study were performed at pH 7.4, 37 °C.
Not determined.
Figure 5.
Initial rates for inactivation of sACE-1 by M-chelate-lisinopril complexes with the coreactants 1 mM ascorbate and 1 mM H2O2 (A + H), 1 mM ascorbate (A), 1 mM H2O2 (H), or no coreactant (None).
Characterization of Inactivated sACE-1
Incubation of sACE-1 with M-chelate-lisinopril complexes and coreactants typically reduced enzyme activity to a non-zero activity; residual activity was observed even after several hours of incubation and appeared to be the result of either quantitative conversion of sACE-1 to forms displaying reduced activity, through either modification of amino acid side chains or cleavage of the protein backbone (or both) that diminished, but did not completely abolish, substrate binding and hydrolysis,15 or in part through depletion of redox coreactants, since addition of fresh ascorbate and peroxide resulted in further modest reductions in activity. Following inactivation with each M-chelate-lisinopril complex and coreactants, the substrate cleavage activity of inactivated sACE-1 was monitored using a range of substrate concentrations, and the percentage of full length sACE-1 remaining was independently quantified by SDS-PAGE with silver staining (Figure SM37, Table SM6). The Michaelis-Menten kinetic parameters kcat, KM, and kcat/KM for substrate cleavage by inactivated sACE-1 as well as the percentage of full length sACE-1 remaining (% that was not cleaved) after preincubation are listed in Table 3. The predominant mechanism of sACE-1 inactivation for each catalyst (side chain modification vs protein cleavage) was assessed by comparison of the remaining percentage of kcat/KM and the percentage of full length sACE-1 remaining following preincubation. Inactivation of sACE-1 appeared to occur primarily as a result of side chain modification, rather than backbone cleavage, since the observed decrease in full length sACE-1 remaining was typically not significant enough to account for even half of the larger overall decreases in kcat/KM; this result is consistent with the fact that higher initial rates were observed for inactivation than for cleavage. Additionally, it is possible that certain backbone cleavage events could arise only as a result of initial side chain oxidation. The enzyme efficiency (kcat/KM) of inactivated sACE-1 was observed to decrease by as much as 12-fold (for Cu-GGH-lisinopril) relative to unmodified sACE-1, and this decreased enzyme efficiency resulted from an increased KM, a decreased kcat, or both, depending on the complex used, and conversion from active to inactivate enzyme required the presence of both M-chelate-lisinopril and coreactants (Figures SM35–SM36). The reaction pathway for sACE-1 inactivation appeared to vary depending on the M-chelate-lisinopril complex used. Increased KM values, which most likely reflect oxidation of sACE-1 residues that contribute to substrate binding by sACE-1, were observed for all complexes except for Co- and Ni-GGH-lisinopril and Ni- and Cu-DOTA-lisinopril, and increases in KM were highest for M-EDTA-lisinopril, Co-NTA-lisinopril, and Fe-DOTA-lisinopril complexes. Decreases in kcat, reflecting either protein cleavage or modification of active site residues directly involved in catalytic hydrolysis of substrate were observed for all complexes, with the exception of Fe-, Co-, and Ni-NTA-lisinopril and Fe- and Ni-EDTA-lisinopril. The magnitude of the decrease in kcat was greatest for M-GGH-lisinopril, M-DOTA-lisinopril, and Cu-chelate-lisinopril complexes. The relative changes in the kcat and KM of sACE-1, as well as the relative level of sACE-1 cleavage, that occurred upon inactivation by each M-chelate-lisinopril complex provide useful information regarding which type of active site residues (substrate binding vs substrate hydrolysis) were modified, or alternatively, the degree to which the backbone of sACE-1 was cleaved. Michaelis-Menten parameters for inactivated sACE-1, after inactivation with each M-chelate-lisinopril complex and coreactants, are summarized in Table 3 and are complimentary to the results for time-dependent inactivation of sACE-1.
Table 3.
Characterization of inactivated sACE-1. Michaelis-Menten kinetic parameters for cleavage of the substrate Mca-RPPGFSAFK(Dnp)-OH by inactivated sACE-1, and percentage of full length sACE-1 remaining, following preincubation of sACE-1 with M-chelate-lisinopril complexes and coreactants
| Complex a | Kinetic parameters for substrate cleavage by inactivated sACE-1 b | % full length sACE-1 remaining after incubation c |
||
|---|---|---|---|---|
| kcat (min−1) | KM (μM) | kcat/KM (min−1μM−1) | ||
| Fe-NTA-lisin | 1,180 ± 80 | 9 ± 2 | 130 ± 30 (48%) | 86 ± 2 |
| Co-NTA-lisin | 1,500 ± 200 | 13 ± 4 | 110 ± 40 (41%) | 91 ± 3 |
| Ni-NTA-lisin | 1,200 ± 100 | 8 ± 2 | 150 ± 40 (56%) | 96 ± 1 |
| Cu-NTA-lisin | 910 ± 10 | 7.5 ± 0.3 | 122 ± 6 (45%) | 90 ± 20 |
| Co-GGH-lisin | 750 ± 20 | 4.6 ± 0.5 | 160 ± 20 (59%) | 80 ± 20 |
| Ni-GGH-lisin | 840 ± 30 | 6.3 ± 0.6 | 130 ± 10 (48%) | 70 ± 20 |
| Cu-GGH-lisin | 150 ± 10 | 7 ± 1 | 22 ± 4 (8.1%) | 50 ± 40 |
| Fe-EDTA-lisin | 1,350 ± 40 | 10.9 ± 0.9 | 120 ± 10 (44%) | 80 ± 30 |
| Co-EDTA-lisin | 780 ± 10 | 11.8 ± 0.3 | 66 ± 2 (24%) | 89 ± 7 |
| Ni-EDTA-lisin | 1,510 ± 50 | 11 ± 1 | 140 ± 10 (52%) | 85 ± 1 |
| Cu-EDTA-lisin | 218 ± 3 | 11.1 ± 0.4 | 19.7 ± 0.9 (7.3%) | 40 ± 30 |
| Fe-DOTA-lisin | 540 ± 20 | 15 ± 1 | 36 ± 4 (13%) | 87 ± 1 |
| Co-DOTA-lisin | 840 ± 10 | 8.7 ± 0.3 | 97 ± 3 (36%) | 91 ± 9 |
| Ni-DOTA-lisin | 610 ± 30 | 5.6 ± 0.7 | 110 ± 20 (41%) | 90 ± 30 |
| Cu-DOTA-lisin | 520 ± 20 | 6.0 ± 0.8 | 90 ± 10 (33%) | 74 ± 1 |
| None | 1,220 ± 50 | 4.5 ± 0.6 | 270 ± 40 (100%) | 100 ± 8 |
lisin = lisinopril.
kcat values below background, KM values above background, and kcat/KM values below background are shown in bold. The % kcat/KM remaining after each preincubation, relative to control, are shown in parentheses to allow comparison with % full length sACE-1 remaining. The concentration of each complex used for preincubation corresponded to 20% saturation of sACE-1, and ascorbate and H2O2 were each present at 1 mM.
