Abstract
The homologous cellular coactivators p300 and CBP contain intrinsic lysine acetyl transferase (termed HAT) activity. This activity is responsible for acetylation of several sites on the histones as well as modification of transcription factors. In a previous study, we found that HBZ, encoded by the Human T-cell Leukemia Virus type 1 (HTLV-1), binds to multiple domains of p300/CBP, including the HAT domain. In this study, we found that HBZ inhibits the HAT activity of p300/CBP through the bZIP domain of the viral protein. This effect correlated with a reduction of H3K18 acetylation, a specific target of p300/CBP, in cells expressing HBZ. Interestingly, lower levels of H3K18 acetylation were detected in HTLV-1 infected cells compared to non-infected cells. The inhibitory effect of HBZ was not limited to histones, as HBZ also inhibited acetylation of the NF-κB subunit, p65, and the tumor suppressor, p53. Recent studies reported that mutations in the HAT domain of p300/CBP that cause a defect in acetylation are found in certain types of leukemia. These observations suggest that inhibition of the HAT activity by HBZ is important for the development of adult T-cell leukemia associated with HTLV-1 infection.
INTRODUCTION
In mammalian cells, the coactivators p300 and CBP, also called KAT3B and KAT3A, respectively, play an essential role in transcription. These ubiquitously-expressed proteins are highly homologous and frequently referred to singularly as p300/CBP. They are recruited to promoters or enhancers through interactions with numerous transcription factors where they engage additional regulators and bridge transcription factors to the general transcription machinery (1). The widespread use of p300 and CBP in transcription is due to the presence of multiple, independent domains in these coactivators (2) that, together, contact more than 400 transcriptional regulators in the cell (3). p300/CBP also carries a lysine acetyl transferase activity (classically designated histone acetyl transferase or HAT activity) that acetylates both histones (4,5) and transcription factors (6). Acetylation of lysine residues within the N-terminal tails as well as the globular domains of the histones is generally linked to active transcription (7,8). In contrast, acetylation of transcription factors produces both positive and negative effects on activity by influencing such properties as cellular localization, stability and molecular interactions (9). p300/CBP is capable of acetylating several core histone lysine residues (10), many of which are also targeted by other proteins with acetyl transferase activity (10,11). However, recent data indicate that lysines 18 and 27 of histone H3 (H3K18ac and H3K27ac) are distinctly acetylated by p300/CBP, as depletion of both coactivators in mouse embryonic fibroblasts leads to a reduction in these modifications (12). In addition, p300/CBP also specifically acetylates lysine 56 of H3 during the DNA damage response (13).
Accumulating evidence indicates that CBP and, to a lesser extent p300, function as tumor suppressors. Mutations in p300 and CBP have been identified in many types of cancer (14). In mice, deletion of a single allele of the CBP gene produces defects in hematopoietic differentiation and an increased incidence of hematologic malignancies (15), while homozygous deletion of the gene causes embryonic lethality (16), In humans, deletions or mutations within a single allele of the CREBBP or EP300 gene is sufficient to cause Rubenstein–Taybi syndrome, which is associated with a high frequency of tumor development among other clinical manifestations (17). Therefore, in mice and humans, p300 and CBP appear limiting in the cell, and a reduction in their functional activities may lead to transformation.
Recent studies show that disruption of p300/CBP HAT activity, specifically, may play a primary role in certain hematological transformation events. Indeed, mutations in CBP and p300 that disrupt HAT activity were found to be prevalent in cases of diffuse large B-cell lymphoma (18,19) and follicular carcinoma (18–20). In relapsed acute lymphoblastic leukemia, the CREBBP gene is also frequently mutated or deleted such that HAT activity is repressed (21). Interestingly, for some types of cancer, a global reduction in the level of H3K18ac serves as a prognostic indicator of a poor clinical outcome (22–28). These observations indicate that p300/CBP HAT activity is targeted during transformation.
Human T-cell Leukemia Virus type 1 (HTLV-1) is a complex retrovirus that causes adult T-cell leukemia/lymphoma (ATL), a malignancy characterized by the abnormal proliferation of mature CD4+ cells (29,30). ATL is a heterogeneous disease with different clinical stages. The acute and lymphoma subtypes are the most aggressive forms of ATL, and their prognosis is poor, with less than 1 year survival for patients diagnosed with these subtypes (31). HTLV-1 encodes several unique proteins that participate in viral replication, viral infectivity, persistence and transformation (32). Among these proteins, HTLV-1 basic leucine zipper factor (HBZ) was recently shown to induce T-cell lymphoma in transgenic mice as well as skin inflammation similar to that observed in HTLV-1 infected patients (33).
HBZ is localized to the nucleus where it functions as a transcriptional regulator/deregulator. Through its C-terminal leucine zipper (ZIP) domain, HBZ is able to form heterodimers with cellular basic leucine zipper (bZIP) transcription factors of the AP-1 (34–36) and ATF/CREB families (37,38). Due to the abnormal basic region of its bZIP domain, HBZ sequesters these factors from their consensus DNA-binding sites, thereby repressing transcription mediated through CRE and AP-1 promoter elements. JunD represents an exception to this model, as its transcriptional activity is enhanced rather than inhibited by HBZ (35). HBZ also contains two LXXLL motifs in an N-terminal activation domain that are responsible for formation of a high-affinity interaction with the KIX domain of p300/CBP (39,40). The KIX domain harbors two distinct binding surfaces that are generally contacted by separate sets of transcription factors (34,37,38). Through its interaction with a single surface, HBZ is able to inhibit interactions involving that surface and, alternatively, enhance the binding of factors to the other surface (40). This dual effect on the KIX domain correlates with specific examples of transcriptional repression and activation by HBZ (39,41,42).
In addition to its interaction with the KIX domain, HBZ was found to bind two partially overlapping regions encompassing the HAT and C/H3 domains of p300/CBP (39). However, the significance of these latter interactions has been unclear. In the this study, we further dissected the binding of HBZ to the HAT and C/H3 domains and found that HBZ contacts separate sites within each domain: one located at the C-terminus of the HAT domain and the other encompassing a transcription factor docking site within the C/H3 domain termed TAZ2 (43). By binding to the HAT domain, HBZ inhibited p300/CBP HAT activity. This effect was mediated by the bZIP domain of HBZ, supporting previous data that the N-terminal LXXLL motifs are not involved in this interaction (39). Cells expressing HBZ exhibited a deficiency in p300/CBP-targeted H3K18ac. Importantly, this mark was reduced in HTLV-1-infected T-cells compared to uninfected T-cells. In addition to the core histones, HBZ also inhibited p300/CBP-mediated acetylation of the NF-κB factor, p65, and the tumor suppressor, p53. Together, these observations link certain molecular effects of HBZ to an emerging prognostic indicator for many cancers, that of disrupted p300/CBP HAT activity.
MATERIALS AND METHODS
Plasmids and antibodies
All GST fusions proteins were derived from CBP. Escherichia coli expression plasmids for GST, GST-HAT (1096–1757 amino acids) and GST-C/H3 (1514–1894 amino acids) have been described (39). GST-N-HAT (1096–1514 amino acids), GST-C-HAT (1514–1679 amino acids), GST-ΔC-HAT (1514–1723 amino acids) and GST-TAZ2 (1758–1894 amino acids) were constructed by PCR-amplification of pRc/RSV-CBP (44) and cloning products into pGEX-2T (GE Healthcare) at the BamHI/EcoRI sites. GST-HBZ and GST-HBZ-Mut(LXXAA)2 (39), HBZ-ΔbZIP (1–122 amino acids) and HBZ-bZIP(120–206 amino acids) (38), pRSETA-p53 (45) have been described. c-Jun bZIP (257–334 amino acids) was PCR-amplified from pBiFC-bJunVN173 (46) and cloned into pRSETA at the BamHI/EcoRI sites. Mammalian expression plasmids for p300-Flag and p300-ΔHAT-Flag (deletion of amino acids 1472–1522) and pcDNA-HBZ-His-Myc have been described (38,47). Flag-p65 was a gift from Dr Baldwin (University of North Carolina at Chapel Hill). pNF-κB-Luc was purchased from Stratagene. Pan acetyl lysine (#9441), acetyl K382 p53 (#2525), acetyl K320 p65 (#3045), p65 (#3034), acetyl H3 K18 (#9675), trimethyl H3K27 (C36B11, #9733) and NUP98 (C39A3, #2598) antibodies were from Cell Signaling. Acetyl K9K14 H3 (06-599) and Myc (clone 4A6, 05-724) antibodies were from Millipore. p53 (DO-1, sc-126), CBP (A-22, sc-369) and p300 (C-20, sc 585) antibodies were from Santa-Cruz. Histone H3 (Ab 1791) and 6× histidine (Ab 9108) antibodies were from Abcam. The actin (MAB1501R) and histone H4 (39270) antibodies were from Chemicon and Active Motif, respectively. The 6× histidine (R930) antibody used for immunofluorescence was from Invitrogen. FITC-conjugated Flag (M2, F4049) and Flag (F-1804) antibodies were from Sigma.
