Abstract
Biogenesis of trans-acting siRNAs (tasiRNAs) is initiated by miRNA-directed cleavage of TAS gene transcripts and requires RNA-dependent RNA polymerase 6 (RDR6) and Dicer-like 4 (DCL4). Here, we show that following miR173 cleavage the entire polyadenylated parts of Arabidopsis TAS1a/b/c and TAS2 transcripts are converted by RDR6 to double-stranded (ds)RNAs. Additionally, shorter dsRNAs are produced following a second cleavage directed by a TAS1c-derived siRNA. This tasiRNA and miR173 guide Argonaute 1 complexes to excise the segments from TAS2 and three TAS1 transcripts including TAS1c itself to be converted to dsRNAs, which restricts siRNA production to a region between the two cleavage sites. TAS1c is also feedback regulated by a cis-acting siRNA. We conclude that TAS1c generates a master siRNA that controls a complex network of TAS1/TAS2 siRNA biogenesis and gene regulation. TAS1/TAS2 short dsRNAs produced in this network are processed by DCL4 from both ends in distinct registers, which increases repertoires of tasiRNAs.
INTRODUCTION
Micro RNAs (miRNAs) regulate gene expression by silencing their target genes through mRNA cleavage and/or translational repression (1,2). In plants, some miRNA targets spawn 21-nt secondary siRNAs (3,4), a subset of which, called trans-acting siRNAs (tasiRNAs), can also target genes in trans. The tasiRNA-targeted genes can in turn spawn tasiRNAs (5), thus leading to silencing cascades which are predicted to form complex networks with distinct regulatory functions (6). miRNAs, tasiRNAs and secondary siRNAs produced from natural antisense transcripts are involved in regulation of plant development, physiology and stress responses (7–9).
The biogenesis of tasiRNAs requires RNA-dependent RNA polymerase 6 (RDR6) which converts a portion of target RNA upstream or downstream of the miRNA/siRNA cleavage site to double-stranded RNA (dsRNA). The resulting dsRNA is then processed by Dicer-like 4 (DCL4) from the miRNA-cleavage end into 21-nt in-phase siRNA duplexes with 2-nt 3′-overhangs (4,10,11).
The Arabidopsis thaliana genome contains four families of tasiRNA-generating TAS genes, classified based on miRNA-dependence, sequence similarity and target gene identity. miR173-dependent TAS1a/b/c and TAS2 tasiRNAs and miR828-dependent TAS4 tasiRNAs are encoded downstream of the miRNA cleavage site and target the genes of pentatricopeptide repeat and MYB transcription factor families, respectively (3,4). miR390-dependent TAS3a/b/c tasiRNAs are encoded upstream of the miRNA-cleavage site and target the genes of auxin response factor family (4,10).
One of the open questions concerning secondary siRNA biogenesis is which factors route some (but not all) miRNA/tasiRNA target transcripts into the RDR6/DCL4 pathway. For the Arabidopsis TAS3 family, a second, non-cleavable, miR390-binding site located upstream of the miR390 cleavage site is essential for tasiRNA biogenesis (12) and the region of TAS3a/b/c transcripts between the two miR390 sites spawns most of secondary siRNAs including tasiRNA species (4). Interestingly, TAS3 transcripts from moss and pine are cleaved at both miR390-target sites (12). These results, together with a finding that secondary siRNAs more often emanate from the genes with two known miRNA/tasiRNA target sites, suggested a two-hit trigger model for secondary siRNA biogenesis (12,13). However, this model left unexplained a robust production of siRNAs from miR173-dependent TAS genes. Efforts to identify a second hit by a known small RNA or some other transcript feature required for tasiRNA production have failed so far (14).
miRNAs and siRNAs exert their silencing functions in association with Argonaute (AGO) proteins that sort sRNAs based on size and 5′-terminal nucleotide (13,15,16). Thus, AGO1 binds mostly 21-nt RNAs with 5′-terminal uridine (5′U), while AGO2 binds mostly 21-nt RNAs with 5′-terminal adenine (5′A). Interestingly, both miR173 and miR390 that initiate tasiRNA biogenesis deviate from this general rule. miR390 is a 21-nt 5′A-RNA but it is specifically associated with AGO7 that plays an essential role at both cleavable and non-cleavable TAS3 target sites (13,17). miR173 is a 22-nt 5′U-RNA that associates with AGO1 to guide the cleavage of TAS1a/b/c and TAS2 transcripts (14). The 22-nt size of miRNA is sufficient to trigger RDR6-dependent siRNA production from miRNA target transcripts including miR173 targets (18,19). A more recent study, however, indicates that asymmetrically positioned bulged bases in a miRNA:miRNA* duplex are more important than the miRNA length for the initiation of RDR6-dependent siRNA biogenesis (20).
Here we used blot hybridization and circularization-reverse transcription-PCR (cRT-PCR) to characterize the structures of dsRNAs originating from Arabidopsis miR173-dependent TAS genes and immunoprecipitation (IP) to identify tasiRNAs associated with AGO complexes. Our study uncovers a complex sRNA network connecting TAS2 and TAS1 genes which is controlled by miR173 and a TAS1c-derived tasiRNA.
MATERIALS AND METHODS
Arabidopsis wild-type, rdr6-15 and dcl mutant lines used in this study, their growth conditions and infection with CaMV, and the detailed protocols for total RNA preparation and high-resolution blot hybridization have been described earlier (21,22). ago1-25 and ago2-1 were kindly provided by Dr Baulcombe. Seeds of tas1c-1 (SAIL_242_C04) were obtained from the Arabidopsis Biological Resource Center and the homozygous individuals were identified by genotyping. Probes used for RNA blot hybridization and genotyping are listed in Experimental Procedures in Supplementary Data. cRT-PCR mapping of TAS RNAs was performed as described earlier [(23); for details, see Methods in Supplementary Data].
The IP experiment with native AGO1-antibodies (Agrisera) and blot hybridization analysis of AGO1-associated sRNAs were performed as we described recently (24). The sRNA probes are listed in Methods in Supplementary Data.
Transient expression experiments using Nicotiana benthamiana were performed as described previously (10). The 35S:TAS1c construct was mutagenized by replacing the region between Pfo I and Pac I sites with synthetic gene fragments carrying the corresponding mutations. For further details, see Methods in Supplementary Data.
RESULTS
Long and short co-terminal dsRNAs originate from all miR173-targeted TAS genes
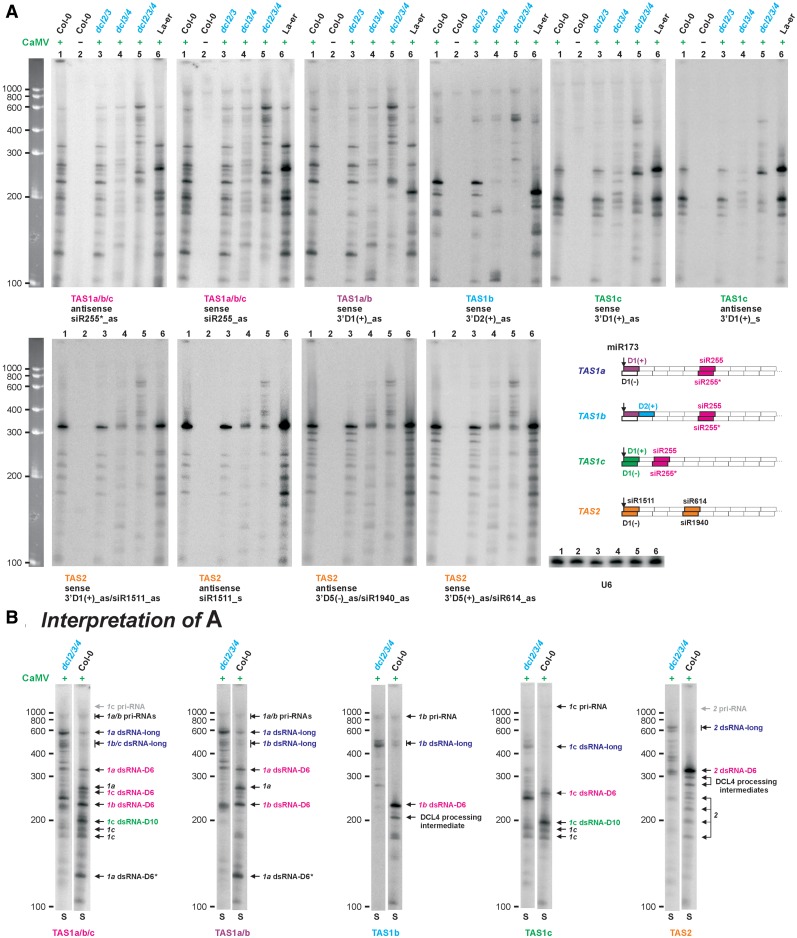
Previously we found that infection with Cauliflower mosaic virus (CaMV) leads to stabilization of RDR6-dependent precursors of tasiRNAs in Arabidopsis (21). Using high-resolution gel separation of total RNA followed by blot hybridization with DNA oligonucleotide probes specific for TAS1 and TAS2 loci, we further characterized the stabilized sense (S) and antisense (AS) precursors (Figure 1). To investigate DCL requirements for dsRNA biogenesis, we compared CaMV-infected wild-type Arabidopsis (Col-0 and La-er) with double (dcl2/3 and dcl3/4) and triple (dcl2/3/4) DCL-mutant lines [all in Col-0 background; (22)]. All the mutants still express DCL1 that produces miRNAs.
