Abstract
Toxin–antitoxin systems are widespread in bacteria and archaea. They perform diverse functional roles, including the generation of persistence, maintenance of genetic loci and resistance to bacteriophages through abortive infection. Toxin–antitoxin systems have been divided into three types, depending on the nature of the interacting macromolecules. The recently discovered Type III toxin–antitoxin systems encode protein toxins that are inhibited by pseudoknots of antitoxic RNA, encoded by short tandem repeats upstream of the toxin gene. Recent studies have identified the range of Type I and Type II systems within current sequence databases. Here, structure-based homology searches were combined with iterative protein sequence comparisons to obtain a current picture of the prevalence of Type III systems. Three independent Type III families were identified, according to toxin sequence similarity. The three families were found to be far more abundant and widespread than previously known, with examples throughout the Firmicutes, Fusobacteria and Proteobacteria. Functional assays confirmed that representatives from all three families act as toxin–antitoxin loci within Escherichia coli and at least two of the families confer resistance to bacteriophages. This study shows that active Type III toxin–antitoxin systems are far more diverse than previously known, and suggests that more remain to be identified.
INTRODUCTION
Bacteria are constantly faced with environmental stresses and the threats of viral predation. Through adaptive evolution they have developed multiple systems to ensure survival, including the toxin–antitoxin (TA) systems (1–4). TA systems are near-ubiquitous throughout the plasmids and chromosomes of bacteria and archaea and usually comprise bicistronic operons encoding two small genes; one for a toxic component and a second for a cognate antitoxin (1,2,5). Though originally identified in 1983 as plasmid maintenance systems (6), TA systems have been attributed to many physiological roles, including formation of persister cells (7), stress resistance (8), protection from bacteriophages (phages) (9) and regulation of biofilm formation (10), among others (11).
TA systems have been divided into three Types, depending on the nature of the interacting toxin and antitoxin macromolecules (4). Within Type I systems, an RNA antitoxin interacts with the toxin transcript and inhibits translation of the toxic protein (12). The toxins and antitoxins of Type II systems interact as proteins (5). Both Type I and Type II systems were originally identified through their role in plasmid maintenance (5,12). The recently discovered Type III systems, first identified by an ability to abort phage infections (9), rely upon the direct interaction of an RNA antitoxin with the toxin protein (13).
Recent studies have identified an ever-increasing number of experimentally defined, or putative, Type I and Type II TA systems (5,12,14–16). Type I toxins are generally small proteins of <60 amino acids. Type I antitoxins are usually encoded as an antisense RNA product, but they can be transcribed divergently from the toxin (12). By combining iterative psi-BLAST searches with Type I-specific parameters (such as the presence of tandem copies of the full loci, the free-energy minima of the antitoxin and the presence of transmembrane domains), the authors were able to detect multiple copies of known Type I loci within new hosts across 774 bacterial genomes (12). The authors were also able to identify and experimentally validate novel Type I TA systems (12). In some cases, many Type I TA system families, and multiple members thereof, were identified in the same host; in the extreme case, Escherichia coli O157:H7 str. Sakai contained 26 Type I loci (12). By comparing the phylogenetic tree of these identified Type I loci with the host taxonomy, it appeared that Type I loci have not been freely disseminated by horizontal gene transfer but may have a common ancient ancestor (12).
Global identifications of Type II systems have often relied on identifying putative toxin and antitoxin genes by distant sequence similarity, followed by ‘guilt-by-association’, where the homologue pairs must cluster into a putative bicistron (5,16). Type II systems previously have been grouped according to the toxin genes present in the locus (2), though they could be re-classified at the level of toxin structure (4), which correlates well with distant sequence similarity (3,5). The most recent global bioinformatic search identified 12 toxin and 20 antitoxin super-families within 2181 prokaryotic genomes (5). This recent study also highlighted the mosaic nature of Type II systems; whereas, previously, specific toxin genes were thought to be associated with specific antitoxin genes, there is clear heterogeneity and interchangeability within Type II systems (5). Type II loci were observed in even greater numbers than Type I loci, with the maximum peaking at 97 Type II loci within a single genome (5). Unlike Type I systems, there appears to have been significant horizontal gene transfer of Type II systems (5). Type II systems are also highly abundant in mobile genetic elements, which may relate to their ‘addictive’ nature and ability to maintain these replicons (17). Web-based tools, such as the search engine RASTA (18) and the database TADB (19), have been developed to assist in identification and cataloguing of the Type II systems.
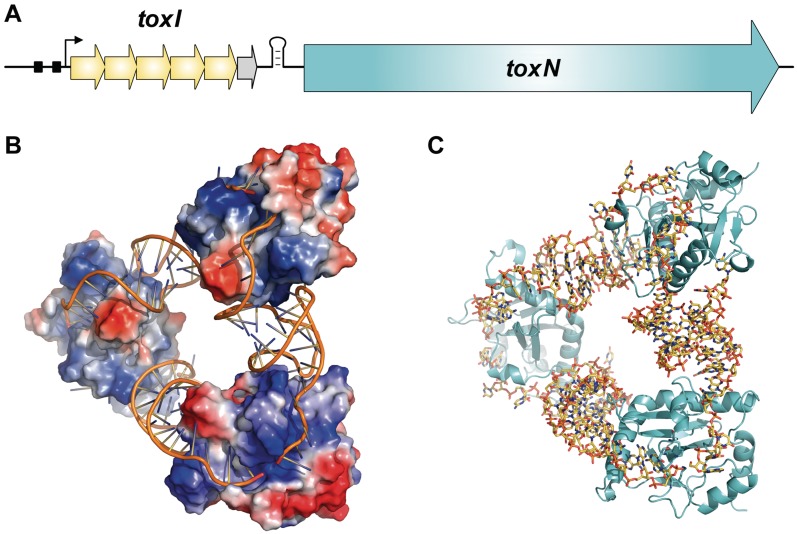
The recently identified Type III TA loci were originally isolated and defined as abortive infection systems, protecting bacterial populations from bacteriophage (phage) assault (9,20,21). Within each Type III locus, a toxin gene is preceded by a short palindromic repeat, which is itself preceded by a tandem array of nucleotide repeats (Figure 1A). The short palindromic repeat acts as a transcriptional terminator, regulating the relative levels of antitoxic RNA and toxin transcript (9,21). The first Type III TA system, ToxIN, was encoded on plasmid pECA1039 of the Gram-negative phytopathogen, Pectobacterium atrosepticum. This locus encodes a 19.7-kDa toxic protein, ToxN, and upstream there is a repetitive array containing 5.5 tandem repeats of a 36 nt sequence, collectively known as the ToxI antitoxin. Through genetic studies, it was predicted that each 36 nt ToxI RNA repeat was able to inhibit the activity of ToxN (9). The crystal structure of the ToxIN complex revealed a heterohexameric triangular assembly of three ToxN proteins interspersed by three, 36 nt, ToxI RNA pseudoknots (4,13) (Figure 1B and C). ToxN was demonstrated to be an endoribonuclease, related in structure to the endoribonucleases Kid and MazF (13).
Figure 1.
Overview of Type III toxin–antitoxin loci. (A) Schematic of a Type III toxin–antitoxin locus. The paradigmatic Type III system, ToxIN from P. atrosepticum, is depicted. The toxin gene, ToxN, is preceded by a stem–loop structure formed from a palindromic repeat. This is itself preceded by a set of tandem nucleotide repeats which act as antitoxic RNA pseudoknots. In the case of ToxIN, the antitoxin, ToxI, is encoded by 5.5, 36 nt tandem repeats (yellow arrows for a full repeat, grey arrow for the half repeat). The locus is transcribed from a constitutive promoter, with the −35 and −10 elements shown as black boxes and the transcriptional start site shown by a black arrow. (B) ToxIN trimer (PDB: 2XDD), with ToxI monomers shown as cartoons and ToxN monomers shown with electrostatic surfaces, where red represents electronegative potential and blue is electropositive. Three monomers of ToxN are held at respective corners of a heterohexameric triangular assembly, formed entirely through protein–RNA interactions with the interspersed pseudoknots of 36 nt ToxI RNAs. (C) ToxIN trimer (PDB: 2XDD) with ToxI shown as yellow sticks and ToxN as cyan cartoons. Figure and legend adapted with the author’s permission from Ref. (4).
Following characterization of ToxIN, simple BLAST (22) searches using the ToxN amino acid sequence-identified multiple homologues (9). These were found on the plasmids and chromosomes of both Gram-negative and Gram-positive species, within human and animal pathogens, oceanic and soil bacteria and extremophiles. Though these were identified by shared identity with ToxN, the cognate ToxI varied greatly in terms of the number of repeats, the length of repeats and the underlying sequence of repeats (9). These related ToxN proteins are therefore likely to bind to very different RNA sequences, thereby providing a whole new subset of complexes upon which to study protein–RNA interactions. Judging from the macromolecular structure of ToxIN and the offset nature of the ToxI pseudoknot sequence (13), the specific RNA sequence that will physically bind to each ToxN cannot be trivially deduced from the pattern of DNA repeats; it is more likely that each pseudoknot RNA is generated across the DNA repeat boundaries.