Expression and purification of recombinant proteins and in vitro translation
GST-HBZ and GST- HBZ-Mut(LXXAA)2 E. coli expression plasmids were transformed into BL21-codon plus (DE3) cells (Agilent Technologies); other plasmids were transformed into BL21(DE3) pLysS cells (Agilent Technologies). Proteins were expressed and purified by glutathione–agarose affinity chromatography or Ni–NTA agarose (Qiagen) as described previously (40). Flag-CBP and 6× His-p300 were expressed from recombinant baculoviruses in Sf9 cells and purified as described (39). Purified proteins were dialyzed against HM 0.1 M [100 mM KCl, 50 mM HEPES (pH 7.9), 12.5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.025% (v/v) Tween 20 and 20% (v/v) glycerol], aliquoted, and stored at −80°C. Recombinant p65 was purchased from Active Motif.
GST pull-down assay
Glutathione–agarose beads (20 µl of a 50% slurry) were equilibrated in 0.5× Superdex buffer [12.5 mM HEPES (pH 7.9), 6.25 mM MgCl2, 5 µM ZnSO4, 75 mM KCl, 20% [vol/vol] glycerol, 0.05% Nonidet P-40, 0.5 mM EDTA, 1 mM DTT and 1 mM PMSF] and combined with GST fusion proteins for 1 h at 4°C. Bead–protein complexes were washed twice with 0.5× Superdex buffer. HBZ was prepared by in vitro transcription/translation using the TNT translation system (Promega) in the presence of [35S] methionine according to the manufacturer’s protocol. Radiolabeled HBZ was combined with bead-protein complexes, and reactions were incubated overnight at 4°C. Beads were washed four times with 0.5× Superdex buffer, and bound proteins were eluted with SDS sample dye and resolved by SDS–PAGE. HBZ was detected using a Typhoon 9410 Imager and ImageQuant TL software (GE Healthcare).
In vitro HAT assays
Recombinant proteins were combined in HAT buffer [50 mM Tris (pH 8), 10% glycerol, 15 µM valproate, 1 mM DTT, 1 mM PMSF and 5 µM acetyl-CoA] and incubated at 30°C for 60 min. Proteins were resolved by SDS–PAGE (14% gels or 4–20% gradient gels), and acetylation was detected by western blot using ECL Plus (GE Healthcare). Nitrocellulose filters were scanned with a Typhoon 9410 Imager (GE Healthcare) and analyzed using ImageQuant TL software. Curcumin (D3420) was purchased from LKT Laboratories Inc. and reconstituted with DMSO prior to each use. Protein concentrations are indicated in the figure legends.
Cell culture, transfections and luciferase assays
HeLa-HBZ (41), HeLa-pcDNA (41) and HeLa A57 (plasmid 3Enh-κB-conA-Luc integrated, 48) cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, 50 μg/ml streptomycin and 0.5 mg/ml G418. T-cell lines and HTLV-1 infected cells were cultured in Iscove’s modified Dulbecco medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine and penicillin–streptomycin. Jurkat stable cell lines were established by electroporation of pMACS Kk.II (Miltenyi Biotec) and pcDNA-HBZ-SP1-Myc (36) or pcDNA3.1 (Invitrogen), followed by pMACS purification of transfected cells (41) and selection with 1.5 mg/ml G418. Jurkat-HBZ clonal cell lines were obtained by initiating cultures of <1 cell/well in a 96-well plate. For transient expression of E1A or HBZ, HeLa cells were electroporated and transfected cells were pMACS-purified as described (49). For immunofluorescence analysis, 1.2 × 105 HeLa cells were seeded onto coverslips one day prior to transfection. Cells were transfected with 4 μg of expression plasmid using Turbofect (Fermentas) according to the manufacturer’s instructions. Cells were fixed 24 h after transfection. Luciferase assays were performed using Jurkat cells as described (40). Firefly luciferase activity was normalized to Renilla luciferase activity from pRL-TK-Luc (Promega).
Cell extracts
Core histones were extracted from the indicated cell lines using the EpiQuik Total Histone Extraction Kit (Epigentek) with buffers containing 400 nM of trichostatin A (TSA; Cayman Chemical). For the experiment shown in Figure 3E, equal quantities of cells were directly resuspended in SDS-loading dye. To prepare nuclear extracts, cells were resuspended in hypotonic buffer [20 mM HEPES (pH 7.6), 20% glycerol, 10 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.1% Igepal, 400 nM TSA, 2 μg/ml leupeptin, 5 μg/ml aprotinin and 1 mM PMSF], ice-chilled for 10 min, and centrifuged at 800 g and 4°C for 5 min. Pelleted nuclei were lysed in RIPA buffer (50 mM Tris [pH 8], 1% Triton X-100, 100 mM NaCl, 1 mM MgCl2, 400 nM TSA, 2 μg/ml leupeptin, 5 μg/ml aprotinin, 1 mM PMSF and 1 mM benzamidine) for 15 min on ice. Lysates were centrifuged at 15 000 g and 4°C for 15 min. Supernatants were aliquoted and stored at −80°C. Whole-cell extracts were prepared with RIPA buffer as described above. Immunoprecipitations were performed as published (39). Etoposide was purchased from Sigma and MG132 from EMD.
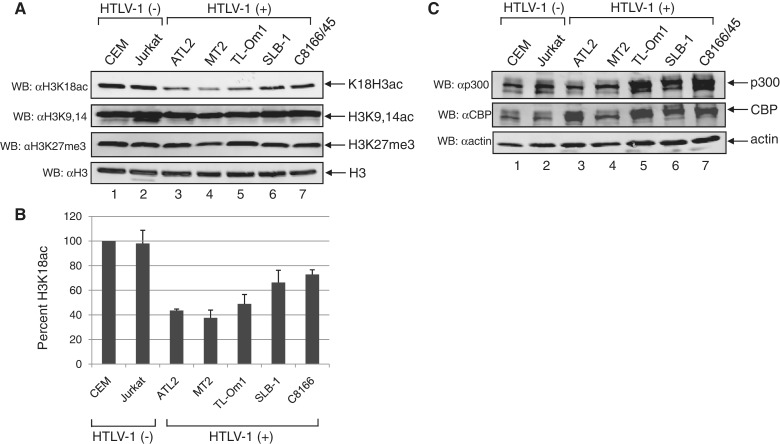
Figure 3.
HBZ reduces H3K18ac in cells. (A) The level of H3K18ac is reduced in HeLa cells expressing HBZ compared to control cells. HeLa cell lines carrying the empty or the HBZ expression vector, as indicated, were analyzed by indirect immunofluorescence confocal microscopy (magnification 40×) to compare levels of H3K18ac. (B) A graphical representation of immunofluorescence data. The mean signal density per cell for H3K18ac or H3 was determined for HBZ-expressing or control cells. Data are the average values (± S.E.M.) obtained from >100 cells. The asterisk denotes a P-value <0.0001 for a two-tailed Student t-test. (C) Jurkat cell lines were established that express HBZ with Myc and 6×His epitope tags or carry the empty vector. HBZ was immunoprecipitated from whole-cell extracts using an antibody against the 6×His tag and then detected by western blot using an antibody against the Myc tag. (D) HBZ is localized to the nucleus in Jurkat cells stably expressing HBZ. The indicated Jurkat cells were analyzed by indirect immunofluorescence confocal microscopy (magnification ×100/zoom × 2) for HBZ expression. NUP98 was stained to mark the nuclear membrane. (E) The level of H3K18ac is reduced in Jurkat cells expressing HBZ compared to control cells. The indicated Jurkat cell lines were analyzed by immunofluorescence confocal microscopy (magnification ×100/zoom × 2) to compare levels of H3K18ac. (F) Histones extracted from HeLa and Jurkat cells stably expressing HBZ exhibit less H3K18ac than control cells. Histones were resolved by SDS–PAGE and analyzed by western blot using the antibodies indicated. (G) Transient expression of HBZ reduces H3K18ac. Histones (2 µg) from HeLa cells transfected with E1A, HBZ or the empty pcDNA vector were analyzed by western blot with the antibodies indicated.