Figure 1.
Blot hybridization analysis of dsRNA precursors of TAS1a/b/c and TAS2 tasiRNAs. (A) The panel shows blot hybridization analysis (5% PAGE) of total RNA from CaMV-infected (+) or mock-inoculated (−) wild-type (Col-0 and La-er), double (dcl2/3 and dcl3/4) and triple (dcl2/3/4) DCL-deficient mutants. The scans represent the same blot successively hybridized with probes specific for TAS1a/b/c-, TAS1a/b-, TAS1b-, TAS1c- and TAS2-derived 21-nt siRNAs and U6 RNA as a loading control. Single-stranded RNA markers are shown on the left. The scheme shows location and polarity of the probed siRNAs (colored boxes with their names indicated) with respect to the miR173 cleavage site (indicated by arrow) on hypothetical dsRNAs from the four TAS genes. Identical siRNAs/probes, i.e. siR255/siR255* duplex encoded by TAS 1a, 1b and 1c and 3′D1(−) siRNA of TAS 1a and 1b, are indicated in pink and purple, respectively. Cyan, blue and orange indicate siRNAs/probes specific for TAS1b, TAS1c and TAS2, respectively. (B) Interpretation of the analysis shown in panel A. The lanes corresponding to Col-0 and dcl2/3/4 samples were cropped from the above blots of sense (S) polarity. Positions of pri-RNAs and dsRNAs are indicated by arrows for each TAS gene (1a, 1b, 1c and 2).
Given that tasiRNA siR255 is encoded in sense strands of all three TAS1 genes (1a, 1b and 1c) the probe complementary to siR255 would detect their primary Pol II transcripts (pri-RNAs) and the sense strands of RDR6-dependent dsRNAs. The probe complementary to the star strand of siR255/siR255* duplex would detect only the antisense strand of RDR6-dependent dsRNAs. The two probes gave identical hybridization patterns in the range of 100–600 nt (Figure 1A), indicating that the RNAs detected within this range can potentially be sense and antisense strands of distinct dsRNA precursors of siR255/siR255*. To further classify these RNAs, we used the probes specific to the 21-nt sequences adjacent to the miR173 cleavage site (identical in both TAS1a and TAS1b but not TAS1c) and the probe specific to TAS1b (Figure 1A). The resulting hybridization patterns allowed to identify the pri-RNAs and putative dsRNAs produced from each of the three TAS1 loci (Figure 1B). Likewise, probes specific for the TAS2 sequences identified one major and several minor sense/antisense RNA pairs (Figure 1).
Comparison of the hybridization patterns in wild-type versus dcl-mutant lines indicated that most of the TAS1 and TAS2 putative dsRNAs, except for the longest species, require DCL4 activity for their biogenesis. The longest sense/antisense pair derived from each locus over-accumulated in dcl2/3/4 (Figure 1A). The longest and shorter pairs from each locus, except for one TAS1b and two TAS2 non-abundant pairs (Figure 1B; designated DCL4 processing intermediates), all contain sequences adjacent to the miR173 cleavage site. Thus, most of the shorter dsRNAs are not intermediates of DCL4 processing of the longest dsRNAs from the miR173 end. The results described below are consistent with this conclusion.
Confirming our previous results (22), CaMV-infected rdr6-15 mutant plants, which lack RDR6 activity, did not accumulate any TAS1/TAS2-derived sense/antisense RNA pairs. Only sense strands of the longest dsRNAs, i.e. miR173-cleavage products of TAS1/TAS2 pri-RNAs (see below), could be detected by blot hybridization in rdr6-15 (Supplementary Figure S1).
Polyadenylated 3′-products of miR173-cleaved TAS pri-RNAs are entirely converted by RDR6 to dsRNAs
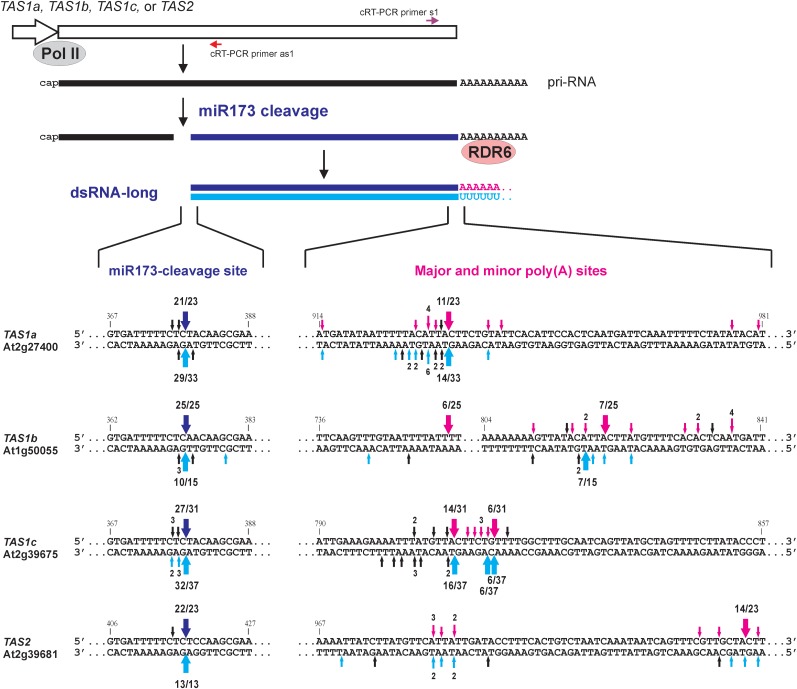
To precisely map TAS dsRNAs we used cRT-PCR that allows simultaneous sequencing of 5′- and 3′-termini of a given RNA [(23), Supplementary Figure S2A]. Pyrophosphatase treatment of total RNA, prior to RNA ligation, allowed circularization of transcripts not only with a 5′-monophosphate present on 3′-fragments of miRNA-cleaved TAS pri-RNA, but also with a 5′-triphosphate likely present on an RDR6-synthesized complementary RNA. cRT-PCR primers proximal to the termini of a given sense or antisense RNA (Figures 2 and 3; Supplementary Figure S2) were designed based on the sizes of putative dsRNAs estimated by blot hybridization (Figure 1B). cRT-PCR products were gel-purified, cloned and sequenced (∼10–30 individual clones for each).
Figure 2.
The entire, polyadenylated 3′-products of miR173-cleaved pri-RNAs of TAS1a, TAS1b, TAS1c and TAS2 genes are converted by RDR6 to dsRNAs. Diagrammatic representation of TAS genes transcribed by Pol II into 5′-capped and 3′-polyadenylated pri-RNAs which are then cleaved by miR173-AGO1 complexes. The 3′-products are converted by RDR6 to long dsRNAs. The regions of the miR173 cleavage site and the poly(A) sites of each TAS pri-RNA are enlarged. Arrows above the upper strand indicate positions of the 5′-terminus (miR173 site) and the 3′-terminus of at least one cloned cRT-PCR product representing the sense strands of TAS dsRNAs; the 3′-termini to which poly(A) is added are indicated in pink. Arrows below the complementary strand indicate the termini of one or more cloned cRT-PCR product representing the corresponding antisense strands of dsRNAs. Termini of the antisense RNA that begin with a poly(U) stretch are indicated in cyan. Thick arrows indicate the termini of the major sense and antisense RNAs; the number of specific cRT-PCR products/total products sequenced for each strand are indicated. Positions of cRT-PCR primers used for long dsRNA mapping are indicated above and below of the TAS gene body.
