Abstract
While most siRNAs induce sequence-specific target mRNA cleavage and degradation in a process mediated by Ago2/RNA-induced silencing complex (RISC), certain siRNAs have also been demonstrated to direct target RNA reduction through deadenylation and subsequent degradation of target transcripts in a process which involves Ago1/RISC and P-bodies. In the current study, we present data suggesting that a third class of siRNA exist, which are capable of promoting target RNA reduction that is independent of both Ago and RISC. These siRNAs bind the target messenger RNA at the polyA signal and are capable of redirecting a small amount of polyadenylation to downstream polyA sites when present, however, the majority of the activity appears to be due to inhibition of polyadenylation or deadenylation of the transcript, followed by exosomal degradation of the immature mRNA.
INTRODUCTION
RNA interference (RNAi) is a mechanism by which double-stranded RNA triggers the inhibition of target gene expression by inducing sequence-specific target mRNA degradation (1). Short interfering RNAs (siRNAs), exogenously administered RNA duplexes of 21–23 nt, lead to degradation of specific mRNAs through the RNA-induced silencing complex (RISC) (2). After cellular delivery of synthetic siRNAs, the double-stranded molecules are incorporated into the RISC-Loading Complex (RLC), which consists of Ago2, Dicer and TRBP (the HIV trans-activating response RNA-binding protein) (3). Prior to target mRNA recognition, an siRNA duplex goes through an ATP-dependent unwinding process and one of the two siRNA strands, referred to as the passenger strand, is released, while the other strand (guide strand) remains bound to the Argonaut protein, Ago2 (4,5). This strand then facilitates interaction of RISC with its complementary target RNA, which is finally cleaved at the site opposite the 10th and 11th positions of the siRNA guide strand by the RNase activity located in the Ago2 protein, triggering its destruction (6–11).
Recent studies suggest that siRNA-mediated off-target mRNA reduction is the result of Ago2- independent degradation processes (12,13), and like miRNAs (8), direct target RNA reduction through deadenylation and subsequent degradation of target transcripts in a process which likely involves P-bodies (14–16). More recently, it has been demonstrated that at least a portion of some siRNA on-target activity is also Ago2 independent (17).
In the current study, a series of siRNAs was screened for activity in Ago2−/− cells. Several siRNA that exhibited significant activity were identified. One siRNA, targeted to Il4R-α, was found to be equally active in wild-type and Ago2−/− cells. This siRNA was determined to bind the target at an upstream poly A site and was found to be active independent of any Ago or other RISC associated proteins. We present data demonstrating that this siRNA is capable of redirecting a small amount of polyadenylation to a downstream polyA site; however, degradation of the message appears to be the result of inhibition of polyadenylation or deadenylation of the transcript. siRNAs targeted to the polyA sites of certain other RNAs were also capable of promoting target degradation in an Ago and P-body independent manner.
MATERIALS AND METHODS
Preparation of antisense oligonucleotides and siRNAs
Synthesis and purification of phosphorothioate/2′-MOE oligonucleotides was performed using an Applied Biosystems 380B automated DNA synthesizer as described previously (18). All RNaseH-dependent antisense oligonucleotides (ASOs) used for target mRNA reduction were 18–20 bases in length, full phosphorothioate with 2′-O-methoxyethyl substitutions at the positions indicated by underlined type. The sequences of the ASOs and siRNA guide strands can be found in Table 1. MOE modified IL4-Ra siRNAs were synthesized as previously described (19). The positions of the MOE substituted nucleotides on the antisense strand are underlined. The siRNA sense strand is unmodified RNA.
Table 1.
Sequences of siRNAs and antisense oligonucleotides used in study
| ISIS No. | Target | Sequence | Chemistry |
|---|---|---|---|
| 383266 | Il4R-α | ACCTCAGCAACAACAGCAC | siRNA |
| 383268 | Il4R-α | GTGAAGGATCTGCTCCCAG | siRNA |
| 383270 | Il4R-α | CACAGACCTCAGCAACAAC | siRNA |
| 383273 | Il4R-α | TTCTGGGAAACCTGCCAGC | siRNA |
| 383275 | Il4R-α | GTTCTCAGGTGACATGCTC | siRNA |
| 383281 | Il4R-α | TTTATTGACATAAAGCTCC | siRNA |
| 383281-2MM | Il4R-α | TTTATTGACATAtAGCTgC | siRNA |
| 383281-4MM | Il4R-α | TTaATTcACATAtAGCTgC | siRNA |
| 383281-SCR | Il4R-α | agatcTctacgtctataat | siRNA |
| 459728 | Il4R-α | TAAAGACTTTATTGACATAA | siRNA |
| 459730 | Il4R-α | GACAAGATAAAGACTTTATT | siRNA |
| 459732 | Il4R-α | TATTGACATAAAGCTCCATG | siRNA |
| 451522 | Il4R-α | TTTATTGACATAAAGCTCC | siRNA |
| 455185 | Il4R-α | TTTATTGACATAAAGCTCC | siRNA |
| 29589 | PTEN | TTAAATTTGGCGGTGTCA | siRNA |
| 29592 | PTEN | TGTCTCTGGTCCTTACTT | siRNA |
| 29593 | PTEN | ACATAGCGCCTCTGACTG | siRNA |
| 29600 | PTEN | TGCTAGCCTCTGGATTTG | siRNA |
| 29604 | PTEN | GTGTCAAAACCCTGTGGA | siRNA |
| 371864 | Eg5 | CAACAAGGATGAAGTCTAT | siRNA |
| 549896 | WAF1 | TTTATTGAGCACCAGCTTTG | siRNA |
| 549894 | GATA3 | TTTAATAAATACCATCAAAA | siRNA |
| 549900 | STAT2 | TTTATTATGACGTACGAAGG | siRNA |
| 308299 | Exosc9 | TGCCAAACCTTTTCACCAGC | RNaseH |
| 454373 | SKIV2l | GCCAGCCTCATCCACATAGC | RNaseH |
| 395392 | Ago1 | CTGGACTATTTGCATAGCAA | RNaseH |
| 399560 | Ago3 | GTTTGCCCATAGTGCCATAC | RNaseH |
| 395927 | Ago4 | GGCTAACAGTCGAATTGGTT | RNaseH |
| 368473 | Dcp1a | CACCATGTTGCTCAGGGAGG | RNaseH |
| 365128 | DCP2 | CTGCATGTAGAAATCCAAGT | RNaseH |
| 443335 | GW182 | TCCTCAGTAGATTTCCATCC | RNaseH |
| 339800 | DDX6 | CTCGGGTAAACAGATCAGTG | RNaseH |
All siRNA are full RNA in both sense and antisense strand except underlined bases which indicate 2′-O-methoxyethyl substitutions (2′MOE). All RNaseH-dependent ASOs are full phosphorothioate with 2′-O-methoxyethyl substitutions at the positions indicated by underline. Nucleotide mismatches are represented by lower case.
