Abstract
The unusual cyclin-dependent protein kinase 5 (CDK5) was discovered based on its sequence homology to cell cycle regulating CDKs. CDK5 was found to be active in brain tissues, where it is not involved in cell cycle regulation but in the regulation of neuronal cell differentiation and neurocytoskeleton dynamics. An aberrant regulation of CDK5 leads to the development of various neurodegenerative diseases including Alzheimer’s disease. Although CDK5 is not regulated by cyclins, its activity does depend on the association with a protein activator and the presence or absence of further inhibitory factors. Recently, CDK5RAP1 was discovered to inhibit the active CDK5 kinase. Here, we show that CDK5RAP1 is a radical SAM enzyme, which postsynthetically converts the RNA modification N6-isopentenyladenosine (i6A) into 2-methylthio-N6-isopentenyladenosine (ms2i6A). This conversion is surprisingly not limited to mitochondrial tRNA, where the modification was known to exist. Instead, CDK5RAP1 introduces the modification also into nuclear RNA species establishing a link between postsynthetic kinase-based protein modification and postsynthetic RNA modification.
INTRODUCTION
Cyclin-dependent protein kinase 5 (CDK5) is an atypical CDK which has no function in cell-cycle progression and does not require cyclin interaction for its activity. Instead, CDK5 has its major function in the organization of the cytoarchitecture of the central nervous system (CNS) (1). The kinase activity is regulated by the proteins p35 (CDK5R1) and p39 (CDK5R2) or their truncated forms p25 and p29, respectively (2). These regulatory subunits are mainly expressed in brain tissue, while CDK5 itself plays a role in various other tissues as well. In the brain, CDK5 is involved in neuronal development and neurogenesis (3). Its aberrant regulation leads to neurodegenerative diseases like Alzheimer’s and Parkinson’s disease. In Alzheimer’s disease, for example, CDK5 is hyperactivated by tight binding to a proteolytically truncated version of the regulator CDK5R1, p25. This in turn leads to aberrant phosphorylation of the tau protein, which then undergoes detrimental fibrillation (4). The tight regulation of CDK5R1 is consequently of paramount importance. While three interaction partners of CDK5R1 were identified in a yeast-two-hybrid screen in 2000 (5), only one of these proteins (C42) was shown to inhibit CDK5 activity by preventing the formation of the activated complex (6). Recently, it was suggested that C42 (later renamed CDK5 regulatory subunit associated protein 1, CDK5RAP1) is a radical SAM enzyme (7) with homology to the bacterial MiaB protein (8), which introduces a methylthio-group into N6-isopentenyladenosine in tRNA at position A37, next to the anticodon (9,10) (Supplementary Scheme S1). The resulting hypermodified adenine, 2-methylthio-N6-isopentenyladenosine (ms2i6A) functions in codon suppression (11) and stabilization of the codon/anticodon interaction (12). The biochemical link established by CDK5RAP1 between the enzymatic modification of tRNA anticodon loops and CDK5 kinase activity is highly unusual, particularly because the modified base ms2i6A is known to exist in tRNA of procaryotic origin (10). In mammals it is hence expected to occur only in mitochondrial tRNA, where it is found in tRNATrp, tRNAPhe and tRNATyr (13). Indeed, we recently confirmed that only traces of ms2i6A are present in cytosolic tRNA, while it is very abundant in mitochondrial tRNA (14).
MATERIALS AND METHODS
Subcellular localization
GFP-reporter plasmids for the subcellular localization of the human CDK5RAP1 variants were obtained from Origene (RG216600 and RG208724). HeLa cells were grown at 37°C and 5% CO2 in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum and 0.1% antibiotic/antimycotic (Gibco). Splitting was performed at 80% confluence. For transfection, 1.5 × 104 cells/300 µl were seeded in an 8-well plate (µslide, IBIDI) with 1 cm2 surface area. After 24 h, the cells were transfected with 25 µl jetPrime reaction mix (Peqlab) containing 250 ng plasmid DNA. After 4 h, the medium was exchanged. Forty-eight hours after transfection, the cells were visualized on a Leica TCS SPE confocal microscope at 488- (GFP) and 532 nm excitation for MitoTracker Red (Molecular Probes) to visualize the mitochondria.