Indirect immunofluorescence analysis
HeLa cells were seeded onto coverslips one day prior to staining. Jurkat cells were allowed to adhere to Poly-d-Lysine (Sigma) coated coverslips for 45 min prior staining. Transfected HeLa cells were stained 24 h after transfection. Cells were washed twice with PBS (137 mM NaCl, 2.7 mM KCl, 3 mM Na2HPO4 and 1.5 mM KH2PO4), fixed with 4% paraformaldehyde at room temperature for 15 min, washed three times with PBS, and permeabilized with PBS/0.1% Triton X-100/0.3% BSA for 10 min. Antibodies were diluted in PBS/0.1% Triton X-100/0.3% BSA and incubated with coverslips overnight at 4°C. Coverslips were washed three times with PBS then incubated with secondary goat anti-rabbit Alexa Fluor 488 or 633, or goat anti-mouse Texas Red conjugated antibodies (Invitrogen) at 37°C for 1 h. Coverslips were then washed three times with PBS and DNA was counterstained with TOPRO3 (Molecular Probes/Invitrogen) at 37°C for 30 min. Excess TOPRO3 was removed with two PBS washes, and coverslips were mounted onto microscope slides using Prolong Antifade Gold (Invitrogen) according to the manufacturer’s instructions. Flag-p300 was detected by additional incubation of coverslips at 37°C for 1 h with the FITC-conjugated Flag antibody diluted in PBS/0.1% Triton X-100 followed by three washes with PBS. Fluorescence images were acquired by confocal microscopy as indicated either at a magnification of 40× or 100× using a LSM 510 confocal microscope (Zeiss), and images were acquired using Zen software (Zeiss). In Figure 3B, Image-Pro Plus (version 4.5) software (Media Cybernetics) was used to calculate the mean density of the fluorescence signal for individual cells. The mean densities for more than 100 cells for each antibody/cell line were averaged and analyzed using a two-tailed Student t-test.
Chromatin immunoprecipitation assay
HeLa A57 cells were electroporated as described (50). Twenty four hours after electroporation cells were treated with 5 µM MG132 (EMD) and harvested 5 h later. Chromatin immunoprecipitation (ChIP) assays were performed as described (51) using 5 μg of Flag antibody for immunoprecipitation. Coimmunoprecipitated DNA was purified using Chelex-100 (BioRad) as described (52). Real-time PCR was performed and data were analyzed as described (50). The following primers were used: 3kB-CONA-F, ATCGGTGATGTCGGCGATATAGG; 3kB-CONA-R, CCTGGCTGTGTTTGCAGAAGCAAT; VCAM1kB-F, TGGAACTTGGCTGGGTGTCTGTTA; VCAM1kB-R, TATTTGTGTCCCACCTGTGTGTGC; VCAM1+2644F, GCAATGGCCACGTGAAGTAGTGTA; VCAM1+2644R, TGCCTCTGACAGGAATTCATCCAC.
RESULTS
HBZ interacts with the p300/CBP HAT domain in vitro
We previously showed that HBZ directly interacts with three domains in p300/CBP: the KIX domain, the HAT domain, and the C/H3 domain (39). The HAT and C/H3 domains are adjacent, partially overlapping domains that share a common zinc-binding sub-domain defined as the ZZ domain (43) (Figure 1A). The C/H3 region also contains a unique sub-domain known as TAZ2 that folds independently (53) of, and does not interact with the ZZ domain (54). While the functional significance of the ZZ domain is unclear, TAZ2 serves as docking site for a number of transcription factors (55–59). To identify the precise HBZ-binding sites within the HAT and C/H3 domains, we performed GST pull-down assays with HBZ and various truncations of the HAT and C/H3 domains. We used the splice 1 isoform of HBZ in all experiments, which is the major isoform expressed in HTLV-1-infected cells (60–62). In these binding assays, GST was fused to the p300/CBP polypeptides, including the HAT domain with a C-terminal truncation that also lacked ZZ (GST-N-HAT), the HAT domain with an N-terminal truncation (GST-C-HAT), and a polypeptide encompassing only the TAZ2 domain (GST-TAZ2) (Figure 1A). We confirmed specific binding of HBZ to the HAT domain (Figure 1B, lane 3); however, this interaction was dramatically reduced using N-HAT (lane 4). The interaction was similarly reduced with ΔC-HAT, which lacks the ZZ domain (Figure 1C, lane 5). In contrast, HBZ bound specifically to C-HAT, suggesting that HBZ targets the C-terminal region of the HAT domain (Figure 1C, lane 4). Surprisingly, HBZ also bound independently to the TAZ2 domain (lane 6).
Figure 1.
HBZ binds the HAT domain of p300/CBP. (A) Schematic representation of CBP and the CBP fragments used in GST pull-down assays. (B) C-terminal truncation of the HAT domain disrupts HBZ-binding. HBZ was incubated with 10 pmol of GST, GST-HAT or GST-N-HAT. Proteins retained on glutathione beads were analyzed by SDS–PAGE and autoradiography. (C) HBZ interacts with a region of p300/CBP encompassing the C-terminal portion of the HAT domain and the ZZ domain, and separately with the TAZ-2 domain. HBZ was incubated with 10 pmol of the indicated GST fusion proteins. Proteins retained on the glutathione beads were analyzed by SDS–PAGE and autoradiography.
HBZ inhibits histone acetylation by p300/CBP in vitro
The observation that HBZ binds to the HAT domain of p300/CBP prompted us to test whether HBZ affects this enzymatic activity of the coactivator, first using in vitro HAT assays. For these assays recombinant, purified proteins were combined with acetyl–CoA, and protein acetylation was evaluated by western blot with an antibody against acetylated lysine. Recombinant proteins used in these assays are shown in Supplementary Figure S1. Using the core histones as substrates for acetylation, we found that GST-HBZ caused a dramatic reduction in p300 HAT activity, while GST alone did not produce such an effect (Figure 2A). In this experiment, the histones were detected as a single band due to the gel-migration conditions. To determine whether HBZ inhibits acetylation of both histones H3 and H4, western blots were also probed for total H4 (Figure 2B). As expected, HBZ inhibited histone H4 acetylation. In these assays detection of histone H4 by western blot was reduced by acetylation; however, SYPRO Ruby-staining confirmed the presence of equivalent histone quantities in each reaction. In agreement with the fact that the HAT domains of p300 and CBP are highly conserved (>90% identity) (63), HBZ also inhibited CBP-mediated acetylation of histones (Figure 2C, lane 5). In addition to the histones, p300 and CBP were autoacetylated; however, autoacetylation was not significantly inhibited by HBZ. This trend is similar for other proteins known to inhibit HAT activity such as E1A (64).
Figure 2.
HBZ inhibits acetylation of histones by p300/CBP. (A) HBZ inhibits p300 HAT activity. HAT assays were performed using recombinant histones (2 µM), p300 (4 nM), GST (0.4 µM) and GST-HBZ (0.24 µM) as indicated. Reactions were analyzed by western blot using antibodies against acetylated lysine and histone H3 as indicated. (B) HBZ inhibits acetylation of histone H3 and H4. HAT assays were performed using recombinant histones (2 µM), p300 (4 nM) and GST-HBZ (0.3 µM) as indicated. Reactions were analyzed by western blot using antibodies against acetylated lysine, histone H3 and histone H4 as indicated. Proteins from a portion of each reaction were also stained with SYPRO Ruby (bottom panel). (C) HBZ inhibits CBP HAT activity. HAT assays were performed with histones (2 µM), p300 (4 nM) or CBP (4 nM), and GST-HBZ (0.24 µM) as indicated. (D) HBZ inhibits histone acetylation by the HAT domain alone. HAT assays were performed with histones (2 µM), p300 or GST-HAT (0.06 µM) and GST (0.4 µM) or GST-HBZ (0.24 µM) as indicated. Each lower panel shows the membrane from the middle panel reprobed for histone H3.
A small group of proteins have been shown to repress p300/CBP HAT activity though their interactions with the C/H3-TAZ2 domain (65,66). To test whether this property applies to HBZ, we substituted the full-length coactivators with GST-HAT, which lacks TAZ2. Interestingly, HBZ retained the ability to repress histone acetylation mediated by the HAT domain alone (Figure 2D), suggesting that the interaction between HBZ and C-HAT is sufficient to inhibit p300/CBP HAT activity. In these assays the ZZ domain appeared to be important for HAT activity, as deletion of this domain (GST-ΔHAT) severely impaired acetylation (4).
HBZ inhibits histone H3K18 acetylation, a p300/CBP HAT target, in cells
To test whether HBZ represses p300/CBP HAT activity in vivo, we first compared levels of H3K18ac in HeLa clonal cell lines that express HBZ or carry the empty pcDNA expression vector (41). H3K18 is known to be a specific target of p300/CBP HAT activity in vivo (12,67,68). Relative levels of H3K18ac were determined by indirect immunofluorescence microscopy, using an antibody against the modification for staining (green). In addition, cellular DNA was stained with TOPRO3 (blue) and HBZ was stained with an antibody against its C-terminal 6× histidine tag (red). In experiments H3K18ac was monitored with the same laser intensity between cells lines in order to directly compare levels of the modification. Figure 3A shows that H3K18ac is attenuated in the cells expressing HBZ. Quantification of the data shows that the level of H3K18ac is 3-fold lower in these cells compared to the cells with the empty vector (Figure 3B).