Monitored by SDS-PAGE.
Kinetic Stability of M-Chelate Complexes
Of potential concern was the degree to which the stability of each M-chelate-lisinopril complex was affected by the mandatory presence of Zn2+, because sACE-1 is a zinc-dependent enzyme. The ability of Zn2+ to exchange with Fe3+, Co2+, Ni2+, and Cu2+ for the complexes studied was evaluated by incubating Zn2+ and each M-chelate (both present at 100 µM) at 37 °C. Exchange was monitored by UV/vis spectroscopy by following the change in absorbance at the specified wavelengths (Figure SM40) for each M-chelate over time. All complexes were effectively stable in the presence of Zn2+, although very slow exchange was observed for only Fe-EDTA, Co-EDTA, and Fe-DOTA, with second order rate constants for exchange of 350 ± 10, 480 ± 20, and 3.1 ± 0.1 M−1min−1, respectively (Table SM7). Under the conditions used for sACE-1 binding and catalytic inactivation experiments, exchange with Zn2+ for all species was insignificant. For example, displacement of iron from 1 µM Fe-EDTA by 10 µM Zn2+ would occur at a calculated initial rate of only 3.5 × 10−9 M/min, or 0.35 % of the metal complex per minute. The fact that both sACE-1 binding and sACE-1 inactivation by Fe-DOTA-, Fe-EDTA-, and Co-EDTA-lisinopril followed expected trends further supports the observation that these complexes were effectively stable under the conditions used.
DISCUSSION
Concentration-Dependent Inactivation of sACE-1
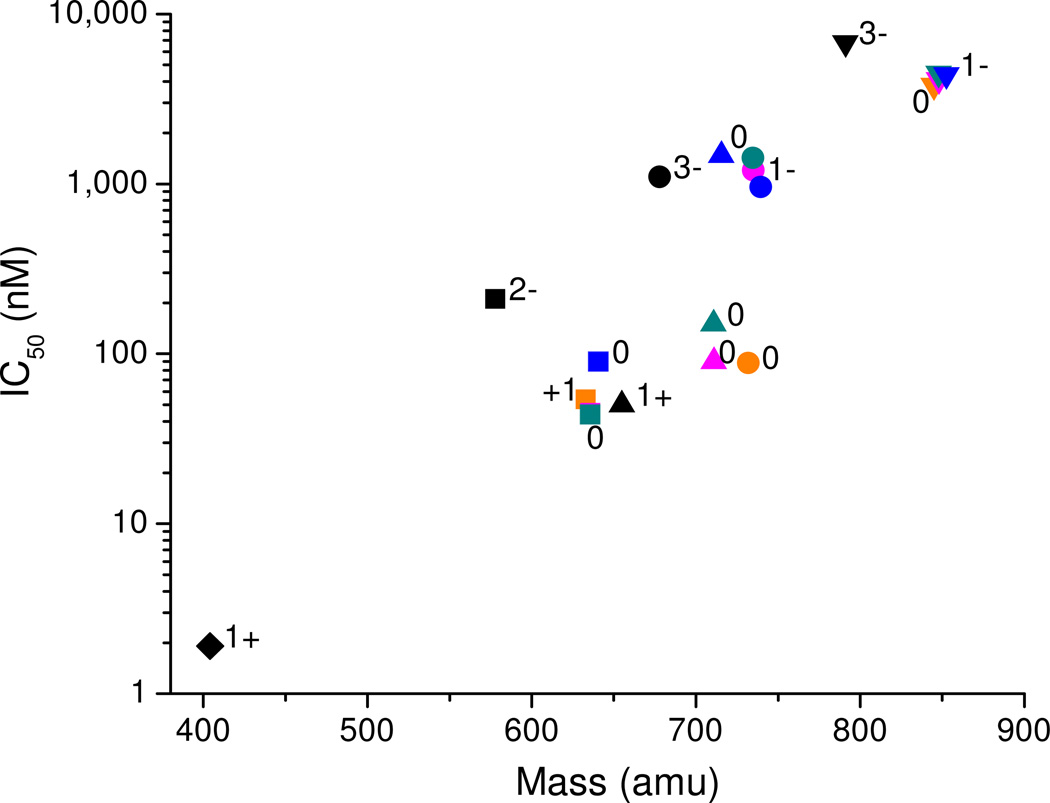
The ability of each M-chelate-lisinopril complex to catalytically inactivate sACE-1 was dependent upon both sACE-1 binding-affinity and the kinetics of irreversible sACE-1 inactivation by M-chelate-lisinopril complexes following active site binding. Prior to studying the kinetics of irreversible sACE-1 inactivation by the M-chelate-lisinopril complexes, the relative affinities of each M-chelate-lisinopril complex for sACE-1 were determined. Each M-chelate-lisinopril complex was tested for the ability to reversibly inhibit sACE-1 activity, and for each M-chelate-lisinopril species the concentration-dependence was expressed as the half-maximal inhibitory concentration (IC50). The sACE-1 binding-affinities of M-chelate-lisinopril species were greatly enhanced relative to the respective M-chelates lacking lisinopril, as expected, and the sACE-1 binding-affinities of the M-chelate-lisinopril species were found to be inversely correlated with both the size and negative charge of the attached M-chelates (Figure 6), whereas the degree of coordination unsaturation of the attached M-chelate appeared to variably affect the sACE-1 binding-affinity.
