Abstract
Variation in the number of individual chromosomes (chromosomal aneuploidy) or chromosome segments (segmental aneuploidy) is associated with developmental abnormalities and reduced fitness in all species examined; it is the leading cause of miscarriages and mental retardation and a hallmark of cancer. However, despite their documented importance in disease, the effects of aneuploidies on the transcriptome remain largely unknown. We have examined the expression effects of seven heterozygous chromosomal deficiencies, both singly and in all pairwise combinations, in Drosophila melanogaster. The results show that genes in one copy are buffered, i.e. expressed more strongly than the expected 50% of wild-type level, the buffering is general and not influenced by other monosomic regions. Furthermore, long genes are significantly more highly buffered than short genes and gene length appears to be the primary determinant of the buffering degree. For short genes the degree of buffering depends on expression level and expression pattern. Furthermore, the results show that in deficiency heterozygotes the expression of genes involved in proteolysis is enhanced and negatively correlates with the degree of buffering. Thus, enhanced proteolysis appears to be a general response to aneuploidy.
INTRODUCTION
Aneuploidy, i.e. variation in the number of individual chromosomes (chromosomal aneuploidy) or chromosome segments (segmental aneuploidy) is associated with developmental abnormalities and reduced fitness in all species examined (1–7). At the organism level the amount of genetic material gained (as in trisomies) or lost (as in segmental monosomies) is correlated to the severity of the associated defects (1,3). In general, segmental trisomies are better tolerated than segmental monosomies and in fruit flies heterozygous deficiency in up to 1% of the genome, on average, is tolerated (3). Aneuploidy is found in ∼90% of human cancers and is the leading cause of miscarriages and mental retardation (2,8). Furthermore, despite the proliferation disadvantage it confers, aneuploidy is a hallmark of cancer, a disease characterized by the proliferative potential of affected cells (9). Clearly, therefore, regardless of whether aneuploidy is a cause or consequence of tumorigenesis, which is debated (1), it may be tolerated if an aneuploid cell has acquired proliferative potential under conditions that prevent the division of surrounding (euploid) cells.
Although aneuploidy is a well-known hallmark of cancer until recently we have had very little insight into how changes in numbers of chromosomes or parts of chromosomes affect gene-expression levels and the responses aneuploidies induce. Early investigations of the relations between chromosomal dose and expression responses have mainly relied on assays of enzymes encoded by a small number of genes (10–12), with only a few studies examining corresponding RNA-levels (13,14). Accurate measurement of genome-wide expressional effects of segmental aneuploidies is difficult, because in many cases they are weak, and few attempts have been made as yet to evaluate the effects of copy number variation on transcription levels in any detail. In general, previous genome-wide studies addressing this question have confirmed the existence of functional autosomal dosage compensation of gene-expression, but the magnitude of the effect has sometimes been overestimated (15–18). These studies generally examined changes in the expression of all genes with altered copy levels. However, genes that are inactive under the test conditions and genes with expression levels below the detection level will appear to be unaffected by the copy number, complicating attempts to assess the variations and their biological significance. More recently it has been confirmed that organisms have intrinsic mechanisms that compensate for aneuploidy at the level of transcription output (18–21). It has been shown that both monosomic and trisomic genomic regions are expressed more closely to wild-type (diploid) levels than would be expected if mRNA levels correlated perfectly to gene dose (19,20) and this effect is called ‘buffering’. Monosomic regions are more strongly buffered than trisomic regions and, interestingly, differentially expressed genes are more strongly buffered in monosomic condition than ubiquitously expressed genes (19). In one case the mechanism for autosomal buffering has been identified, the compensation for monosomic condition in the fourth chromosome in D. melanogaster, mediated by the protein Painting of Fourth (POF) (19,22–24). These findings further imply that aneuploid regions are buffered at the RNA level by general buffering systems rather than by feedback regulation of a few individual genes. To explore in more detail the factors that affect degrees of buffering at regional and individual gene levels, and transcription responses to aneuploidies, we have analysed effects of seven deletions, both singly and in all pairwise combinations, within the aneuploid regions and across the D. melanogaster genome.
Our results confirm that genes in monosomic condition are buffered and indicate both that this effect is general and that the buffering effect at one region is not altered by combinatorial effects. In addition, long genes are better buffered than short genes. For short genes only differentially expressed genes are buffered and genes with low expression, while the buffering of long genes is not affected by either the ubiquity or strength of their expression. We have observed no signs of spreading of the buffering effect. Furthermore, our results suggest that aneuploidy induces enhanced proteolysis.
MATERIALS AND METHODS
Fly stocks and genetic crosses
The DrosDel isogenic deficiency strains Df(3R)ED10953, Df(2L)ED4559, Df(2L)ED1770, Df(2L)ED1612, Df(2L)ED3, Df(3R)ED5071, Df(3R)ED7665, Df(2L)ED748 and Df(3R)ED10946 were obtained from the Drosophila Genetic Resource Center (DGRC) in Kyoto, Japan. The isogenic background stock w1118iso; 2iso; 3iso was used as wild-type. All deficiency strains were crossed to w1118iso; 2iso; 3iso to generate Df/+ or crossed pairwise to create combinations of two deficiency regions. Two deficiencies, Df(2L)ED748 and Df(3R)ED10946, were incorrect, i.e. they had no deleted region, therefore these strains were treated as wild-type. Two of the combinations, Df(3R)ED5071/Df(3R)ED7665 and Df(2R)ED1770/+; Df(3R)ED10953/+, caused lethality, thus there are no expression results for these combinations. In all cases, flies were cultivated and crossed at 25°C in vials containing potato mash-yeast-agar. All crosses are listed in Supplementary Table S1.