Because T-cells represent a major target of HTLV-1 infection, we established Jurkat cell lines that stably express HBZ or carry the empty vector (Figure 3C). As in other cells, HBZ was distributed throughout the nucleus as evident by staining within the perimeter of the nucleus, which was marked by staining of NUP98, a component of the nuclear pore complex (69) (Figure 3D). Similar to HeLa cells, the Jurkat cells expressing HBZ exhibited less H3K18ac than the cells without the viral protein (Figure 3E). Interestingly, the pattern of H3K18ac staining diverged between HeLa and Jurkat cells, with a diffuse signal detected throughout the nucleus of HeLa cells and a signal that was more pronounced at the nuclear periphery in Jurkat cells. The latter pattern of H3K18ac staining was observed using another T-cell line, HUT78 (Supplementary Figure S2), suggesting that this phenomenon occurs specifically in T-cell lines. The same laser intensity was used to compare HBZ negative and positive cells, producing an appearance of almost complete loss of H3K18ac in HBZ-expressing Jurkat cells. However, with higher laser intensity, a more substantial signal was detected (data not shown), suggesting HBZ reduces but does not completely eliminate H3K18ac in these cells.
Microscopy results were confirmed by western blot analysis of H3K18ac levels following cellular extraction of histones. Using this approach we again detected less H3K18ac from the clonal HeLa and Jurkat cells stably expressing HBZ than from the cells with the empty vector (Figure 3F, top panels). Quantification of the western-blots showed 1.5- and 2-fold reductions in H3K18ac in HeLa and Jurkat cells, respectively. We also noted that the level of H3K56ac, another p300/CBP modification, was also diminished in HBZ-expressing Jurkat cells (Supplementary Figure S3). In contrast, dual acetylation of lysines 9 and 14 on histone H3 (H3K9,14ac) remained constant between cells lacking or expressing HBZ (Figure 3F, middle panels). This histone mark arises from the redundant functions of multiple proteins that carry HAT activity, and therefore, does not explicitly involve p300/CBP (12,70).
Interestingly, H3K18ac was reduced to a similar extent in HeLa and Jurkat cells expressing HBZ even though the Jurkat cell line expressed less HBZ (Supplementary Figure S4A). Our data indicate that this observation is partly due to lower levels of p300 and CBP in Jurkat cells compared to HeLa cells (Supplementary Figure S4A). Furthermore, Jurkat cells are known to carry a frame-shift mutation in the sequence encoding the HAT domain of one CREBBP allele (21), which is expected to reduce the enzymatic activity of CBP. The activity of p300 may be similarly reduced, as we observed less acetylation using p300 immunoprecipitated from Jurkat than from HeLa cells in in vitro HAT assays (Supplementary Figure S4B). In these experiments we used saturating quantities of extract to ensure equal immunoprecipitation of p300. Similar to results using recombinant proteins, immunoprecipitates from HBZ-expressing cell lines exhibited less HAT activity than their control-cell counterparts. Finally, it is important to note that HeLa cells are infected with human papillomavirus, which expresses the viral protein E6. Because E6 has been reported to bind the C/H3 domain of p300/CBP and inhibit HAT activity, it may limit the accessibility and the effect of HBZ on the coactivator (71–73). Therefore, although the cellular level of HBZ is important, it is likely that repression of HAT activity by HBZ is also dictated by the ratio between the viral protein and active and/or accessible p300/CBP.
We also transiently transfected HeLa cells with the HBZ expression vector to show that the reduction in H3K18ac was due to HBZ and not a non-specific event arising during the creation of the clonal cell lines (the typical level of HBZ after transfection is shown in Supplementary Figure S5). As a positive control for the H3K18ac decrease, cells were separately transfected with an expression vector for the adenovirus protein, E1A, which is known to directly inhibit p300 HAT activity (64,74,75). As with constitutive expression of HBZ in the clonal cell lines, transient expression of HBZ also led to a decrease in H3K18ac, while no apparent change in the level of H3K9,14ac was observed (Figure3G). However, HBZ did not reduce H3K18ac to the same extent as E1A.
To correlate the reduction in H3K18ac with inhibition of p300 HAT activity by HBZ, we tested whether overexpression of p300 would restore H3K18ac in cells expressing HBZ. An expression vector for p300 carrying a C-terminal Flag epitope tag (p300-Flag) was transiently transfected into HeLa cells stably expressing HBZ. Indirect immunofluorescence was then used to identify cells expressing p300-Flag (green) and to assess relative levels of H3K18ac (blue). Figure 4 shows that overexpression of p300 augmented H3K18ac in both cells expressing HBZ and cells carrying the empty vector. Similar levels of H3K18ac were observed in HBZ-expressing cells with p300-Flag and in untransfected cells lacking HBZ. This result indicates that the increased abundance of p300 overwhelmed the ability of HBZ to inhibit HAT activity and reestablished H3K18ac. In contrast to wild-type p300, overexpression of p300 carrying a deletion in the HAT domain did not restore H3K18ac in the presence of HBZ (data not shown).
Figure 4.
Overexpression of p300 restores H3K18ac in HeLa cells stably expressing HBZ. Immunofluoresence confocal microscopy (magnification ×100/zoom × 1) was used to detect ectopic expression of p300 (denoted by arrows) and to compare levels of H3K18ac in the indicated HeLa cell lines.
HTLV-1-infected T-cell lines exhibit low levels of H3K18ac
Based on the cellular reduction in H3K18ac by HBZ, we were interested in evaluating levels of this modification in HTLV-1-infected cells. We therefore extracted histones from five infected T-cell lines and two uninfected T-cell lines, and performed a western blot analysis of H3K18ac. Interestingly, levels of H3K18ac appeared lower in all of the HTLV-1-infected cells compared to the uninfected T-cells (Figure 5A, upper panel). In contrast, levels of H3K9,14ac and H3 lysine 27 methylation (H3K27me3) remained relatively constant (Figure 5A). Quantification of the H3K18ac relative to total H3 revealed that ATL-2, MT-2 and TL-OmI contained the lowest levels of H3K18ac (Figure 5B). Importantly, the combined abundance of p300 and CBP in infected cell lines was equal to or greater than the combined abundance of these proteins in the uninfected cell lines (Figure 5C), indicating that the reduction in H3K18ac was not due to lower levels of p300/CBP. Our previous data show that, among the cell lines tested, HBZ expression is highest in ATL-2 and MT-2 cells (41). These results suggest that HBZ inhibits p300/CBP HAT activity in the context of HTLV-1 infection.
Figure 5.
The level of H3K18ac is reduced in HTLV-1-infected cells. (A) Infected cell lines contain lower levels of H3K18ac than uninfected T-cell lines. Histones (400 ng) extracted from five HTLV-1-infected cell lines and two uninfected T-cell lines were analyzed by western blot with the antibodies indicated. The lower panel shows the membrane from the top panel reprobed for histone H3. (B) A graphical representation of the western blot data. The band density for H3K18ac was divided by that for H3, and values were normalized to that for the CEM cell line, which was set to 100%. Data are the average (±S.E.M.) of three independent extractions from CEM, Jurkat and MT2 cells, and two independent extractions from the other cell lines. (C) The combined levels of p300 and CBP in the HTLV-1-infected T-cell lines are equal to or greater than those in the uninfected cell lines. Levels of p300 and CBP in 50 µg of whole-cell extracts were analyzed by western blot.
The bZIP domain of HBZ is sufficient for inhibition of p300/CBP HAT activity
We were next interested in determining which region of HBZ mediates the inhibition of p300/CBP HAT activity. Our previous data showed that two LXXLL motifs in the activation domain of HBZ form a strong interaction with the coactivator KIX domain, but are not necessary for binding to the HAT domain (39,40). Based on those observations, we tested whether an HBZ mutant with LL to AA substitutions in both LXXLL motifs (GST-HBZ-MutAD) retained the ability to inhibit p300/CBP HAT activity. This mutant is known to exhibit a defect in binding to the KIX domain (39,40). Figure 6A shows that GST-HBZ-MutAD inhibited p300 HAT activity to a similar extent as the wild-type viral protein in vitro (compare lanes 4 and 5), suggesting that the LXXLL motifs in HBZ are not involved in inhibition.
Figure 6.