Immunocytochemistry
The growth conditions were essentially the same as described earlier, except that a 16-well plate (Chamber slide, Thermo Fisher Scientific) was seeded with 1.0 × 103 cells/150 µl. After 24 h, the cells were fixed for 15 min in 4% fresh PFA solution [4% (w/v) paraformaldehyde dissolved at 70°C in PBS]. After three 10-min washing steps in PBS, the cells were incubated over night with rabbit anti-CDK5RAP1 (SAB2105127, Sigma-Aldrich) 1:300 in PBS supplemented with 0.3% Triton X-100 and 5% Chemiblocker (Millipore). The cells were washed three times 5 min in PBS and incubated with Alexa488-conjugated anti-rabbit IgG F(ab′)2 fragment (Cell Signaling) 1:750 in PBS containing 2% Chemiblocker for 1.5 h. After two washing steps, the cells were immersed for 5 min in PBS containing 0.4 µg/ml propidium iodide (Sigma-Aldrich). For co-staining of CDK5, mouse anti-CDK5 (C6118, Sigma-Aldrich) was added at 1:300 dilution and visualized with Alexa555-conjugated anti-mouse IgG F(ab′)2 fragment (Cell Signaling). The slides were mounted in Fluoromount G (Beckman Coulter) and analyzed on a Leica TCS SPE confocal microscope.
Cytochrome C oxidase assay
The respiratory activity of mitochondria in whole cell lysates of pork tissue was measured using the Cytochrome C Oxidase Assay Kit of Sigma-Aldrich. The tissue samples (200 mg) were homogenized with 450 µl ice-cold PBS at 30 Hz for 2 min with a tissue lyser (Qiagen). Subsequently, 50 µl of 10 mM maltoside was added and the lysis solution was shaken at 4–9°C for at least 15 min. The mixture was centrifuged at 4°C, 16 000 × g for 10 min and the supernatant was diluted 10-fold in enzyme dilution buffer. Measurements and analysis of the data took place according to the manufacturer. The mean values in nmol/(min × ml) (=1 mU) were eventually divided by the total protein concentration of the cell lysates determined using the Bradford assay. The enzyme activity is shown in nmol/(min × mg protein).
Complementation assay
The bacterial expression construct of the shorter CDK5RAP1 variant 2 was obtained by PCR amplification from the human cDNA ORF Clone (Origene, RG216600) using Phusion High-Fidelity DNA polymerase (NEB) with the two primers 5′-CACCATGATGGATGAACTTCTTGGAAGGC-3′ and 5′-TCAGCAATATGCAGAAGAGTCCCTCAGAG-3′. The 1498-bp PCR product was cloned in a two-step Gateway reaction via the vector pENTR/TEV/D-TOPO (Invitrogen) into the expression vector pDEST007 (15), which results in the addition of an N-terminal Strep-Tag II (IBA). To exclude effects of protein overexpression and antibiotic selection, all strains were transformed with the same plasmid backbone, encoding either human CDK5RAP1-v2 or the unrelated protein BstPOL I (control). We used the wild-type strain BW25113 as well as the deletion mutant ΔmiaB (JW0658) from the Keio Collection, NBRP (NIG, Japan) (16). All deletions were PCR-verified. The cells were grown under 100 µg/ml carbenicillin selection and the deletions were further selected with 50 µg/ml kanamycin at 37°C, while shaking at 200 rpm in 1 l of LB-medium until OD600 = 0.6 was reached. The expression of CDK5RAP1-v2 and the control protein was induced by addition of 200 µg anhydrotetracycline and the incubation was continued at 16°C for 12 h. Cells were harvested (10 000 × g, 8 min, 4°C) and stored at −20°C until RNA preparation.
siRNA knockdown
The growth conditions were essentially the same as described for the subcellular localization, except that a 75-cm2 tissue culture flask was seeded with 1.5 × 106 cells. After 24 h, the cells were transfected with 4 µg of endonuclease-derived siRNA (esiRNA, esiMission CDK5RAP1, Sigma-Aldrich) or the same amount of an unrelated esiRNA (esiMission EGFP, Sigma-Aldrich). Twenty-four hour post-transfection, the medium was exchanged and HeLa cells were transfected again as described earlier. Twenty-four hours after the second transfection, the medium was exchanged. Total RNA was extracted after 72 h as described below.