Microarray analysis
For microarray analysis total RNA was isolated using TRIzol or TRI Reagent (Invitrogen, Ambion) followed by purification using an RNeasy kit (Qiagen) according to the manufacturer’s instructions. Fifteen adult females (0–24 h) were used for each replicate; 2–3 replicates for each single Df/+ genotype (seven different Df), 19 pairwise Df combinations and six replicates of the w1118iso; 2iso; 3iso wild-type control. The labelled cDNA probes (44 in total) were then hybridized to an Affymetrix Drosophila gene chip (version 2) and the intensity values were normalized and summarized using robust multi-array analysis (RMA) in R (www.R-project.org) and the Bioconductor package (25). The resulting data are available at http://www.ncbi.nlm.nih.gov/geo/ (Accession number: GSE34400). Based on expression array data in the FlyAtlas database (26) (Geo accession number: GSE7763), ubiquitously expressed genes were defined as genes showing expression levels of at least 6 in all of the 12 examined tissues after RMA normalization, while all other genes were defined as differentially expressed genes. After removing all genes expressed at levels <6 or >2 in wild-type or with a standard deviation in the six wild-type replicates >1 the data were scaled, by adding an array specific constant (positive or negative; on average −0.0089; average absolute value 0.032) to all the mutant array expression values so that the total genomic expression on all mutant arrays matched that of the wild-type. The average expression relative to wild-type was then measured for all of the expressed genes. The expression ratios obtained in the different experiments were calculated as follows. To analyse the expression ratio for each Df in different combinations, the mean expression ratio of all genes for each region and each replicate were calculated, giving 57 different ratios. To analyse the effects of different deletion criteria on buffering, the mean ratio of all genes in all replicates for each deficiency region were calculated, giving a total of seven different ratios. To analyse buffering at the gene level, the median expression ratio for each gene in all 7-9 replicates, giving a total of 320 genes, was calculated.
To calculate combinatorial effects the mean expression ratio for each region and each replicate were calculated as described above. The mean expression ratios for each of the 2–3 single replicates were then combined to calculate a single median expression ratio per Df. Then all deficiency regions were sorted in descending order of their buffering effect: ED4559>ED1612>ED10953>ED3>ED1770>ED5071>ED7665. Next, we calculated the difference between the mean expression ratio for the Df when single and the mean expression ratio of the same Df when combined with a Df with a lower expression ratio [e.g. Df(2L)ED4559/+ versus Df(2L)ED4559/Df(2R)ED1612, Df(2L)ED4559/+ versus Df(2L)ED4559/+; Df(3R)ED10953/+ etc.] This was done for all Df and the values were then summed and a mean value for the directional effect of each Df was calculated based on data obtained for the 19 combinations. Reciprocal calculations were applied to see if more strongly buffering regions affect regions with a weaker buffering effect.
Gene criteria definitions
To identify the gene features that affect a specific gene’s buffering we defined 16 gene criteria (Supplementary Table S2). Gene length was defined as the length of the longest annotated transcript for each gene. CDS length was defined as the sum of all non-overlapping coding sequences annotated for all transcripts of the corresponding gene. Where overlapping features were found, only the longest was included in the sum. Relative CDS length was calculated as the ratio between CDS length and gene length. Lengths of 5′ UTRs, 3′ UTRs and introns were calculated in the same way. Median WT expression was defined as the median expression value of the six wild-type replicates. FlyAtlas expression pattern is the total number of FlyAtlas-defined tissue types (12 tissue types) in which the gene has an expression > 6, for details see (19). Standard deviation FlyAtlas expression pattern is the standard deviation of expression levels for each gene in the 12 tissue types. Standard deviation of 6 WT is the standard deviation of the mean for the six wild-type replicates. Buffering level of neighbouring gene was defined as the buffering level of the gene closest (in start or stop position) to the gene of interest (start or stop position). Distance to 5′ neighbouring gene is the distance between the start coordinate for the gene of interest and the stop or start coordinate of the neighbouring gene upstream of the gene of interest. Distance to 5′ expressed gene is defined as above, but the distance is calculated to the closest expressed gene upstream of the gene of interest.
Breakpoint effects analysis
Diploid genes outside Df breakpoints from all Df were grouped so that all genes within 0–5000 bp from a breakpoint, all genes within 0–10 000 bp from a breakpoint etc., up to 0–1 280 000 bp from a breakpoint, were grouped. In addition all diploid genes on the same chromosome arm as a Df were included as a group and finally all monosomic genes from each Df were included as a reference group. A mean breakpoint effect for each bin was then calculated based on the median expression ratio of each gene and all values were combined in a mean plot.
Statistical analysis
All statistical analyses were performed on log2-scaled data using Statsoft Statistica 10.0 or Microsoft Excel 2010. Bonferroni correction was applied by multiplying all P-values with the number of tests performed.
Pathway analysis
To determine if (and if so which) biological pathways are induced by aneuploidy we calculated the mean expression ratio (mean of all 38 datasets) for all diploid genes (excluding genes encompassed by one or more Df ). The 100 most strongly up-regulated genes were then submitted to the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources v.6.7 hosted by the National Institute of Allergy and Infectious Diseases (NIAID/NIH) (http://david.abcc.ncifcrf.gov/) (27,28). We used the whole Drosophila genome gene list as background and included GOterm level 5.