Just as the number and diversity of identified Type I and Type II loci continues to increase (5,12,16), it follows that the current list of Type III TA loci must be hugely under-representative. When the structure of ToxN was first obtained, a comparative search of entries within the PDB identified Type II toxin proteins Kid, MazF, CcdB and YdcE as high-scoring structural homologues of ToxN (4,13). The structures of members of this Kid/MazF/CcdB superfamily overlay well with the core regions of ToxN, though ToxN has specific additional folds that appear to allow binding of the antitoxic RNA pseudoknot (4,13). While these comparisons relied upon known structures, it was also possible to perform structure-based homology searches to identify novel Type III systems, de novo. Initial searches using FUGUE (23) generated a list of 880 putative ToxN homologues. Due to the complexity of the Type III locus, an algorithm was not available with which to sort these putative hits. Each hit was therefore assessed on a case-by-case basis, identifying those hits that match the criteria of a Type III TA locus. Having identified putative loci, iterative BLAST searches were performed to expand the list of potential homologues. In this manner, 125 putative Type III TA loci were identified. These hits were further divided into three families, according to protein sequence homology. Phylogenetic analyses were performed to assess the distribution of these Type III loci in relation to the taxonomy of their hosts. For the first time, we were able to identify putative Type III loci in both an archaeal species and a bacteriophage. The findings were validated experimentally by taking exemplars of each family and testing for toxic and antitoxic effects within a reconstructed E. coli over-expression model. Each locus tested was confirmed as an active TA system. Furthermore, these loci were tested for their ability to protect from infection by coliphages. One of the newly identified loci conferred phage resistance. This global analysis using structure-based homology models has greatly increased our knowledge of the prevalence and diversity of Type III TA systems within sequenced prokaryotic genomes.
MATERIALS AND METHODS
Bioinformatic approach
Homology-based searches
Based on the crystal structure of ToxIN (PDB: 2XDD), an environment-specific substitution profile was created by using JOY (24) and FUGUE (23), with additional homologous sequences collected from the NCBI nr database. Sequences of bacterial and archaeal proteomes were downloaded from integr8 (25) (http://www.ebi.ac.uk/integr8/), using a custom script. The fugueprf programme in the FUGUE suite was used to perform structure-based homology searches against these sequences with the ToxN profile as a query. All the hits with Z-scores ≥3.5 were collected. FUGUE has been widely used for detecting remote homologues and previous benchmark results suggest that Z-scores ≥ 6.0, ≥ 4.0 and ≥ 3.5 would correspond to confidence levels of 99, 95 and 90%, respectively.
Manual searches
The first 270 FUGUE hits, with Z-scores ranging from 27.4 to 4.0, were analysed. For each protein sequence, the coding sequence and 1 kb up- and downstream were extracted from the NCBI database. This extracted locus sequence was then analysed using Tandem Repeat Finder (26); default settings were used for the initial searches (match, mismatch, indels = 2, 7, 7; min score = 50), followed by less stringent searches for those homologues with more variable antitoxin sequences (match, mismatch, indels = 2, 3, 5; min score = 50). If repeats were identified, the locus sequence was then examined for a palindromic repeat and an E. coli consensus promoter sequence. Toxin amino acid sequences from the resulting positive hits were then aligned against each other using BLASTp (22), in order to group them by families of related sequences. Examples from each family were then used in iterative rounds of BLASTp searches until all homologues had been identified from the NCBI database (current as of July 2011). Information about each identified hit, including sequence information, is stored within a searchable spreadsheet, as Supplementary Table S1. Full details of the 880 FUGUE hits, their analysis and the subsequent BLAST searches are stored within a second spreadsheet, as Supplementary Table S2.
Phylogenetics
Phylogenetic analysis was performed on 69 toxin sequences which were part of putative Type III TA loci unambiguously containing all the required sequence elements, (as outlined in the ‘Results’ section), together with sequences of the Type II toxins Kid, MazF, YdcE, CcdB and RelE. The alignment of these 74 protein sequences was performed using Clustal Omega (27). Additionally, 16S rDNA sequences were retrieved from the Ribosomal Database Project for the strains encoding the corresponding putative Type III TA loci (28). Maximum Likelihood phylogenetic analysis was performed using TREEFINDER (http://www.treefinder.de) (29). TREEFINDER was first used to select an appropriate model to analyse the aligned datasets. Following the Akaike information criterion, the VT substitution model was chosen for the ToxN sequences, while the GTR model was used for the 16S sequences. Phylogenies were reconstructed using the default settings of TREEFINDER. LR-ELW (Local Rearrangement-Expected-Likelihood Weights) edge support (using 1000 replicates) was enabled to provide approximate bootstrap analysis.
Alignment of ToxI
Single repeat consensus sequences of ToxI homologues were manually aligned with ToxI from P. atrosepticum, according to sequence and on the length and spacing of potential base-pairing regions. Nested base-pairing regions in each ToxI sequence were identified using pknotsRG (30). The start and end of each predicted ToxI pseudoknot was assigned from the relative position of structural features and is not anchored to the ToxI DNA repeat.
Bacterial strains, bacteriophages and media
Bacterial strains
All functional assays were performed within E. coli strain DH5α (Gibco/BRL). Genomic DNA was extracted from 10 ml overnight cultures of Photorhabdus luminescens subsp. laumondii TT01 (31). For further details about each strain, see Supplementary Table S3.
Bacteriophages
All bacteriophages used were isolated from treated sewage effluent, collected from a river Cam outlet at Milton, near Cambridge, UK (Supplementary Table S3).
Media
E. coli strains were grown at 37°C and P. luminescens was grown at 30°C, in Luria broth (LB) at 250 rpm or on LB-agar (LBA) containing 1.5% w v−1 or 0.35% w v−1 agar, to make LBA plates or top-LBA, respectively. Growth (OD600) was measured using a Heλios α spectrophotometer set to 600 nm. When required, media was supplemented with ampicillin (Ap) at 10 µg ml−1, spectinomycin (Sp) at 50 µg ml−1, d-glucose (glu) at 0.2% w v−1, l-arabinose (L-ara) at 0.1% w v−1 and isopropyl-β-d-thiogalactopyranoside (IPTG) at 1 mM. Bacteriophages were stored over chloroform in phage buffer; 10 mM Tris–HCl, pH 7.4, 10 mM MgSO4 and 0.01% w v−1 gelatin.
Cloning of Type III toxin–antitoxin loci
Molecular biology techniques were performed as described previously (32). All primers were obtained from Sigma-Genosys and are listed in Supplementary Table S4. All plasmids constructed and/or used in this study are listed in Supplementary Table S5. The molecular nature of each recombinant plasmid was verified by DNA sequencing. Genomic DNA was obtained from P. luminescens using an extraction kit (Qiagen). The genomic DNA from other strains was kindly provided to us from other researchers (see ‘Acknowledgements’ section).
Toxin cloning
Each toxin sequence was amplified by PCR using genomic DNA as a template and cloned into pBAD30 (33), using the designated primers and restriction enzymes (Supplementary Tables S4 and S5). Each toxin was cloned such that the protein was translated using the native ribosome binding site. Transformants of pBAD30-based vectors were selected on LBA supplemented with ampicillin and glucose.
Antitoxin cloning
An expression vector carrying a single repeat of the P. luminescens tenpI antitoxin was generated by first amplifying the required insert using primers TRB214 and PF185, with pQE-80L as a template. The resulting amplicon was then cloned into pTA100, using PstI and XhoI. For all other antitoxins, the sequence was amplified by PCR using genomic DNA as a template and cloned into pTA100 (9), using the designated primers and restriction enzymes (Supplementary Tables S4 and S5).
Type III toxin–antitoxin locus cloning
Each Type III toxin–antitoxin locus was cloned into pBR322 (34), using amplicons generated by PCR amplification from genomic DNA, with the designated primers and restriction enzymes (Supplementary Tables S4 and S5). The region cloned for each locus includes up to 500 bp upstream of the toxin start codon and <100 bp downstream of the toxin stop codon.
Bacteriophage isolation
A 10 ml sample of treated sewage effluent was shaken vigorously with 500 µl of chloroform for 1 min. A 200-µl aliquot of this treated sample was mixed with 200 µl of a DH5α overnight culture and 3 ml of top-LBA, and then poured as an overlay on an LBA plate. These plates were incubated overnight at 37°C and the resulting phage plaques were picked with sterile toothpicks into 50 µl of phage buffer, which was then treated with 20 µl of chloroform.