RNA isolation
Total RNA was prepared with the Trifast reagent from Peqlab. tRNA was isolated according to our established method (17). The isolation of mRNA followed the mTRAP Maxi Kit from Active Motif. rRNA was obtained via the enrichment of ribosomes. For the preparation of small RNA, the mirPremier-Kit (Sigma Aldrich) was used. For detailed information, see Supplementary Methods.
RESULTS
Subcellular localization of CDK5RAP1
In order to unravel the unusual link between CDK5 kinase activity and RNA modification chemistry, we investigated the catalytic properties and the cellular distribution of CDK5RAP1, which exists in different splice variants (18). Variant 1 contains an N-terminal extension that is by computational methods predicted to comprise a mitochondrial import signal (score = 0.647, TargetP1.1) (19), while variant 2 lacks this extension (Figure 1a). To analyze the subcellular distribution of both splicing variants, we transfected HeLa cells with GFP fusions of these two CDK5RAP1 splice variants. The confocal analysis clearly shows a mitochondrial localization of variant 1. In order to secure the interpretation we performed a colocalization with a mitochondrial counterstain and indeed observed the colocalization of variant 1 and the MitoTracker (Figure 1b). Variant 2, which is lacking the putative import signal, we found distributed in both cytoplasm and nucleus, respectively (Figure 1b), with the stronger signal appearing in the cytoplasm. This distribution was confirmed by immunocytochemical (ICC) staining of CDK5RAP1, however with a stronger preference for nuclear localization (Figure 1c). Interestingly, in both live-cell imaging and ICC methods, CDK5RAP1 appeared to be excluded from the nucleoli. This is a somehow surprising observation since the nucleoli are the compartment where RNA processing mainly occurs. ICC staining with an antibody against CDK5 showed that there is no apparent colocalization of CDK5 with CDK5RAP1 (Figure 1c). This is in accordance with CDK5RAP1s mode of CDK5-inhibition by preventing formation of the activated CDK5 complex. In some cells, CDK5RAP1 shows an association with the mitotic spindle (Figure 1d). Such localization was previously shown also for CDK5RAP2 (20), the second CDK5R1-binding protein identified in the yeast two hybrid screen. This result suggests that CDK5RAP1 might be interacting with cell cycle regulating mitotic CDKs as well. The mitochondrial localization of CDK5RAP1 variant 1 is in full agreement with the suggested MiaB-like function in ms2i6A biosynthesis in mitochondrial tRNA.
Figure 1.