The bZIP domain of HBZ is responsible for inhibition of p300/CBP HAT activity. (A) Mutation of the LXXLL motifs in HBZ does not impair inhibition of HAT activity. HAT assays were performed using recombinant histones (2 µM), p300 (4 nM) and GST (0.3 µM), GST-HBZ (0.25 µM) or GST-HBZ-MutAD (0.45 µM) as indicated. (B) The bZIP domain of HBZ inhibits HAT activity. A schematic of the HBZ constructs tested in HAT assays. Asterisks denote the positions of the LXXLL motifs. HAT assays were performed using recombinant histones (2 µM), p300 (4 nM) and either GST (0.7 µM), GST-HBZ (0.24 µM), HBZ1–122 (2.3 µM) or HBZ-bZIP (2.6 µM) as indicated. (C) Inhibition of HAT activity is retained with different purified preparations of the bZIP domain of HBZ. HAT assays were performed using recombinant histones (2 µM), p300 (4 nM) and either GST-HBZ (0.24 µM), GST-HBZ-bZIP (0.33 µM) or one of three different HBZ-bZIP preparations (1 µM) as indicated. (D) Inhibition of HAT activity by HBZ-bZIP is blocked by the bZIP domain of c-Jun. HAT assays were performed using recombinant histones (2 µM), p300 (4 nM) and either HBZ-bZIP (0.3 µM), c-Jun-bZIP (0.3 µM), or HBZ-bZIP preincubated with c-Jun-bZIP (0.3 µM of each) as indicated. (E) The bZIP domain of HBZ does not function as a deacetylase. HAT assays were performed using recombinant histones (2 μM), p300 (4 nM) and HBZ-bZIP (0.3 μM) as indicated. In lane 5, HBZ-bZIP was added to the post-acetylation reaction, which was incubated for an additional 30 min. In all experiments reactions were analyzed by western blot using the antibodies indicated, and each lower panel shows the membrane from the middle panel reprobed for histone H3.
We then compared the inhibitory effects of two truncation mutants of HBZ. The first mutant consists of the activation domain and the two central basic regions of HBZ, BR1 and BR2 (1–122 amino acids), and the second encompasses only the bZIP domain (amino acids 120–206) (Figure 6B). In an in vitro HAT assay, the bZIP domain blocked histone acetylation, while HBZ1–122 did not (Figure 6B, compare lanes 4 and 5). These results indicate that the bZIP domain of HBZ is responsible for inhibition of p300/CBP HAT activity.
To corroborate this finding, we tested three separate protein preparations of the bZIP domain. HBZ-bZIP#1 and HBZ-bZIP#3 were prepared by distinct heat treatments of the cellular lysate followed by heparin-agarose chromatography. HBZ-bZIP#2 was purified using Ni2+-affinity chromatography. Like full-length HBZ, each preparation of the bZIP domain inhibited histones acetylation (Figure 6C, lanes 3–5). However, inhibition was blocked by fusion of GST to the N-terminus of the bZIP domain (Figure 6C, Lane 2). Interestingly, loss of inhibition also occurred through the formation of a heterodimer between the bZIP domain the HBZ and that of c-Jun (Figure 6D). These latter experiments imply that the GST fusion and the bZIP domain of c-Jun obstruct binding site on HBZ that is required for the interaction with the HAT domain of p300/CBP.
It was also necessary to evaluate the bZIP domain of HBZ for histone deacetylase activity. Accordingly, we performed a HAT assay in which the core histones were acetylated by p300 prior to the addition of the HBZ bZIP domain to the reaction (Figure 6E, lane 5). As expected, this approach did not lead to the reduction in histone acetylation that was observed when the bZIP domain was added at the initial stage of the reaction (Figure 6E, lane 4). This result confirms that the bZIP domain does not carry deacetylase activity.
HBZ is more potent than curcumin for inhibition of p300/CBP HAT activity in vitro
To gauge the efficiency with which HBZ inhibits histone acetylation, we compared its inhibitory effect to that of curcumin, which is known to specifically target p300/CBP HAT activity (76). We performed a parallel set of in vitro HAT assays, in which reactions were supplemented with increasing concentrations of full-length HBZ, HBZ-bZIP, or curcumin. Results from these experiments show a reduction in histone acetylation at lower concentrations of full-length HBZ or HBZ-bZIP than curcumin (compare Figure 7A and B to C). Quantification of the data revealed an IC50 of 0.1 µM for full-length HBZ and the bZIP domain and an IC50 of 25 µM for curcumin (Figure 7D). Importantly, the curcumin value is similar to that reported previously (76). These results demonstrate that HBZ is a more potent inhibitor of p300/CBP HAT activity than curcumin. Figure 7A shows acetylation of HBZ by p300. This modification was consistently detected with the full-length viral protein in in vitro HAT assays (Figure 8 and data not shown); however, its significance remains undefined. Importantly, acetylation of HBZ is not inconsistent with its effect on HAT activity, as E1A is similarly acetylated by p300, while functioning to inhibit the HAT activity of the coactivator (64,74,75).
Figure 7.
HBZ is a potent inhibitor of p300/CBP HAT activity in vitro. HAT assays were performed using recombinant histones (2 µM, lanes 1–7), p300 (4 nM, lanes 2–7) and increasing concentrations of GST-HBZ [(A), HBZ-bZIP (B), or curcumin (C)]. Reactions were analyzed by western blot using the antibodies indicated. In each lower panel, the membrane showing acetylated histones was reprobed for histone H3. (D) The graph shows the decrease in histone acetylation in response to increasing concentrations of GST-HBZ, HBZ-bZIP or curcumin. Western blot data were used to quantify levels of histone acetylation and data were plotted relative to the concentration of the inhibitor. The graph shows data averaged from two independent experiments.
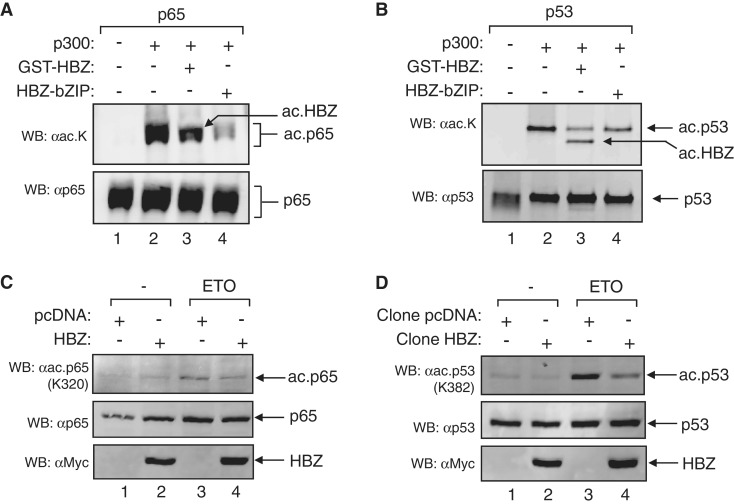
Figure 8.
HBZ inhibits p65 and p53 acetylation by p300/CBP. (A) HBZ inhibits p65 acetylation by p300 in vitro. HAT assays were performed using recombinant p65 (80 nM), p300 (4 nM) and GST-HBZ (0.3 µM) or HBZ-bZIP (0.3 µM) as indicated. (B) HBZ inhibits p53 acetylation by p300 in vitro. HAT assays were performed using recombinant p53 (50 nM), p300 (4 nM) and GST-HBZ (0.5 µM) or HBZ-bZIP (0.5 µM) as indicated. In (A) and (B), the lower panel shows the membrane from the top panel reprobed for total p65 and p53, respectively. Reactions were analyzed by western blot using the antibodies indicated. (C) HBZ inhibits p65 acetylation in vivo. Nuclear extracts (40 µg) were prepared from HeLa cell lines expressing HBZ or containing the empty pcDNA vector. Cells used in these experiments were stimulated with 50 µM etoposide (ETO) for 5 h. (D) HBZ inhibits p53 acetylation in vivo. Experiments were performed as described in (C). In (C) and (D) the middle panel shows the membrane from the upper panel reprobed for total p65 and p53, respectively. Extracts were analyzed by western blot using the antibodies indicated.
HBZ inhibits the acetylation of transcription factors by p300/CBP
Certain proteins known as INHATs (inhibitors of histone acetyltransferase activity) inhibit histone acetylation by binding to histone tails and preventing p300/CBP from accessing these substrates (77–79). Our in vitro results indicated that HBZ did not function in this manner, as concentrations of HBZ lower than that of the histones were capable of inhibiting acetylation. To provide further evidence that HBZ targets the HAT domain and not the histone tails, we used HAT assays to analyze acetylation of the NF-κB transcription factor, p65, and the tumor-suppressor transcription factor, p53. These proteins are known substrates of p300/CBP HAT activity: p65 is acetylated specifically by p300/CBP at K320 (80), and p53 is acetylated at K382 (81,82). The overall effect of these modifications is to increase the transcriptional activity of the factors (80–82). We found that both full-length HBZ and HBZ-bZIP efficiently inhibited p300-mediated acetylation of p65 in vitro (Figure 8A). A similar effect was observed with p53 (Figure 8B), confirming that HBZ inhibits the HAT activity of p300/CBP through its interaction with the HAT domain. In our experiments, HBZ consistently induced only a modest reduction in p53 acetylation, with the full-length viral protein serving as a more potent inhibitor than the bZIP domain. These observations may stem from the ability of p53 to contact several domains of p300/CBP, including the KIX and TAZ2 domains that are targeted by HBZ (57,83). Because the bZIP domain of HBZ does not form a high-affinity interaction with the KIX domain, it may be less effective in competing with p53 for binding to p300/CBP than full-length HBZ. As shown in Figure 7 above, p300 also acetylated HBZ in these assays.