siRNA/ASO treatment
Tissue culture medium, trypsin, Lipofectamine RNAiMAX and Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA). Wild-type and Ago2 knockout mouse embryonic fibroblasts (MEF) were a kind gift of Dr Gregory J. Hannon. Cells were cultured in DMEM supplemented with 10% fetal calf serum, streptomycin (0.1 µg/ml) and penicillin (100 U/ml). For the knockdown of RISC or P-Body associated genes, cells were treated at 30–100 nM with the indicated ASO/siRNA in Opti-MEM media (Invitrogen) containing 6 µg/ml Lipofectamine 2000 or Lipofectamine RNAiMAX for 4 h at 37°C, as described previously (19). Cells were split 24–72 h after the start of ASO treatment and seeded in 96-well plates at 3500 cells/well. The following day, wild-type and/or Ago/RISC-reduced cells were treated with 3–300 nM siRNA to generate of IC50 curves (N = 3–4/concentration). The next day total RNA was purified from 96-well plates using an RNeasy 3000 BioRobot (Qiagen, Valencia, CA) and target mRNA levels assessed by qRT/PCR. For siRNA competition experiments, cells were co-transfected at the indicated concentrations as previously described (20).
Reduction of target mRNA expression was determined by qRT-PCR performed essentially as described elsewhere (21). Briefly, total RNA was analyzed in a final volume of 30 µl containing 200 nM gene-specific PCR primers, 0.2 mM of each dNTP, 75 nM fluorescently labeled oligonucleotide probe, 5 µl RT-PCR buffer, 5 mM MgCl2, 2 U of Platinum Taq DNA Polymerase (Invitrogen) and 8 U of RNase inhibitor. Reverse transcription was performed for 30 min at 48°C followed by PCR: 40 thermal cycles of 30 s at 94°C and 1 min at 60°C using an ABI Prism 7700 Sequence Detector (Applied Biosystems). To avoid artifacts based upon well to well variation in cell number, mRNA levels were normalized to the total amount of RNA present in each reaction as determined by Ribogreen assay (22) (Invitrogen). IC50 curves and P-values were generated using Prism 4 software (GraphPad). Sigmoidal dose response was calculated according to Y = Bottom + (Top − Bottom)/(1 + 10((LogEC50 − X))); where X is the logarithm of concentration and Y is the response. The sequence for the Il4R-α primer/probe set used in the RT-PCR reaction is TCCCATTTTGTCCACCGAATA for the forward primer, GTTTCTAGGCCCAGCTTCCA for the reverse primer and TGTCACTCAAGGCTCTCAGCGGTCC for the probe. Where indicated, a second Il4R-α primer/probe set downstream of polyA site 1 was used: (FP-CCTTCCACTGCTTATACCCG, RP-GCTTGTACCTTAGTTCCCTCAG, Probe-TTACCACCCTCTCCCATCTCCT). The sequence for the PTEN primer/probe set is ATGACAATCATGTTGCAGCAATTC for the forward primer, CGATGCAATAAATATGCACAAATCA for the reverse primer, and CTGTAAAGCTGGAAAGGGACGGACTGGT for the probe. The sequence for the GAPDH primer/probe set is GGCAAATTCAACGGCACAGT for the forward primer, GGGTCTCGCTCCTGGAAGAT for the reverse primer, and AAGGCCGAGAATGGGAAGCTTGTCATC for the probe. The sequence for the cyclophilin A primer/probe set is TCGCCGCTTGCTGCA for the forward primer, ATCGGCCGTGATGTCGA for the reverse primer, and CCATGGTCAACCCCACCGTGTTC for the probe. The sequence for the Eg5 primer/probe set used in the RT-PCR reaction is GACTTGTATTGGTGAATGCCATCT for the forward primer, AGGAGTCGAAGACAAACTAGAAAAGG for the reverse primer and TCTGGCCAAGGCTGTTTCCCTACCTCTAAC for the probe.
Primer probe sets used to analyze the levels of targeted RISC/P-body genes are as follows. Mouse Ago1: AGTGATGCCAACACTCCATCTG (forward primer), AGGGTGATGGAACACGAAGTAGA (reverse primer) and CCTGTCCATCGCCTTCATAGACA (probe). Mouse Ago2: CCATCTAGCTGTGAAGGCTCTGA (forward primer), TTCTTAGGGCCAGGCTTTAAAA (reverse primer) and CATGAAAGCCACACCATTTCCATTGGG (probe). Mouse DDX6: GCAAAAGCGGTGCCTACCT (forward primer), CACCATTGCTTGAATATTGTCCTT (reverse primer) and TCCCCTTACTTGAAAGGCTGGACCTGA (probe). Mouse GW182: GACAATGTCATGCCCCACACT (forward primer), CAAGCCTATAGGAAAGTTGCTGAAA (reverse primer) and CACCCGAGCTGCAGAAAGGGCC (probe).
Western blots were performed as described elsewhere (20). The rabbit polyclonal antibody to Il4R-α was obtained from Santa Cruz Biotechnology (sc-686) and mouse Anti-γ-Tubulin from Sigma-Aldrich (T5326).
RLM-RACE mapping of polyadenylation sites
Ago2−/− MEF cells were treated with Il4R-α siRNA 383281 or the corresponding full MOE ASO at a concentration of 100 nM. 3′ RACE was performed using 1 µg total RNA purified from the cells the following day using a First Choice RLM-RACE kit according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA). The sequence of the Il4R-α forward primer for the 3′RACE PCR reaction was GGAAGCTGGGCCTAGAAACT. Following 35 cycles of PCR, 10 µl was run on a 2% agarose gel in 1 × TBE buffer and visualized by ethidium bromide staining. The gel was then transferred to nylon membrane essentially as described previously (23). Hybridization was performed using 5′-end-labeled oligonucleotide probes, as described previously (24). Probe 1: CATCTGCTATCCCTACCACCTG (short) and Probe 2: TCAATGCCTTGACACATTCAAC (long). Eithidium bromide stained PCR products were also gel purified and cloned into pCR4-TOPO (Invitrogen) for sequence analysis.
To quantitate polyA site usage total RNA was also reverse transcribed using oligo dT primer. qPCR was then performed cells using primer probe sets upstream or downstream of PA site #1 as described above for Il4R-α. The WAF1 primer/probe set was FP-CAGGCACCATGTCCAATCCT, RP-GAGACAACGGCACACTTTGCT, Probe-TGATGTCCGACCTGTTCCGCACA, and the 3′RACE forward primer was CATTCTATGGTGTGTGGTGGTG.