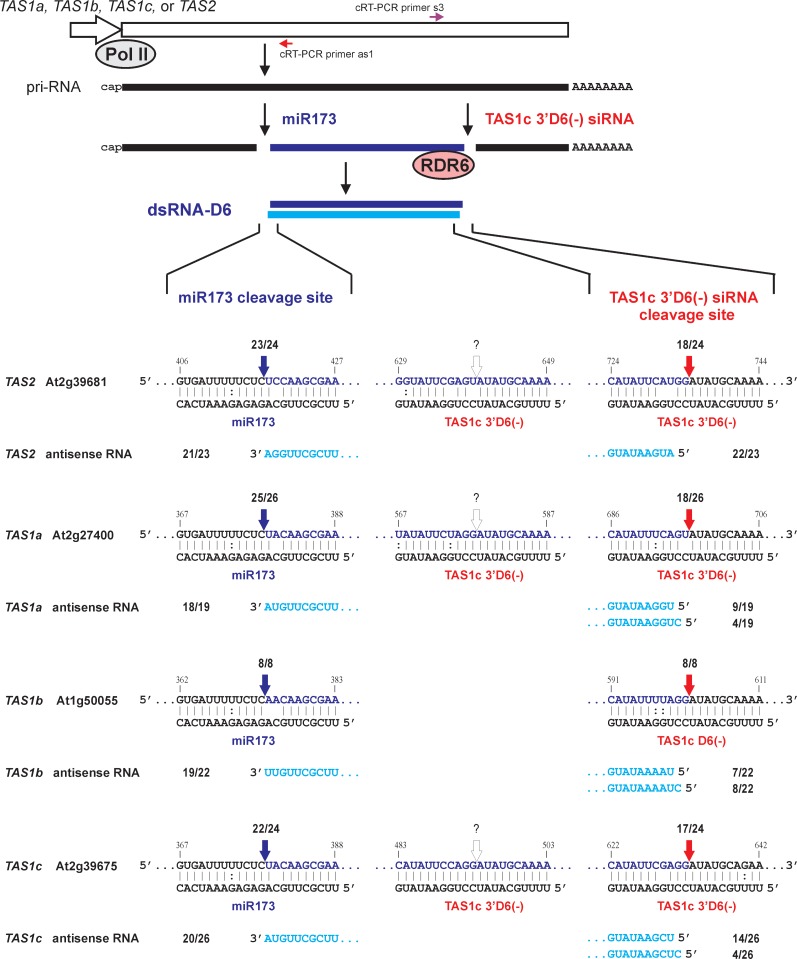
Figure 3.
Two-cleavage mechanism for biogenesis of TAS1a-, TAS1b-, TAS1c- and TAS2-derived dsRNAs. Diagrammatic representation of TAS genes transcribed by Pol II into pri-RNAs which are then cleaved at the two sites by miR173- and TAS1c 3′D6(−) siRNA-AGO1 complexes, respectively. The excised fragment between the two-cleavage sites is converted by RDR6 to dsRNA. The cleavage sites for each pri-RNA and the siRNA/target pri-RNA duplexes are shown in the expanded regions. Additional complementary sites for TAS1c 3′D6(−) siRNA in the intervening regions of TAS2 and TAS1a pri-RNAs as well as the origin site of TAS1c 3′D6(−) are also shown. Colored arrowheads indicate positions of the 5′-terminus (miR173 site; in blue) and the 3′-terminus [TAS1c 3′D6(−) site; in red] of cloned cRT-PCR products representing the sense strands of TAS dsRNAs. The 5′- and 3′-terminal sequences of cRT-PCR clones representing the corresponding antisense strands of TAS dsRNAs are shown below the duplexes and highlighted in cyan. The number of specific cRT-PCR products/total products sequenced for each strand is indicated. Positions of potential cleavage sites for 3′D6(−) siRNA are indicated by open arrowheads. Positions of the cRT-PCR primers used for the short dsRNA mapping are indicated above and below of the TAS gene body.
For each TAS, we first mapped the longest sense/antisense RNA pair from wild-type (Col-0) and dcl2/3/4 plants. Typically, no abundant products could be obtained using total RNA from mock-inoculated Arabidopsis, whereas the CaMV-infected material-yielded abundant cRT-PCR products of expected size (Supplementary Figure S2). Along with our blot hybridization (Figure 1), this indicates that tasiRNA precursors are efficiently processed by DCLs and do not accumulate at high levels, unless they are stabilized [in our case by the CaMV transactivator protein; (21)]. cRT-PCR analysis revealed that most of the TAS1a longest sense RNAs are the entire 3′-products of miR173-directed cleavage of the pri-RNA: they start with U379 (numbered from the cap site) and terminate with a poly(A) tail of up to 24 nt attached at one major site (A933) and several minor sites (Figure 2 and Supplementary Figure S2A). One to three terminal uridines were attached to some poly(A) tails. Most of the TAS1a longest antisense RNAs (RDR6 products) terminate with A379 and a large fraction of those start with a poly(U) stretch of up to 26 nt attached to U933 or at nearby positions. Some antisense RNAs have short A-rich stretches at the very 5′-end (Supplementary Figure S2A). Thus, the antisense RNAs are essentially complements of the sense RNAs. These results demonstrate that RDR6 copies the entire 3′-product of miR173-cleaved TAS1a pri-RNA into a complementary RNA.
cRT-PCR analysis of the longest dsRNAs from TAS1b, TAS1c and TAS2 genes (Supplementary Figure S2B, S2C and S2D, respectively) led us to similar conclusions with some peculiarities in each case. For TAS1b, two major (U753 and A822) and several minor poly(A) sites were identified (Figure 2 and Supplementary Figure S2B). The longest antisense RNAs from TAS1b have very short U-stretches, UA-rich stretches or just a single uridine at the 5′ termini, suggesting that long poly(A) tails are almost entirely removed from the 3′-cleavage product before its conversion to dsRNAs. For TAS1c, two major poly(A) sites were mapped (A810 and G816), one of which (A810) was reported earlier (14). TAS1c pri-RNAs of both types are used as templates for RDR6 following miR173-cleavage, since the counterpart antisense RNAs with long U-stretches at the 5′-end were found (Figure 2 and Supplementary Figure S2C). The longest TAS2 dsRNAs were composed of miR173-cleaved pri-RNA terminating at several minor and one major (A1031) poly(A) sites and their antisense counterparts (Figure 2 and Supplementary Figure S2D). Notably, in this case the longest dsRNAs could be detected by cRT-PCR only in dcl2/3/4 but not in wild-type plants, which is consistent with the blot hybridization analysis (Figure 1).
A second cleavage of miR173-targeted TAS pri-RNAs generates an additional template for RDR6
cRT-PCR results summarized in Figure 3 show that sense RNA of the most abundant sense/antisense pair derived from TAS2 in wild-type plants starts with U418 and ends predominantly with G734. Thus, this RNA is a derivative of the miR173-cleaved pri-RNA 3′-product truncated by a second, downstream cleavage. The corresponding antisense RNA starts with A732 and ends with A418 (Figure 3). Thus, RDR6 synthesis of a complementary RNA begins at the third nucleotide of the sense template, which creates a 2-nt 3′-overhang at the end of the resulting dsRNA, and stops at the template’s last nucleotide. A large fraction of the antisense RNAs contained one to three non-template nucleotides, predominantly UU or UA (Supplementary Figure S2D). Since a tomato RDR activity was reported to add one non-template nucleotide to a fraction of the run-off antisense transcripts in vitro (25), we assume that the Arabidopsis RDR6 has a similar property in vivo. cRT-PCR analysis of the shorter TAS2 dsRNAs in dcl2/3/4 plants which lack DCL4 activity revealed heterogeneous populations of sense and antisense RNAs with the sense RNAs starting at the miR173 cleavage site but ending at various positions and the antisense RNAs mirroring their sense counterparts (Supplementary Figure S2D). We conclude that TAS2 pri-RNA is cleaved twice, first at the miR173 site and second at the downstream site by a cleavage indirectly dependent on DCL4. The 317-nt RNA fragment between the two cleavage sites is converted by RDR6 to a 315-bp dsRNA (Figure 1). This dsRNA has a 2-nt 3′-overhang at the second cleavage end and an up to 3-nt 3′-overhang at the miR173 end.
cRT-PCR analysis of TAS1b short sense RNA also revealed a second, DCL4-dependent, cleavage site downstream of the miR173 site: all the cloned RNAs start with A374 and end with G601 (Figure 3 and Supplementary Figure S2B). The corresponding antisense RNA starts either with C600 or U599 and ends predominantly with U374 followed in 50% cases by one or two non-template uridines (Supplementary Figure S2B). Thus, the most abundant precursor of TAS1b tasiRNAs is 227- or 226-bp dsRNA with a 2-nt 3′-overhang at the second cleavage end and an up to 2-nt 3′-overhang at the miR173 end.