Real-time PCR
Total RNA from four Df/+ replicates, Df(3R)ED7665 or Df(3R)ED3, was reverse-transcribed using an iScript™ cDNA Synthesis Kit and amplified by real-time PCR using iQ™SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. Primer pairs used are listed in Supplementary Table S4. The expression levels were normalized to the amount of Rpl32 mRNA in each replicate.
RESULTS
Gene expression is buffered when genes are in monosomic condition
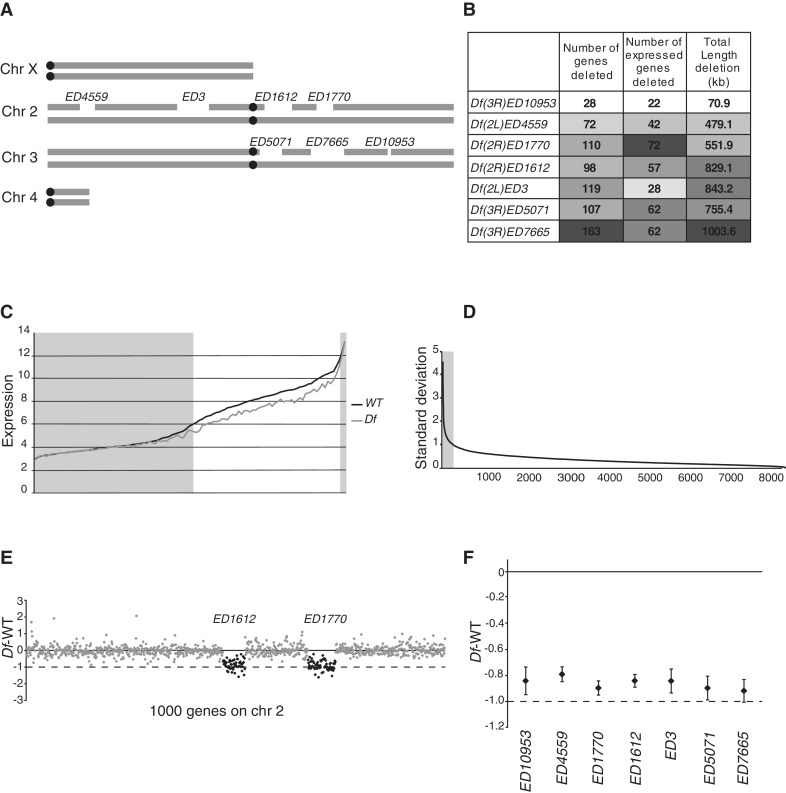
To study the buffering of genes in monosomic condition we chose seven segmental deficiencies (Df) differing in length, number of uncovered genes and number of uncovered expressed genes (Figure 1A and B). To explore their effects we outcrossed strains, each heterozygously harbouring one of the deficiencies, to wild-type (to make single, Df/+, deletion strains) and created all combinations of pairwise crosses (Supplementary Table S1). We then analysed adult female expression levels in the wild-type (six biological replicates) and each Df/+ strain, both single-deletion (2–3 biological replicates) and all non-lethal pairwise combinations (single replicates) by Affymetrix microarray analysis. To minimize the effect of genetic background we used isogenic DrosDel strains in all our crosses (29,30). Two of the 21 Df combinations were lethal. Since genes with non- or sub-detectable expression and genes with very high expression (which tend to reach saturation on the arrays) will behave as fully compensated in expression microarray studies, we plotted transcript levels in each Df/+ against wild-type transcript levels for Affymetrix probe sets (hereafter called genes) within the affected regions. The resulting plot showed that that transcript levels of genes with an expression level <6 (log2-scale) and >12 did not differ between wild-type and any Df/+ (Figure 1C). This implies that genes with transcript levels below or above these thresholds are either fully buffered or their transcript levels cannot be reliably measured. These genes were therefore removed from all further analysis. We have previously noticed that expression levels of a subset of genes recorded in all microarray analyses are highly variable due to technical reasons and/or the developmental stage analysed. We therefore used our six wild-type replicates to identify genes with highly variable expression and excluded all genes from further analysis for which the standard deviation of expression level for the wild-type replicates exceeded 1 in log2 scale (corresponding to a ±2-fold variation in expression) (Figure 1D). In summary, out of the 18 769 genes (probe sets) on the arrays we removed 52% due to low expression, 2.6% due to very high expression and 1.4% due to high variability between replicates. All further analysis was performed on the 8255 remaining genes, of which 320 were uncovered by a deficiency. The expression values of these 8255 genes correlates very well between the wild-type replicates (average rs = 0.93, for all pairwise comparisons).
Figure 1.
Segmental monosomic regions are buffered. (A) Schematic illustration of the seven Drosophila deficiency regions included in the study. (B) Heatmap table showing the total number of uncovered genes (number of Affymetrix probe sets), total number of uncovered expressed genes (i.e. probe sets with a wild-type expression level >6) and the total length of the deficiency regions. (C) Absolute expression level (in log2 scale) after RMA normalization of all genes uncovered by a deficiency. The black line represents the median expression of six wild-type replicates (diploid gene copy number) sorted by expression levels. The grey line represents expression of the deficiency strains (monosomic gene copy number). The deficiency data were plotted as running averages of 50 genes. Genes with a wild-type expression level <6 or >12 were excluded from further analysis (grey boxes). (D) Standard deviation (in log2 scale) of the mean calculated from the six wild-type replicates for all expressed genes (after removing <6; >12), sorted in descending order. A total of 259 genes with SD>1 are defined as unstably expressed and were thus excluded from further analysis (black line). (E) Expression ratios (log2 scale) of 1000 expressed genes along chromosome 2, showing a representative dataset containing two adjacent deficiency regions (in black). (F) The mean expression ratio (log2 scale) for each deletion (diamonds indicate mean values for all genes from the two or three single replicates). Whiskers indicate 95% CIs. Black lines represent 0 in log2 scale or 100% of wild-type expression level and dashed lines represent −1 in log2 scale or 50% of wild-type expression level (i.e. the expected expression level in the absence of a buffering effect).