Figure 6.
sACE-1 binding-affinity of M-chelate-lisinopril complexes was inversely correlated with the size and negative charge of the species attached to the lysine sidechain of lisinopril. Lisinopril and all M-chelate-lisinopril species (metal-bound and metal-free) are shown: ♦ = lisinopril; ■ = NTA-lisinopril; ▲ = GGH-lisinopril; ● = EDTA-lisinopril; ▼ = DOTA-lisinopril. Orange = Fe; pink = Co; cyan = Ni; blue = Cu; black = no metal. The charge of the modified lysine sidechain of lisinopril is listed for each attachment; the charge for each Fe3+ complex was 1+ higher than for each corresponding M2+ complex.
Attachment of smaller M-chelates to lisinopril, as in the case of M-NTA-lisinopril, resulted in tighter binding to the active sites of sACE-1 (lower IC50 values), whereas increases in size of the attached M-chelate (M-NTA < M-GGH < M-EDTA < M-DOTA) incrementally reduced the binding affinity for sACE-1. The M-DOTA-lisinopril complexes, which contain the largest M-chelates, had the weakest ACE-affinity among the M-chelate-lisinopril complexes tested. Structural modeling confirmed this correlation, since lisinopril-attached M-DOTA had difficulty entering the active site of both N-domain ACE and C-domain ACE during energy minimization, whereas lisinopril-attached M-NTA, M-GGH, and M-EDTA all preferred localization within the active site binding pockets of ACE. Additionally, the planar profile of the attached M-chelate appeared to contribute to ACE-affinity. M-GGH-lisinopril complexes that contain relatively low-profile square-planar M-GGH chelates tended to have lower IC50 values. The sole exception was Cu-GGH-lisinopril, possibly reflecting a distinct structure compared to other M-GGH-lisinopril complexes in the unbound state as a result of intramolecular coordination of one of the two lisinopril carboxylates to an axial coordination site of the Cu-GGH-lisinopril complex. Indeed, the square-planar Cu-GGH complex is known to weakly coordinate a 5th oxygen ligand at one of the two axial coordination sites, probably as a result of Jahn-Teller distortion of the d9 Cu2+ center,33 and energy-minimized 3D models of Cu-GGH-lisinopril are consistent with the required geometry. Cu-GGH-lisinopril could alternatively accommodate intermolecular axial coordination to a nearby amino acid side chain from sACE-1, such as D140 from the N-terminal domain or E162 or D377 from the C-terminal domain, although this is less likely, as evidenced by the higher IC50 value. The M-NTA-lisinopril complexes similarly possess two empty metal-coordination sites, which also have the potential for either intramolecular coordination to lisinopril carboxylates or intermolecular coordination to sACE-1 amino acid sidechains. The latter most likely contributes to the observed high-affinity binding to sACE-1 by the M-NTA-lisinopril complexes. M-NTA-lisinopril complexes differ from Cu-GGH-lisinopril in that the empty metal-coordination sites of M-NTA-lisinopril appear to favor sACE-1 binding through coordination of sACE-1 residues, whereas sACE-1 binding appears to be abrogated by empty coordination sites in the case of Cu-GGH-lisinopril, most likely through distinct modes of intermolecular and intramolecular metal-coordination, respectively. Another key difference between the metal-coordination environments of M-NTA- and M-GGH-lisinopril is that the two empty coordination sites of each exist in cis and trans orientations, respectively, and this difference could partially contribute to their distinct binding affinities for sACE-1.
In addition to steric and metal-coordination effects, the charge of the M-chelates attached to lisinopril appeared to affect the affinity of the M-chelate-lisinopril complexes toward sACE-1. The charge (z) of the M-chelates coupled to the lysine sidechain of lisinopril increased from negative to positive in the order M-DOTA = M-EDTA (z = −1) < M-GGH = M-NTA (z = 0) < uncoupled amine (z = +1) for non-iron complexes, and the observed sACE-1 binding-affinity followed the same trend. Metal-bound chelators typically had a more positive charge than metal-free chelators (except for GGH-lisinopril) as a result of the positive charge of the bound metal, and these followed the same trend. Similarly, iron complexes of EDTA-lisinopril and DOTA-lisinopril were observed to possess significantly higher sACE-1 binding-affinity than the analogous complexes with cobalt, nickel, and copper, as a result of the greater 3+ oxidation state relative to the other metals. Indeed, X-ray crystal structures of the lisinopril/ACE co-complex show a close proximity between the negatively-charged ACE residues (D140 for N-domain ACE; D377 and E162 for tACE) and the positively-charged lysine sidechain of ACE-bound lisinopril,5,6 and the resulting electrostatic attraction most likely contributes to the high-affinity binding of sACE-1 by lisinopril. Conversion of the positively-charged lysine sidechain of lisinopril to a negative or neutral M-chelate is expected to negatively impact the sACE-1 binding affinity to some degree, as is observed. Lisinopril had the highest affinity for sACE-1 (IC50 = 1.9 nM) of all species studied, which punctuates the inverse correlations between sACE-1 binding-affinity and both size and negative charge of attached M-chelates. However, the binding interaction between lisinopril and sACE-1 was sufficiently tolerant of attachment of M-chelates to lisinopril that high-affinity binding was retained for each of the M-chelate-lisinopril species, and this high-affinity binding is a prerequisite for targeted catalytic inactivation of sACE-1 by M-chelate-lisinopril complexes.