The TAS1a sense/antisense pair of ∼320 bp is more abundant than the TAS1a longest dsRNA but less abundant than two shorter pairs derived from this locus (Figure 1B). cRT-PCR mapping showed that this dsRNA is produced by the two-cleavage mechanism involving miR173-directed and DCL4-dependent cleavages. These cleavages excise a 318-nt fragment from TAS1a pri-RNA (from U379 to U696) which is copied by RDR6 to the complementary RNA starting either with U694 or C695 and ending with A379, often followed by one or two non-template nucleotides (Figure 3 and Supplementary Figure S2A). Thus, this dsRNA resembles the most-abundant TAS1b dsRNA.
TAS1c-derived tasiRNA directs a second cleavage of TAS2, TAS1a and TAS1b pri-RNAs
Our inspection of the second cleavage sites mapped on TAS2, TAS1b and TAS1a pri-RNAs revealed a remarkable similarity: all these sites are near-perfectly complementary to the TAS1c-derived siRNA 3′D6(−); this siRNA is encoded at the sixth processing cycle from the miR173-cleavage site in the antisense orientation (4,10). In all three cases, the cleavage occurs within a target pri-RNA/tasiRNA duplex between positions 10 and 11 from the tasiRNA 5′-end (Figure 3), thus representing a conserved, AGO-mediated cleavage. Inspection of sRNA deep-sequencing data [http://asrp.cgrb.oregonstate.edu; (4)] showed that this 21-nt RNA is one of the most abundant siRNAs derived from TAS1c. It is shifted forward by 1 nt with respect to the 21-nt phase set by miR173 cleavage, because a 22-nt siRNA/siRNA* duplex is generated at the fourth or the fifth processing cycle (Supplementary Figure S3A). The 1-nt phase forward drift was also found to occur in TAS3a and may result from misprocessing by DCL4 (4).
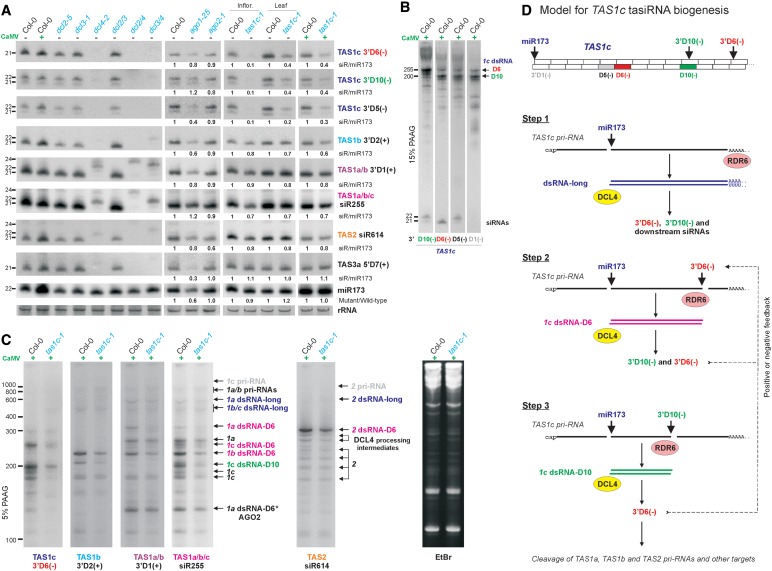
Consistent with the deep-sequencing data, our blot hybridization showed that TAS1c 3′D6(−) appears as one 21-nt class, while 3′D5(−) accumulates as minor (21 nt) and major (22 nt) classes (Figure 4A). Genetic analysis revealed that the biogenesis of both 3′D6(−) and 3′D5(−) requires DCL4, with no substantial contribution of DCL2, the dicer normally involved in generating 22-nt RNAs (Figure 4A).
Figure 4.
TAS1c is a master tasiRNA-generating unit regulated by feedback loops. (A) Genetic requirements for the biogenesis of TAS1/2-derived siRNAs. The panel shows blot hybridization analysis (15% PAGE) of total RNA from CaMV-infected (+) or mock-inoculated (−) wild-type Arabidopsis (Col-0), single (dcl2-5, dcl3-1 and dcl4-2) and double (dcl2/3, dcl2/4 and dcl3/4) DCL null mutants, AGO mutants (ago1-25 and ago2-1) and TAS1c siRNA-deficient mutant (tas1c-1) plants. The cropped scans represent blots successively hybridized with probes specific for TAS1c-, TAS1b-, TAS1a/b-, TAS1a/b/c-, TAS2- and TAS3a-derived siRNAs and miR173. EtBr-stained rRNA is shown as a loading control. The relative levels of TAS siRNA accumulation in a mutant line versus Col-0 were calculated using the miR173 level as an internal control for each sample; for each blot, siRNA levels in Col-0 were set to 1. (B) Blot hybridization analysis of TAS1c-derived dsRNA-D6 and dsRNA-D10. The cropped scans are from the blot shown in panel (A) (the second lane from left). (C) Blot hybridization analysis of TAS1c-, TAS1b-, TAS1a-, TAS2-derived dsRNAs in CaMV-infected Col-0 and tas1c-1 plants using 5% PAGE. The total RNA samples analyzed are the same as in panel A (the two from right). EtBr-stained RNA is shown as a loading control. Positions of dsRNAs derived from TAS1c, TAS1b, TAS1a and TAS2 are indicated by arrows (same as in Figure 1B). TAS1c 3′D6(−) tasiRNA-dependent dsRNAs are highlighted in pink, TAS1c 3′D10(−)-dependent dsRNA is in green and tasiRNA-independent longest dsRNAs are in blue. (D) Model for TAS1c tasiRNA biogenesis and feedback regulation.
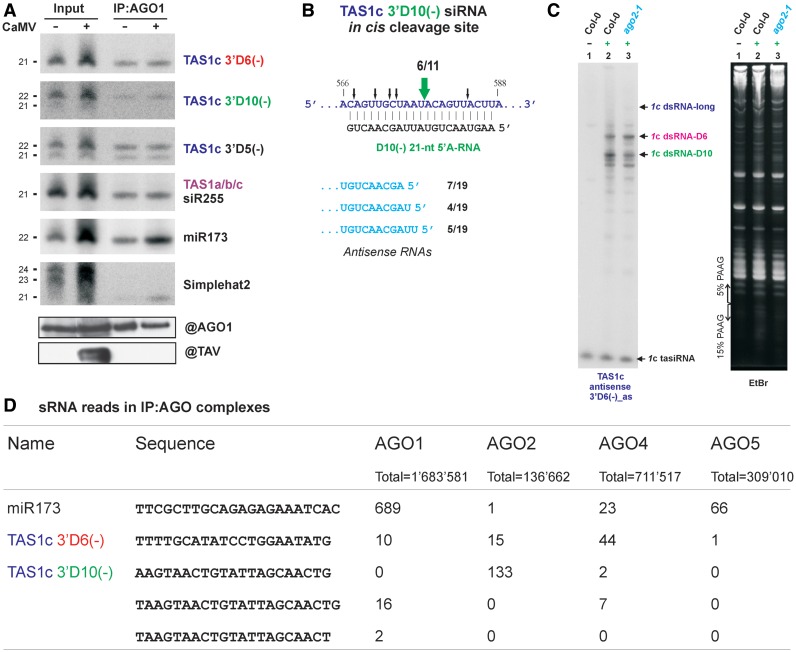
The 3′D6(−) is a 21-nt 5′U-RNA which would predict its association with AGO1 (15). Indeed, our IP experiment using native AGO1-antibodies revealed that AGO1 protein binds this siRNA as tightly as miR173 (Figure 5A). We conclude that both miR173 (18) and TAS1c 3′D6(−) tasiRNA guide AGO1-mediated cleavage of TAS2 and TAS1a/b pri-RNAs at their respective target sites.
Figure 5.