RESULTS AND DISCUSSION
Identification of Ago2 cleavage independent siRNAs
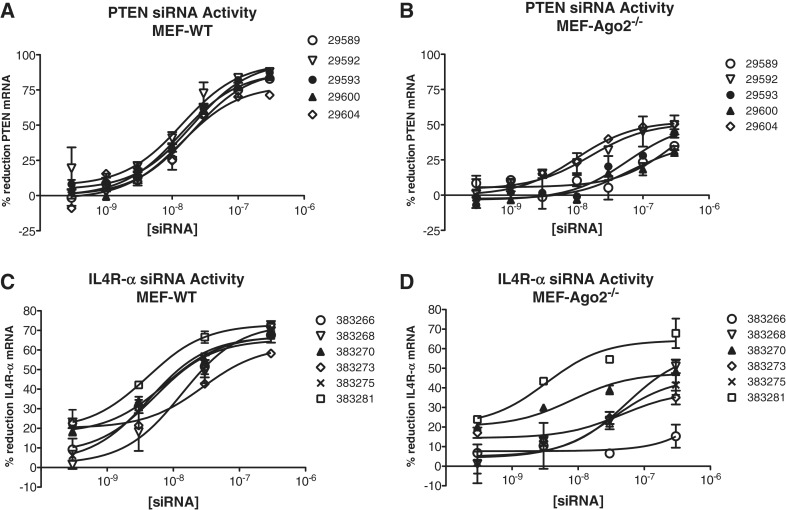
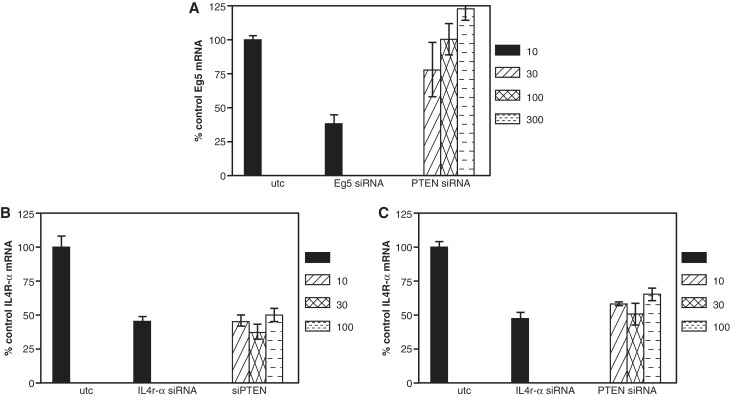
We have previously shown that a significant portion of on-target siRNA activity mediated by an siRNA directed against PIK3CB is maintained in mouse Ago2−/− cells and is Ago2 cleavage independent (17). To identify other Ago2 cleavage independent siRNAs, a series of siRNAs directed against Il4R-α or PTEN was screened in wild-type and Ago2−/− mouse fibroblasts. All PTEN siRNAs evaluated had similar activity in wild-type cells each with an IC50 of ∼10 nM (Figure 1A). In Ago2−/− cells, the activity of the same set of siRNAs was more variable (Figure 1B), and the potency and efficacy (maximum target reduction) were much lower than observed in the wild type cells. The siRNAs targeted to Il4R-α displayed a greater variation in activity in the wild-type cells with IC50’s ranging from ∼5–50 nM (Figure 1C). While all, but one Il4R-α siRNA (383266) exhibited some activity in the Ago2−/− cells (Figure 1D), one siRNA, 383281, had similar efficacy and an IC50 of <10 nM in both Ago2- and wild-type cells.
Figure 1.
Multiple siRNAs demonstrate significant Ago2-independent activity. Wild-type and Ago2−/− mouse embryonic fibroblasts (MEFs) were transfected with siRNAs at concentrations ranging from 300 pM to 300 nM as detailed in ‘Materials and Methods’ section. The following day target mRNA reduction was measured by qRT/PCR. Percent inhibition at each treatment concentration is shown. (A) PTEN siRNA activity in WT cells. (B) PTEN siRNA activity in Ago2−/− cells. (C) Il4R-α siRNA activity in WT cells. (D) Il4R-α siRNA activity in Ago2−/− cells.
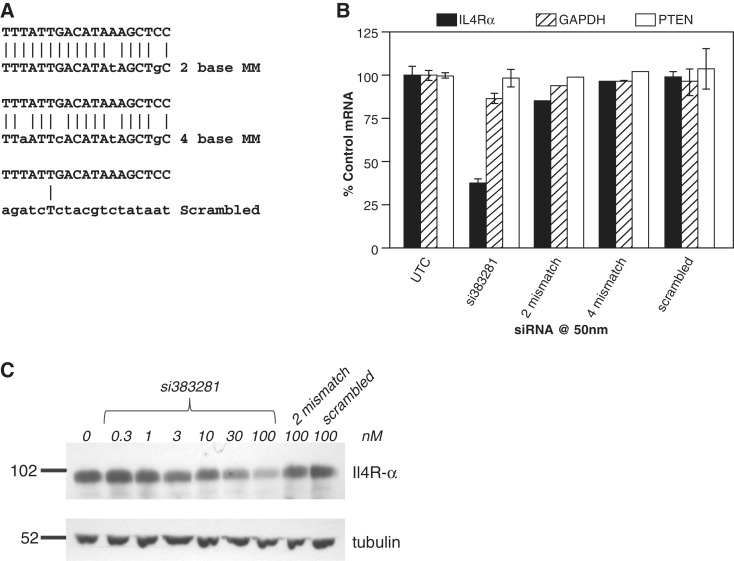
To confirm that the observed inhibition was sequence specific, two and four base mismatches and a scrambled control were designed based upon the siRNA 383281 sequence (Figure 2A). Ago2−/− cells were treated with siRNA 383281 and control siRNAs at a concentration of 50 nM. The following day levels of Il4R-α mRNA as well as G3PDH and PTEN were evaluated by qRT/PCR. Treatment with siRNA 383281 resulted in ∼60% reduction of Il4R-α mRNA, however no reduction of the Il4R-α mRNA was observed with the four base mismatch or scrambled control and only ∼10% reduction was observed with the two base mismatch (Figure 2B). No significant reduction of non-target mRNAs was observed with any siRNA. To confirm that protein levels were also reduced, Ago2−/− fibroblasts were transfected with siRNA at concentrations ranging from 300 pM to 100 nM. Western blot analysis was performed 24 h post-transfection. Il4R-α protein was significantly reduced by treatment with siRNA 383281 with an IC50 of ∼10 nM (Figure 2C), a concentration comparable to the IC50 for RNA reduction. At the highest concentration, mismatch and scrambled controls had little effect on Il4R-α protein expression.
Figure 2.