Domain Selectivity
A possible consequence of attachment of M-chelates to lisinopril is that the modest selectivity of lisinopril for the C-domain of sACE-1, which is thought to result from the higher negative charge density within the C-domain active site that lies in close proximity to the lysine sidechain of lisinopril,6 could become reversed, so that binding to the N-domain is more favored. Keeping in mind that previous studies have demonstrated the N-domain to be a slightly more efficient catalyst of Angiotensin I hydrolysis than the C-domain (kcat/KM = 13 µM−1min−1 for N-domain vs 9.6 µM−1min−1 for C-domain and 11 µM−1min−1 for sACE-1), despite similar substrate binding affinities (KM values ranging approximately 17 to 19 µM),37 and that there is much evidence for unique critical roles of the N-domain in hematopoietic stem cell regulation,38 the inversion of inhibitor selectivity in favor of binding to the N-domain could be beneficial for certain therapeutic uses. The inverse correlation between negative charge and binding-affinity most likely applies to both active sites. However, our experiments with somatic ACE-1 and the substrate Mca-RPPGFSAFK(Dnp)-OH, which is derived from bradykinin (RPPGFSPFR), a C-domain selective substrate,39 most likely reflect binding/inactivation of the C-domain and therefore provide no assessment of domain selectivity. Consistent with this fact, reported kcat/KM values for hydrolysis of a fluorogenic substrate similar to that used herein (Abz-LFK-Dnp-OH) by N-domain ACE, tACE (identical to C-domain), and sACE-1 are 10 µM−1min−1 (kcat = 60 min−1), 380 µM−1min−1 (kcat = 1,240 min−1), and 220 µM−1min−1 (kcat = 798 min−1), respectively,40 although the exact domain selectivity of the substrate Mca-RPPGFSAFK(Dnp)-OH is unknown. Clarification of the domain selectivity of the M-chelate-lisinopril complexes will be the focus of future studies.
Kinetics and Mechanism of Catalytic Inactivation
Catalytic inactivation of sACE-1 was observed for most of the M-chelate-lisinopril complexes investigated in this work, and these complexes provided a remarkable range of reactivity. Catalytic inactivation of sACE-1 by M-chelate-lisinopril complexes was found to be dependent upon the ability of lisinopril to localize each attached M-chelate to the active sites of sACE-1 and the specific properties of each M-chelate, including the reduction potential, coordination environment, and identity of the transition metal present within each M-chelate. M-chelate-lisinopril reactivity was amplified by the presence of physiologically available redox coreactants, which provided the driving force responsible for the observed reactivity. The presence/absence of the coreactants ascorbate and H2O2, which have reported physiological concentrations ranging from µM-mM and pM-µM, respectively, were varied to evaluate redox mechanism and coreactant selectivity.41,42 We believe the catalytic reaction to be similar for ascorbate/O2 and ascorbate/peroxide systems, with the introduction of peroxide providing a mechanistic “shunt”, with cytochrome P450 providing good precedent for formation of peroxide at a catalytic redox center from O2 and an electron source.43
The ability of each M-chelate-lisinopril complex to catalyze irreversible inactivation of sACE-1 was attributed to the known ability of M-chelates to facilitate conversion of the oxidants O2 or H2O2 to reactive oxygen species (ROS) through single electron transfer,44–46 followed by the reaction of nascent ROS with nearby sACE-1 residues, resulting in active site destruction. Meanwhile, ascorbate, a single electron reductant, is known to function as a pro-oxidant by re-reducing oxidized metal centers following generation of ROS.46–48 Therefore, in the presence of both oxidant and reductant, M-chelate complexes possess the ability to undergo multiple turnovers of oxidation/reduction, catalyzing production of multiple ROS per metal center in the process (Figures SM41–SM43). This ability of M-chelates to function with multiple turnovers was previously observed to be optimal when the M-chelate reduction potential is poised between the reduction potentials of the reductant and oxidant half reactions, since both reduction and oxidation of the M-chelate are thermodynamically favored in this range—a similar theme is employed by native superoxide dismutase (SOD), in which the reduction potential of SOD is poised between those of the two relevant half reactions.49,50 The reduction potentials for ascorbyl radical/ascorbate and H2O2/OH• at neutral pH are −66 mV and 380 mV,51,52 respectively, and we demonstrated previously that M-chelates with reduction potentials within this range provide the fastest rates of consumption of redox coreactants, ROS generation, and multiple-turnover redox cycling of the M-chelate. As a result, M-chelate-mediated cleavage of both DNA and RNA was observed to be optimal within this range, while reduction potentials outside this range provided very mild levels of reactivity.12,26 However, a key difference between nucleic acid and protein targets, such as sACE-1, is that proteins tend to be more stable than nucleic acids toward oxidative insults.36,53 Therefore, nucleic acid cleavage by M-chelates appears to be optimized by selection of M-chelates with reduction potentials that lie close to those of relevant coreactants, with facile subsequent cleavage of nucleic acids by nascent ROS. By contrast, the efficiency of enzyme inactivation by M-chelates is more likely to require optimization of catalyst reduction potentials to match specific amino acid and/or protein backbone oxidation requirements, which are more demanding than for nucleic acids. For example, previously determined reduction potentials for Tyr•+/Tyr, Trp•+/Trp, and His•+/His at pH 7 are 930 mV, 1015 mV, and 1170 mV, respectively,36,53 and these residues each exist within the active sites of sACE-1.5,6 Indeed the fastest rates of sACE-1 inactivation were observed for Cu-GGH-lisinopril, and the Cu-GGH chelate displays the highest reduction potential of the M-chelates tested, E° = 1038 mV. Accordingly, inactivation of sACE-1 appears to be mediated by oxidation of active site residues, and to a lesser extent, cleavage of the protein backbone (Table 2), consistent with our previous observations for metallopeptide-mediated inactivation of human carbonic anhydrase,15 as well as the reduced catalytic efficiency of inactivated sACE-1.