Loading of TAS1c siRNAs into AGO1 and AGO2 complexes that mediate in trans and in cis cleavages. (A) The upper panel shows blot hybridization analysis of total sRNAs (input) and sRNAs associated with AGO1 protein in mock-inoculated (−) or CaMV-infected (+) plants following IP with AGO1-antibodies (IP:AGO1). The scans represent the same blot successively hybridized with probes specific for TAS1c siRNAs, miR173 and, as a negative control, the transposon SIMPLEHAT2-derived 24-nt siRNA. The lower panel shows Western blot analysis of AGO1 protein accumulation in the input and the IP:AGO1 fractions using AGO1-antibody (@AGO1). Underneath is the same blot stained with antibodies specific for CaMV TAV protein (@TAV); (B) The in cis cleavage site for TAS1c-derived siRNA 3′D10(−) at TAS1c pri-RNA (the numbering is from the 5′ cap-site). Thick and thin arrows indicate the major and minor cleavage products, respectively, determined by cRT-PCR; the number of clones is given when more than one clone had the same 3′-terminus. The sequence of 3′D10(−) siRNA guiding the cleavage is shown under the pri-RNA sequence. The 5′-termini of predominant antisense RNAs of 1c dsRNA-D10 are shown, with corresponding numbers of specific cRT-PCR products/total products indicated; (C) Blot hybridization analysis of TAS1c-derived dsRNAs in CaMV-infected Col-0 and ago2-1 plants using 5% PAGE. The blot was hybridized with TAS1c 3′D6(−) siRNA-specific probe. EtBr-stained RNA is shown as a loading control. Positions of TAS1c-derived dsRNAs and dsRNA-D10 are indicated by arrows; (D) sRNA sequencing reads for TAS1c-derived siRNAs and miR173 in sRNA libraries prepared from AGO:IP complexes [(15); GEO accession: GSE10036].
We also identified additional target sites for TAS1c 3′D6(−) on TAS2 and TAS1a pri-RNAs, each located between the two mapped cleavage sites (Figure 3).
TAS1c 3′D6(−) directs in trans cleavage of TAS1c pri-RNA downstream of the origin site, which leads to its shorter dsRNA precursor
TAS1c 3′D6(−) has the potential to cleave its own precursor (TAS1c pri-RNA) in cis, i.e. at the site of its origin (Figure 3), which together with miR173-cleavage would give rise to a 115-nt fragment. However, we did not detect any abundant sense or antisense RNA of this size with TAS1c probes (Figures 1 and 4B). Instead, we identified another complementary site for TAS1c 3′D6(−) in a downstream position of TAS1c pri-RNA, and confirmed by cRT-PCR that it is cleaved at the expected position (Figure 3 and Supplementary Figure S2C). The 255-nt fragment (U378 to G632) excised from TAS1c pri-RNA by miR173 and TAS1c 3′D6(−) is converted to dsRNA with 1 or 2-nt 3′-overhangs on both ends, since the termini of the corresponding antisense RNA were mapped to positions C631 or U630 (5′ end) and A378 (3′-end); two non-template nucleotides were added in most cases (Figure 3 and Supplementary Figure S2C). A sense/antisense RNA pair of the corresponding size is the second most abundant TAS1c pair detected by blot hybridization (Figures 1 and 4B; Supplementary Figure S2C; designated 1c dsRNA-D6). This finding is indicative of a feedback loop regulating TAS1c 3′D6(−) biogenesis (Figure 4D).
The most abundant precursor of TAS1 3′D6(−) is generated via in cis cleavage
The most abundant sense/antisense RNA pair derived from TAS1c is shorter than the earlier described 1c dsRNA-D6 (Figures 1 and 4B; designated 1c dsRNA-D10). cRT-PCR (Supplementary Figure S2C) revealed that it is generated by a two-cleavage mechanism involving miR173 and TAS1c-derived siRNA 3′D10(−) which appears to cleave TAS1c pri-RNA in cis at the sites of its origin (Figure 5B). The sense strand of the resulting dsRNA starts with U379 and terminates predominantly with U577. The antisense strand 5′-ends map predominantly to U576, U575 and A574, thus resulting in 1- to 3-nt 3′-overhang (Figure 5B). The corresponding 3′-ends map to A379 followed with a few non-template nucleotides (Supplementary Figure S2C).
The 3′D10(−) itself can be generated from both TAS1c longest dsRNA and 1c dsRNA-D6 (Figure 4C) by DCL4 processing at the tenth cycle from the miR173 end. On blots, the 3′D10(−) appears as 21-nt and, more abundant, 22-nt classes (Figure 4A). The mapped cleavage site on TAS1c pri-RNA would predict that both size-classes should have 5′-terminal A587 to direct an AGO-mediated cleavage at the mapped position (Figure 5B).
Genetic analysis showed that both 21-nt and 22-nt classes of TAS1c 3′D10(−) require DCL4 for their biogenesis. The 21-nt class is selectively reduced in the null mutant ago2-1 (15), whereas the 22-nt class is selectively reduced in the hypomorphic mutant ago1-25 (26) (Figure 4A). This suggests that TAS1c 3′D10(−) exists as 21-nt 5′A587-RNA bound to AGO2 and 22-nt 5′U588-RNA bound to AGO1. Our AGO1-IP and other AGO-IP studies support this conclusion (Figure 5A and D).
Taken together, our findings suggest that the 21-nt 5′A-form of TAS1c 3′D10(−) may guide AGO2-mediated cleavage of TAS1c pri-RNA in cis. AGO2 is known to be associated with 21-nt 5′A-RNAs (15) but a slicer activity of AGO2 was not demonstrated so far.
In ago2-1 mutant, apart from the reduced accumulation of 21-nt 3′D10(−), accumulation of other TAS1c-derived tasiRNAs including 3′D6(−) is not drastically affected (Figure 4A). This can be explained by increased (compensatory) production of tasiRNAs from 1c dsRNA-D6 and also from TAS1c longest dsRNA. Indeed, accumulation of 1c dsRNA-D6 is increased ∼2- to 3-fold in ago2-1 compared to wild-type plants, concomitant with ∼2- to 3-fold decrease in accumulation of 1c-dsRNA-D10; the longest dsRNA remains at a similar low level (Figure 5C and Supplementary Figure S4). Thus, in the absence of AGO2, 1c dsRNA-D6 becomes the most abundant precursor of TAS1c-derived tasiRNAs.
Surprisingly, in dcl2/3/4 plants that do not produce any tasiRNA, in addition to the longest TAS1c dsRNA, one shorter dsRNA accumulates to high levels (Figure 1). This dsRNA has 3′-overhangs of one to several nucleotides and is generated by a mechanism involving the miR173 cleavage and a precise cleavage that occurs predominantly 7-nt upstream of the 3′D6(−) cleavage site (Supplementary Figure S2C). It remains to be investigated which DCL1-dependent sRNA directs the second cleavage in this case.
The majority of TAS1/TAS2 siRNAs are produced from the region between the cleavage sites
Our inspection of the deep-sequencing data (4) revealed that for each miR173-dependent TAS gene, most of unique sRNAs and the overwhelming majority of sRNA reads originate from the region between miR173 and TAS1c 3′D6(−) cleavage sites (Supplementary Figure S3B). Thus, the short dsRNAs (dsRNA-D6) produced by the two-cleavage mechanism are likely the major precursors of TAS1/TAS2 tasiRNAs. The region of TAS1a/b/c and TAS2 genes downstream of the TAS1c 3′D6(−) cleavage site also generates sRNAs, albeit at much lower levels (Supplementary Figure S3B). These sRNAs are likely processed from the longest, poly(A)/poly(U) duplex-containing dsRNAs.
TAS2- and TAS1a/b/c-derived short dsRNAs are processed from both ends
The dsRNAs derived from TAS2, TAS1a, TAS1b and TAS1c can potentially be processed from both ends. If this is true, then in addition to miR173-phased tasiRNAs, siRNAs in phase with the TAS1c 3′D6(−) cleavage site should also accumulate. Inspection of the deep-sequencing data (4) revealed that each of the four miR173-dependent TAS loci spawns 21-nt RNAs upstream of and in phase with the 3′D6(−) cleavage site (Supplementary Figure S3A; highlighted in pink). Also, more abundant 21-nt RNAs are produced in a 1-nt-forward-drifted phase caused by generation of 22-nt RNAs at the first (TAS1a and TAS1b) or the seventh processing cycle (TAS2) (Supplementary Figure S3A; purple). Thus, similar to miR390-directed cleavage for TAS3 family pri-RNAs (12), the 3′D6(−) cleavage of TAS1a/b/c and TAS2 pri-RNAs sets a register for production of phased siRNAs upstream of the cleavage site. This second register is shifted with respect of the miR173 register either forward (by 2 nt in TAS2 and TAS1c and 3 nt in TAS1a) or backward (by 3 nt in TAS1b). Interestingly, an expected cleavage of TAS1a and TAS2 pri-RNAs at the additional target sites for TAS1c 3′D6(−) (Figure 3) also appears to initiate processing of phased siRNAs from the upstream region (Supplementary Figure S3A; cyan and blue).