To verify the presence and genomic position of all Df, results of all expression array analyses were first plotted as the Df/+ expression level divided by wild-type expression level (designated Df – WT) for all genes along the chromosome arms carrying the respective deletion. The Df regions could be easily detected at the correct positions in all cases (Figure 1E). We next calculated the average expression ratio (Df –WT) of the seven single Df/+, and found their average expression ratio varied from −0.9 to −0.79 (Figure 1F). We conclude that in all cases there is a weak buffering of average gene expression in the monosomic region.
The buffering level is not affected by combining Df
An important objective of the study was to determine whether there are any combinatorial effects, i.e. whether buffering effects of one Df influence buffering effects of a second Df when combined. We addressed three hypotheses: (i) the buffering effects of a Df may depend on whether it is present singly or in combination with another Df at a different genomic position; (ii) a Df may enhance or reduce buffering of another Df with weaker buffering; (iii) a Df may have directional (positive or negative) influence on the buffering of all other Df when combined, irrespective of whether the others have stronger or weaker buffering.
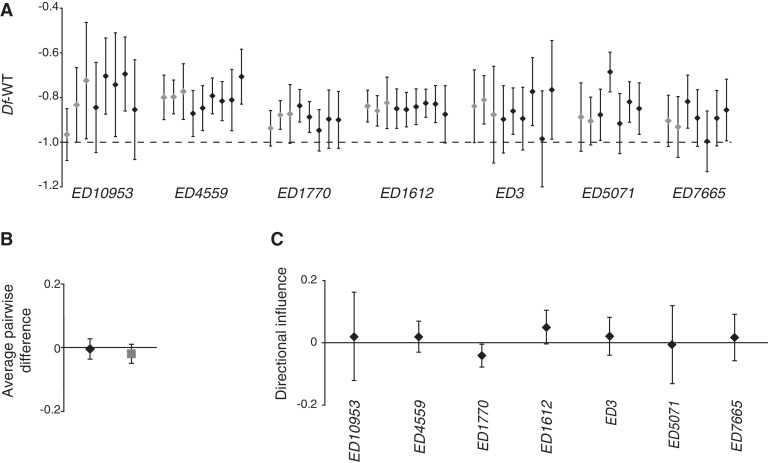
The expression ratios for each Df, both singly and in combination with each of the other Df, are shown in Figure 2A. No clear combinatorial effects were detected, and the variation in buffering of each Df was similar among single-deletion replicates and all combinations including it. A Kruskal–Wallis test on all groups in Figure 2A (the single replicates and all combinations involving each Df) detected only one significant effect (a significant difference in expression levels of single and paired replicates of ED5071, P = 0.049, due to a deviation in buffering associated with the ED5071/ED10953 combination).
Figure 2.
Buffering is not affected by combinatorial effects. (A) Mean expression ratio (log2 scale) for each deficiency region and replicate. Each deficiency region was present in 7–9 replicates and the replicates of the Df/+ when single and when paired with another Df/+ are indicated in grey and black, respectively. The dashed line represents −1 in log2 scale or 50% of wild-type expression level. (B) Average directional effects on expression levels when more strongly buffered Df/+ were combined with less buffered Df/+ (black diamonds) and when less buffered Df/+ were combined with more strongly buffered Df/+ (grey square); mean values for 19 combinations. (C) Influence on expression ratio of single Df. The plot shows the influential effect of each Df when paired (none of the directional influences are significantly different from 0, Bonferroni-corrected Wilcoxon matched pair test). All y-axis values are in log2 scale. Whiskers indicate 95% confidence intervals (CIs).
We next assessed whether Df have any mutual influence, by ordering them in terms of buffering effect and calculating the effect on the buffering of each Df of combining it with (i) each more weakly buffering Df and (ii) each more strongly buffering Df (i.e. calculating differences in expression level ratios when the focal Df was alone/in combination). In total there were 19 combinations since two combinations were lethal: Df(3R)ED5071/Df(3R)ED7665 and Df(2R)ED1770/+; Df(3R)ED10953/+. The average effects were close to zero and no significant effects were detected (Figure 2B). Finally, we assessed if individual Df show consistent directional effects when combined with another Df, for example if a combination with Df1 will always lead to an increase or decrease of the buffering of the other Df in a pairwise combination. No significant directional effect was detected, that is, none of the Df affected its partners significantly (Wilcoxon matched pair test, all Bonferroni corrected P-values > 0.3, Figure 2C). Based on our combinatorial analysis we conclude that the average buffering of a Df is unaffected by the presence of another Df.