We also evaluated levels of p65 and p53 acetylation in HeLa cells stably expressing HBZ or carrying the empty pcDNA expression vector. Cells were treated with etoposide to induce acetylation of p65 or p53, as etoposide causes genotoxic stress, thereby activating p53 and NF-κB signaling (84,85). The modified factors were detected in nuclear extracts by western blot using antibodies against the p65 K320 and p53 K382 acetylation marks. Following treatment with etoposide, we observed lower levels of p65 acetylation in the cells expressing HBZ compared to the cells carrying the empty vector (Figure 8C). Similarly, in etoposide-treated cells, p53 acetylation was diminished in the presence of HBZ (Figure 8D). These data further support a role for HBZ in the inhibition of p300/CBP HAT activity in vivo.
We further explored the effect of HBZ on p65 acetylation. HBZ was recently reported to inhibit the transcriptional activity of p65 by stimulating the degradation of this factor (86). To determine whether the reduction in acetylation of p65 by HBZ also participates in this repression, we performed luciferase assays using Jurkat cells transiently transfected with the pNF-kB-Luc reporter plasmid. Cotransfection of an expression vector for p65 significantly increased luciferase activity (Figure 9A, lane 2), and as reported, additional cotransfection of HBZ abrogated this effect (Figure 9A, lane 3) as well as reduced the level of p65 (Figure 9B, lane 3). Interestingly, HBZ retained the ability to repress luciferase activity in cells treated with the proteosome inhibitor, MG132 (Figure 9A), despite stabilization of p65 (Figure 9B, lane 7). Furthermore, cotransfection of p300 partially restored luciferase activity. To directly assess DNA binding by p65, we performed ChIP assays using HeLa cells carrying integrated copies of a reporter construct regulated by a promoter with three κB sequences (48). We found that HBZ reduced enrichment of p65 at this site in the presence of MG132 (Figure 9C). Because cells were treated with MG132, levels of p65 were similar in the presence and absence of HBZ (Figure 9D). Similar results were observed at the endogenous VCAM-1 promoter that is activated by NF-κB and was previously found to be repressed by HBZ (86). Enrichment of p65 was significantly lower at a site downstream of the VCAM-1 promoter. Overall, these results suggest that inhibition of p65 acetylation by HBZ contributes to repression of the classic NF-kB pathway.
Figure 9.
HBZ reduces p65 transcriptional activity. (A) Jurkat cells were transfected with pNF-κB-Luc (100 ng) alone or in combination with Flag-p65 (50 ng), pcDNA-HBZ (400 ng) or pCI-p300 (400 ng) as indicated. Twenty four hours after transfection, one set of cells was treated with MG132 (5 µM) for 5 h. The reported values are the average luminescence ± SE from one experiment performed in duplicate and are representative of four independent experiments. (B) Whole-cell extracts from transfected Jurkat cells were resolved by SDS–PAGE, and proteins indicated were detected by western blot. Asterisks denote non-specific bands. (C) HeLa A57 cells were electroporated with Flag-p65 (4 µg) alone or in combination with pcDNA-HBZ (16 µg); cells were treated with MG132 (5 µM) 5 h prior to ChIP assays. Real-time PCR was used to quantify levels of Flag-p65-enrichment at the synthetic NF-κB sequence, the VCAM-1 promoter, and within the VCAM-1 gene (position +2644 relative to start site) by comparing amplification of coimmunoprecipitated DNA with that of a fraction of the total input DNA. The graph shows the average fold enrichment (± SE) relative to cells electroporated with pcDNA3.1 (set to 1) from two independent ChIP assays. (D) Flag-p65 and HBZ are expressed in transfected cells. Western blot analysis was performed on whole-cell extracts from electroporated cells treated with MG132 as above. The membrane was probed with the indicated antibodies.
DISCUSSION
In this study we found that HBZ inhibits p300/CBP HAT activity by binding directly to the coactivator HAT domain. This is one of three conserved domains within p300/CBP that is contacted directly by HBZ. The others are the TAZ2 domain, shown in this study, and the KIX domain (39). Interestingly, like HBZ, the viral transactivator Tax, also targets multiple p300/CBP domains, including the KIX, C/H1 and CR2 domains (87–89). Therefore, modulation of p300/CBP activity by HTLV-1 appears to play a prominent role in viral infection. Although HBZ is capable of inhibiting p300/CBP HAT activity in vivo, our previous data suggest that HBZ has a lower affinity for the HAT domain than it does for the KIX domain (39). Such properties are not unique, as E1A utilizes one domain to bind stably to the TRAM motif within the C/H3 domain and inhibits HAT activity though a separate, weaker interaction (74,90).
In vitro, the bZIP domain of HBZ was necessary and sufficient for inhibition of HAT activity. Results from previous studies demonstrate that this domain allows HBZ to form heterodimers with certain cellular bZIP transcription factors within the ATF/CREB and AP1 families and modulate their activity (34,35,37,38). It is likely that repression of p300/CBP HAT activity and interaction with bZIP factors are mutually exclusive functions of HBZ, as we found that c-Jun-bZIP blocks inhibition of acetylation by HBZ. Similar non-overlapping functions are associated with the DNA-binding domain of early B-cell factor, which directly represses acetylation by p300/CBP in solution, but not when the factor is bound to DNA (66). Repression of HAT activity by the bZIP domain of HBZ may be dependent on the N-terminal activation domain of the viral protein in vivo. This premise is based on a model in which the LXXLL motifs in the activation domain of HBZ form a high-affinity interaction with the coactivator KIX domain, potentially placing the C-terminal bZIP domain in a favorable position to repress HAT activity. Future experiments will clarify such aspects of the HBZ-p300/CBP complex.
Within cells, we found that direct repression of p300/CBP HAT activity by HBZ leads to a global reduction of H3K18ac. Strikingly, low H3K18ac was also observed in T-cells chronically infected with HTLV-1. H3K18 is known to be specifically targeted for acetylation by p300/CBP (12,67). Interestingly, reductions of this and a subset of other histone modifications have been documented in a variety of cancers and are frequently correlated with poorer clinical outcomes (22). Therefore, such epigenetic changes may play a role in transformation. With respect to H3K18ac, the HAT activity of CBP and, to a lesser extent p300, were recently found to be impaired in a significant number of diffuse large B-cell lymphoma and follicular lymphoma cases through mutations and/or monoallelic deletions in their genes (18). CBP was similarly affected in a number of relapsed acute lymphoblastic leukemia cases (21).
The immediate effects of disrupted p300/CBP HAT activity relating to transformation are not fully defined. One important substrate of p300/CBP is the tumor suppressor p53 (81), which is inactive in HTLV-1-transformed cells through mechanisms reported to involve the viral protein Tax (91–95). Acetylation of p53 is essential for its ability to block cell proliferation and induce apoptosis in response to genotoxic stress that would otherwise lead to the accumulation of mutations and potentially transformation (96). We found that HBZ reduces p53 acetylation in vitro and following genotoxic stress, suggesting that HBZ may contribute to the inactivation of this factor in HTLV-1-infected cells. In addition to p53, HBZ inhibited p65 acetylation by p300/CBP, also an activating modification (80). Recently, HBZ was shown to repress the classical NF-kB pathway by targeting p65 (86). Although attributed to protein turnover, we found that reduced acetylation of p65 also contributes to this effect.
The transforming viral proteins e1A (splice variant of E1A) and SV 40 large T antigen are also capable of reducing the global level of H3K18ac (67). For E1A, this effect is correlated with direct inhibition of HAT (64,74,75). Interestingly, transformation of mouse embryonic fibroblasts by E1A requires its interaction with p300/CBP (97). HBZ has not been reported to exhibit the same in vitro transforming properties as E1A, and we found HBZ is less potent than E1A in reducing global H3K18ac. Based on these observations, it is possible that transformation occurs when p300/CBP HAT activity is decreased below a certain threshold, and inhibition of HAT activity by HBZ does not surpass this threshold under normal circumstances. However, during the course of persistent HTLV-1 infection, rare cellular events may cooperate with HBZ to augment inhibition of HAT activity, in turn contributing to the development of ATL.