Likewise, TAS1c 3′D10(−) cleavage of TAS1c pri-RNA sets a register for 21-nt RNAs from the upstream region (Supplementary Figure S3A; green). Notably, the 22-nt TAS1c D10(−) species itself can be produced by in-phase processing of 1c dsRNA-D6 from either end.
Using a miRNA target prediction software [http://bioinfo3.noble.org/psRNATarget/; (27)] we identified potential target genes for several siRNAs encoded in the registers set by the 3′D6(−) on TAS1a, TAS1b and TAS2 and the 3′D10(−) on TAS1c (Supplementary Table S1).
Taken together, we conclude that the short dsRNAs derived from TAS1a, TAS1b, TAS1c and TAS2 are processed by DCL4 not only from the miR173 end but also from the opposite end defined by TAS1c 3′D6(−) cleavage and, in the case of TAS1c, also by TAS1c 3′D10(−) cleavage. The processing from both ends results in production of siRNAs in two different registers, thus increasing the coding capacity of these tasiRNA-generating loci.
It should be noted that the deep-sequencing data used for the above analyses of tasiRNAs were obtained using non-infected A. thaliana plants (4). Thus, the short dsRNA precursors of tasiRNAs identified here in CaMV-infected plants are likely present and processed from both ends in non-infected plants.
miR173-dependent tasiRNA production from TAS1a/b and TAS2 genes is not drastically affected by TAS1c gene knockdown
Our results so far demonstrated that TAS1c-derived tasiRNA 3′D6(−) can target in trans TAS1a/b and TAS2 pri-RNAs to initiate the biogenesis of abundant dsRNAs that can be processed by DCL4 from both ends. To address the question whether this second cleavage is required for efficient production of TAS1a/b and TAS2 tasiRNAs in the miR173 register, we searched A. thaliana mutant collections for TAS1c gene mutants. By genotyping combined with RNA blot hybridization, we identified a homozygous mutant line (designated tas1c-1) with T-DNA insertion between the two major poly(A) sites of TAS1c (Supplementary Figure S2C). In tas1c-1, accumulation of TAS1c siRNAs including 3′D6(−) and 3′D10(−) was reduced to ∼40% of the wild-type level in rosette leaves and to ∼10% in inflorescence tissues (Figure 4A). This knockdown of TAS1c gene expression strongly reduced accumulation of the 3′D6(−)-dependent short dsRNAs derived from TAS1a, TAS1b and TAS2 (Figure 4C; see ‘dsRNA-D6’ species highlighted in pink), but did not affect the accumulation the 3′D6(−)-independent, longest dsRNAs derived from TAS1a and TAS1b (Figure 4C; highlighted in blue) as well as less abundant, shorter RNAs derived from TAS2 (Figure 4C; indicated by four connected arrows). The knockdown had only a slight impact on accumulation of TAS1a/b and TAS2 tasiRNAs encoded in the miR173 register, which was reduced only to 70–80% of the wild-type levels in both tissues (Figure 4A). We conclude that TAS1c gene expression does not substantially affect the biogenesis of TAS1a, TAS1b and TAS2-derived siRNAs encoded in the miR173 register. Our findings are consistent with the notion that tasiRNA production from TAS1a/b and TAS2 loci can proceed through a single miR173-directed cleavage event (18,19), which leads to the production of the longest, poly(A)/poly(U) duplex-containing dsRNAs.
TAS1c 3′D6(−) tasiRNA-directed cleavage regulates TAS1c 3′D10(−) siRNA biogenesis and precludes TAS2 siRNA production downstream of the target site
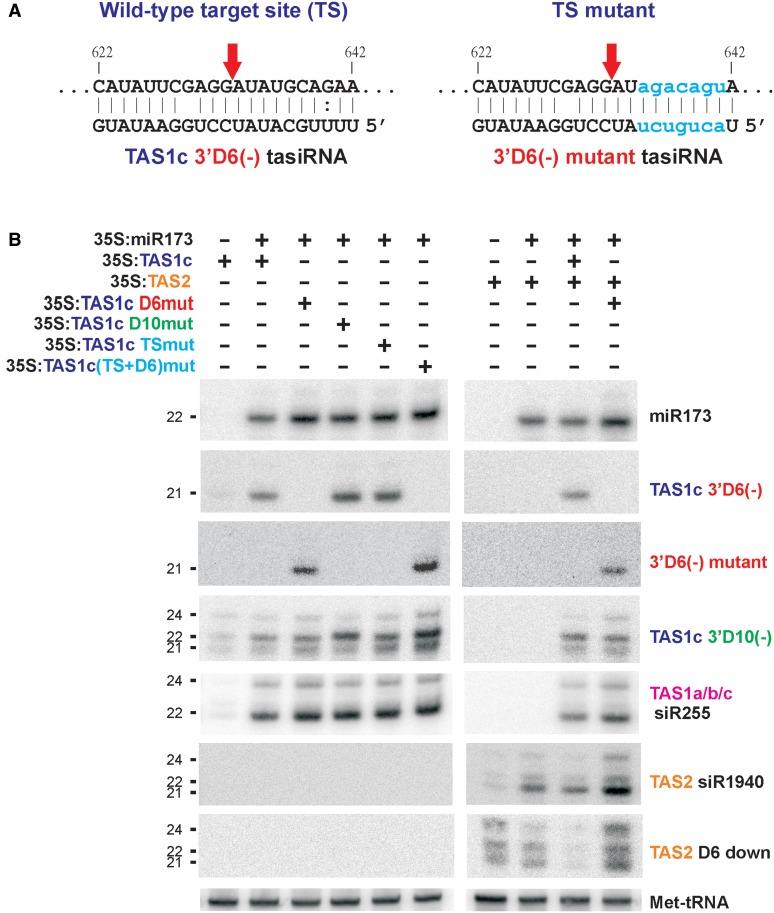
To further investigate a role of the second cleavage of TAS pri-RNAs, we used a previously established transient expression assay using N. benthamiana leaves which recapitulates miR173-dependent tasiRNA biogenesis from the TAS1 and TAS2 transcripts driven by the CaMV 35S promoter (10). Confirming the previous results, robust production of siR255 from 35S:TAS1c and siR1940 from 35S:TAS2 could be achieved only by co-expression of miR173 from a 35S:miR173 construct. We then co-expressed the TAS2 construct with 35S:TAS1c or its derivative in which the seed sequence of the 3′D6(−) tasiRNA had been mutated to prevent the second cleavage of TAS2 pri-RNA. Co-expression of 3′D6(−) species or its seed-mutant form only slightly affected miR173-dependent production of siR1940 from TAS2 (Figure 6), supporting the above conclusions. However, siRNA production from the region downstream of the 3′D6(−) cleavage site was nearly abolished in the presence of 3′D6(−) species but not its mutant form (see ‘TAS2 D6 down’ in Figure 6). Thus, the second cleavage of TAS2 pri-RNA restricts siRNA production to the region between the miR173 and 3′D6(−) cleavage sites. This is consistent with our above analysis of the sRNA deep-sequencing data in A. thaliana. Interestingly, the region downstream of the 3′D6(−) cleavage site generated siRNAs of three size-classes and their production was detected both in the presence and absence of miR173.
Figure 6.
TAS1c 3′D6(−) tasiRNA-directed cleavage regulates TAS1c 3′D10(−) siRNA biogenesis and precludes TAS2 siRNA production downstream of the target site. (A) The wild-type and mutant targets sites (TS) of the wild-type TAS1c pri-RNA and its TS mutant version, respectively. Mutagenized positions are in lower case and highlighted in cyan. Position of the cleavage directed by the wild-type TAS1c 3′D6(−) tasiRNA or its seed-mutant version is indicated by red arrow. (B) Reconstruction of TAS1c and TAS2 tasiRNA biogenesis in a transient expression assay using N. benthamiana. 35S:TAS1c constructs with mutagenized 3′D6(−), 3′D10(−) siRNAs, the 3′D6(−) target site sequences, or both 3′D6(−) and its target site sequences were expressed or coexpressed with 35S:miR173 and also with 35S:TAS2 constructs as indicated above the blot panels. The siRNAs detected by successive hybridization of the blots are shown to the right of each panel.