The number of deleted genes affects the average buffering
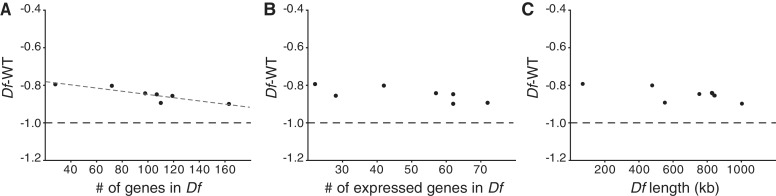
Since we did not detect any significant effect of combinations we can treat a combination of Df1 and Df2 as an additional replicate of both the single Df1 and the single Df2. For the following analyses we therefore had 7–9 biological replicates of each Df. Our seven different deletions yielded slightly different average expression ratios, and hence displayed slightly different buffering effects (Figure 1D). We therefore assessed whether these differences are correlated with any of the Df characteristics listed in Figure 1B. The results showed a small but significant correlation between expression ratio and the total number of deleted genes, so the fewer deleted genes the larger the buffering effect (rs = −0.964, P = 4.5 × 10−4). The total number of expressed genes and deletion length displayed the same trend although not significant (Figure 3).
Figure 3.
The number of deleted genes affects the average buffering. Correlation plots of mean expression ratio (log2 scale) for each deficiency region (calculated for all expressed genes and all replicates for each Df) plotted against total number of genes included in the deficiencies (A) against the total number of expressed genes included in the deficiencies (B) and the total length (in kb) of the deficiencies (C). The dashed line represents −1 in log2 scale or 50% of wild-type expression level. The Spearman’s correlation co-efficient is significant for (A) (rs = −0.964, P = 4.5 × 10−4).
Longer genes are more strongly buffered
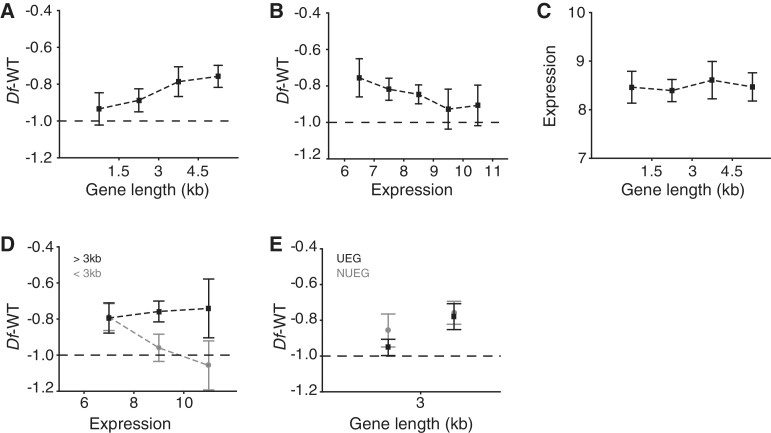
We next evaluated buffering at the gene level, starting by defining 16 gene criteria (Supplementary Table S2). We asked which of these criteria significantly correlate with the buffering of individual genes (median buffering of 7–9 replicates) when present in Df/+ condition. The results show that length criteria correlated positively with buffering, whereas expression correlated negatively (Supplementary Table S2). As expected, the buffering levels associated with all of the different length criteria correlated very well, and we decided to use the criteria gene length for further analysis. Therefore, we plotted the gene length versus expression ratio (Figure 4A). The results showed a clear correlation, i.e. the longer the gene the better it is buffered. We next plotted the transcript level in wild-type versus expression ratio and detected a negative correlation (Figure 4B). Since both gene length and expression are correlated to buffering we next assessed if there is a correlation between gene length and expression (Figure 4C). The results show that the correlations observed for gene length and expression are independent. However, by separating the genes into long genes (>3 kb, n = 132) and short genes (<3 kb, n = 188) we found that expression only correlates to buffering of short genes (Figure 4D).
Figure 4.
Gene length is the primary determinant of buffering. Expression ratio for monosomic genes (median of all replicates) plotted against (A) bins of gene length (kb, n = 71, 117, 44 and 88, respectively) and (B) median wild-type expression (median of six wild-type expression sets, n = 49, 70, 96, 57 or 48). (C) Wild-type expression level plotted against bins of gene length. (D) Expression ratio plotted against median wild-type expression of short (<3 kb) genes (shown in grey; n = 74, 89 or 25) and long (>3 kb) genes (shown in black; n = 45, 64 or 23). (E) Expression ratio plotted against binned lengths of: genes with >6 expression levels in all 12 FlyAtlas-specified tissue types, defined as housekeeping genes (UEG), shown in black (n = 96 or 57); and genes with >6 expression levels in 11 or fewer tissue types, defined as differentially expressed genes (NUEG), shown in grey (n = 92 or 75). Black lines represent −1 in log2 scale or 50% of wild-type expression. All y-axis values are in log2 scale. Squares indicate mean values and whiskers indicate 95% CIs. For correlation co-efficients, see Supplementary Table S2.
We have previously shown that buffering depends on whether genes are ubiquitously expressed [defined as genes expressed at levels >6 in all 12 tissues present in the FlyAtlas database; (26)] or differentially expressed (19). We therefore included this classification when examining the gene length versus buffering relationship (Figure 4E). The results show that differentially expressed short genes are more strongly buffered than ubiquitously expressed short genes, but long genes are buffered to the same extent independent of expression pattern. Taken together, our results show that gene length is the primary determinant for buffering, i.e. longer genes are more strongly buffered, regardless of both expression level and whether the genes are ubiquitously or differentially expressed.