Protection assays
Strains of E. coli, containing both antitoxin and toxin (or control) expression plasmids, were obtained by either sequential or co-transformation. Single colonies of the resulting strains were grown as 10 ml overnight cultures and these were used to inoculate 25 ml of LB, Ap and glu in 250 ml flasks, then grown at 37°C and 250 rpm in an orbital shaker, from a starting OD600 of ∼0.04, until exponential phase [∼1 × 108 colony forming units (cfu) ml−1]. At this end point, samples were removed, washed with phosphate buffered saline, serially diluted and plated for viable counts at 37°C on LBA, Ap, Sp plates containing either (i) glu, so neither toxin or antitoxin is expressed; (ii) glu and IPTG, to express the antitoxin; (iii) L-ara, to express the toxin; or (iv) L-ara and IPTG to express both the toxin and antitoxin. The data presented are the mean viable counts from triplicate data, with error bars representing the standard deviation.
Efficiency of Plating assays
Isolated phage plaques were repeatedly re-plated until plaque pure and homogeneous plaque morphologies were reproducibly obtained. Lysates of the phage were then made by scraping the top-LBA from a plate with a confluent lawn of phage plaques into a glass universal, adding 3 ml of phage buffer and vortexing vigorously with 500 µl of chloroform for 1 min. After standing at room temperature for 30 min, the agar mix was centrifuged at 2200g for 20 min at 4°C. The supernatant was decanted into a glass bijou for phage lysate storage at 4°C, over 100 µl chloroform. Phage lysates were then titrated on the E. coli pBR322 control strain, and test strains. The resulting Efficiency of Plating (EOP) was calculated as (number of plaque-forming units on test strain/number of plaque-forming units on control strain). Each EOP value was calculated as the mean value from triplicate data.
RESULTS
Structure-based homology searches and identification of Type III toxin–antitoxin loci
To date, there has been only one Type III TA locus characterized in detail; the ToxIN system, isolated from plasmid pECA1039 of P. atrosepticum SCRI1039 (9). This publication also reported, as proof-of-principle, that a homologous ToxIN system from Bacillus thuringiensis also acted as a Type III TA system in E. coli (9). A catalogue of 19 other homologues of ToxN was identified from the NCBI database using BLASTp searches (9,22). The current study has attempted to significantly extend this minimal list of Type III TA systems.
The structure of ToxIN was solved by crystallographic analysis (13), and so it was possible to use the program FUGUE (23) to perform structure-based homology searches with ToxN as the search model. FUGUE searches were performed in June 2010, against 1852 bacterial and archaeal proteomes taken from integr8 (25) (http://www.ebi.ac.uk/integr8/). The total number of sequences scanned was >6 million. This generated an initial list of 880 putative ToxN homologues, with Z-scores ranging from 27.4 to 3.5 (Supplementary Table S2).
To identify the subset of putative ToxN homologues which are likely to come from true Type III TA loci, a list of criteria was set, based on the required features of a Type III TA locus as determined experimentally (9,13,21). Namely, the putative toxN homologue should be preceded by a short palindromic repeat to act as a transcriptional terminator, which should, in turn, be preceded by a tandem array of nucleotide repeats to act as the antitoxic toxI. Preferentially, this locus should then be preceded by −35 and −10 promoter elements, with the position defined by the start of toxI. As it became more difficult to predict a promoter in bacteria more distantly related from E. coli, the presence of an obvious promoter was excluded as a strict requirement. No algorithm was immediately available to allow rapid processing of the FUGUE hits, so the analysis was undertaken on a case-by-case basis. Each nucleotide sequence of the identified ToxN homologues was taken together with 500 bp upstream and downstream, and attempts were made to identify the cognate toxI using Tandem Repeat Finder (26). The default settings of Tandem Repeat Finder proved adequate to accurately predict the ToxI for each of the previously identified homologues (9), so these settings were used for all initial searches. Where a suitable toxI-like sequence was identified, the sequence was then screened for a putative transcriptional terminator and promoter by visual inspection. If a toxI-like sequence was not identified, the sequence was not examined any further and these negative hits are recorded within Supplementary Table S2. In this manner, we examined the first 270 FUGUE hits. At this point, the Z-score had reached 4.0 and was slowly decreasing towards 3.5 (Supplementary Figure S1). There were rapidly diminishing returns in identifying Type III loci from the FUGUE hits as we progressed to Z-scores <5.0 (Supplementary Figure S1). Analysis of the raw FUGUE hits was therefore ended after the first 270 hits. At this point, 37 putative Type III loci were identified.
Three families of Type III toxin–antitoxin loci were identified
A BLASTp matrix was formed to compare the amino acid sequences of the 37 ToxN structural homologues with every other sequence; it became immediately clear from the returned E values that the 37 hits could be divided into three families (Supplementary Figure S2). Though all the hits by definition share the same overall tertiary structure, members of the second and third family toxins had low detectable amino acid sequence identity shared with either ToxN or between each other (Supplementary Figure S2), which supports the decision to classify them into three independent families. The first family contained ToxIN from P. atrosepticum, B. thuringiensis and all homologues thereof. When naming the families, it was decided to maintain the ‘IN’ nomenclature, wherein every antitoxin has the suffix ‘I’ for inhibitor and each toxin is denoted ‘N’, as for the toxIN family. This standardization, if used universally, would ensure that when Type III TA systems are identified in future, the reader should be able to conclude readily which component is the antitoxin and which component is the toxin. The second family contained a locus from Coprococcus catus GD/7, so the family was named cptIN (CoPrococcus Type III Inhibitor/toxiN; suggested pronunciation, ‘cap-tin’). The third family contained a locus from P. luminescens subsp. laumondii TT01, so this family was named tenpIN (Type III ENdogenous to Photorhabdus Inhibitor/toxiN).
Having divided the 37 hits into three families, we took examples from each family and performed exhaustive BLASTp searches. The results from these searches were again assessed for hits that represented putative Type III TA systems, following the same criteria as described previously for the FUGUE hits (Supplementary Table S2). Negative hits were then re-visited using less stringent settings of tandem repeat finder to identify Type III loci with greater variability between their antitoxin repeat sequences. A final list of 125 putative Type III TA systems was generated. A consolidated version of this list is presented in Table 1. A more comprehensive, searchable, spreadsheet of these hits, including all the nucleotide and protein sequences, is available as Supplementary Table S1.
Table 1.
Distribution of three identified Type III toxin–antitoxin families
| Strain | Plasmid | Abbreviation | Taxonomy (Phylum, Class, Order) | toxIN loci | cptIN loci | tenpIN loci |
|---|---|---|---|---|---|---|
| Abiotrophia defectiva ATCC 49176 | Ade | Firmicutes, Bacilli, Lactobacillales | 1 | |||
| Acetivibrio cellulolyticus CD2 | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Acidobacterium sp. MP5ACTX9 | pACIX905 | Acidobacteria, Acidobacteria, Acidobacteriales | 1 | |||
| Actinobacillus ureae ATCC 25976 | Proteobacteria, Gammaproteobacteria, Pasteurellales | 1 | ||||
| Alkaliphilus oremlandii OhILAs | Aor | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Anoxybacillus flavithermus WK1 | Afl | Firmicutes, Bacilli, Bacillales | 1 | |||
| Bacillus cereus Rock1-15 | Bce | Firmicutes, Bacilli, Bacillales | 1 | |||
| Bacillus thuringiensis serovar konkukian str. 97-27 | pBT9727 | Firmicutes, Bacilli, Bacillales | 1 | |||
| Bacillus thuringiensis serovar kurstaki pAW63 | pAW63 | Bthsk | Firmicutes, Bacilli, Bacillales | 1 | ||
| Bacillus thuringiensis serovar pondicheriensis BGSC 4BA1 | Bthsp | Firmicutes, Bacilli, Bacillales | 1 | |||
| Bacillus weihenstephanensis KBAB4 | pBWB402 | Bwe | Firmicutes, Bacilli, Bacillales | 1 | ||
| Bryantella formatexigens DSM 14469 | Bfo | Firmicutes, Clostridia, Clostridiales | 2 | 3 | ||
| Caldicellulosiruptor bescii DSM 6725 | Cbe | Firmicutes, Clostridia, Thermoanaerobacterales | 1 | |||
| Caldicellulosiruptor kristjanssonii 177R1B | Ckr | Firmicutes, Clostridia, Thermoanaerobacterales | 1 | |||
| Caldicellulosiruptor lactoaceticus 6A | Firmicutes, Clostridia, Thermoanaerobacterales | 1 | ||||