For TAS1c itself, the mutation of the 3′D6(−) seed sequence did not alter miR173-dependent accumulation of siR255. However, it reduced accumulation of 22-nt 3′D10(−) and increased accumulation of 21-nt 3′D10(−) species, thus resembling the effect observed in Arabidopsis ago1-25 plants. Mutation of the 3′D6(−) target site (TS) in 35S:TAS1c, in such a way that the mutant TS sequence could be cleaved only by the D6 seed-mutant siRNA but not the wild-type siRNA, led to a similar effect on accumulation of the 3′D10(−) size-classes. A combination of the two mutations restored the wild-type ratio of 3′D10(−) size-classes. Furthermore, an A-to-G mutation of the 5′-terminal nucleotide of the 21-nt 3′D10(−) in 35S:TAS1c increased the ratio of the 22-nt class to the 21-nt class, which is reminiscent of the effect observed in Arabidopsis ago2-1 plants. Taken together, these results confirm feed-back regulation of TAS1c siRNA biogenesis by the 3′D6(−)-directed in trans cleavage and the 3′D10(−)-directed in cis cleavage of TAS1c pri-RNA.
DISCUSSION
Structures of RDR6-dependent dsRNA precursors of tasiRNAs
In this work we demonstrate that RDR6 converts the entire, polyadenylated, 3′-products of miR173-cleaved TAS1/TAS2 pri-RNAs to dsRNA. However, the resulting longest dsRNAs accumulate at much lower levels than the shorter dsRNAs produced from all the four loci by a two-cleavage mechanism. In fact, TAS2-derived longest, poly(A)/poly(U) duplex-containing dsRNA could be detected only in dcl2/3/4 plants lacking DCL2, DCL3 and DCL4 activities, while TAS1 loci-derived longest dsRNAs accumulated at much higher levels in dcl2/3/4 than in wild-type plants. Thus, the polyadenylated 3′-fragment of miR173-cleaved TAS pri-RNAs is rapidly truncated by the second cleavage directed by TAS1c-derived tasiRNA 3′D6(−) which generates a shorter, poly(A)-less template for RDR6. We cannot exclude, however, that the longest dsRNA is more efficiently processed by DCL4 than the shorter dsRNA.
Our cRT-PCR analysis also suggests that the poly(A) tail is shortened and, in some cases, fully removed prior to RDR6 binding to the long template. The shortened poly(A) tails are often tagged by short UA-rich sequences. The RNA templates with both tagged and non-tagged poly(A) tails of up to 37 nt are copied by RDR6 to dsRNAs. This finding is supported by the in vitro studies of an affinity-purified A. thaliana RDR6 protein showing that the RDR6 activity is not inhibited by addition of a 40-nt poly(A) tail to the template (28).
The structures of more abundant, short TAS1/TAS2 dsRNAs identified here reveal that RDR6 initiates complementary RNA synthesis preferentially from the third nucleotide of the template’s 3′-end, thus creating a 2-nt 3′-overhang on the resulting dsRNA. Less frequently, the synthesis is initiated from the second or the fourth nucleotide, resulting in 1-nt or 3-nt 3′-overhangs, respectively. In our parallel work, precise mapping of the TAS3a-derived dsRNA revealed that RDR6 initiates complementary RNA synthesis predominantly at the third nucleotide of the miR390-cleaved TAS3a pri-RNA (17). This confirms our findings for miR173-dependent TAS1/TAS2 dsRNAs. Furthermore, we demonstrate here that RDR6-mediated synthesis terminates at the 5′-terminal nucleotide of the template and frequently one or two non-template nucleotides are added to the 3′-end of the complementary RNA. Similar biochemical properties have been reported for an RDR activity isolated from tomato: this RDR copied an artificial single-stranded RNA template from the second or the third nucleotide of the template’s 3′-terminus carrying the hydroxyl group and added one non-template nucleotide to 50% of the complementary run-off transcripts (25). Thus, production of dsRNAs with 1-nt or 2-nt 3′ overhangs at both termini appears to be a common property of plant RDRs. In vitro studies have shown that the dsRNA with a 2-nt 3′-overhang is an ideal substrate of human dicer (29). It remains to be investigated whether or not a short 3′ overhang is absolutely required for DCL4. Notably, a substantial fraction of miR173-dependent dsRNAs derived from TAS1a, TAS1b, TAS1c or TAS2 does not possess any overhang at the miR173 cleavage end.
Our data also support the in vitro results showing that Arabidopsis RDR6 does not require a primer (28).
Role of the second cleavage of pri-RNA in tasiRNA production from TAS1/TAS2 genes
We demonstrate here that miR173-initiated biogenesis of tasiRNAs from Arabidopsis TAS2 and TAS1 genes involves a second cleavage of TAS pri-RNA downstream of the miR173 site. In all cases, the short segment of pri-RNA between the two-cleavage sites is converted by RDR6 to dsRNA. The resulting dsRNA spans the region giving rise to the majority of siRNAs derived from each of the four loci. Hence, the two-cleavage mechanism generates the major dsRNA precursors of tasiRNAs. However, our results also demonstrate that the second cleavage is not required for production of tasiRNAs in the miR173 register. This is consistent with the previous studies showing that a single miR173-dependent cleavage is sufficient to initiate tasiRNA biogenesis (18,19). What could be the role of the second cleavage? Our results in N. benthamiana indicate that the second cleavage directed by TAS1c 3′D6(−) tasiRNA strongly reduces TAS2 siRNA production from the region downstream of the cleavage site. Thus, the two-cleavage mechanism appears to restrict siRNA production to the region between the miR173 and 3′D6(−) cleavage sites. Additionally, identification of 21-nt phased siRNAs in the register set by TAS1c 3′D6(−) cleavage in the sRNA deep-sequencing database shows that the second cleavage could also be to increase the tasiRNA coding capacity of the TAS1/TAS2 genes.
Our findings extend the two-hit trigger model originally proposed for miR390-dependent TAS3 genes (12) to miR173-dependent TAS1/TAS2 genes. But unlike TAS3a/b/c pri-RNAs targeted at two sites by a single miRNA (miR390), TAS1a/b/c and TAS2 pri-RNAs are targeted by miR173 only at one site, while the second hit is guided by TAS1c-derived tasiRNA that cleaves not only TAS1a, TAS1b and TAS2 pri-RNAs but also its own precursor (TAS1c pri-RNA). Previous studies have identified several Arabidopsis genes that have target sites for two or more different miRNAs and/or tasiRNAs; most of these genes generate secondary siRNAs (4,12). We demonstrate here that the miRNA- and the tasiRNA-directed cleavages at one target RNA can recruit RDR6 activity that converts the RNA segment between the two-cleavage sites to a dsRNA substrate for DCL4.
Feedback regulation of TAS1c gene
We propose a feedback regulation model in which TAS1c gene codes for two siRNAs, 3′D6(−) and 3′D10(−), which control tasiRNA production from this gene (Figure 4D). These siRNAs are encoded in antisense orientation at the processing cycles 6 and 10, in phase with the miR173 register, and can initially be generated from the poly(A)/poly(U) duplex-containing longest dsRNA, produced by RDR6 following a single miR173-cleavage. Once generated, each of these siRNAs together with miR173 directs cleavage of TAS1c pri-RNA at the respective target site to initiate RDR6-catalyzed synthesis of two distinct dsRNAs. The most abundant of the two (1c dsRNA-D10) is generated by the 3′D10(−) cleavage in cis and can potentially serve as a precursor of 3′D6(−) siRNA but not of 3′D10(−) itself. Less-abundant dsRNA (1c dsRNA-D6) generated by the 3′D6(−) cleavage in trans can potentially spawn both species (Figure 4D).