Buffering level of one gene does not affect the buffering of neighbouring genes
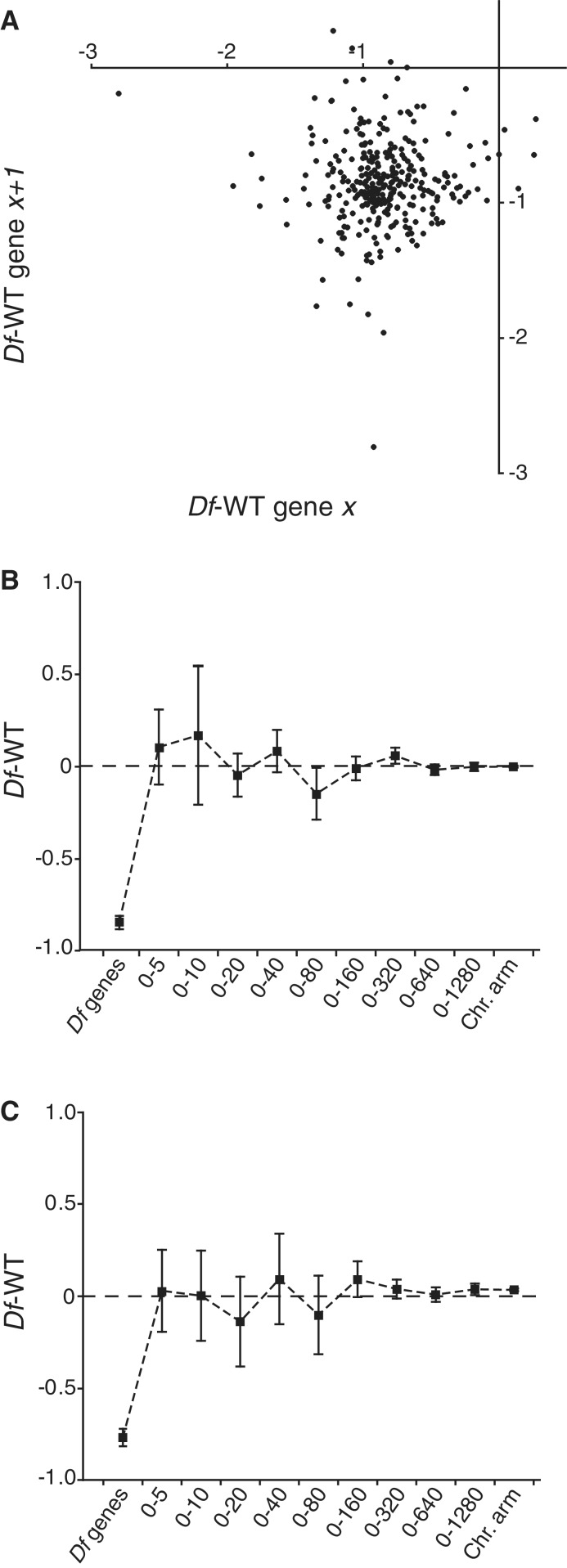
The results obtained from analysis of our large current data set corroborate our previous conclusion that the average calculated buffering is better explained as a general mechanism than by feed-back regulation of individual genes, since the expression differences between strains carrying deficiencies and wild-type are approximately normally distributed [Supplementary Figure S1 and (19)]. However, an alternative hypothesis is that buffering may be caused by feed-back regulation of individual genes and a local spreading of this feed-back generates the observed general effect. To address this possibility we tested for any correlation between the buffering of a given gene and its closest neighbouring gene (Figure 5A). No such spreading effect was detected. We also hypothesized that a spreading effect may occur outside deficiency regions, but close to the breakpoints and therefore calculated average expression ratios as a function of increasing distance from a Df breakpoint (Figure 5B). Since buffering is stronger for long genes we repeated the calculation, this time using only long genes (gene length >3 kb, Figure 5C). We detected no signs of spreading effects in either case. We conclude that the observed buffering is best explained as a general increase in expression mainly acting on long genes and that it shows no detectable spreading.
Figure 5.
Buffering level of one gene does not affect buffering of neighbouring genes. (A) Correlation plot of median expression ratio for each monosomic gene against the median expression ratio of neighbouring expressed monosomic genes. (B,C) Expression ratios of diploid genes within different interval distances (in kb) from a deficiency breakpoint. ‘Df genes’ represents the mean expression ratio of all genes uncovered by any deficiency. ‘Chr. arm’ represents the mean expression ratio of all diploid genes on the same chromosome arm as the deficiency. In (B) and (C) the mean expression ratio for each group was calculated using data for all diploid genes present within the interval and all long genes (>3 kb) within the interval, respectively. The dashed line represents 0 in log2 scale or no change from wild-type expression level. Squares indicate mean values and whiskers indicate 95% CI.
Aneuploidy induces proteolysis
In addition to the effects observed on genes within a deficiency region, we wanted to determine if any general effects on transcription are induced elsewhere in the genome by aneuploidy, e.g. if certain gene networks are activated or inhibited as a response to aneuploidy. To address this possibility, we calculated the mean expression ratio (mean of all 38 datasets) for all diploid genes (excluding all genes uncovered by a deficiency region). The most upregulated gene in the aneuploid samples compared to wild-type is the white gene. Since a functional white gene is an outcome of the deficiency induction and a marker for DrosDel deficiencies this was expected and the white gene was thus excluded from further analysis. The 100 most strongly up-regulated genes were then submitted to the DAVID (27,28). By using the Drosophila genome gene list as background we found that genes involved in peptidase and proteolysis activity were highly over-represented among the top 100 up-regulated genes (Table 1 and Supplementary Table S3). To rule out the possibility that the observed up-regulation of proteolytic genes could be caused by the presence of the white gene, we examined two DrosDel derived Df strains that contain no monosomic region but still carry the white gene [Df(2L)ED748 and Df(3R)ED10946, see ‘Materials and Methods’ section]. As expected, these two strains showed no significant up-regulation of the proteolytic genes.