Ago2-independent activity is sequence specific. (A) two and four base mismatches or scrambled control siRNAs were designed based upon the sequence of siRNA 383281 (upper strand). Mismatches are depicted in lower case on the bottom strand. (B) Levels of Il4R-α (solid bars), cyclophilin (open bars), and GAPDH (striped bars) mRNA were measured by qRT/PCR 48 h post-transfection with 100 nM 383281 and control siRNAs. (C) Ago2−/− fibroblasts were transfected with siRNA 383281 at concentrations ranging from 300 pM to 100 nM and with mismatch and scrambled controls at 100 nM. Western blot analysis was performed 24 h post-transfection as detailed in ‘Materials and Methods’ section. A total of 50 µg protein was loaded in each lane.
siRNA 383281 is active in the absence of Ago and P-body proteins
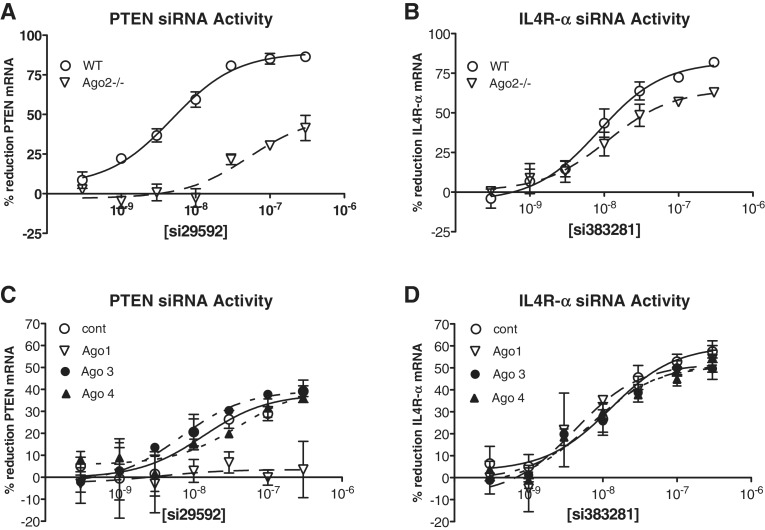
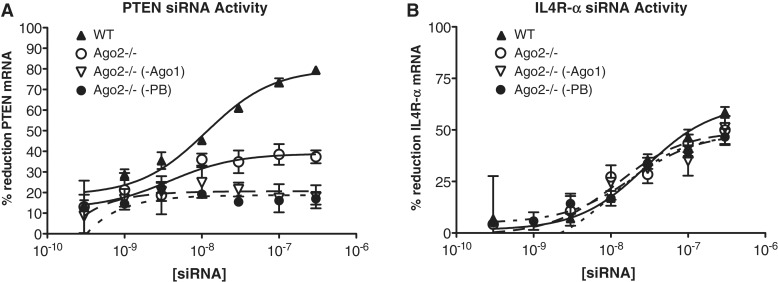
Since we had previously shown that Ago2 independent siRNA activity may be mediated by Ago1, the effect of Ago1 reduction on the activity of siRNA 383281 was next evaluated. Ago2−/− fibroblasts were treated with ASOs targeting Ago1, Ago3 or Ago4 as detailed in ‘Materials and Methods’ section. After 48 h, Ago-reduced and wild-type cells were treated with 3–300 nM siRNA. As observed previously, Il4R-α siRNA 383281 was found to have comparable activity in Ago2−/− and wild-type cells (Figure 3B), while PTEN siRNA 29592 had a potency similar to that of si383281 (Il4R-α) in wild-type cells, but was much less active in the Ago2−/− cells (Figure 3A). The Ago2-independent activity of si29591 (PTEN) was not affected by reducing Ago3 or Ago4, however reduction of Ago1 in the Ago2−/− cells resulted in a complete ablation of activity (Figure 3C). In contrast, the activity siRNA 383281 was not affected by reduction of any of the Ago proteins (Figure 3D). Simultaneous reduction of Agos 1, 3 and 4 in the Ago2−/− cells also had no effect on the activity of this siRNA (Supplementary Figure S1). Reduction of each of the Ago RNAs was greater than 75% as estimated by qRT/PCR which we have previously demonstrated is correlated with similar reduction in protein levels (17).
Figure 3.
Il4R-α siRNA activity is also independent of Ago 1, 3, and 4. Wild-type and Ago2−/− MEFs were treated with ASOs targeting Ago1, Ago3, or Ago4. The following day Ago-reduced cells were seeded in 96-well plates then transfected with PTEN or Il4R-α siRNA at concentrations ranging from 300 pM to 300 nM. Messenger RNA reduction was measured after 24 h by qRT/PCR. IC50 curves are plotted versus no siRNA control for each treatment condition. (A) Activity of PTEN siRNA 341391 in wild-type (WT) and Ago2−/− (KO) cells. (B) Activity of Il4R-α siRNA 383281 in WT and KO MEF cells. (C) Activity of PTEN siRNA 341391 in KO cells (circle), and KO cells reduced in Ago 1 (inverted triangle), Ago 3 (filled circle), or Ago 4 (filled triangle). (D) Activity of Il4R-α siRNA 383281 in KO cells (circle), and KO cells reduced in Ago 1 (inverted triangle), Ago 3 (filled circle), or Ago 4 (filled triangle).
Since degradation of a siRNA targeted message may occur in P-bodies even in the absence of Ago2-mediated cleavage (25), the effect of P-body reduction on the activity of the PTEN and Il4R-α siRNAs was next evaluated. Ago2−/− fibroblasts were treated with ASOs targeting Ago1 or with P-body associated proteins GW182 and DDX6. Reduction of the targeted RNAs was confirmed to be >80% by qRT/PCR (data not shown). After 48 h, Ago1/P-body reduced and control Ago2−/− MEF cells were treated with 3–300 nM siRNA. Reduction of either Ago1 or the P-body proteins resulted in a reduction in Ago2-independent activity of PTEN siRNA 29592 (Figure 4A), suggesting that the Ago2-independent activity of this siRNA is RISC dependent and mediated via a P-body dependent degradation pathway. In contrast, the activity of Il4R-α siRNA 383281 neither was affected by Ago1 reduction nor was activity diminished in the P-body reduced cells (Figure 4B). Together, these data suggest a novel siRNA-mediated degradation pathway independent of Ago1-4, RISC, and associated P-body degradation.
Figure 4.