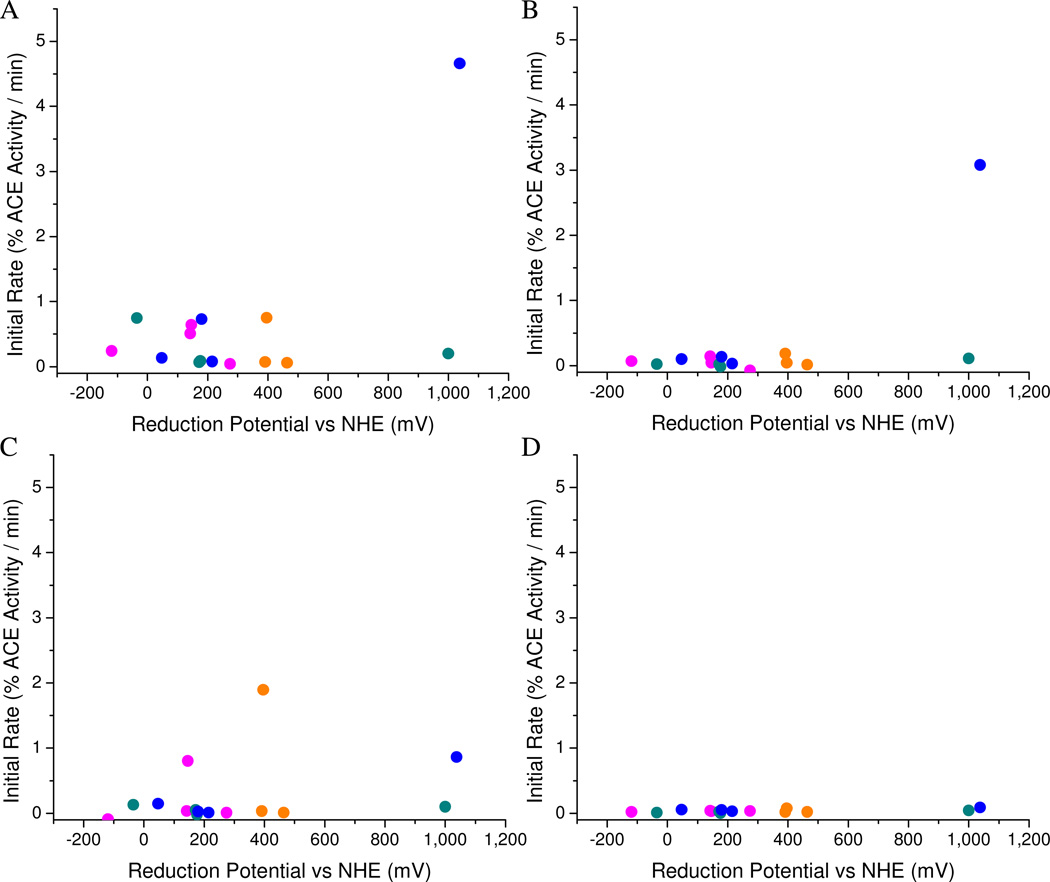
Two hot spots were observed in the relationship between rate of sACE-1 inactivation and M-chelate reduction potential; namely, a low potential region and a high potential region (Figure 7). The low potential region, which ranged from approximately −100 mV to 400 mV, provided relatively mild levels of sACE-1 inactivation, and M-chelate reactivity in this range was attributed to the redox cycling described above, where H2O2 is converted to hydroxyl radicals, and ascorbate re-reduces the oxidized metal center, allowing thermodynamically favored multiple turnovers and ROS generation. Indeed activity in this range was highest when both H2O2 and ascorbate were present. The high potential region, which corresponded to M-chelate reduction potentials near 1000 mV, provided the most efficient catalytic inactivation of sACE-1. M-chelates in this range (Cu-GGH and Ni-GGH) have greater oxidizing power, although prerequisite generation of the oxidized metal is less thermodynamically favored and probably requires an alternate mechanism. The most likely mechanism involves (1) metal-mediated reduction of O2 by ascorbate to form metal-bound superoxide, (2) single electron oxidation of the reduced metal center by metal-bound superoxide, forming H2O2 (possibly metal-bound) with the possibility of subsequent Fenton chemistry resulting in a metal-associated reactive oxygen species, and (3) reaction of this activated species with nearby sACE-1 residues, resulting in sACE-1 inactivation. Each step of this mechanism is supported, respectively, by the facts that (1) Ni-GGH-lisinopril and Cu-GGH-lisinopril are reactive in the presence of the combination of O2 and ascorbate, the addition of H2O2 further stimulates reactivity (mechanistic shunt), and the lack of diffusion of superoxide radicals in our previous work suggests that the superoxide intermediate is metal-bound (Figure SM42);26 (2) the reduction potential of the superoxide/H2O2 half reaction (E° = 890 mV) lies closest to 1000 mV of any of the possible single-electron half reactions between O2, superoxide, H2O2, and hydroxyl radical,51 and superoxide is therefore the most likely species to oxidize the metal center; and (3) these M-chelate-lisinopril complexes catalyzed irreversible inactivation of sACE-1 with high efficiency. Interestingly, in our previous work on M-chelate promoted nucleic acid cleavage we observed that activity was centered in the same two regions (low and high potential range of reduction potential), although nucleic acid cleavage was significantly faster for the low potential region than for the high potential region. This pattern now appears to be inverted in the case of protein (sACE-1) inactivation and most likely reflects the different chemical makeup of the targets (protein vs nucleic acid) and relative difficulty of oxidation of protein sidechains and/or backbone versus (deoxy)ribose rings.
Figure 7.
Initial rates of aerobic sACE-1 inactivation by M-chelate-lisinopril complexes and added coreactants: (A) H2O2/ascorbate, (B) ascorbate, (C) H2O2, (D) none. Orange = Fe; pink = Co; cyan = Ni; blue = Cu. Rates depended on the coreactants present and were highest for M-chelates with reduction potentials near 1000 mV (high region); and modest rates were seen for reduction potentials (low potential region) between those of H2O2/hydroxyl radical (E° = +380 mV) and ascorbyl radical/ascorbate (E° = −66 mV). Rates of catalysis were negligible in the absence of added coreactant.