| Clostridium botulinum BKT015925 | p1BKT015925 | Cbo | Firmicutes, Clostridia, Clostridiales | 1 | ||
| Clostridium cellulovorans 743B | Cce | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Clostridium hiranonis DSM 13275 | Chi | Firmicutes, Clostridia, Clostridiales | 1 | 1 | ||
| Clostridium nexile DSM 1787 | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Clostridium phage D-1873 | CloΦD | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Clostridium sp. HGF2 | Clh | Firmicutes, Clostridia, Clostridiales | 1 | 1 | ||
| Coprobacillus sp. 29_1 | Firmicutes, Erysipelotrichi, Erysipelotrichales | 1 | ||||
| Coprococcus catus GD/7 | Cca | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Coprococcus sp. ART55/1 | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Eubacterium rectale ATCC 33656 | Ere33656 | Firmicutes, Clostridia, Clostridiales | 2 | |||
| Eubacterium rectale DSM 17629 | Ere17629 | Firmicutes, Clostridia, Clostridiales | 1 | 1 | ||
| Eubacterium rectale M104/1 | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Eubacterium saburreum DSM 3986 | Esab | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Eubacterium saphenum ATCC 49989 | Esap | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Eubacterium ventriosum ATCC 27560 | Eve | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Eubacterium yurii subsp. margaretiae ATCC 43715 | Eyu | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Fibrobacter succinogenes subsp. succinogenes S85 | Fsu | Fibrobacteres, Fibrobacteres, Fibrobacterales | 1 | |||
| Finegoldia magna BVS033A4 | Fma | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Fusobacterium nucleatum subsp. nucleatum ATCC 23726 | Fnun | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | |||
| Fusobacterium nucleatum subsp. polymorphum ATCC 10953 | Fnup | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | 1 | ||
| Fusobacterium nucleatum subsp. vincentii ATCC 49256 | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | ||||
| Fusobacterium periodonticum ATCC 33693 | Fpe | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | |||
| Fusobacterium sp. 2_1_31 | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | ||||
| Fusobacterium sp. 3_1_33 | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | 1 | |||
| Fusobacterium sp. 3_1_36A2 | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | ||||
| Fusobacterium sp. 3_1_5R | Fus | Fusobacteria, Fusobacteria, Fusobacteriales | 3 | |||
| Fusobacterium sp. 4_1_13 | Fus4 | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | 1 | ||
| Fusobacterium sp. 7_1 | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | ||||
| Fusobacterium sp. D11 | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | ||||
| Fusobacterium sp. D12 | FusD12 | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | |||
| Fusobacterium ulcerans ATCC 49185 | Ful | Fusobacteria, Fusobacteria, Fusobacteriales | 1 | |||
| Gemella moribillum M424 | Firmicutes, Bacilli, Bacillales | 1 | ||||
| Geobacillus thermoglucosidasius C56-YS93 | Firmicutes, Bacilli, Bacillales | 1 | ||||
| Haemophilus influenzae biotype aegyptius | pF1947 | Hin | Proteobacteria, Gammaproteobacteria, Pasteurellales | 1 | ||
| Haemophilus influenzae biotype aegyptius BPF | pF3028 | Proteobacteria, Gammaproteobacteria, Pasteurellales | 1 | |||
| Haemophilus parasuis | pHPS5839 | Hpa | Proteobacteria, Gammaproteobacteria, Pasteurellales | 1 | ||
| Haemophilus parasuis SH0165 | Hpa0165 | Proteobacteria, Gammaproteobacteria, Pasteurellales | 1 | |||
| Haemophilus somnus 2336 | Hso | Proteobacteria, Gammaproteobacteria, Pasteurellales | 1 | 1 | ||
| Lachnospiraceae bacterium 2_1_46FAA | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Lachnospiraceae bacterium 3_1_46FAA | Lba | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Lachnospiraceae bacterium 4_1_37FAA | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Lachnospiraceae bacterium 5_1_63FAA | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Lachnospiraceae bacterium 8_1_57FAA | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Lachnospiraceae bacterium 9_1_43BFAA | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Lachnospiraceae oral taxon 107 str. F0167 | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Lactobacillus gasseri JV-V03 | Lga | Firmicutes, Bacilli, Lactobacillales | 1 | |||
| Lactobacillus helveticus DSM 20075 | Firmicutes, Bacilli, Lactobacillales | 1 | ||||
| Lactobacillus helveticus MTCC 5463 | Firmicutes, Bacilli, Lactobacillales | 1 | ||||
| Lactobacillus jensenii 1153 | Lje1153 | Firmicutes, Bacilli, Lactobacillales | 1 | |||
| Lactobacillus jensenii 208-1 | Lje208 | Firmicutes, Bacilli, Lactobacillales | 2 | |||
| Lactobacillus jensenii 269-3 | Lje269 | Firmicutes, Bacilli, Lactobacillales | 1 | |||
| Lactobacillus jensenii 27-2-CHN | LjeCHN | Firmicutes, Bacilli, Lactobacillales | 1 | |||
| Lactobacillus jensenii SJ-7A-US | LjeUS | Firmicutes, Bacilli, Lactobacillales | 1 | |||
| Lactobacillus kefiranofaciens ZW3 | Lke | Firmicutes, Bacilli, Lactobacillales | 1 | |||
| Lactococcus lactis subsp. lactis CV56 | pCV56A | Lla | Firmicutes, Bacilli, Lactobacillales | 1 | ||
| Lactococcus lactis W-37, protein ‘AbiQ’ | pSRQ900 | LlaW37 | Firmicutes, Bacilli, Lactobacillales | 1 | ||
| Leptotrichia goodfellowii F0264 | Lgo | Fusobacteria, Fusobacteria, Fusobacteriales | 4 | 1 | 1 | |
| Leptotrichia hofstadii F0254 | Lho | Fusobacteria, Fusobacteria, Fusobacteriales | 2 | 1 | ||
| Mahella australiensis 50-1 BON | Mau | Firmicutes, Clostridia, Thermoanaerobacterales | 1 | |||
| Methanococcus vannielii SB | Euryarchaeota, Methanococci, Methanococcales | 1 | ||||
| Pectobacterium atrosepticum SCRI1039 | pECA1039 | Pba | Proteobacteria, Gammaproteobacteria, Enterobacteriales | 1 | ||
| Peptoniphilus duerdenii ATCC BAA-1640 | Pdu | Firmicutes, Clostridia, Clostridiales | 2 | |||
| Peptoniphilus harei ACS-146-V-Sch2b | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Phascolarctobacterium sp. YIT 12067 | Pha | Firmicutes, Negativicutes, Selenomonadales | 1 | |||
| Photorhabdus luminescens subsp. laumondii TTO1 | Plu | Proteobacteria, Gammaproteobacteria, Enterobacteriales | 1 | |||
| Pyramidobacter piscolens W5455 | Synergistetes, Synergistia, Synergistales | 1 | ||||
| Roseburia intestinalis M50/1 | Rin | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Roseburia intestinalis XB6B4 | Firmicutes, Clostridia, Clostridiales | 1 | ||||
| Ruminococcus lactaris ATCC 29176 | Rla | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Ruminococcus sp. 5_1_39B_FAA | Rum | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Ruminococcus torques ATCC 27756 | Rto | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Ruminococcus torques L2-14 | Rto14 | Firmicutes, Clostridia, Clostridiales | 1 | |||
| Shewanella putrefaciens 200 | Proteobacteria, Gammaproteobacteria, Alteromonadales | 1 | ||||
| Staphylococcus aureus HUNSC491 | pPR9 | Firmicutes, Bacilli, Bacillales | 1 | |||
| Staphylococcus aureus A9754 | Sau | Firmicutes, Bacilli, Bacillales | 1 | |||
| Staphylococcus aureus subsp. aureus Mu50 | pVRSA | Firmicutes, Bacilli, Bacillales | 1 | |||
| Streptococcus sanguinis SK72 | Ssa | Firmicutes, Bacilli, Lactobacillales | 1 | |||
| Taylorella equigenitalis MCE9 | Teq | Proteobacteria, Betaproteobacteria, Burkholderiales | 1 | |||
| Thermosinus carboxydivorans Nor1 | Tca | Firmicutes, Negativicutes, Selenomonadales | 1 | |||
| Treponema succinifaciens DSM 2489 | Tsu | Spirochaetes, Spirocahetes, Spirochaetales | 1 | |||
| Treponema vincentii ATCC 35580 | Spirochaetes, Spirocahetes, Spirochaetales | 1 | ||||
| Veillonella atypica ACS-134-V-Col7a | Vat | Firmicutes, Negativicutes, Selenomonadales | 1 | |||
| Veillonella parvula ACS-068-V-Sch12 | Vpa | Firmicutes, Negativicutes, Selenomonadales | 1 | |||
| Vibrio cholerae MZO-3 | Proteobacteria, Gammaproteobacteria, Vibrionales | 1 | ||||
| Vibrio cholerae O395 | Vch | Proteobacteria, Gammaproteobacteria, Vibrionales | 1 | |||
| Xenorhabdus bovienii SS-2004 | Xbo | Proteobacteria, Gammaproteobacteria, Enterobacteriales | 1 | |||
| Yersinia pseudotuberculosis IP 31758 | p153kb | Yps | Proteobacteria, Gammaproteobacteria, Enterobacteriales | 1 |
Dividing the 125 hits back into families, we had 67 examples of toxIN loci, 33 of cptIN and 25 of tenpIN. We found, for the first time, that multiple copies of the same family can exist within one host, such as within Eubacterium rectale ATCC 33656, Fusobacterium sp. 3_1_5R, Lactobacillus jensenii 208-1 and Peptoniphilus duerdenii ATCC BAA-1640 (Table 1). Furthermore, multiple families may be represented within a single host, such as is the case for Bryantella formatexigens DSM 14469, two clostridia, E. rectale DSM 17629, three fusobacteria, Haemophilus somnus 2336, Leptotrichia hofstadii F0254 and Leptotrichia goodfellowii F0264. Leptotrichia goodfellowii F0264 contained the highest number of Type III TA loci, six in total, with at least one representative from each of the three families (Table 1).