Il4R-α siRNA activity does not require P-body associated proteins. Ago2−/− MEFs were treated with ASOs targeting Ago1 or with P-body (PB) associated proteins GW182 and DDX6. The following day Ago1 or P-body reduced cells were seeded in 96-well plates then transfected with PTEN or Il4R-α siRNA at concentrations ranging from 300 pM to 300 nM. Messenger RNA reduction was measured after 24 h by qRT/PCR. IC50 curves are plotted versus no siRNA control for each treatment condition. (A) Activity of PTEN siRNA 29592 in WT cells (filled triangle), KO cells (circle), or KO cells reduced in Ago 1 (inverted triangle) or PB (filled circle). (B) Activity of Il4R-α siRNA 383281 in WT cells (filled triangle), KO cells (circle), or KO cells reduced in Ago 1 (inverted triangle) or PB (filled circle).
siRNA 383281 does not compete with other siRNAs for free RISC
Previous work has shown that co-transfection of two siRNAs can lead to a reduction in activity as a result of competition for the limited amount free RISC available in the cell (20,26). To confirm the RISC independence of siRNA 383281, we performed experiments in which this Il4R-α siRNA was co-transfected with a RISC-dependent siRNA to a different target to evaluate whether the siRNAs compete for RISC. Wild-type MEFs were treated with a siRNA targeting Eg5 at a concentration of 10 nM. In the absence of a second siRNA, ∼60% reduction in Eg5 mRNA was observed (Figure 5A). A concentration dependent loss in Eg5 siRNA activity was observed when the PTEN siRNA was co-transfected at 10, 30 or 100 nM. In the absence of competition, Il4R-α siRNA 383281 reduced Il4R-α mRNA in both wild-type and Ago2−/− cells by ∼50% at 10 nM (Figures 5B and C). However, no loss in activity was observed even when PTEN siRNA was co-transfected at concentration of up to 100 nM. When Il4R-α siRNA 383281 was used as a competitor to either Eg5 or PTEN siRNA, no competition was observed even at a 10-fold excess over the target siRNA (Supplementary Figure S2). Once again, these data suggest a mechanism for siRNA 383281 that is RISC independent.
Figure 5.
Il4R-α siRNA activity does not compete for free RISC with other siRNAs. (A) WT MEF cells were transfected 10 nM Eg5 siRNA alone or in the presence of 10, 30, or 100 nM PTEN siRNA for 4 h. Reduction of Eg5 mRNA was assessed the following by qRT/PCR and plotted as percent control in untreated cells. (B) WT MEF cells were transfected 10 nM Il4R-α siRNA 383281 alone or in the presence of 10, 30, or 100 nM PTEN siRNA for 4 h. Reduction of Il4R-α mRNA was assessed the following by qRT/PCR and plotted as percent control in untreated. (C) Ago2−/− MEF cells transfected with 10 nM Il4R-α siRNA 383281 alone or in the presence of 10, 30, or 100 nM PTEN siRNA for 4 h.
Nucleotide modifications that prevent Ago binding do not ablate activity of siRNA 383281
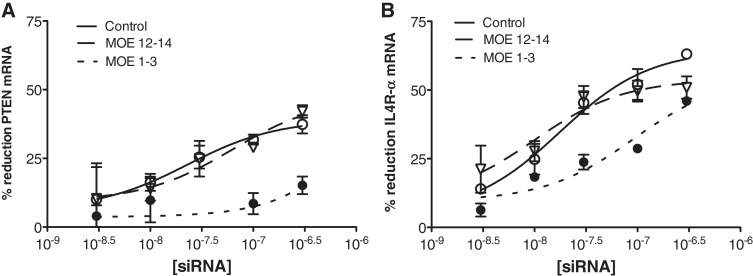
Modified siRNAs substituted with 2′-MOE nucleotides at various positions have been shown to result in reduction of siRNA activity (27). Inserting 2′-MOE nucleotides in positions 1–3 of the guide strand reduces the affinity for Ago and results in ablation of both specific and off-target siRNA activity, while 2′-MOE substitutions at positions 12–14 of the siRNA have essentially no effect on binding affinity, but result in a ∼60-fold reduction in Ago2 cleavage activity (17). Ago2−/− MEFs were transfected with unmodified PTEN siRNA 29591 and Il4R-α siRNA 383281 or with corresponding siRNAs substituted with 2′-MOE nucleotides at positions 1–3 or 12–14. Both siRNAs with 2′-MOE substitutions at positions 12–14 resulted in activity equivalent to the unmodified siRNAs (Figure 6) confirming that cleavage is not required for Ago2-independent activity of either siRNA. No activity was observed in cells treated with PTEN siRNA 29592 with MOE substitution at 1–3. In these cells, which lack Ago2, any activity of this siRNA is likely the result of Ago1 binding and subsequent degradation of the message in P-bodies (Figures 3 and 4) and preventing Ago1 binding with 2′MOE base substitutions eliminates activity. In contrast, significant activity was maintained for the Il4R-α 383281 siRNA with MOE substitution at positions 1–3. This again suggests that a substantial portion of the activity of this siRNA is RISC independent. Although the diminished activity relative to the unmodified siRNA might suggest that some fraction of the activity may be associated with RISC, we think that a more likely explanation is that unwinding of the duplexes is required and the 2′MOE substitution inhibits helicase activity (28). It is also possible that the 2′MOE substitution may have an effect on the mechanism by which this siRNA exerts its activity.
Figure 6.
MOE modification at positions 1–3 does not eliminate activity of Il4R-α siRNA. PTEN and Il4R-α siRNAs were synthesized with 2′MOE modified nucleotides at positions 1–3 (MOE 1–3) or 12–14 (MOE 12–14) of the guide strand. (A) Ago2−/− MEF cells were treated with unmodified or MOE-substituted PTEN siRNAs at concentrations ranging from 300 pM to 300 nM. The following day mRNA reduction was evaluated by qRT/PCR. Control siRNA (solid lines), MOE1-3 siRNA (dotted lines), MOE12–14 siRNA (dashed lines). (B) Ago2−/− MEF cells were treated with unmodified or MOE-substituted Il4R-α siRNAs at concentrations ranging from 300 pM to 300 nM.
siRNA 383281 binds near polyA signal of Il4R-α
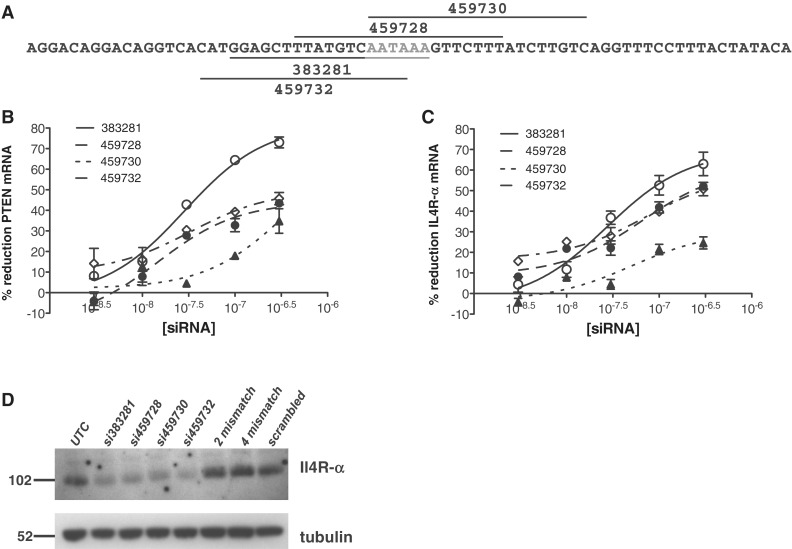
Il4R-α siRNA 383281 binds at position 3555–3575 of the Il4R-α mRNA sequence (ACC. NM_001008700). Since this target site overlaps a putative polyA signal (position 3568–3573), we hypothesized that this siRNA may be interfering with polyadenylation of the pre-mRNA. The siRNA target site was shifted relative to this polyA site as depicted in Figure 7A. Activity of the shifted siRNAs was assessed by qRT/PCR in wild-type and Ago2−/− MEFs as previously detailed. In wild-type cells, siRNAs 459728 and 459732 were less active than the parent siRNA 383281 (Figure 7B), however a comparable level of activity was maintained in the Ago2−/− cells (Figure 7C), suggesting that in both cell types the activity is primarily Ago2 independent. siRNA 459730 was significantly less active, even though it covers the entire polyA signal. This may reflect a directionality requirement for hybridization at the site or may be the result of the reduced binding affinity of si459730 relative to si383281. Ago2−/− MEFs were also treated at a single concentration of 100 nM of the same series of siRNAs, followed 24 h later by western blot analysis. Each of these siRNAs also effectively reduced expression of Il4R-α protein, although there was less discrimination in activity at the concentration used in this experiment. Still, as for the reduction of RNA, si459730 appears to be less active than the others.