The coreactant dependence of the catalytic activity of each M-chelate-lisinopril species appeared to be related to the reduction potential, coordination environment, and electronic properties of each attached M-chelate. For instance, Cu-GGH-lisinopril and Ni-GGH-lisinopril each possess reduction potentials near 1000 mV. However, observed reactivity was much higher for Cu-GGH-lisinopril than for Ni-GGH-lisinopril. Several factors could contribute to the enhanced reactivity of the copper site. In particular, the d9 configuration of Cu2+ is subject to the Jahn-Teller effect that will labilize the non-chelating axial sites and facilitate coreactant and reactant binding and product release, whereas Ni2+ is a relatively stable square planar d8 system. Also, oxidation of Cu2+ yields the electronically stable square planar d8 Cu3+ center, relative to Ni2+ oxidation where it is lost, and so the former reaction is expected to have lower activation barrier and be kinetically favored. Similarly, the iron complexes Fe-DOTA-lisinopril, Fe-EDTA-lisinopril, and Fe-NTA-lisinopril each possess reduction potentials near 400 mV, but show different coreactant selectivities. Fe-DOTA-lisinopril (and Co-EDTA-lisinopril) preferred reaction with the co-oxidant H2O2, with no obvious enhancement following addition of ascorbate, whereas Fe-EDTA-lisinopril preferred reaction with O2 and ascorbate; Fe-NTA-lisinopril surprisingly had no apparent reactivity.45 We have previously observed that both Fe-DOTA and Co-EDTA produce hydroxyl radicals in the presence of H2O2, but not O2 (Figure SM42), and that these complexes do not readily consume ascorbate (Figure SM41), consistent with the lack of enhancement of catalytic activity with added ascorbate for these two complexes.26 We also previously observed that Fe-EDTA reacts with either O2/ascorbate or H2O2/ascorbate to efficiently produce superoxide radicals or hydroxyl radicals, respectively (Figure SM42).26 The fact that inactivation of sACE-1 by Fe-EDTA-lisinopril was more dependent on O2 than H2O2 suggests that superoxide radicals more effectively inactivated sACE-1 than hydroxyl radicals. It was surprising that no catalytic activity was observed for Fe-NTA-lisinopril, since Fe-NTA was also previously found to react rapidly with both H2O2/ascorbate and O2/ascorbate to produce ROS, and so the lack of reactivity of M-NTA-lisinopril complexes in general may reflect the fact that the two cis empty coordination sites of M-NTA-lisinopril are actually occupied by an active site sACE-1 residue, blocking reaction with coreactants,44 consistent with the observed high-affinity binding of sACE-1 by the M-NTA-lisinopril species.
The nature of the ROS produced by each M-chelate-lisinopril complex (superoxide, H2O2, or hydroxyl radical) and whether the ROS was metal-bound or diffusible were likely to be key factors in determining the catalytic efficiency of sACE-1 inactivation for each M-chelate-lisinopril complex, since the most critical contribution to catalysis, other than binding-affinity, appears to be the step in which ROS or ROS-activated M-chelates react directly with active site sACE-1 residues. Our data show no correlation between reactivity and the rate of production of diffusible hydroxyl radical or superoxide species, as reflected by rates of ascorbate consumption (Figure SM41) or reaction with TEMPO and rhodamine B radical traps (Figures SM42–SM43).26 Cu-GGH-lisinopril, the most effective catalyst, appears most likely to inactivate sACE-1 through an activated metal-bound ROS intermediate, rather than through the production of diffusible radicals, and so the most effective M-chelate-lisinopril intermediates were likely to be metal-bound ROS, since a metal-bound ROS would be held in close proximity to the active site for an extended period of time. Similarly, the complexes Ni-GGH, Co-GGH, Co-DOTA, Ni-DOTA, Cu-DOTA were each found to effectively inactivate sACE-1, but none of these complexes was found to produce diffusible radicals in our previous studies (Figures SM42–SM43).26 Even for complexes known to produce diffusible radicals, such as Fe-EDTA, Fe-NTA, Fe-DOTA, Co-EDTA, the ROS produced are likely to remain bound to the metals where they originated for a significant period of time before diffusing, and this metal-bound state is most likely responsible for most of the observed catalytic inactivation of sACE-1 for Fe-EDTA-, Fe-DOTA-, and Co-EDTA-lisinopril. We have also observed this proximity effect in previous studies involving targeted cleavage of HIV RRE RNA by Rev-coupled M-chelates, in which the most effective M-chelates were those not observed to produce diffusible radicals.12 Those M-chelate-lisinopril complexes, such as Cu-GGH-lisinopril, that are effective catalysts but do not produce a damaging abundance of diffusible ROS, are likely to possess the best characteristics for physiological use due to their selective nature of targeted catalytic inactivation. In fact, the in vivo abundance of native ATCUN (Amino Terminal Cu- and Ni-binding) motifs, such as GGH, in albumins,33,34,54,55 histatins,56,57 and neuromedin C,58 typically used for Cu- and Ni-transport, as well as selective oxidation chemistry, most likely reflects both the placement of the Cu-ATCUN and Ni-ATCUN reduction potentials outside of the typical physiological window and the lack of diffusible ROS produced by these complexes. In this study, we have effectively hijacked the natural Cu-ATCUN complex for the purpose of targeted inactivation of sACE-1.
To elucidate the overall catalytic efficiencies of irreversible sACE-1 inactivation and cleavage by the synthesized M-chelate-lisinopril complexes, second order rate constants were established by use of equation (2) and are summarized in Table 4. M-chelate-lisinopril complexes were found to inactivate sACE-1 with a remarkable range of second order rate constants, as high as 150,000 M−1min−1, and many were able to do so under physiologically relevant conditions. Attachment to lisinopril was necessary for targeted inactivation of sACE-1 by M-chelate-lisinopril complexes, as evidenced by the large increase in second order rate constants for inactivation of sACE-1 upon attachment of M-chelates to lisinopril. The magnitude of each second order rate constant was dependent primarily on the relative ability of each M-chelate-lisinopril catalyst to both bind sACE-1 with high-affinity and irreversibly modify active site residues of sACE-1 following binding.
CONCLUSIONS
Catalytic metallodrugs possess the potential for highly efficient multiple-turnover, irreversible inactivation of therapeutic targets. Here we have demonstrated that through attachment of a variety of reactive metal chelates to a targeting ligand (lisinopril), a remarkable range of catalytic activities toward a targeted enzyme (sACE-1) can be achieved, and further, that it is possible to tune this reactivity to physiological optimums by varying parameters of the attached metal complex, including size, charge, coreactant selectivity, coordination environment, and reduction potential. We have illustrated that the size and charge of the attached metal complex greatly affect the relative ability of each catalyst to bind sACE-1 with high affinity, and therefore, to efficiently catalyze inactivation of sACE-1, and these results are likely to prove useful in further development of ACE inhibitors, while similar trends are likely to apply in the development of metallodrugs for other targets. We found that catalytic sACE-1 inactivation was most effective when the catalyst reduction potential was near 1000 mV, in contrast to previous studies of nucleic acid cleavage, where highest activity was observed when the catalyst reduction potential was poised between those of the milder physiological coreactants;12,26 the relative extremity of the catalyst reduction potential favored for sACE-1 inactivation most likely reflects the increased stability of enzymes relative to nucleic acids toward oxidation and the increased oxidative power required to modify most amino acids. The ability of each catalyst to inactivate sACE-1 appeared to depend to a great extent upon the mechanism employed, and catalysts that inactivated sACE-1 by a metal-bound ROS appeared to function more efficiently than those known to produce diffusible ROS. Catalysts that proceed through metal-bound, rather than diffusible, ROS are likely to prove more valuable for physiological use, since diffusible ROS would be hazardous to non-targeted tissues. This class of compounds possesses a significant advantage over the current reversible inhibitor lisinopril, in that irreversible inactivation of sACE-1 is achieved, and this work marks significant progress toward the ultimate goal of the design of highly-efficient multiple-turnover metallodrugs.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health [HL093446 and AA016712]. Jeff Joyner was supported by an NIH Chemistry/Biology Interface training grant (T32 GM08512). Lalintip Hocharoen was supported by a fellowship from the Ministry of Science and Technology, Thailand. Dr. Michael Freitas kindly allowed use of his LCQ-Deca mass spectrometer, and the Bruker MicroTOF instrument used for all other mass analysis was provided by a grant from the Ohio BioProducts Innovation Center. We thank Jingwei Li for his kind assistance with NMR collection and analysis.