The majority of hits were found on bacterial chromosomes and plasmids, though this analysis identified a toxIN locus encoded on a plasmid prophage; Clostridium phage D-783. This converting phage carries the neurotoxin cluster encoding a major pathogenicity determinant of Clostridium botulinum. Type I and Type II loci also have been identified on phages, such as the Hok/Gef Type I system of the enterobacterial phage 933W (12) and the Type II phd/doc locus of P1 (35).
Each of the hits contained in Table 1 is unique, in that the individual locus is contained within a unique host. However, there are cases where exact copies of the same Type III TA locus have been detected in multiple host strains. This was seen with the toxIN locus of Ruminococcus torques ATCC 27756 and Lachnospiraceae bacterium 8_1_57FAA. Similarly, Lactobacillus jensenii 208-1 contains two toxIN loci; one is found duplicated in Lactobacillus jensenii 27-2-CHN, while the other is duplicated in both Lactobacillus jensenii 269-3 and Lactobacillus jensenii 1153. The tenpIN locus identified within the two Vibrio species, O593 and MZO-3, is also identical except for one, silent, base substitution in the toxin-coding sequence.
The criteria for identification of each putative Type III TA locus required a toxin gene to be preceded by a terminator and, further upstream, a set of tandem repeats. While the majority of hits have a relatively small distance between the repeats, the terminator and the toxin gene (around 1–50 bp between each component), there were some examples with larger intervening distances. The toxIN locus of Actinobacillus ureae ATCC 25976 has a gap of 340 bp between toxI and toxN. The toxIN loci of Roseburia intestinalis M50/1 and Roseburia intestinalis XB6B4 have similar such gaps, of 232 and 292 bp, respectively. There were no ORFs or other features of note detected within these gaps.
Whereas many Type II TA loci have been identified in Archaea (5), previous studies have not identified any examples of either Type I or Type III TA loci in this third superkingdom (9,12). Our new analysis has identified one putative cptIN locus within the Archaeon, Methanococcus vannielii SB. This system also has a larger than expected gap of 242 bp separating the antitoxic repeats and toxin gene. These hits were retained within the consolidated list of putative systems (Table 1 and Supplementary Table S1), as items of interest for further study and validation.
The toxIN locus of Lactobacillus jensenii SJ-7A-US follows the Type II higBA TA locus (36), in that the canonical arrangement of toxin to antitoxin genes is swapped. It was observed that the antitoxic toxI repeats and terminator appear to be downstream of the toxN gene. Though there is a predicted promoter downstream of this toxN homologue potentially driving transcription of the cognate toxI, using the current model, it is unclear how this locus might successfully regulate the levels of the two species. This will be a focus of future study.
There are several hits within our analysis that have toxin sequences representing partial sequences of other, longer, toxins. These smaller toxins have full sets of antitoxic repeats and terminators, though only shorter versions of the toxin genes. It is unclear whether these partial proteins are still toxic or possess a different cellular activity. Two of the three cptIN loci of Bryantella formatexigens DSM 14469 encode toxins of 66 and 45 amino acids, respectively, and the singular cptIN locus of the Lachnospiraceae bacterium 9_1_43BFAA (54 amino acids), Methanococcus vannielii SB (101 amino acids) and Pyramidobacter piscolens W5455 (81 amino acids) also encode shorter toxins, all of which share sequence similarity with the 162 amino acids EUBREC_0659 protein from E. rectale ATCC 33656. The toxin from the singular tenpIN locus of Leptotrichia hofstadii F0254 (70 amino acids) is a shortened version of the TenpN toxin from P. luminescens (143 amino acids). These short proteins could represent non-toxic forms of the longer homologues, or, they could have also arisen through sequencing errors within draft genomes that would result in premature termination of the predicted proteins.
Analysis of the three Type III toxin–antitoxin families
Selected features of the sequences within each of the three families of Type III TA loci are summarized in Table 2. The summary shows that, although the two original toxIN loci (from P. atrosepticum and B. thuringiensis), were found on plasmids (9), the majority of the Type III TA loci are encoded in the host chromosome. This may be a reflection of sequencing projects focusing on chromosomes rather than extrachromosomal genomes. In the case of cptIN, however, there were no examples on any other replicon but the chromosome.
Table 2.
Overview of Type III toxin–antitoxin families
| Family name | Total members of family | Replicon |
Antitoxin repeat length (nucleotides) |
Number of tandem antitoxic repeats |
Toxin length (amino acids) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Plasmid | Phage | Mean | Range | Mean | Range | Mean | Range | ||
| toxIN | 67 | 57 | 9 | 1 | 38.1 | 31–62 | 2.8 | 1.9–5.6 | 168.0 | 70–288 |
| cptIN | 33 | 33 | 0 | 0 | 44.8 | 40–48 | 2.4 | 1.9–3.4 | 148.6 | 45–213 |
| tenpIN | 25 | 20 | 5 | 0 | 50.9 | 39–57 | 2.2 | 1.9–3.0 | 164.2 | 70–259 |
Comparing antitoxin sequences of the toxIN family with those of cptIN and tenpIN, it seems there is a general shift to progressively fewer copies, but longer lengths, of antitoxic repeats. The toxin size, however, stays approximately the same. The smaller value for mean toxin size in cptIN is skewed by the presence of multiple small putative toxins, as discussed above.
Phylogeny of ToxN proteins
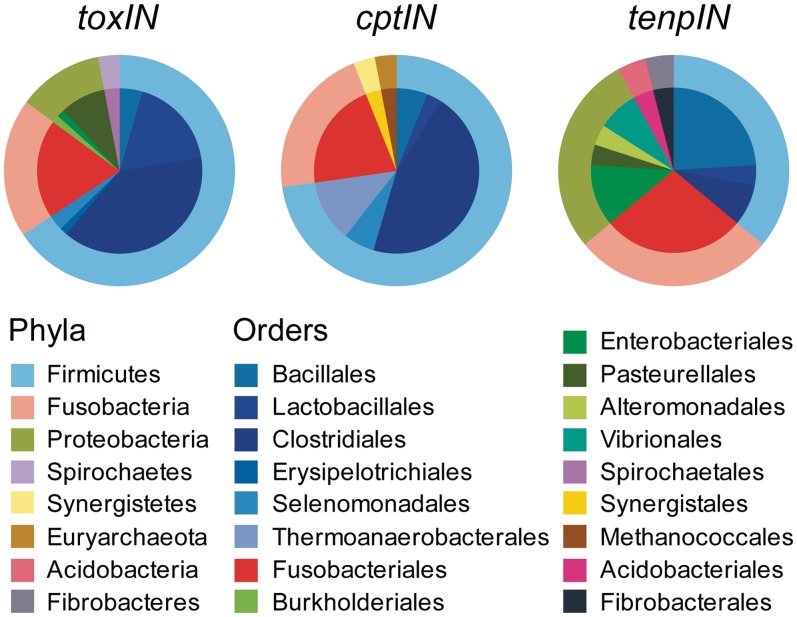
A recent global analysis of Type I TA systems indicated that there was little horizontal gene transfer contributing to dissemination of these loci (12). In contrast, Type II loci have been more freely distributed among evolutionarily unrelated species (5). Table 1 lists the taxonomy of each host organism. This distribution can also be viewed in Figure 2. It appears that the vast majority of Type III TA loci are found within either the Firmicutes, (mainly Orders Bacillales, Lactobacillales and Clostridiales), or the fusobacteria (Figure 2). In the case of the toxIN and tenpIN families, a substantial proportion of these loci are found in the Proteobacteria, while none of the cptIN loci were found in this Phylum. Though the original toxIN locus was found in the enterobacterium, Pectobacterium, the other toxIN loci within the Proteobacteria were found in either Pasteurellales or (as for one example) within Burkholderiales. In contrast, the tenpIN loci identified within the Proteobacteria included examples from several enteric bacteria, such as Photorhabdus, Xenorhabdus and Yersinia, along with loci in the Pasteurellales, Alteromonadales and Vibrionales.
Figure 2.
Taxonomy of Type III toxin–antitoxin loci. The taxonomic distribution of the identified members from each Type III toxin–antitoxin family is shown as a pie chart, with the outer ring representing the different Phyla and the inner portion representing the respective subdivisions of each Phylum into Orders. Class has been omitted for clarity. Colours are as indicated in the inset key.