Table 1.
Ontology groups enriched within the set of the 100 most up-regulated diploid genes identified using the Functional Annotation Chart in NIH DAVID, and level 5 GOterms
| Category | Term | Count | Fold Enrichment | Bonferroni |
|---|---|---|---|---|
| GOTERM_MF_5 | serine-type peptidase activity | 17 | 7.54 | 2.76E-09* |
| GOTERM_MF_5 | serine-type endopeptidase activity | 16 | 7.81 | 8.20E-09* |
| GOTERM_BP_5 | proteolysis | 22 | 4.74 | 6.31E-08* |
| GOTERM_MF_5 | endopeptidase activity | 18 | 5.38 | 1.11E-07* |
| GOTERM_BP_5 | chitin metabolic process | 6 | 8.28 | 0.12 |
| GOTERM_BP_5 | aminoglycan metabolic process | 6 | 6.65 | 0.28 |
| GOTERM_MF_5 | exopeptidase activity | 5 | 6.45 | 0.16 |
| GOTERM_CC_5 | integral to membrane | 10 | 2.29 | 0.13 |
| GOTERM_CC_5 | intrinsic to membrane | 10 | 2.25 | 0.15 |
| GOTERM_BP_5 | positive regulation of translation | 2 | 31.47 | 1.00 |
| GOTERM_MF_5 | serine-type exopeptidase activity | 2 | 24.85 | 0.87 |
Significantly enriched (Bonferroni-adjusted, P < 0.001) ontology groups are indicated in bold. For a detailed list of genes, see Supplementary Table S3.
To test the stability of this finding we applied an alternative approach for sorting out up-regulated genes by sorting genes based on the number of Df replicates that displayed an expression ratio of more than one in log2 scale (two times higher expression in Df compared to wild-type). The top 100 gene list extracted using this criterion was compared to the top 100 list extracted using the mean expression ratio obtained from all data sets, and found that 69 genes were identical in the two groups, verifying the robustness of the gene list. Reciprocally, the 100 most strongly down-regulated diploid genes were examined using DAVID, but in this case no significantly overrepresented gene ontology group was found.
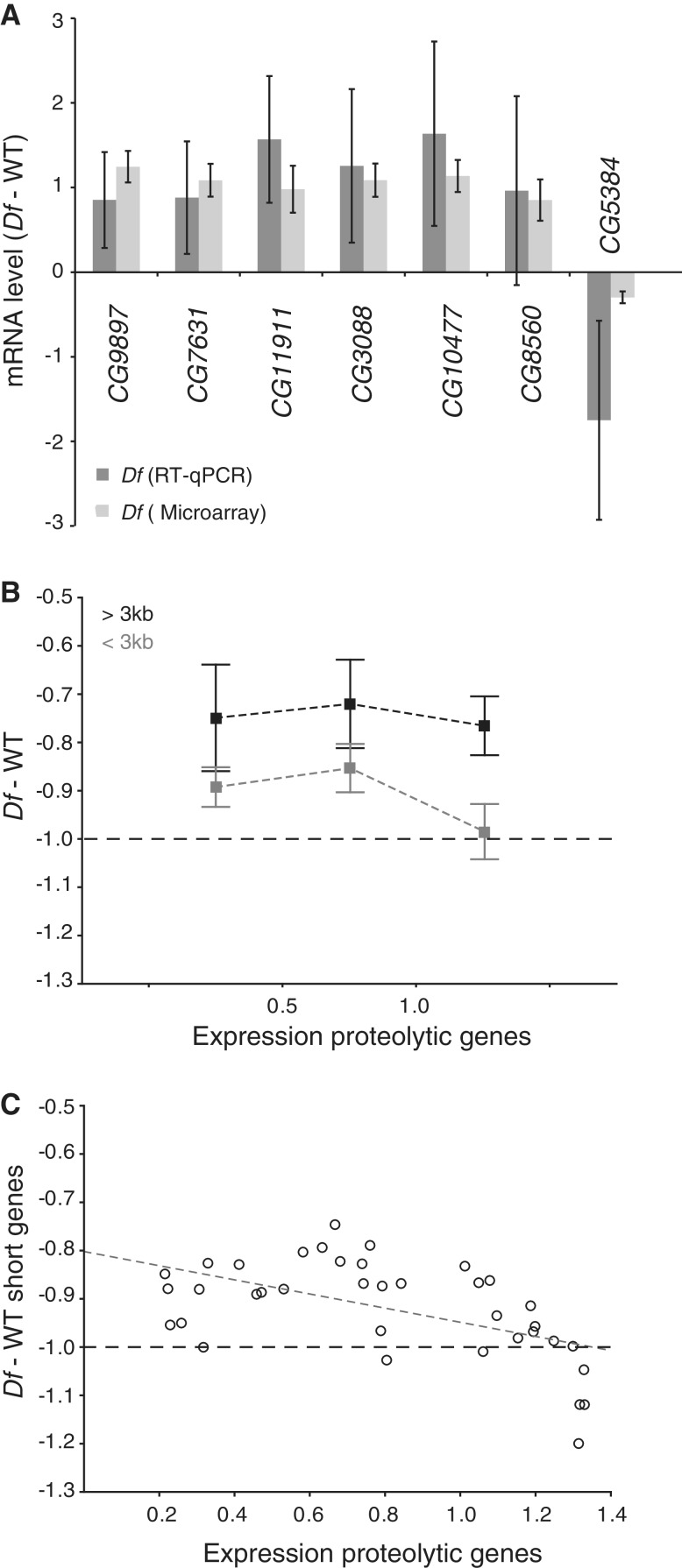
To confirm that proteolysis genes are transcriptionally up-regulated in Df/+ flies, six of the up-regulated genes defined as being involved in proteolytic activity were examined by RT–qPCR (Figure 6A) using four replicates of Df/+ and four replicates of wild-type. The results from the qPCR were compared to the mean expression ratios of all 38 microarray datasets. The RT–qPCR results support the conclusion that genes involved in proteolysis are up-regulated when parts of the genome are in monosomic condition.