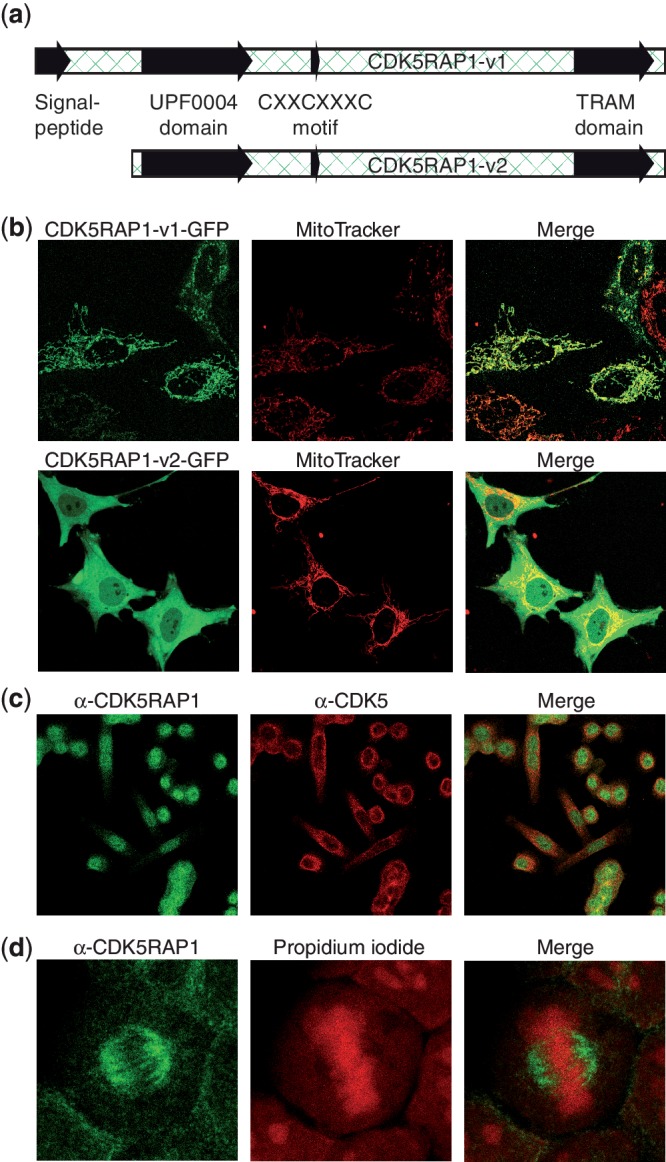
Subcelluar distribution of CDK5RAP1 variants. (a) Representation of the two investigated CDK5RAP1 variants with mitochondrial import sequence and catalytic domain. (b) Live-cell images of HeLa cells transfected with C-terminally GFP-tagged CDK5RAP1-v1 and CDK5RAP1-v2. For the counterstain of mitochondria, cells were incubated with MitoTracker Red dye. The merge shows that CDK5RAP1-v1 colocalizes with mitochondria, while CDK5RAP1-v2 is distributed in both cytoplasm and nucleus. (c) Formaldehyde-fixed HeLa cells were stained with an antibody against the N-terminal region of CDK5RAP1-v2 (Alexa-488, green) and against CDK5 (Alexa-555, red). The localization of CDK5RAP1 is predominantly nuclear with little overlap to CDK5. (d) Again, fixed HeLa cells were stained with anti-CDK5RAP1 (green). For localization of nucleic acids, propidium iodide (red) was used. CDK5RAP1 shows an association with the mitotic spindle.
Occurrence of ms2i6A in mitochondrial tRNA
The presence of ms2i6A in mitochondrial tRNA was shown previously (21), and we have further verified its localization. In order to investigate the role of ms2i6A for mitochondrial activity, we isolated tRNA from various pork tissues, fully digested these RNA fractions and analyzed the nucleoside mixture by LC-MS (Supplementary Methods). Precise quantification of ms2i6A in these lysates was enabled by adding a synthetic, isotope-labeled ms2i6A standard to the mixture as described recently by us in detail (17). As a marker for mitochondrial activity, we quantified the activity of cytochrome C oxidase (COX). The results summarized in Figure 2 clearly show a strong correlation with mitochondrial activity. Tissues with a high-energy demand like muscle and brain tissue feature also the highest concentrations of ms2i6A. We further enriched mitochondria from porcine heart and quantified ms2i6A in both mitochondrial and cytosolic tRNA. A strong enrichment was achieved in preparations of mitochondria (80 modifications per 1000 tRNA) as compared to cytosolic tRNA (6 per 1000). Residual ms2i6A in these preparations likely originates from ruptured mitochondria. This data is taken from Ref. (14).
Figure 2.
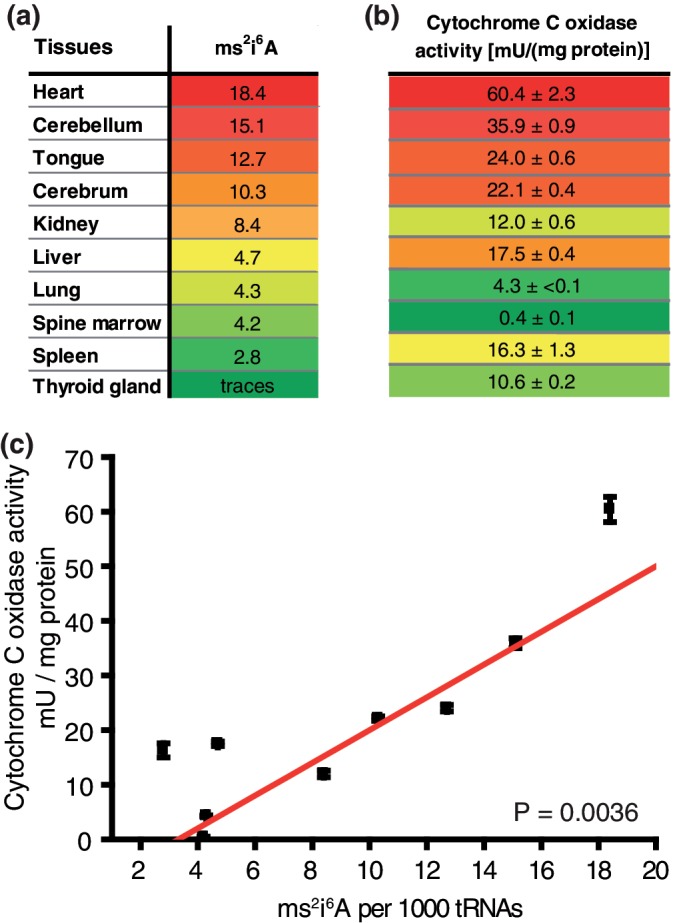
Correlation of ms2i6A content with mitochondrial activity. (a) Content of ms2i6A in tRNA of various pork tissues, sorted by descending amount. Highest values in red, lowest in green. (b) Cytochrome C oxidase activity in the same tissues. For comparison, the same color code was used. (c) Correlation plot of COX activity versus ms2 i 6A content.