Figure 7.
siRNA microwalk around Il4R-α polyA site. (A) Position of siRNAs relative to Il4R-α upstream poly A site. PolyA site is shown in grey. Wild-type (B) and Ago2−/− (C) MEF cells were transfected with siRNAs at concentrations ranging from 300 pM to 300 nM as detailed in ‘Materials and Methods’ section. The following day target mRNA reduction was measured by qRT/PCR. Percent inhibition at each treatment concentration is shown. (D) Ago2−/− fibroblasts were transfected with siRNAs 100 nM. Western blot analysis was performed 24 h post-transfection as detailed in ‘Materials and Methods’ section. A total of 50 µg protein was loaded in each lane.
Binding of siRNA 383281 to the polyA signal results in deadenylation or inhibition of polyadenylation of Il4R-α mRNA
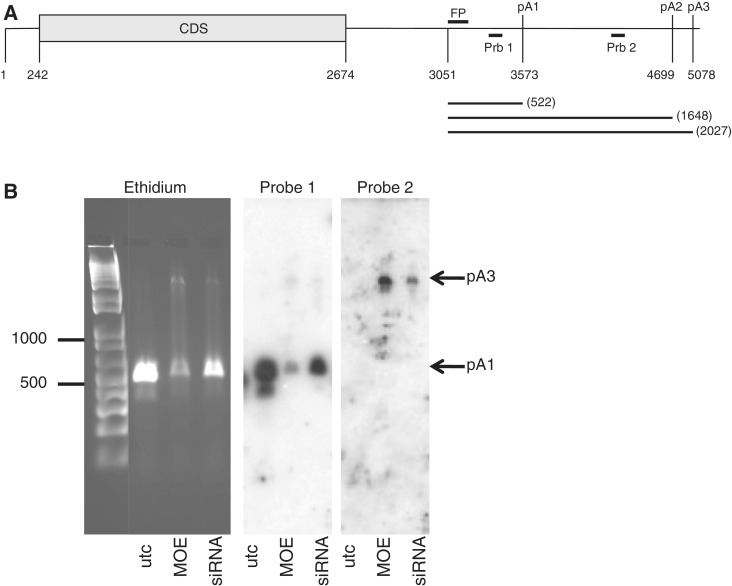
Studies based on Expressed Sequence Tags (ESTs) have determined that ∼29% of human mRNAs have multiple polyadenylation sites (29) producing mature mRNAs with 3′ regions of variable lengths. As UTRs may contain regulatory elements affecting mRNA stability or translation efficiency, the choice of alternate polyadenylation sites may strongly affect the final expression of the gene. Indeed, differential polyadenylation has been shown repeatedly to occur in a tissue- or disease-specific manner (30). We next performed 3′RACE to determine if siRNA treatment did, in fact, result in a reduction of polyadenylated mRNA. Wild-type and Ago2−/− MEFs were treated with Il4R-α siRNA 383281 or with ASO 473768, a full 2′MOE/P = S oligonucleotide targeting the same site. Previous work has shown that ASOs with this chemistry are capable of inhibiting polyadenylation when targeted to the polyA signal or site (31). Three potential polyA signals were identified on Il4R-α mRNA at positions 3573, 4699 and 5078 (Figure 8A). The first two sites are the canonical AAUAAA, while the third signal is the AUUAAA variant, which has been shown to be used in ∼10% of transcripts evaluated (32). While there is no empirical evidence that the second signal is used, accessions with validated 3′ ends have been identified for both the first and third signal (33). In untreated Ago2−/− MEF cells, a single band of ∼520 bp, consistent with usage of the first polyA signal (pA1), was identified by 3′ RACE (Figure 8B). Subsequent sequencing of this fragment confirmed polyadenylation at this site. Treatment with either the siRNA or ASO resulted in a pronounced decrease in this RACE product. However, the MOE ASO appeared to more effectively reduce the polyadenylated mRNA as compared with the siRNA. In MOE ASO-treated cells, a second larger band was also observed with a size consistent with usage of the third polyA signal (pA3). To confirm the identity of the bands, Southern blots of the RACE gel were performed using probes targeted to the sequence upstream of pA1 (Probe 1) or downstream of pA1 and upstream of pA3 (Probe 2). With Probe 1, the majority of the signal was in the lower band corresponding to pA1 and the intensity of the band was reduced by treatment with either the siRNA or the MOE ASO. In addition, a weak signal was observed at a position consistent with usage of pA3. These data indicate that only a small fraction of the long transcript is present in siRNA treated cells. Probe 2 would be expected to hybridize only with mRNA polyadenylated at pA2 or pA3. Indeed, no pA1 signal was observed using Probe 2, but a signal consistent with the longer transcript was observed in cells treated with the MOE ASO and, to a lesser extent, with the siRNA. Taken together these data primarily support a mechanism by which this siRNA either deadenylates or prevents polyadenylation of the message presumably by blocking the binding of CPSF to this signal (34). A fraction of the activity may also be related to redirection of a small, but significant amount of polyadenylation to pA3. Consistent with this, mRNA half lives for transcripts polyadenylated at pA1 and pA3 were virtually identical (Supplementary Figure S3).
Figure 8.
Treatment Il4R-α siRNA 383281 results in a small increase in polyadenylation at a downstream polyA site. (A) PolyA site and RACE primer localization on the Il4R-α transcript. Nucleotide position of 3′ RACE forward primer (FP), and polyA sites is indicated. Expected length of RACE products is given below the transcript. (B) Ago2−/− MEF cells were treated with Il4R-α siRNA 383281 or the corresponding full MOE ASO at a concentration of 100 nM. 3′ RACE was performed using total RNA purified from the cells the following day. Left Panel, ethidium bromide stained gel. Center panel, Southern blot of RACE gel probed with Prb1. Right panel, Southern blot of RACE gel probed with Prb2.