Although histone acetylation is associated with transcriptionally active regions of chromatin, a global reduction in H3K18ac may not be sufficient to cause an overall decrease in gene expression. Indeed, several proteins have been identified with HAT activity (11), and acetylation at other lysine residues of histone H3 and the other core histones may provide a redundant role for the H3K18ac mark (98). Furthermore, there are multiple examples of p300/CBP participating in transcriptional activation through a HAT-independent mechanism (68,99,100). These observations support gene-expression microarray data for HBZ that show an even distribution of up and downregulated genes (62). Therefore, similar to e1A, the global reduction in H3K18ac caused by HBZ appears to diverge from its role in deregulating gene expression (22,101). It is possible that a decrease in H3K18ac by HBZ may hinder repair of DNA double-strand breaks. Acetylation of histones at DNA damage sites by p300/CBP is an important step in the repair process, and H3K18ac represents one of the marks that accumulate at sites of damage (13,102). Whether HBZ affects repair of double-strand breaks is under investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
National Institutes of Health [CA 128800 to I.L.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Joan Boyles for the gift of the p300-Flag and p300-ΔHAT-Flag plasmids, Dr Baldwin for the Flag-p65 plasmid, Dr M. Matsuoka for the ATL-2 and TL-OmI cell lines and Dr F. Bex for the HeLa A57 cells.
REFERENCES
- 1.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell. Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 2.Arany Z, Sellers WR, Livingston DM, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 3.Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Tang Y, Cole PA, Marmorstein R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr. Opin. Struct. Biol. 2008;18:741–747. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brownell JE, Allis CD. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 9.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 10.Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 11.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 15.Kung AL, Rebel VI, Bronson RT, Ch'ng LE, Sieff CA, Livingston DM, Yao TP. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc. Natl Acad. Sci. USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roelfsema JH, Peters DJ. Rubinstein-Taybi syndrome: clinical and molecular overview. Expert. Rev. Mol. Med. 2007;9:1–16. doi: 10.1017/S1462399407000415. [DOI] [PubMed] [Google Scholar]
- 18.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerchietti LC, Hatzi K, Caldas-Lopes E, Yang SN, Figueroa ME, Morin RD, Hirst M, Mendez L, Shaknovich R, Cole PA, et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J. Clin. Invest. 2010;120:4569–4582. doi: 10.1172/JCI42869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurdistani SK. Histone modifications in cancer biology and prognosis. Prog. Drug Res. 2011;67:91–106. doi: 10.1007/978-3-7643-8989-5_5. [DOI] [PubMed] [Google Scholar]
- 23.Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. Am J. Pathol. 2009;174:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 25.Manuyakorn A, Paulus R, Farrell J, Dawson NA, Tze S, Cheung-Lau G, Hines OJ, Reber H, Seligson DB, Horvath S, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J. Clin. Oncol. 2009;28:1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 27.Mosashvilli D, Kahl P, Mertens C, Holzapfel S, Rogenhofer S, Hauser S, Buttner R, Von Ruecker A, Muller SC, Ellinger J. Global histone acetylation levels: prognostic relevance in patients with renal cell carcinoma. Cancer Sci. 2010;101:2664–2669. doi: 10.1111/j.1349-7006.2010.01717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puppin C, Passon N, Lavarone E, Di Loreto C, Frasca F, Vella V, Vigneri R, Damante G. Levels of histone acetylation in thyroid tumors. Biochem. Biophys. Res. Commun. 2011;411:679–683. doi: 10.1016/j.bbrc.2011.06.182. [DOI] [PubMed] [Google Scholar]
- 29.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particle from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl Acad. Sci. USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl Acad. Sci. USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goncalves DU, Proietti FA, Ribas JG, Araujo MG, Pinheiro SR, Guedes AC, Carneiro-Proietti AB. Epidemiology, treatment, and prevention of human T-cell leukemia virus type 1-associated diseases. Clin. Microbiol. Rev. 2010;23:577–589. doi: 10.1128/CMR.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kannian P, Green PL. Human T lymphotropic virus type 1 (HTLV-1): molecular biology and oncogenesis. Viruses. 2010;2:2037–2077. doi: 10.3390/v2092037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satou Y, Yasunaga J, Zhao T, Yoshida M, Miyazato P, Takai K, Shimizu K, Ohshima K, Green PL, Ohkura N, et al. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 2011;7:e1001274. doi: 10.1371/journal.ppat.1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basbous J, Arpin C, Gaudray G, Piechaczyk M, Devaux C, Mesnard JM. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 2003;278:43620–46327. doi: 10.1074/jbc.M307275200. [DOI] [PubMed] [Google Scholar]
- 35.Thebault S, Basbous J, Hivin P, Devaux C, Mesnard JM. HBZ interacts with JunD and stimulates its transcriptional activity. FEBS Lett. 2004;562:165–170. doi: 10.1016/S0014-5793(04)00225-X. [DOI] [PubMed] [Google Scholar]
- 36.Hivin P, Basbous J, Raymond F, Henaff D, Arpin-Andre C, Robert-Hebmann V, Barbeau B, Mesnard JM. The HBZ-SP1 isoform of human T-cell leukemia virus type I represses JunB activity by sequestration into nuclear bodies. Retrovirology. 2007;4:14. doi: 10.1186/1742-4690-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard JM. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 2002;76:12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemasson I, Lewis MR, Polakowski N, Hivin P, Cavanagh MH, Thebault S, Barbeau B, Nyborg JK, Mesnard JM. Human T-cell leukemia virus type 1 (HTLV-1) bZIP protein interacts with the cellular transcription factor CREB to inhibit HTLV-1 transcription. J. Virol. 2007;81:1543–1553. doi: 10.1128/JVI.00480-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clerc I, Polakowski N, Andre-Arpin C, Cook P, Barbeau B, Mesnard JM, Lemasson I. An interaction between the human T cell leukemia virus type 1 basic leucine zipper factor (HBZ) and the KIX domain of p300/CBP contributes to the down-regulation of tax-dependent viral transcription by HBZ. J. Biol. Chem. 2008;283:23903–23913. doi: 10.1074/jbc.M803116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cook PR, Polakowski N, Lemasson I. HTLV-1 HBZ protein deregulates interactions between cellular factors and the KIX domain of p300/CBP. J. Mol. Biol. 2011;409:384–398. doi: 10.1016/j.jmb.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polakowski N, Gregory H, Mesnard JM, Lemasson I. Expression of a protein involved in bone resorption, Dkk1, is activated by HTLV-1 bZIP factor through its activation domain. Retrovirology. 2010;7:61. doi: 10.1186/1742-4690-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao T, Satou Y, Sugata K, Miyazato P, Green PL, Imamura T, Matsuoka M. HTLV-1 bZIP factor enhances TGF-beta signaling through p300 coactivator. Blood. 2011;118:1865–1876. doi: 10.1182/blood-2010-12-326199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponting CP, Blake DJ, Davies KE, Kendrick-Jones J, Winder SJ. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci. 1996;21:11–13. [PubMed] [Google Scholar]
- 44.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 45.Van Orden K, Yan JP, Ulloa A, Nyborg JK. Binding of the human T-cell leukemia virus Tax protein to the coactivator CBP interferes with CBP-mediated transcriptional control. Oncogene. 1999;18:3766–3772. doi: 10.1038/sj.onc.1202703. [DOI] [PubMed] [Google Scholar]
- 46.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques. 2006;40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 47.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez MS, Thompson J, Hay RT, Dargemont C. Nuclear retention of IkappaBalpha protects it from signal-induced degradation and inhibits nuclear factor kappaB transcriptional activation. J. Biol. Chem. 1999;274:9108–9115. doi: 10.1074/jbc.274.13.9108. [DOI] [PubMed] [Google Scholar]
- 49.Lemasson I, Polakowski NJ, Laybourn PJ, Nyborg JK. Tax-dependent displacement of nucleosomes during transcriptional activation of human T-cell leukemia virus type 1. J. Biol. Chem. 2006;281:13075–13082. doi: 10.1074/jbc.M512193200. [DOI] [PubMed] [Google Scholar]
- 50.Lemasson I, Polakowski N, Laybourn PJ, Nyborg JK. Transcription regulatory complexes bind the human T-cell leukemia virus 5' and 3' long terminal repeats to control gene expression. Mol. Cell. Biol. 2004;24:6117–6126. doi: 10.1128/MCB.24.14.6117-6126.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemasson I, Polakowski N, Laybourn PJ, Nyborg JK. Transcription factor binding and histone modifications on the integrated proviral promoter in HTLV-I-infected T-cells. J. Biol. Chem. 2002;277:49459–49465. doi: 10.1074/jbc.M209566200. [DOI] [PubMed] [Google Scholar]