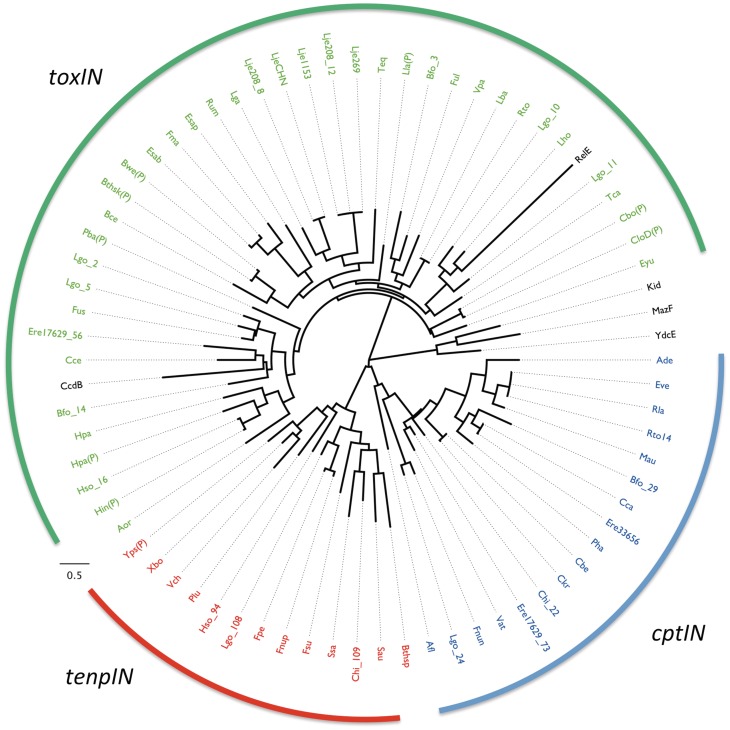
To assess the impact of horizontal gene transfer on the spread of Type III TA loci, 69 of the 125 toxin sequences were aligned and then analysed to construct a phylogenetic tree using Maximum Likelihood (Figure 3). Previous results identified the Type II toxins MazF (E. coli), Kid (E. coli plasmid R1), YdcE (Bacillus subtilis) and CcdB (E. coli F plasmid) as structural homologues of ToxN (13). These were therefore also included in the analysis, as well as RelE (E.coli), the principal member of a Type II toxin family that has not been identified as similar in structure to ToxN or Kid/MazF. The resulting dendrogram shows clear separation between toxins of the three predicted Type III TA families (Figure 3). Furthermore, it shows that the Type III toxins CptN and TenpN share a common route of divergence away from ToxN (Figure 3). The Type II endoribonucleases Kid, MazF and YdcE formed a clade independent of the three Type III families. It is interesting to note, however, that within this analysis CcdB, a topoisomerase inhibitor, and RelE were grouped with the ToxN family of proteins, albeit with large evolutionary distances.
Figure 3.
Phylogeny of selected toxin sequences. Sixty-nine toxin sequences from loci unambiguously containing all features of a Type III system (presence of putative antitoxic repeats, promoter, terminator) were aligned, together with five Type II toxins, and then analysed with TREEFINDER (29). In the case where a certain species has more than one Type III TA system selected in this manner, the number following the underscore (e.g. Lgo_14) refers to the reference number for that system (Table 1 and Supplementary Table S1). A ‘P’ in parentheses implies that the source TA system is encoded upon a plasmid, rather than within the chromosome. Entries from the toxIN family are coloured green, cptIN are blue and tenpIN are red, while Type II toxins are in black. The scale bar represents the approximate number of changes per amino acid position as the tree expands radially.
When a second phylogenetic tree was constructed using the 16S rRNA sequences from 44 of the host bacteria, we saw that these were now not tightly grouped, suggesting horizontal movement of similar Type III TA loci between unrelated bacteria (Supplementary Figure S3).
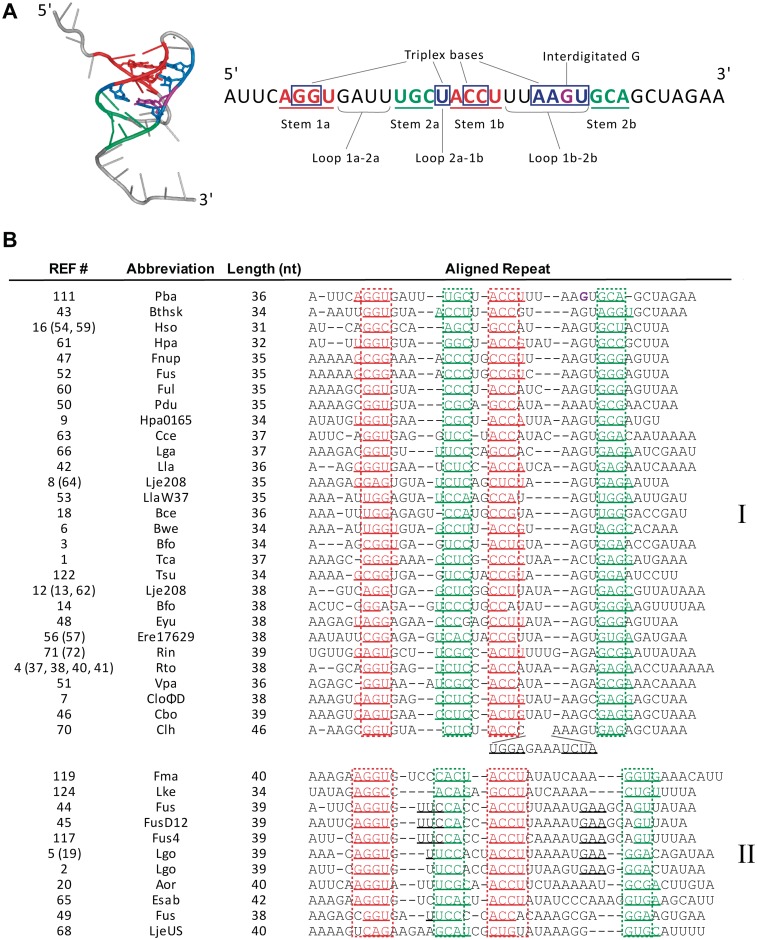
Alignment of ToxI sequences
The ToxI antitoxin of P. atrosepticum folds as a compact, hairpin-type pseudoknot with two single-stranded tails, which binds and inhibits two ToxN monomers at distinct surfaces, such that three molecules of the protein are held in a self-closing, inactive complex by three pseudoknots of ToxI (13). The pseudoknot core comprises two base-paired stems, stabilized by three base triplexes and interdigitation of a guanine between bases of the opposite strand (Figure 4A). To determine whether this structure is conserved within the new toxIN-family antitoxins, which mostly show only limited sequence similarity, attempts were made to align the new ToxI sequences to the structural template of ToxI from P. atrosepticum (Figure 4B). The alignment was performed manually, based on the placement of base-pairing regions and lengths of the interspersing loops. The precise sequence corresponding to one antitoxic RNA repeat is not known for the new homologues, so each consensus DNA repeat sequence was offset to maximize the alignment to ToxI from P. atrosepticum. In this way, 39 unique ToxI sequences, corresponding to entries for 52 toxIN loci, could be aligned to the ToxI structural template. Examination of the alignment (Figure 4B) shows that the pseudoknot structure is predicted to be conserved within this family; each ToxI contains two nested base-pairing regions, typically 3–4 nt in length. The spacing of these elements is also conserved, with a general pattern of a medium-length (3–4 nt) ‘Loop 1a–2a’, a short (1–2 nt) ‘Loop 2a–1b’, and a longer, variable length ‘Loop 1b–2b’ separating the base pairing sequences (Figure 4A and B). A short Loop 2a-1b is a common feature in RNA pseudoknots because it allows coaxial stacking of the two stem-loops in order to form a compact helical core (37). The length of Loop 1b-2b was used to divide the ToxI homologues into two groups; this was defined as ≤7 nt for Group I and >7 nt for Group II, with the exception of entry 70 from Clostridium sp. HGF2, which contains a 12 nt hairpin insertion in this loop. It was not possible to determine whether the triplex base interactions in the ToxI P. atrosepticum pseudoknot are conserved due to the sequence variability of the loop regions, however, over half of the aligned ToxI sequences retained the interdigitated guanine preceding stem region 2b.
Figure 4.