Occurrence of ms2i6A in nuclear RNA
Based on this data, we reasoned that the nucleocytoplasmic localization of variant 2 of CDK5RAP1 causes the transformation of i6A into ms2i6A in nuclear RNA species as well, where ms2i6A was not observed so far. We applied our isotope-based quantification method on preparations of various RNA species (Figure 3a). In agreement with our earlier data and with the Warburg effect, which describes strongly impaired mitochondrial function in cancer cells, we were unable to detect significant amounts of ms2i6A in the total tRNA fraction of HeLa cells (17). Also, the ribosomal RNA, which was obtained from ribosome-enriched preparations, did contain only traces of ms2i6A. In contrast, polyadenylated RNA enriched by oligo-dT hybridization gave a significant signal for ms2i6A (Figure 3b). The amount of the precursor i6A in polyA-RNA is very low, showing that in this RNA species full conversion to ms2i6A occurs (Figure 3c). The highest amount of ms2i6A was found in preparations of small RNA, using a commercial miRNA preparation kit. These preparations contain next to tRNA (∼70–100 nt) also RNA species of a larger size (Supplementary Figure S1). The presence of tRNA is reflected by the high i6A content observed, similar to that seen in preparations of tRNA. By exclusion of ribosomal and transfer RNA as the source of ms2i6A, we reason that ms2i6A is indeed an RNA modification that next to mitochondrial tRNA exists also in a yet unidentified nuclear RNA species. The data prove that ms2i6A is far more wide spread than thought until today.
Figure 3.
ms2i6A/i6A content in different RNA species of HeLa cells. (a) Overview of investigated RNA fractions. (b) ms2i6A content in different RNA species of HeLa cells relative to unmodified adenosine (ppm). tRNA and rRNA contain no or only trace amounts of ms2i6A. However we found ms2i6A to be enriched in polyA-RNA and even higher in miRNA. (c) i6A content in different RNA species of HeLa cells relative to adenosine (ppm). tRNA and miRNA contain high amounts of i6A. In polyA-RNA and rRNA, i6A is only present in traces or low amounts.