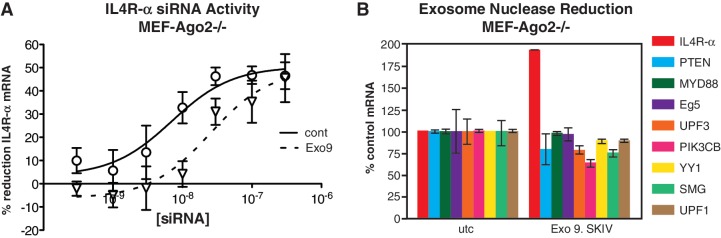
Ago-independent reduction by siRNA 383281 is at least partially mediated by the nuclear exosome
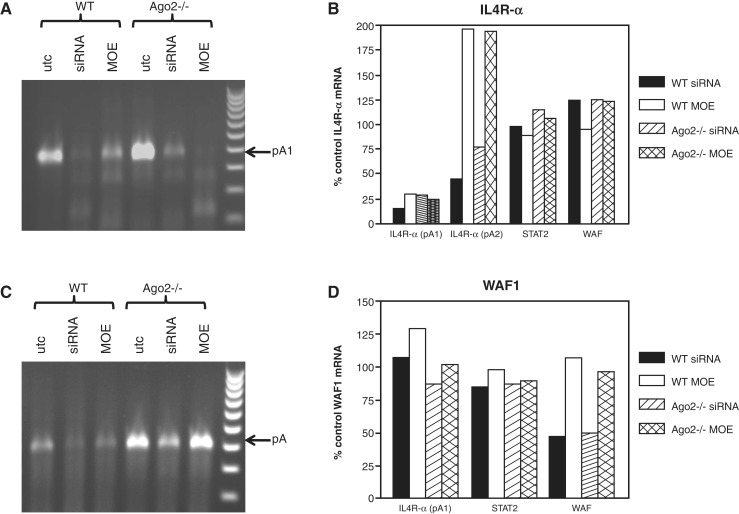
Degradation of messenger RNA initiated by deadenylation has previously been demonstrated to be mediated by the exosome, a 3′-5′-exoribonuclease complex that functions in both the nucleus and cytoplasm (35). In the nucleus, the RNA exosome complex is involved in the elimination of mRNAs with processing defects, thereby limiting or excluding their export to the cytoplasm. We evaluated the effect of exosome reduction on the activity of Il4R-α si383281. MEFs were treated with 75 nM each of ISIS 308299, an ASO targeting PmSCl-75 (Exosc9), a non-catalytic component of the RNA exosome complex which has 3′ → 5′ exoribonuclease activity, and ISIS 454373, an ASO targeting SKIV2L, a putative RNA helicase, which has been shown to mediate 3′–5′ RNA degradation by the exosome. The following day cells were seeded in 96-well plates, then transfected with si383281. Cells were harvested the following day and expression of Il4R-α mRNA evaluated by quantitative RT/PCR. Reduction of these exosomal proteins resulted in a significant decline in the activity of Il4R-α siRNA 383281 (Figure 9A), suggesting that ago-independent reduction by IL4 targeting siRNA is at least partially mediated by the exosome. The expression of several other transcripts was also evaluated in the same cells (Figure 9B). Interestingly, exosome reduction resulted in an increase in the levels of Il4R-α, but not the other transcripts evaluated. However, more extensive screening using PCR arrays of control and exosome reduced cells identified several transcripts also affected by exosome reduction (Supplementary Figure S4). siRNAs were designed to target the poly A sites of three of these transcripts: GATA3, WAF1 and STAT2. Only the siRNA directed to the polyA site of WAF1 demonstrated comparable activity in both wild-type and Ago2−/− MEF cells (Supplementary Figure S5). The targeted polyA signal is the only one known to be used for WAF1 mRNA. We therefore performed 3′ RACE and qRT/PCR to compare the activity to that of siRNA 383281. Wild-type and Ago2−/− MEFs were treated with WAF1 and Il4R-α siRNA and corresponding full MOE ASOs at 100 nM. The following day 3′ RACE was performed using total RNA purified from the treated cells. As previously observed, treatment with both the siRNA and MOE ASO resulted in a significant decrease in polyadenylated Il4R-α mRNA in WT and Ago2−/− cells (Figure 10A). Quantitative PCR was performed using oligo dT-primed cDNA. Amplification with a primer/probe set specific to the region upstream of the targeted polyA site indicated that treatment with both the Il4R-α siRNA and MOE ASO resulted in a strong decrease in mRNA polyadenylated at site 1 (Figure 10B). However, when a primer/probe set specific to the region downstream of the targeted polyA was used, treatment with the MOE ASO resulted in an increase in mRNA polyadenylated at site 2 or 3. In contrast, treatment with the corresponding siRNA resulted in a decrease in both long and short mRNA, suggesting that the full MOE and siRNA work via different mechanisms.
Figure 9.
Exosome reduction leads to specific inhibition of Il4R-α siRNA 383281 activity. Ago2−/− MEFs were treated with antisense ASOs targeting exosomal proteins Exosc9 and SKIV2l. The following day cells were seeded in 96-well plates, then transfected with si383281. Cells were harvested the following day and expression of Il4R-α mRNA evaluated by quantitative RT/PCR. (A) Concentration response curves for si383281 in control (solid line) or exosome-reduced (dashed line) Ago2−/− MEF cells. (B) Gene expression in exosome-reduced relative to untreated control cells at 48 h post-transfection.
Figure 10.
An siRNA targeting the WAF1 poly A site also reduces polyadenylated WAF1 mRNA. Wild-type and Ago2−/− MEF cells were treated with siRNAs and corresponding full MOE ASO as described in ‘Materials and Methods’ section. 3′ RACE was performed using total RNA purified from the cells the following day. Total RNA was also reverse transcribed using oligo dT primer. Oligo dT primed cDNA was then quantitated by qPCR. (A) Ethidium bromide stained gel of Il4R-α RACE products in WT and Ago2−/− cells. (B) qPCR of Il4R-α siRNA/MOE treated cells using primer probe sets upstream or downstream of PA site #1. (C) Ethidium bromide stained gel of WAF1 RACE products in WT and Ago2−/− cells. (D) qPCR of WAF1 siRNA/MOE treated cells. Primer/probe sets used are indicated at the bottom of the graph.