Footnotes
Supporting Information Available. HPLC traces, mass spectra, 1H-NMR spectra, metal ion titrations, and extinction coefficients for synthesized M-chelate-lisinopril complexes, concentration-dependent sACE-1 inhibition plots, time-dependent sACE-1 inactivation and cleavage plots, additional rate constants and rates, complex kinetic stability, energy-minimized structural models of M-chelate-lisinopril complexes bound to N-domain ACE, Michaelis-Menten plots and SDS-PAGE analyses for inactivated sACE-1, and redox reactivity of M-chelates. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Brown NJ, Vaughan DE. Circulation. 1998;97:1411–1420. doi: 10.1161/01.cir.97.14.1411. [DOI] [PubMed] [Google Scholar]
- 2.Deddish PA, Marcic B, Jackman HL, Wang H-Z, Skidgel RA, Erdös EG. Hypertension. 1998;31:912–917. doi: 10.1161/01.hyp.31.4.912. [DOI] [PubMed] [Google Scholar]
- 3.Chappell MC, Pirro NT, Sykes A, Ferrario CM. Hypertension. 1998;31:362–367. doi: 10.1161/01.hyp.31.1.362. [DOI] [PubMed] [Google Scholar]
- 4.Georgiadis D, Beau F, Czarny B, Cotton J, Yiotakis A, Dive V. Circ. Res. 2003;93:148–154. doi: 10.1161/01.RES.0000081593.33848.FC. [DOI] [PubMed] [Google Scholar]
- 5.Natesh R, Schwager SLU, Sturrock ED, Acharya KR. Nature. 2003;421:551–554. doi: 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- 6.Corradi HR, Schwager SLU, Nchinda AT, Sturrock ED, Acharya KR. J. Mol. Biol. 2006;357:964–974. doi: 10.1016/j.jmb.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 7.Deddish PA, Wang J, Michel B, Morris Pw, Davidson NO, Skidgel RA, Erdos EG. Proc. Natl. Acad. Sci. USA. 1994;91:7807–7811. doi: 10.1073/pnas.91.16.7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lonn EM, Yusuf S, Jha P, Montague TJ, Teo KK, Benedict CR, Pitt B. Circulation. 1994;90:2056–2069. doi: 10.1161/01.cir.90.4.2056. [DOI] [PubMed] [Google Scholar]
- 9.Hocharoen L, Cowan JA. Chem. Eur. J. 2009;15:8670–8676. doi: 10.1002/chem.200900821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowan JA. Pure Appl. Chem. 2008;80:1799–1810. [Google Scholar]
- 11.Jin Y, Cowan JA. J Am Chem Soc. 2006;128:410–411. doi: 10.1021/ja055272m. [DOI] [PubMed] [Google Scholar]
- 12.Joyner JC, Cowan JA. J. Am. Chem. Soc. 2011;133:9912–9922. doi: 10.1021/ja203057z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gokhale NH, Cowan JA. J. Biol. Inorg. Chem. 2006;11:937–947. doi: 10.1007/s00775-006-0145-2. [DOI] [PubMed] [Google Scholar]
- 14.Jin Y, Cowan JA. J Biol Inorg Chem. 2007;12:637–644. doi: 10.1007/s00775-007-0221-2. [DOI] [PubMed] [Google Scholar]
- 15.Gokhale NH, Bradford S, Cowan JA. J. Am. Chem. Soc. 2008;130:2388–2389. doi: 10.1021/ja0778038. [DOI] [PubMed] [Google Scholar]
- 16.Lee J, Udugamasooriya DG, Lim H-S, Kodadek T. Nat. Chem. Biol. 2010;6:258–260. doi: 10.1038/nchembio.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher J, Zelenko O, Walts AD, Sigman DS. Biochemistry. 1998;37:2096–2104. doi: 10.1021/bi971565j. [DOI] [PubMed] [Google Scholar]
- 18.Suh J, Yoo SH, Kim G, Chae PS, Lee TY, Lee J, Lee J, Jang YA, Ko EH. Angew. Chem. 2007;119:7194–7197. [Google Scholar]
- 19.Kim MG, Kim M-s, Lee SD, Suh J. J. Biol. Inorg. Chem. 2006;11:867–875. doi: 10.1007/s00775-006-0139-0. [DOI] [PubMed] [Google Scholar]
- 20.Suh J, Chei WS. Curr. Op. Chem. Biol. 2008;12:207–213. doi: 10.1016/j.cbpa.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 21.Jeong K, Chung WY, Kye Y-S, Kim D. Bioorg. Med. Chem. 2010;18:2598–2601. doi: 10.1016/j.bmc.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 22.Chei WS, Ju H, Suh J. J. Biol. Inorg. Chem. 2011;16:511–519. doi: 10.1007/s00775-010-0750-y. [DOI] [PubMed] [Google Scholar]
- 23.Suh J, Chei WS, Lee TY, Kim MG, Yoo SH, Jeong K, Ahn JY. J. Biol. Inorg. Chem. 2008;13:693–701. doi: 10.1007/s00775-008-0354-y. [DOI] [PubMed] [Google Scholar]
- 24.Meggers E. Chem. Commun. 2009:1001–1010. doi: 10.1039/b813568a. [DOI] [PubMed] [Google Scholar]
- 25.Brown KC, Yang S-H, Kodadek T. Biochemistry. 1995;34:4733–4739. doi: 10.1021/bi00014a030. [DOI] [PubMed] [Google Scholar]