CDK5RAP1 is an N6-isopentenyladenosine methylthiotransferase
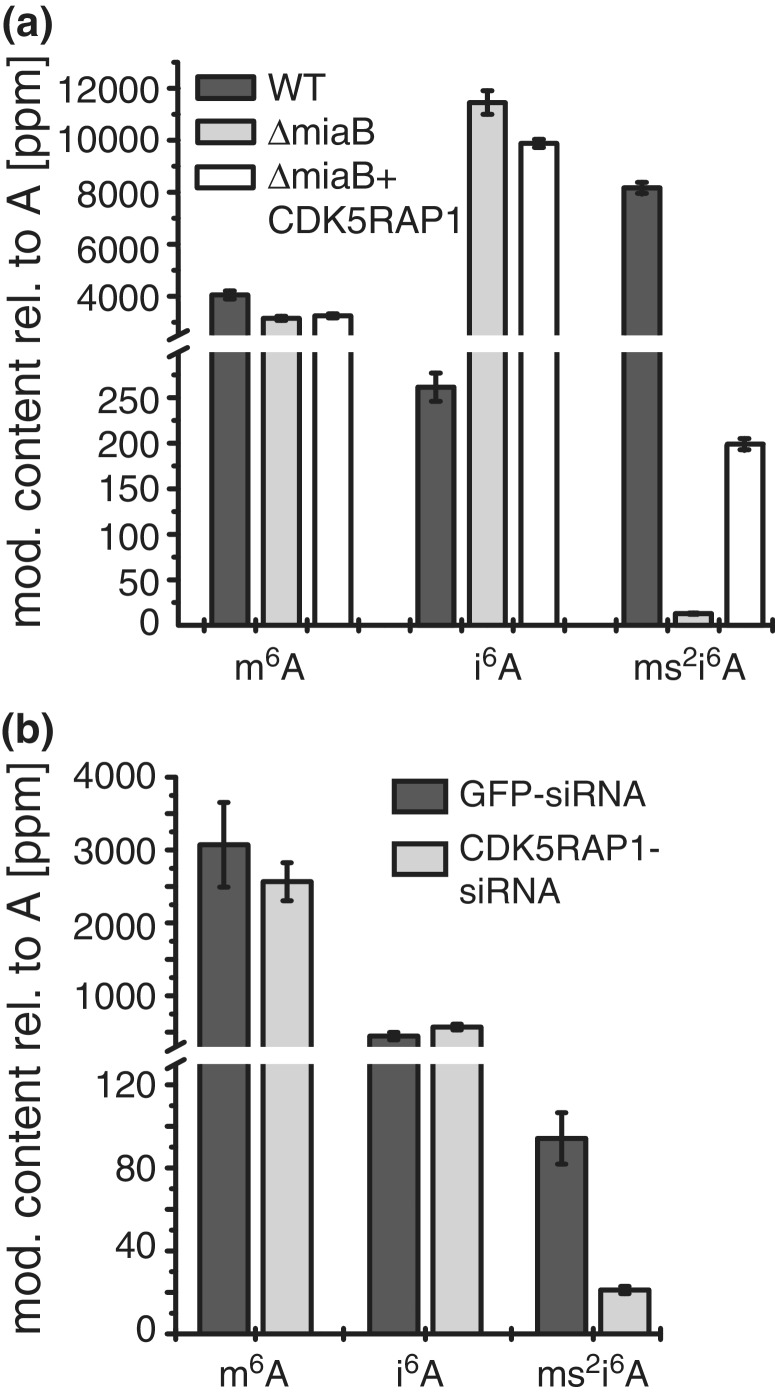
In order to proof that the CDK5 inhibitory protein CDK5RAP1 is responsible for the biosynthesis of ms2i6A in RNA (Supplementary Scheme S1), we expressed variant 2, which is lacking the mitochondrial import sequence, in a MiaB-deficient Escherichia coli strain from the Keio collection (16) and analyzed the ms2i6A content in tRNA by mass spectrometry. Although the protein turned out to be rather insoluble (Supplementary Figure S2), we detected significantly elevated levels of ms2i6A in the complemented cells (Figure 4a), showing that CDK5RAP1 has indeed MiaB-like catalytic properties. To address the possibility that bacterial tRNA is a bad substrate for human CDK5RAP1, we measured also the ms2i6A content of total RNA and received the same results, however with less pronounced differences (Supplementary Figure S3). We could further exclude the possibility that CDK5RAP1 generates ms2t6A. Whereas t6A is present in E. coli in significant amounts, ms2t6A remaines undetectable even after CDK5RAP1 overexpression (Supplementary Figure S4b). To further verify that CDK5RAP1 is the responsible enzyme for the synthesis of the detected ms2i6A in RNA extracts from HeLa cells, we next measured ms2i6A levels in HeLa cells after knockdown of CDK5RAP1 by transfection with endonuclease-derived siRNA (esiRNA), targeting both variants. Transfection with esiRNA resulted indeed in a 78% decrease of ms2i6A content after 72 h in comparison to cells treated with esiRNA against green fluorescent protein (Figure 4b). Again, the content of ms2t6A is not affected by knockdown of CDK5RAP1, proving that t6A is not recognized as a substrate of CDK5RAP1 (Supplementary Figure S4a). The amount of m6A served in this study as an internal reference in both experiments and did not show significant alterations. From this study, we can conclude that the ms2i6A found in RNA of HeLa cells is generated by the action of CDK5RAP1.
Figure 4.