Unlike the Il4R-α target, full MOE ASOs targeted to the WAF1 polyA site had little effect on mRNA levels in either wild-type or Ago2−/− cells (Figure 10C and D). However, the corresponding siRNA did effectively reduce polyadenylated mRNA in both cell types. These data suggest that MOE ASOs can inhibit or redirect polyadenylation when a second signal is present, but that there is little effect on transcripts which have only a single poly A site. On the other hand, siRNAs directed to certain poly A signals are able to reduce targeted message in an Ago-independent manner which is also not dependent on alternative polyadenylation. Our data support a model in which siRNA targeted to the polyA signal results in deadenylation or inhibition of polyadenylation and subsequent degradation of the immature mRNA transcript. This model requires the presence of the siRNA in the nucleus. While it has been reported that active Ago2-dependent siRNAs are excluded from the nucleus (36), other studies demonstrating RNAi in nuclei of Caenorhabditis elegans (37), and siRNA-directed RNA degradation in the nucleus of plants (38), indicate that this is not always the case. Our own studies using fluorescently labeled siRNAs also suggest that siRNA 383281 is present in both the nucleus and cytoplasm of treated cells (Supplementary Figure S6).
Several questions remain to be answered. Foremost, where and how is the Il4R-α siRNA 383281 unwound, and how does the antisense strand get to the nucleus? We reduced several helicases using ASOs and siRNAs (Translin, Trax, C3PO, SKIV2L and MOV10) and none affected the activity of this siRNA, so we cannot comment on this step. We also reduced proteins thought to be involved in nuclear transport of RNAs (CLP1, EXP5, IMP8, NPM1, DCR and Drosha) and again no effects were observed on the activity of Il4R-α siRNA 383281 (data not shown). Finally, it is not clear how important or broadly applicable the proposed mechanism is. We did identify one other mRNA (WAF1) which appears to be degraded by the same mechanism and has equivalent activity in wild-type and Ago2−/− cells (Figure 10, Supplementary Figures S4 and S5). However, targeting of the polyA signal did not result in RISC-independent degradation of the targeted message for two other mRNAs to which we designed similar siRNAs. Whether or not polyA targeted siRNA triggers degradation is likely to depend on the complement of specific proteins associated with a given target and/or on some specific features of the siRNA-binding site and its RNA context (12,14,39). We did explore the possibility that siRNA/ASO binding to the polyA signal might change the structure of 3′UTR of Il4R-α, forming a STAU1-binding site, and leading to subsequent Stau1-mediated mRNA decay (40). However, Stau1 reduction had no effect on the observed activity (data not shown). Clearly, more screening will be required in order to determine if this is a rare process limited to few mRNA targets, or is more ubiquitous.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
Funding for open access charge: ISIS Pharmaceuticals.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Xue-hai Liang for critical reading of this manuscript and for providing expert guidance in confocal imaging and analysis. We also thank Tracy Reigle for help in preparation of the figures.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 3.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC Couples MicroRNA Biogenesis and Posttranscriptional Gene Silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 5.Matranga C, Tomari Y, Shin C, Bartel D, Zamore P. Passenger-Strand Cleavage Facilitates Assembly of siRNA into Ago2-Containing RNAi Enzyme Complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 6.Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 7.Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-Stranded Antisense siRNAs Guide Target RNA Cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 8.Okamura K, Ishizuka A, Siomi H, Siomi M. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 Mediates RNA Cleavage Targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Song J-J, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 11.Rivas FV, Tolia NH, Song J-J, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struc. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 12.Alemán LM, Doench J, Sharp PA. Comparison of siRNA-induced off-target RNA and protein effects. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic Ago proteins for on-target RNA interference. Curr. Biol. 2008;18:1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Rivas FV, Wohlschlegel J, Yates JR, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vickers TA, Lima WF, Wu H, Nichols JG, Linsley PS, Crooke ST. Off-target and a portion of target-specific siRNA mediated mRNA degradation is Ago2 ‘Slicer’ independent and can be mediated by Ago1. Nucleic Acids Res. 2009;37:6927–6941. doi: 10.1093/nar/gkp735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM, Bennett CF. 2′-O-(2-methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 19.Vickers TA, Zhang H, Graham MJ, Lemonidis KM, Zhao C, Dean NM. Modification of MyD88 mRNA splicing and inhibition of IL-1beta signaling in cell culture and in mice with a 2′-O-methoxyethyl-modified oligonucleotide. J. Immunol. 2006;176:3652–3661. doi: 10.4049/jimmunol.176.6.3652. [DOI] [PubMed] [Google Scholar]
- 20.Vickers TA, Lima WF, Nichols JG, Crooke ST. Reduced levels of Ago2 expression result in increased siRNA competition in mammalian cells. Nucleic Acids Res. 2007;35:6598–6610. doi: 10.1093/nar/gkm663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winer J, Kwang C, Jung S, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase ± polymerase chain reaction for monitoring gene erxpression in cardiac myocytes in vitro. Anal. Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto JG, Beadles-Bohling AS, Wiren KM. Comparison of RiboGreen and 18S rRNA quantitation for normalizing real-time RT-PCR expression analysis. BioTechniques. 2004;36:58–60. doi: 10.2144/04361BM06. [DOI] [PubMed] [Google Scholar]
- 23.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 24.Liang X-H, Vickers TA, Vickers TA, Guo S, Crooke ST. Efficient and specific knockdown of small non-coding RNAs in mammalian cells and in mice. Nucleic Acids Res. 2010;39:1–17. doi: 10.1093/nar/gkq1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi JJ. RNAi and the P-body connection. Nat. Cell Biol. 2005;7:643–644. doi: 10.1038/ncb0705-643. [DOI] [PubMed] [Google Scholar]
- 26.Koller E, Propp S, Murray H, Lima W, Bhat B, Swayze EE, Marcusson EG, Dean NM. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prakash TP, Allerson CR, Dande P, Vickers TA, Sioufi N, Jarres R, Baker BF, Swayze EE, Griffey RH, Bhat B. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 28.Lima WF, Wu H, Nichols JG, Sun H, Murray HM, Crooke ST. Binding and cleavage specificities of human Argonaute2. J. Biol. Chem. 2009;284:26017–26028. doi: 10.1074/jbc.M109.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beaudoing E, Freier S, Wyatt J, Claverie J-M, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vickers TA, Wyatt JR, Burckin T, Bennett CF, Freier SM. Fully modified 2′ MOE oligonucleotides redirect polyadenylation. Nucleic Acids Res. 2001;29:1293–1299. doi: 10.1093/nar/29.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 33.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Chen Z-Y, Wang W, Baker CC, Krug RM. The 3′-end-processing factor CPSFis required for the splicing of single-intron pre-mRNAs in vivo. RNA. 2001;7:920–931. doi: 10.1017/s1355838201010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Res. 2006;34:1369–1380. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffer P, Ivashuta S, Pontes O, Vitins A, Pikaard C, Mroczka A, Wagner N, Voelker T. Posttranscriptional gene silencing in nuclei. Proc. Natl Acad. Sci. USA. 2011;108:409–414. doi: 10.1073/pnas.1009805108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3[prime] UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.