- 26.Joyner JC, Reichfield J, Cowan JA. J. Am. Chem. Soc. 2011;133:15613–15626. doi: 10.1021/ja2052599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmier MO, Van Doren SR. Anal. Biochem. 2007;371:43–51. doi: 10.1016/j.ab.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittal RR, Harris L, McKinnon RA, Sorich MJ. J. Chem. Inf. Model. 2009;49:704–709. doi: 10.1021/ci800390m. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein KE, Welsh SL, Inman JK. Biochem. Biophys. Res. Comm. 1990;167:310–316. doi: 10.1016/0006-291x(90)91766-l. [DOI] [PubMed] [Google Scholar]
- 30.Riordan JF, Harper JW, Martin M. J. Cardio. Pharmacol. 1986;8(Suppl. 10):S29–S34. doi: 10.1097/00005344-198600101-00007. [DOI] [PubMed] [Google Scholar]
- 31.Ehlers MRW, Riordan JF. Biochemistry. 1991;30:7118–7126. doi: 10.1021/bi00243a012. [DOI] [PubMed] [Google Scholar]
- 32.Lu J, Stewart AJ, Sadler PJ, Pinheiro TJT, Blindauer CA. Biochem. Soc. Trans. 2008;36:1317–1321. doi: 10.1042/BST0361317. [DOI] [PubMed] [Google Scholar]
- 33.Camerman N, Camerman A, Sarkar B. Can. J. Chem. 1976;54:1309–1316. [Google Scholar]
- 34.Lau S-J, Kruck TPA, Sarkar B. J. Biol. Chem. 1974;249:5878–5884. [PubMed] [Google Scholar]
- 35.Stadtman ER. Annu. Rev. Biochem. 1993;62:797–821. doi: 10.1146/annurev.bi.62.070193.004053. [DOI] [PubMed] [Google Scholar]
- 36.Harriman A. J. Phys. Chem. 1987;91:6102–6104. [Google Scholar]
- 37.Rice GI, Thomas DA, Grant PJ, Turner AJ, Hooper NM. Biochem. J. 2004;383:45–51. doi: 10.1042/BJ20040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaud A, Williams TA, Chauvet MT, Corvol P. Mol. Pharmacol. 1997;51:1070–1076. doi: 10.1124/mol.51.6.1070. [DOI] [PubMed] [Google Scholar]
- 39.Jaspard E, Wei L, Alhenc-Gelas F. J. Biol. Chem. 1993;268:9496–9503. [PubMed] [Google Scholar]
- 40.Woodman ZL, Schwager SLU, Redelinghuys P, Carmona AK, Ehlers MRW, Sturrock ED. Biochem. J. 2005;389:739–744. doi: 10.1042/BJ20050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman DL, Keaney JF, Jr, Biegelsen ES, Duffy SJ, Coffman JD, Vita JA. Hypertension. 2000;35:936–941. doi: 10.1161/01.hyp.35.4.936. [DOI] [PubMed] [Google Scholar]
- 42.Gokce N, Keaney JF, Jr, Frei B, Holbrook M, Olesiak M, Zachariah BJ, Leeuwenburgh C, Heinecke JW, Vita JA. Circulation. 1999;99:3234–3240. doi: 10.1161/01.cir.99.25.3234. [DOI] [PubMed] [Google Scholar]
- 43.Cowan JA. Inorganic Biochemistry. An Introduction. 2nd ed. New York: Wiley-VCH; 1997. pp. 333–337. [Google Scholar]
- 44.Graf E, Mahoney J, Bryant R, Eaton J. J. Biol. Chem. 1984;259:3620–3624. [PubMed] [Google Scholar]
- 45.Inoue S, Kawanishi S. Cancer Research. 1987;47:6522–6527. [PubMed] [Google Scholar]
- 46.Buettner GR, Jurkiewicz BA. Radiat. Res. 1996;145:532–541. [PubMed] [Google Scholar]
- 47.Chiou S-H. J Biochem. 1984;96:1307–1310. doi: 10.1093/oxfordjournals.jbchem.a134951. [DOI] [PubMed] [Google Scholar]
- 48.Jin Y, Cowan JA. J. Am. Chem. Soc. 2005;127:8408–8415. doi: 10.1021/ja0503985. [DOI] [PubMed] [Google Scholar]
- 49.Azab HA, Banci L, Borsari M, Luchinat C, Sola M, Viezzoli MS. Inorg. Chem. 1992;31:4649–4655. [Google Scholar]
- 50.Barrette WCJ, Sawyer DT, Fee JA, Asada K. Biochemistry. 1983;22:624–627. doi: 10.1021/bi00272a015. [DOI] [PubMed] [Google Scholar]
- 51.Wood P. Biochem. J. 1988;253:287–289. doi: 10.1042/bj2530287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borsook H, Keighley G. Proc. Natl. Acad. Sci. USA. 1933;19:875–878. doi: 10.1073/pnas.19.9.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navaratnam S, Parsons BJ. J. Chem. Soc. Faraday Trans. 1998;94:2577–2581. [Google Scholar]
- 54.Bradshaw RA, Peters T. J. Biol. Chem. 1969;244:5582–5589. [PubMed] [Google Scholar]
- 55.Lau S-J, Sarkar B. J. Biol. Chem. 1971;246:5938–5943. [PubMed] [Google Scholar]
- 56.Cabras T, Patamia M, Melino S, Inzitari R, Messana I, Castagnola M, Petruzzelli R. Biochem Biophys Res Commun. 2007;358:277–284. doi: 10.1016/j.bbrc.2007.04.121. [DOI] [PubMed] [Google Scholar]
- 57.Grogan J, McKnight CJ, Troxler RF, Oppenheim FG. FEBS Letts. 2001;491:76–80. doi: 10.1016/s0014-5793(01)02157-3. [DOI] [PubMed] [Google Scholar]
- 58.Harford C, Sarkar B. Biochem. Biophys. Res. Commun. 1995;209:877–882. doi: 10.1006/bbrc.1995.1580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.