CDK5RAP1 is the human homolog of the bacterial protein MiaB. (a) Complementation assay in E. coli. Quantification of m6A, i6A and ms2i6A in tRNA of wild-type E. coli, a MiaB-deficient strain and a MiaB-deficient strain complemented with CDK5RAP1. m6A was used as internal standard and levels did not change during the experiment. i6A level increased in ΔMiaB and compared to WT, because of lack of conversion to ms2i6A. ΔMiaB + CDK5RAP1 shows a decreased i6A level in comparison to ΔMiaB, generated by the complementation. By the expression of CDK5RAP1, the ms2i6A level increases 15-fold in comparison to ΔMiaB. (b) siRNA knockdown in HeLa cells. Cells were transfected twice with esiRNA against CDK5RAP1. Non-coding esiRNA against EGFP served as a control. After 72 h, total RNA was extracted and the modification content was determined by LC-MS. m6A and i6A remain constant, while ms2i6A decreased ∼78%.
DISCUSSION
Our study provides a direct link between posttranslational protein modification processes such as phosphorylations catalyzed by CDK5 and the processes that implement posttranscriptional RNA modifications. Furthermore, we show that nuclear RNA modifications are not limited to rather small modifications such as methylation or A-to-I editing (22), but that the modifications involve also the introduction of unusual hypermodified bases such as ms2i6A. We present the biosynthetic enzyme CDK5RAP1, which has a dual role as RNA methylthiotransferase and inhibitor of CDK5. We further find differential localization of the two splicing variants of CDK5RAP1 to mitochondria and cytosol. Alternative splicing could therefore regulate the dual function of this enzyme. The involvement of RNA modifying enzymes in other developmental or metabolic processes is a new concept that gained recently support by two other observations. First, CDKAL1, which is a long-known factor in typ II diabetes (T2D) and a close homolog of CDK5RAP1, was characterized as an N6-threonyladenosine (t6A) methylthiotransferase. Despite the high homology to CDK5RAP1, which led to its name, CDKAL1 does not accept i6A as a substrate (23). Further it was shown to localize in the endoplasmatic reticulum (ER) with the C-terminus being the determinant for this localization (24). While t6A occurs in mitochondrial tRNA just as i6A, its methylthiolated species ms2t6A was not found in mitochondrial tRNA species and appears to be restricted to the anticodon loop of tRNALys (http://trnadb.bioinf.uni-leipzig.de/) (13). The second example that connects RNA modifications with lifestyle disease, is the fat and obesity associated protein FTO, which was discovered in 2007 to be an iron(II) and 2-oxoglutarate dependent demethylase (25). Just recently it was found that FTO is able to demethylate m6A, which is an important and highly abundant modification in mRNA. FTO co-localizes with the splicing factor SC35 in the nucleus, where it was suggested to have a role in regulation of splicing (26). Our results now show that RNA modification is connected to the regulation of a critical kinase (CDK5) involved in neuronal cell differentiation. We believe that the introduction of ms2i6A into RNA will strongly influence the function of the RNA species. ms2i6A is a modification which strongly alters the RNA structure and duplex stability (27) and influences for example codon recognition by stacking interactions with neighboring RNA duplexes (12). The ability of the base to stabilize loop structures can serve multiple purposes in many RNA species. It can alter for example the specific target binding of miRNA by duplex interaction, influence miRNA processing by perforation of the loop structure in pre-miRNA or it may aid in intron splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4, Supplementary Methods, Supplementary Methods Figures 1–3 and Supplementary Scheme 1.
FUNDING
Center for Integrated Protein Science Munich and Sonderforschungsbereich 749. Funding for open access charge: Center for Integrated Protein Science München.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stylianos Michalakis for his advice in immunocytochemistry and Prof. Beckmann and Thomas Becker for providing us the ribosome isolation methodology.