Alignment of putative pseudoknot elements with toxIN antitoxins. (A). Structure of a single pseudoknot repeat of P. atrosepticum ToxI (left, PDB: 2XDD), and schematic of secondary and tertiary interactions within each ToxI unit (right). (B) Alignment of consensus repeat sequences of the ToxIN family antitoxins. Stem loops 1 and 2 are shown in red and teal, respectively; additional potential base pairing regions are underlined. The intercalated G19 of ToxI is shown in purple. The reference numbers in brackets indicate entries with identical ToxI consensus repeat sequences, as listed in Supplementary Table S1. Entries 10, 11 and 30 could not be aligned because of overall length (∼60 nt). Entries 15 (69), 17 (39), 55, 58, 67, 118, 120 (121), 123 and 125 all contained two nested base pairing regions of >3 nt each, but could not be aligned, because the loop lengths between base pairing regions did not match the pattern of either group I or II. Strain abbreviations can be related back to entries in Table 1 and Supplementary Table S1.
Nine of the ToxI antitoxins could not be aligned because the spacing between their predicted pseudoknot base-pairing regions did not match the overall pattern in the alignment. These putative ToxI sequences may have different pseudoknot structures, or they may not encode functional antitoxins. Another three antitoxins could not be aligned because they are ∼60 nt in length, in contrast to the 31–46 nt of the aligned ToxI sequences.
The cptIN and tenpIN antitoxin sequences are generally longer than the ToxI sequences (Table 2). The CptI and TenpI antitoxins could not be readily aligned with the structural template of ToxI due to their length and sequence divergence from the canonical ToxI antitoxin family. Alignment within the CptI and TenpI families was also not possible as these longer sequences generally contained multiple possible pseudoknot base-pairing regions, and because the offset of the processed antitoxic RNA relative to the DNA repeat is not known for any members of these families.
Functional analysis of putative Type III toxin–antitoxin loci
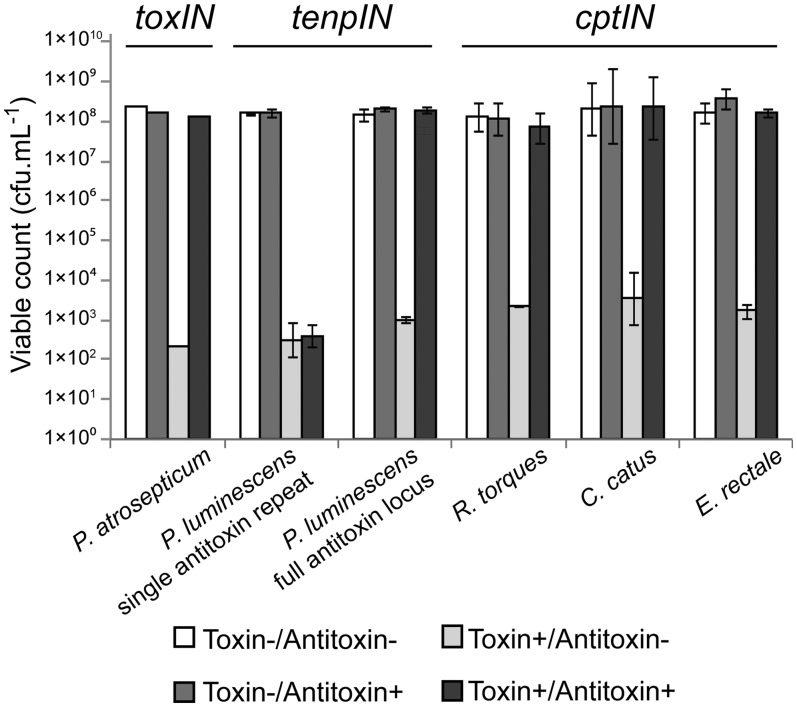
To determine whether the putative systems function as Type III TA loci, we assessed the toxicity of the proposed toxin gene and the ability of the cognate antitoxic repeats to inhibit the lethal effects. The toxin genes from the tenpIN family P. luminescens TT01 locus, and cptIN family C. catus GD/7, R. torques L2-14 and E. rectale DSM 17629 loci were cloned under the control of the l-arabinose inducible promoter in pBAD30 (33), including the native ribosome binding site. The respective antitoxic repeats, either as single repeats or as the full tandem array, were cloned into a spectinomycin-resistant derivative of pQE-80L, pTA100 (9), where they could be over-expressed by addition of IPTG. By co-transforming E. coli DH5α with both toxin and antitoxin plasmids, we were able to assess the toxicity and antitoxicity of each component (Figure 5). In the case of P. luminescens, a single TenpI repeat did not provide antitoxicity, over and above the empty vector control (Figure 5). However, the full TenpI locus of P. luminescens, acted to inhibit the cognate toxin (Figure 5). This implies that the active pseudoknot from this locus is encoded over the DNA repeat boundary, so at least two DNA repeats are required to form at least one active inhibitory pseudoknot. Following the results from P. luminescens, the full antitoxins were cloned from C. catus, R. torques and E. rectale. These loci acted as expected, with the toxin inhibited by the cognate antitoxic RNA. The cloned antitoxin sequences do not encode any predicted, translatable, open reading frames, supporting the model that the repeats encode antitoxic pseudoknots of RNA, rather than any antitoxic peptide.
Figure 5.
Protection of E. coli DH5α from Type III toxins by cognate antitoxins. Protection assays were performed as described in Materials and Methods. Results for the toxIN system of P. atrosepticum have been published previously (9); data from a single toxIN experiment is included for illustrative purposes. Of the four new loci tested, all toxin genes reduced viability of the host E. coli, which could then be restored by the full cognate antitoxin. Data shown are the mean values from triplicate experiments, with standard deviations represented by error bars.
ToxIN was first identified as an abortive infection system and is known to be active in multiple host backgrounds, providing protection against a range of bacteriophages (9,21). To assess whether the newly identified Type III TA families might also cause abortive infection, attempts were made to clone the full tenpIN locus of P. luminescens and the cptIN loci of C. catus, E. rectale and R. torques into pBR322. No recombinants were obtained from the C. catus cloning, and while the other three plasmids were made successfully, on a qualitative level the strain containing pTRB265, encoding the cptIN locus from R. torques, formed smaller colonies than the other recombinant strains.
Strains of E. coli DH5α containing the test plasmids, alongside a vector control, were infected with new environmental coliphages. These coliphages were isolated from treated sewage effluent taken on three independent visits to a sewage treatment plant. Following isolation, individual phages were re-tested against all the cloned Type III TA systems, in order to obtain Efficiency of Plating (EOP) data, as a measure of phage resistance (Table 3). As expected, the toxIN system from P. atrosepticum dramatically reduced the EOP of three phages (Table 3). The tenpIN locus from P. luminescens also reduced the EOP of the same three phages. The cptIN loci from E. rectale and R. torques did not affect the EOPs of any of the six phages tested. This confirms that at least one of the two, new, tenpIN and cptIN Type III TA families has abortive infection capacity and can provide high levels of resistance to bacteriophage infection.
Table 3.
Bacteriophage resistance provided by Type III toxin–antitoxin systems
| Phage | EOP on test DH5α strain (plasmid carried, Type III toxin–antitoxin family encoded, source organism) |
|||
|---|---|---|---|---|
| pTA46 | pFR2 | pFLS112 | pTRB265 | |
| toxIN | tenpIN | cptIN | cptIN | |
| Pectobacterium atrosepticum | Photorhabdus luminescens | Eubacterium rectale | Ruminococcus torques | |
| ΦF6 | 2.6 × 10−7 | 1.8 × 10−4 | 0.9 | 0.8 |
| ΦF8 | 0.6 | 0.5 | 0.9 | 1.0 |
| ΦTB23 | 1.1 | 1.2 | 1.0 | 1.1 |
| ΦTB27 | <6.0 × 10−8 | <6.0 × 10−8 | 0.7 | 0.9 |
| ΦTB28 | <4.3 × 10−9 | <4.3 × 10−9 | 1.1 | 0.9 |
| ΦTB29 | 0.9 | 0.9 | 0.8 | 0.7 |
DISCUSSION
Previous global studies have used and developed automated methods to consolidate and extend lists of Type I and Type II TA loci, while also validating selected new entries (5,12,16). Type III TA loci are a recent discovery and to date only a limited list, containing 19 homologues of one family, has been published (9). The aim of the current study was to search for, catalogue, define and characterize new Type III systems and families.
Identification of Type III toxin–antitoxin systems
Due to the complexity and specific features required of the nucleotide sequences defining a Type III TA locus, there was no readily available method for fully automated screening of the collected sequence databases. Having recently solved the macromolecular structure of ToxIN by crystallography (4), it became possible to begin screening for Type III TA systems using the ToxN structure for structure-based homology searches. Analysis of the resulting structure-based search hits identified 37 putative Type III TA systems, which were divided into three families. Exhaustive sequence homology searches then further increased the numbers included within each of these families, producing a final list of 125 putative Type III TA loci. This marks the first identification of Type III TA loci encoding toxins bearing no significant amino acid sequence similarity with the initial Type III TA locus toxin, ToxN. This confirms the prediction that the ‘Type III’ descriptor extends to many more families and is not an isolated case for ToxN and homologues thereof. This list also greatly extends the known and predicted homologues within the toxIN family. As the results of this study were driven from an initial structure-based homology search, there is clear bias towards a subset of database entries. It is highly likely that there are many more Type III TA families that remain to be identified. An automated system, combining searches for tandem repeats or pseudoknot sequences coupled to palindromic repeats and an ORF, would be a very powerful tool to further expand our understanding of the numbers and spread of Type III TA loci.