Several lines of evidence support our feedback model and allow speculation on potentially positive or negative functions of the two feedback loops. First, we have detected by blot hybridization and sequenced the three TAS1c-derived dsRNAs, namely, the single miR173-dependent longest dsRNA (1c dsRNA-long; Figures 1B and 2), the miR173- and TAS1c 3′D6(−) tasiRNA-dependent dsRNA (1c dsRNA-D6; Figures 1B and 3) and the miR173- and TAS1c 3′D10(−) siRNA-dependent dsRNA (1c dsRNA-D10; Figures 1B, 3 and 5). The terminal sequences of the dsRNA template strands indicate the conserved, AGO-mediated cleavages of TAS1c pri-RNA directed by miR173 on the upstream site and by the 3′D6(−) or 3′D10(−) siRNA at the downstream site. The relative accumulation levels of the three dsRNAs would suggest that DCL4 processing of the two shorter dsRNAs might generate the majority of TAS1c-derived siRNAs. However, we cannot exclude that the longest dsRNA is processed more efficiently than the shorter ones. Second, we provide genetic evidence that reduced accumulation of 21-nt 3′D10(−) siRNA in ago2-1 mutant plants correlates with reduced accumulation of 1c dsRNA-D10 and increased accumulation of 1c dsRNA-D6 (Figure 5). Thus, the 3′D10(−)-directed cleavage required for 1c dsRNA-D10 biogenesis appears to interfere with 1c dsRNA-D6 biogenesis, possibly owing to competition between 3′D10(−) and 3′D6(−) siRNAs for the same target TAS1c pri-RNA. This suggests that the in cis cleavage directed by 3′D10(−) may be involved in negative feedback regulation of TAS1c siRNA biogenesis. Third, our reconstitution of TAS1c siRNA production in the N. benthamiana transient assay shows that the 3′D6(−) tasiRNA-directed cleavage of TAS1c pri-RNA regulates the biogenesis of 3′D10(−) siRNA size-classes by decreasing relative accumulation of the cleavage-competent, 21-nt species (Figure 6), thus suggesting a positive function of this cleavage event. Further experiments are needed to clarify the roles of the two feedback loops in regulation of TAS1c gene expression.
We established that, similar to miR173, TAS1c 3′D6(−) tasiRNA is tightly associated with AGO1 protein to exert its function in TAS1/TAS2 siRNA biogenesis. Interestingly, this 21-nt 5′U-RNA was also co-immunoprecipitated with AGO2 which preferentially binds 21-nt 5′A-RNAs and with AGO4 which preferentially binds 24-nt 5′A-RNAs [(15); Figure 5D]. We speculate that this apparent competition for the 3′D6(−) species may play a role in regulation of TAS1c expression by reducing the levels of functional AGO1::3′D6(−) complexes. Additionally, AGO4::3′D6(−) complex may function in the nucleus to downregulate transcription of TAS1c pri-RNA.
AGO2 was implicated in TAS3 siRNA biogenesis and in antiviral defense since it was found to be associated with miR390 (15) and virus-derived siRNAs (16), respectively. Recently, the antiviral role of AGO2 was confirmed for some RNA viruses that encode suppressors specifically targeting AGO1 (30). However, so far there was no evidence for AGO2 catalytic activity. Our results suggest that this protein binds TAS1c-derived 21-nt siRNA 3′D10(−) to mediate in cis cleavage of TAS1c pri-RNA. Further experiments are needed to confirm that AGO2 is a functional slicer and that its slicer activity is required for AGO2 action in TAS1c tasiRNA biogenesis and antiviral defense.
TAS1/TAS2 gene regulation network
A sRNA interaction network connecting miR173-dependent TAS2, TAS1a and TAS1c genes was recently predicted by a bioinformatic analysis of sRNA populations sequenced from A. thaliana (6). Our data confirms the existence of this network as well as refines and extends the prediction by demonstrating that TAS1c-derived tasiRNA can target all the four miR173-dependent TAS loci. Few unmapped RNAs that we detected by blot hybridization (Figure 1) may arise from additional targeting events predicted by MacLean et al. (6).
The existence of this complex network with a central role for TAS1c is not surprising, given the fact that the four TAS loci are paralogous: TAS2 and TAS1c arose by a direct duplication, while TAS1a and TAS1b arose from TAS1c after it had diverged from TAS2 (11). Interestingly, TAS2 and TAS1 tasiRNAs including TAS1c 3′D6(−) and 3′D10(−) described here target several pentatricopeptide repeat family genes giving rise to secondary siRNAs with the potential to target in trans other genes within and outside of this multigene family (4). This creates another complex interaction network subordinate to the miR173 network. Our findings for TAS1c would also suggest that feedback loops in the hubs of sRNA networks can fine-tune the levels of a master siRNA, such as TAS1c 3′D6(−), required for in trans regulation of other genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1, 2 (A, B, C and D), 3 (A and B) and 4, Supplementary Table 1, Supplementary Methods and Inventory of Supplementary Data.
FUNDING
Swiss National Science Foundation Grants (31003A_127514 to M.M.P. and 31003A_122469 to Thomas Hohn and M.M.P.) and European Commission Grant (a Marie Curie fellowship PIIF-237493-SUPRA to R.R.). Funding for open access charge: University of Basel.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nachelli Malpica for technical assistance, David Baulcombe for ago1-25 and ago2-1 seeds and James Carrington for the 35S:GUS, 35S:TAS1c, 35S:TAS2 and 35S:miR173 constructs. We are grateful to Thomas Hohn for support and critical reading of the manuscript and to Thomas Boller for hosting the group at the Botanical Institute. M.M.P. designed the research; R.R., M.A., A.S.Z., B.B. and E.G.G. performed research; R.R. and M.MP. analyzed data and M.M.P. wrote the article.
REFERENCES
- 1.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 2.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 3.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl Acad. Sci. USA. 2010;107:15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacLean D, Elina N, Havecker ER, Heimstaedt SB, Studholme DJ, Baulcombe DC. Evidence for large complex networks of plant short silencing RNAs. PLoS One. 2010;5:e9901. doi: 10.1371/journal.pone.0009901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 9.Katiyar-Agarwal S, Jin H. Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 2010;48:225–246. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 13.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM, Alexander A, Nguyen G, Allen E, Ahn JH, et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl Acad. Sci. USA. 2008;105:20055–20062. doi: 10.1073/pnas.0810241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 17.Rajeswaran R, Pooggin MM. RDR6-mediated synthesis of complementary RNA is terminated by miRNA stably bound to template RNA. Nucleic Acids Res. 2012;40:594–599. doi: 10.1093/nar/gkr760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 2010;17:997–1003. doi: 10.1038/nsmb.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HM, Li YH, Wu SH. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl Acad. Sci. USA. 2007;104:3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manavella PA, Koenig D, Weigel D. Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl Acad. Sci. USA. 2012;109:2461–2466. doi: 10.1073/pnas.1200169109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shivaprasad PV, Rajeswaran R, Blevins T, Schoelz J, Meins F, Jr, Hohn T, Pooggin MM. The CaMV transactivator/viroplasmin interferes with RDR6-dependent trans-acting and secondary siRNA pathways in Arabidopsis. Nucleic Acids Res. 2008;36:5896–5909. doi: 10.1093/nar/gkn590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Jr, Hohn T, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shivaprasad PV, Akbergenov R, Trinks D, Rajeswaran R, Veluthambi K, Hohn T, Pooggin MM. Promoters, transcripts, and regulatory proteins of Mungbean yellow mosaic geminivirus. J. Virol. 2005;79:8149–8163. doi: 10.1128/JVI.79.13.8149-8163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blevins T, Rajeswaran R, Aregger M, Borah BK, Schepetilnikov M, Baerlocher L, Farinelli L, Meins F, Hohn T, Pooggin MM. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 2011;39:5003–5014. doi: 10.1093/nar/gkr119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiebel W, Haas B, Marinković S, Klanner A, Sänger HL. RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J. Biol. Chem. 1993;268:11858–11867. [PubMed] [Google Scholar]
- 26.Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai X, Zhuang Z, Zhao PX. Computational analysis of miRNA targets in plants: current status and challenges. Brief Bioinform. 2010;12:115–121. doi: 10.1093/bib/bbq065. [DOI] [PubMed] [Google Scholar]
- 28.Curaba J, Chen X. Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J. Biol. Chem. 2008;283:3059–3066. doi: 10.1074/jbc.M708983200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 30.Harvey JJ, Lewsey MG, Patel K, Westwood J, Heimstädt S, Carr JP, Baulcombe DC. An antiviral defense role of AGO2 in plants. PLoS One. 2011;6:e14639. doi: 10.1371/journal.pone.0014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.