REFERENCES
- 1.Dhavan R, Tsai LH. A decade of CDK5. Nat. Rev. Mol. Cell Biol. 2001;2:749–759. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- 2.Liebl J, Fürst R, Vollmar AM, Zahler S. Twice switched at birth: cell cycle-independent roles of the “neuron-specific” cyclin-dependent kinase 5 (Cdk5) in non-neuronal cells. Cell. Signal. 2011;23:1698–1707. doi: 10.1016/j.cellsig.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 3.Jessberger S, Gage FH, Eisch AJ, Lagace DC. Making a neuron: Cdk5 in embryonic and adult neurogenesis. Trends Neurosci. 2009;32:575–582. doi: 10.1016/j.tins.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong CX, Iqbal K. Hyperphosphorylation of microtubule-associated protein tau: a promising therapeutic target for Alzheimer disease. Curr. Med. Chem. 2008;15:2321–2328. doi: 10.2174/092986708785909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ching YP, Qi Z, Wang JH. Cloning of three novel neuronal Cdk5 activator binding proteins. Gene. 2000;242:285–294. doi: 10.1016/s0378-1119(99)00499-0. [DOI] [PubMed] [Google Scholar]
- 6.Ching YP, Pang AS, Lam WH, Qi RZ, Wang JH. Identification of a neuronal Cdk5 activator-binding protein as Cdk5 inhibitor. J. Biol. Chem. 2002;277:15237–15240. doi: 10.1074/jbc.C200032200. [DOI] [PubMed] [Google Scholar]
- 7.Atta M, Mulliez E, Arragain S, Forouhar F, Hunt JF, Fontecave M. S-Adenosylmethionine-dependent radical-based modification of biological macromolecules. Curr. Opin. Struct. Biol. 2010;20:684–692. doi: 10.1016/j.sbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Kaminska KH, Baraniak U, Boniecki M, Nowaczyk K, Czerwoniec A, Bujnicki JM. Structural bioinformatics analysis of enzymes involved in the biosynthesis pathway of the hypermodified nucleoside ms(2)io(6)A37 in tRNA. Proteins. 2008;70:1–18. doi: 10.1002/prot.21640. [DOI] [PubMed] [Google Scholar]
- 9.Pierrel F, Douki T, Fontecave M, Atta M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J. Biol. Chem. 2004;279:47555–47563. doi: 10.1074/jbc.M408562200. [DOI] [PubMed] [Google Scholar]
- 10.Esberg B, Leung HC, Tsui HC, Bjork GR, Winkler ME. Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 1999;181:7256–7265. doi: 10.1128/jb.181.23.7256-7265.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouadloun F, Srichaiyo T, Isaksson LA, Bjork GR. Influence of modification next to the anticodon in tRNA on codon context sensitivity of translational suppression and accuracy. J. Bacteriol. 1986;166:1022–1027. doi: 10.1128/jb.166.3.1022-1027.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 13.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Globisch D, Pearson D, Hienzsch A, Brückl T, Wagner M, Thoma I, Thumbs P, Reiter V, Kneuttinger AC, Müller M, et al. Systems-based analysis of modified tRNA bases. Angew. Chem. Int. Ed. Engl. 2011;50:9739–9742. doi: 10.1002/anie.201103229. [DOI] [PubMed] [Google Scholar]
- 15.Ober M, Müller H, Pieck C, Gierlich J, Carell T. Base pairing and replicative processing of the formamidopyrimidine-dG DNA lesion. J. Am. Chem. Soc. 2005;127:18143–18149. doi: 10.1021/ja0549188. [DOI] [PubMed] [Google Scholar]
- 16.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brückl T, Globisch D, Wagner M, Müller M, Carell T. Parallel isotope-based quantification of modified tRNA nucleosides. Angew. Chem. Int. Ed. Engl. 2009;48:7932–7934. doi: 10.1002/anie.200902740. [DOI] [PubMed] [Google Scholar]
- 18.Zou X, Ji C, Jin F, Liu J, Wu M, Zheng H, Wang Y, Li X, Xu J, Gu S, et al. Cloning, characterization and expression of CDK5RAP1_v3 and CDK5RAP1_v4, two novel splice variants of human CDK5RAP1. Genes Genet. Syst. 2004;79:177–182. doi: 10.1266/ggs.79.177. [DOI] [PubMed] [Google Scholar]
- 19.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 20.Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 21.Sakurai M, Ohtsuki T, Suzuki T, Watanabe K. Unusual usage of wobble modifications in mitochondrial tRNAs of the nematode Ascaris suum. FEBS Lett. 2005;579:2767–2772. doi: 10.1016/j.febslet.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Knoop V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell. Mol. Life Sci. 2011;68:567–586. doi: 10.1007/s00018-010-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arragain S, Handelman SK, Forouhar F, Wei FY, Tomizawa K, Hunt JF, Douki T, Fontecave M, Mulliez E, Atta M. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6- threonylcarbamoyladenosine in tRNA. J. Biol. Chem. 2010;285:28425–28433. doi: 10.1074/jbc.M110.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei FY, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A, Matsui H, Atta M, Michiue H, Fontecave M, et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 2011;121:3598–3608. doi: 10.1172/JCI58056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kierzek E, Kierzek R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003;31:4472–4480. doi: 10.1093/nar/gkg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.