Distribution and abundance of Type III toxin–antitoxin systems
The Type III families identified in this study are dominated by entries from the Firmicutes and Fusobacteria (Figure 2). In the cases of toxIN and tenpIN, this also extends, in part, to the Proteobacteria. The distribution of Type III systems within each of these Phyla does not appear biased by the relative levels of sequenced strains from each Phylum; as of September 2011, considering the total number of bacterial genome projects in the NCBI, 28% of entries were for the Firmicutes, 44% were for the Proteobacteria and <1% were for the Fusobacteria. Both Type I and Type II TA systems also appear over-represented within the Firmicutes and Proteobacteria (5,12).
Type III loci were identified in bacteria with a wide-range of lifestyles (Table 1 and Supplementary Table S1). The most pertinent category may be that of the human pathogens. Bacillus cereus Rock1-15 can cause food poisoning, while several clinical isolates were also identified; Yersinia pseudotuberculosis IP 31758, the two Vibrio strains and the Staphylococcus aureus strains. Perhaps of greatest clinical relevance is the presence of tenpIN loci on conjugative multi-resistance plasmids from S. aureus. Though only plasmid pVRSA (38) is listed in Table 1, many other S. aureus plasmids contain these systems, including pGO1, which is associated with aminoglycoside resistance (39) and the related plasmid pSK41, a member of the β-lactamase-heavy-metal-resistance plasmid family (40).
It is of note that 70 of the 125 hits come from strains that have been sequenced as part of the Human Microbiome Project (HMP) (http://www.hmpdacc.org/) (41). A further eight hits are from strains in the metaHIT project (http://www.metahit.eu/). While metaHIT focuses solely on the Human Intestinal Tract, the HMP samples from many sites around the healthy human body. As of October 2011, the HMP had deposited ∼800 of the ∼12 500 bacterial genome project sequences in the NCBI database. As this small proportion of HMP sequences, ∼6% of the database, contains over half of the putative Type III TA systems, it appears these loci are well represented within human commensals.
The relative numbers of Type III loci present within a strain (maximum of six thus far) are currently low in comparison to the large numbers of Type II systems (up to 97) or medium numbers of Type I systems (up to 26) identified in other strains (5,12). These values for Type I and Type II systems have been steadily increasing since initial discovery of TA systems; further global searches for Type III systems will probably increase these numbers in a similar fashion.
Functional roles of Type III toxin–antitoxin systems
Using co-over-expression assays, members of both the cptIN and tenpIN families were confirmed as active TA systems, in an E. coli model (Figure 5). While ToxIN from P. atrosepticum also acts as an abortive infection system in the endogenous and other host enteric strains (9,21), and it was shown that TenpIN from P. luminescens can abort infection by coliphages in E. coli (Table 3), it cannot be concluded that this is the natural ‘role’ of Type III TA loci. Furthermore, the cptIN loci tested were not able to provide phage resistance against this small subset of phages and in an E. coli model. It is of note that both the toxIN and tenpIN systems were identified in enteric bacteria, so phage resistance may be a specific attribute of Type III TA systems from this host taxon. We also used an enteric model of phage infection; the cptIN loci came from Firmicutes, so the coliphages used may be too distantly related from the phages targeting E. rectale and R. torques to be recognized and aborted, or the cptIN systems may not be correctly expressed in E. coli. Transferred back to their natural hosts, these cptIN loci may be active against cognate phages, though they may also fulfil an entirely different requirement. Further investigations are required to identify cellular roles for cptIN. The original biological role of Type III TA systems is likely to remain under debate. It will be interesting to study the activities of other Type III TA systems, both in model systems and within their endogenous hosts, to investigate their full physiological capabilities and evolutionary significance.
CONCLUSION
The numbers of known Type I and Type II TA systems has greatly increased as methods of identification have been streamlined and the sequence databases swell with entries. In comparison, though this study identifies new Type III TA systems for the first time, the numbers of families and examples within each family are comparatively few in regard to the Type I and II systems. Performing further studies of the full sequence databases in an automated and unbiased fashion is highly likely to prove useful and instructive. It is predicted that there are many more undiscovered Type III TA systems ready to be examined. Doing so will provide greater understanding of their diversity, abundance and biological roles and will provide a useful range of molecular ‘reagents’ with which to study protein–RNA interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S5, Supplementary Figures S1–S3.
FUNDING
Biotechnology and Biological Sciences Research Council (UK); Wellcome Trust and the Marsden Fund, Royal Society of New Zealand; Commonwealth Scholarship awarded from the Commonwealth Scholarship Commission in the United Kingdom and co-funded by the Cambridge Commonwealth Trust (to F.L.S). Work with P. atrosepticum was performed under a plant health license from the Department for Environment, Food and Rural Affairs (UK). Funding for open access charge: BBSRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Keith Turner for providing the genomic DNA of Coprococcus catus GD/7, Ruminococcus torques L2-14 and Eubacterium rectale DSM 17629. Photorhabdus luminescens subsp. laumondii TT01 was kindly provided by Dr David Clarke. We also thank Chen Yi-An of the Mizuguchi lab, for help in constructing the bacterial and archaeal proteome database.
REFERENCES
- 1.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 2.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 3.Hayes F, Van Melderen L. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 2011;46:386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- 4.Blower TR, Salmond GP, Luisi BF. Balancing at survival’s edge: the structure and adaptive benefits of prokaryotic toxin-antitoxin partners. Curr. Opin. Struct. Biol. 2011;21:109–118. doi: 10.1016/j.sbi.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Dréze P, Van Melderen L. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011;39:5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogura T, Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl Acad. Sci. USA. 1983;80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc. Natl Acad. Sci. USA. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal ‘addiction module’ regulated by guanosine [Corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl Acad. Sci. USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl Acad. Sci. USA. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011;77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magnuson RD. Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blower TR, Pei XY, Short FL, Fineran PC, Humphreys DP, Luisi BF, Salmond GPC. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat. Struct. Mol. Biol. 2011;18:185–190. doi: 10.1038/nsmb.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jørgensen MG, Pandey DP, Jaskolska M, Gerdes K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 2009;191:1191–1199. doi: 10.1128/JB.01013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of Type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makarova KS, Wolf YI, Snir S, Koonin EV. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 2011;193:6039–6056. doi: 10.1128/JB.05535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sevin EW, Barloy-Hubler F. RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007;8:R155. doi: 10.1186/gb-2007-8-8-r155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao Y, Harrison EM, Bi D, Tai C, He X, Ou HY, Rajakumar K, Deng Z. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2010;39:D606–D611. doi: 10.1093/nar/gkq908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Blower TR, Fineran PC, Johnson MJ, Toth IK, Humphreys DP, Salmond GP. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin–antitoxin locus of Erwinia. J. Bacteriol. 2009;191:6029–6039. doi: 10.1128/JB.00720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 24.Mizuguchi K, Deane CM, Blundell TL, Johnson MS, Overington JP. JOY: protein sequence-structure representation and analysis. Bioinformatics. 1998;14:617–623. doi: 10.1093/bioinformatics/14.7.617. [DOI] [PubMed] [Google Scholar]
- 25.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, Phan I, et al. Integr8 and Genome Reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jobb G. TREEFINDER version of March 2011. 2011. Munich, Germany. [Google Scholar]
- 30.Reeder J, Steffen P, Giegerich R. pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucleic Acids Res. 2007;35:W320–W324. doi: 10.1093/nar/gkm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- 32.Fineran PC, Everson L, Slater H, Salmond GP. A GntR family transcriptional regulator (PigT) controls gluconate-mediated repression and defines a new, independent pathway for regulation of the tripyrrole antibiotic, prodigiosin, in Serratia. Microbiol. 2005;151:3833–3845. doi: 10.1099/mic.0.28251-0. [DOI] [PubMed] [Google Scholar]
- 33.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 35.Lehnherr H, Maguin E, Jafri S, Yarmolinsky MB. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 1993;233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- 36.Tian QB, Ohnishi M, Tabuchi A, Terawaki Y. A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 1996;220:280–284. doi: 10.1006/bbrc.1996.0396. [DOI] [PubMed] [Google Scholar]
- 37.Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3:e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 39.Caryl JA, O'Neill AJ. Complete nucleotide sequence of pGO1, the prototype conjugative plasmid from the staphylococci. Plasmid. 2009;62:35–38. doi: 10.1016/j.plasmid.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Berg T, Firth N, Apisiridej S, Hettiaratchi A, Leelaporn A, Skurray RA. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 1998;180:4350–4359. doi: 10.1128/jb.180.17.4350-4359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.