- 52.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 53.De Guzman RN, Liu HY, Martinez-Yamout M, Dyson HJ, Wright PE. Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP [In Process Citation] J. Mol. Biol. 2000;303:243–253. doi: 10.1006/jmbi.2000.4141. [DOI] [PubMed] [Google Scholar]
- 54.Legge GB, Martinez-Yamout MA, Hambly DM, Trinh T, Lee BM, Dyson HJ, Wright PE. ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module. J. Mol. Biol. 2004;343:1081–1093. doi: 10.1016/j.jmb.2004.08.087. [DOI] [PubMed] [Google Scholar]
- 55.Burge S, Teufel DP, Townsley FM, Freund SM, Bycroft M, Fersht AR. Molecular basis of the interactions between the p73 N terminus and p300: effects on transactivation and modulation by phosphorylation. Proc. Natl Acad. Sci. USA. 2009;106:3142–3147. doi: 10.1073/pnas.0900383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 2009;28:948–958. doi: 10.1038/emboj.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferreon JC, Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc. Natl Acad. Sci. USA. 2009;106:6591–6596. doi: 10.1073/pnas.0811023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferreon JC, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proc. Natl Acad. Sci. USA. 2009;106:13260–13265. doi: 10.1073/pnas.0906770106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He J, Ye J, Cai Y, Riquelme C, Liu JO, Liu X, Han A, Chen L. Structure of p300 bound to MEF2 on DNA reveals a mechanism of enhanceosome assembly. Nucleic Acids Res. 2011;39:4464–4474. doi: 10.1093/nar/gkr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cavanagh M, Landry S, Audet B, Arpin-Andre C, Hivin P, Pare M-E, Thete J, Wattel E, Marriott SJ, Mesnard JM, et al. HTLV-I antisense transcripts initiate in the 3’LTR and are alternatively spliced and polyadenylated. Retrovirology. 2006;3:15. doi: 10.1186/1742-4690-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murata K, Hayashibara T, Sugahara K, Uemura A, Yamaguchi T, Harasawa H, Hasegawa H, Tsuruda K, Okazaki T, Koji T, et al. A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization. J. Virol. 2006;80:2495–2505. doi: 10.1128/JVI.80.5.2495-2505.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl Acad. Sci. USA. 2006;103:720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cohen I, Poreba E, Kamieniarz K, Schneider R. Histone modifiers in cancer: friends or foes? Genes Cancer. 2:631–647. doi: 10.1177/1947601911417176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Q, Yao H, Vo N, Goodman RH. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc. Natl Acad. Sci. USA. 2000;97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li GD, Fang JX, Chen HZ, Luo J, Zheng ZH, Shen YM, Wu Q. Negative regulation of transcription coactivator p300 by orphan receptor TR3. Nucleic Acids Res. 2007;35:7348–7359. doi: 10.1093/nar/gkm870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao F, McCarrick-Walmsley R, Akerblad P, Sigvardsson M, Kadesch T. Inhibition of p300/CBP by early B-cell factor. Mol. Cell. Biol. 2003;23:3837–3846. doi: 10.1128/MCB.23.11.3837-3846.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kasper LH, Lerach S, Wang J, Wu S, Jeevan T, Brindle PK. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. EMBO J. 2010;29:3660–3672. doi: 10.1038/emboj.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Griffis ER, Xu S, Powers MA. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol. Biol. Cell. 2003;14:600–610. doi: 10.1091/mbc.E02-09-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kouzarides T. SnapShot: Histone-modifying enzymes. Cell. 2007;131:822. doi: 10.1016/j.cell.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 71.Zimmermann H, Degenkolbe R, Bernard HU, O'Connor MJ. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 1999;73:6209–6219. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co- activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas MC, Chiang CM. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol. Cell. 2005;17:251–264. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 74.Chakravarti D, Ogryzko V, Kao HY, Nash A, Chen H, Nakatani Y, Evans RM. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 75.Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 76.Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 77.Macfarlan T, Parker JB, Nagata K, Chakravarti D. Thanatos-associated protein 7 associates with template activating factor-Ibeta and inhibits histone acetylation to repress transcription. Mol. Endocrinol. 2006;20:335–347. doi: 10.1210/me.2005-0248. [DOI] [PubMed] [Google Scholar]
- 78.Hublitz P, Kunowska N, Mayer UP, Muller JM, Heyne K, Yin N, Fritzsche C, Poli C, Miguet L, Schupp IW, et al. NIR is a novel INHAT repressor that modulates the transcriptional activity of p53. Genes Dev. 2005;19:2912–2924. doi: 10.1101/gad.351205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 80.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 82.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc. Natl Acad. Sci. USA. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fritsche M, Haessler C, Brandner G. Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene. 1993;8:307–318. [PubMed] [Google Scholar]
- 85.Kasibhatla S, Brunner T, Genestier L, Echeverri F, Mahboubi A, Green DR. DNA damaging agents induce expression of Fas ligand and subsequent apoptosis in T lymphocytes via the activation of NF-kappa B and AP-1. Mol. Cell. 1998;1:543–551. doi: 10.1016/s1097-2765(00)80054-4. [DOI] [PubMed] [Google Scholar]
- 86.Zhao T, Yasunaga J, Satou Y, Nakao M, Takahashi M, Fujii M, Matsuoka M. Human T-cell leukemia virus type 1 bZIP factor selectively suppresses the classical pathway of NF-kappaB. Blood. 2009;113:2755–2764. doi: 10.1182/blood-2008-06-161729. [DOI] [PubMed] [Google Scholar]
- 87.Kwok RP, Laurance ME, Lundblad JR, Goldman PS, Shih H, Connor LM, Marriott SJ, Goodman RH. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 88.Lemasson I, Nyborg JK. Human T-cell leukemia virus type I Tax repression of p73beta is mediated through competition for the C/H1 domain of CBP. J. Biol. Chem. 2001;276:15720–15727. doi: 10.1074/jbc.M100131200. [DOI] [PubMed] [Google Scholar]
- 89.Scoggin KE, Ulloa A, Nyborg JK. The oncoprotein Tax binds the SRC-1-interacting domain of CBP/p300 to mediate transcriptional activation. Mol. Cell. Biol. 2001;21:5520–5530. doi: 10.1128/MCB.21.16.5520-5530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O'Connor MJ, Zimmermann H, Nielsen S, Bernard HU, Kouzarides T. Characterization of an E1A-CBP interaction defines a novel transcriptional adapter motif (TRAM) in CBP/p300. J. Virol. 1999;73:3574–3581. doi: 10.1128/jvi.73.5.3574-3581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reid RL, Lindholm PF, Mireskandari A, Dittmer J, Brady JN. Stabilization of wild-type p53 in human T-lymphocytes transformed by HTLV-I. Oncogene. 1993;8:3029–3036. [PubMed] [Google Scholar]
- 92.Akagi T, Ono H, Tsuchida N, Shimotohno K. Aberrant expression and function of p53 in T-cells immortalized by HTLV- I Tax1. FEBS Lett. 1997;406:263–266. doi: 10.1016/s0014-5793(97)00280-9. [DOI] [PubMed] [Google Scholar]
- 93.Cereseto A, Diella F, Mulloy JC, Cara A, Michieli P, Grassmann R, Franchini G, Klotman ME. p53 functional impairment and high p21waf1/cip1 expression in human T- cell lymphotropic/leukemia virus type I-transformed T cells. Blood. 1996;88:1551–1560. [PubMed] [Google Scholar]
- 94.Gartenhaus RB, Wang P. Functional inactivation of wild-type p53 protein correlates with loss of IL-2 dependence in HTLV-I transformed human T lymphocytes. Leukemia. 1995;9:2082–2086. [PubMed] [Google Scholar]
- 95.Takemoto S, Trovato R, Cereseto A, Nicot C, Kislyakova T, Casareto L, Waldmann T, Torelli G, Franchini G. p53 stabilization and functional impairment in the absence of genetic mutation or the alteration of the p14(ARF)-MDM2 loop in ex vivo and cultured adult T-cell leukemia/lymphoma cells. Blood. 2000;95:3939–3944. [PubMed] [Google Scholar]
- 96.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rasti M, Grand RJ, Mymryk JS, Gallimore PH, Turnell AS. Recruitment of CBP/p300, TATA-binding protein, and S8 to distinct regions at the N terminus of adenovirus E1A. J. Virol. 2005;79:5594–5605. doi: 10.1128/JVI.79.9.5594-5605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl Acad. Sci. USA. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 100.Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ, Kurdistani SK. Epigenetic reprogramming by adenovirus e1a. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, Yasui A, Yokota J, Kohno T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011;30:2135–2146. doi: 10.1038/onc.2010.592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.