Figure 6.
Aneuploidy induces proteolysis. (A) Comparison of the mean levels of mRNA (log2 scale) from six genes encoding proteolytic proteins and one encoding a deubiquitinating protein (CG5384) determined by RT–qPCR (dark grey bars). The corresponding mean expression ratio for all 38 Df/+ microarray replicates is shown in light grey. Note that a reduction in CG5384 expression is linked to increased proteolysis (31). The mRNA levels measured by qPCR were normalized against that of the Rpl32 gene in each replicate and 100% was set as the mean expression in wild-type. Error bars represent 95% CIs of four biological replicates (qPCR) and 38 biological replicates (microarray). (B) Expression ratios for all long genes (>3 kb, shown in black) within each deficiency region (each replicate represented by a separate data point) and for all short genes (<3 kb, shown in grey) plotted against binned values for the mean expression ratio of the top 20 up-regulated proteolysis genes. Squares indicate mean values and whiskers indicate 95% CIs of n = 10, 12 and 15 for each bin, respectively. (C) Scatter plot of expression ratio for all short genes within each deficiency region plotted against mean expression ratio of the 20 most up-regulated proteolysis genes. The Spearman’s correlation co-efficient is significant (rs = −0.529, p = 7.5 × 10−4). All y-axis values are in log2 scale. Dashed lines represent −1 in log2 scale or 50% of wild-type expression.
It has previously been shown that disomic yeast strains acquired loss-of function mutations in the gene encoding the deubiquitinating enzyme Ubp6 and it was showed that this was an aneuploidy tolerating mutation (31). We therefore examined the Drosophila ortholgue to Ubp6, i.e. CG5384. Interestingly, since UBP6 is a deubiquitinating enzyme loss of UBP6 leads to increased protein degradation, and in accordance with the increased expression of proteolysis genes we detected down-regulation of CG5384 in Df/+ flies (Figure 6A).
The level of induction of the proteolytic genes varies between the different Df strains. We next investigated whether this variation was related to the variation in buffering of the Df. For this analysis we calculated the average expression change of the top 20 up-regulated proteolytic genes displayed in our Df datasets. Interestingly, all but one of the 38 Df datasets showed that the presence of a Df increased average expression of proteolytic genes. We then plotted the average expression change versus the average buffering of the long and short genes displayed in the 37 Df datasets showing induced expression of proteolytic genes (Figure 6B). Interestingly, the induction of proteolytic genes appears to be strongest when the short genes within a Df are poorly buffered. Indeed, we observed a significant correlation between the average buffering of short genes and induction of genes involved in proteolysis (rs = −0.53, P = 7.5 × 10−4, Figure 6C). We conclude that increased proteolysis is a consequence of the genomic imbalance caused by aneuploidy and will also be induced by monosomic regions. In addition, buffering dampens the induced proteolysis.
DISCUSSION
Compensation of expression output in response to dosage differences is a well-known and thoroughly described phenomenon in dosage compensation systems acting on the sex-chromosomes (32–34). These dosage compensation mechanisms are required to equalize the X-linked gene expression between males and females and to ensure balance between X-chromosomal and autosomal gene products (18,32,35). The relations between doses of autosomal chromosomes or chromosome segments and their expression output have until recently been much more elusive. However, the existence of autosomal buffering has been demonstrated in several comparisons of gene expression to copy number (15,17,19–21,36). A generalized model that may account for autosomal dosage compensation, the ‘inverse dosage effect’ has also been proposed by Birchler and colleagues (37,38). For a segmental monosomy, this model predicts that the reduced expression caused by the loss of one gene copy is compensated or ‘buffered’ by the loss of negative regulators nearby. Compensation via an inverse effect basis will occur in segmental monosomies if negative regulators and target genes are simultaneously changed in dosage.
It is important to stress that the final expression output in response to dosage changes will be dependent on their effects at several levels and in the current study we only measured transcript levels using expression microarrays. Nevertheless, our previous results and those presented here confirm the existence of buffering of segmental monosomic states. To measure dosage compensation and buffering robustly it is essential to exclude all genes with expression levels outside the dynamic range of the method applied, since these genes will appear to be buffered, regardless of whether they are or not, leading to overestimates of the buffering effect. In the present study we therefore removed genes with expression levels above or below the dynamic range of the array method. We also removed genes with highly variable expression in wild-type to improve the reliability of the data. Although it is likely that our calculated buffering effects and those of the gene criteria affecting buffering could be extrapolated to the excluded genes, it should be stressed that we have made no attempt to determine the buffering of genes with expression levels below and above our cut-offs. Generally, deep sequencing (RNA-seq) techniques are considered to provide a wider dynamic range when determining transcript levels than expression microarrays. However, similar considerations should also be applied in the interpretation of RNA-seq data, as recently exemplified by conflicting reports on up-regulation of the X-chromosome in mammals (39,40).
Buffering is not affected by combinatorial effects
Cell lines and cancer cells are known to accumulate multiple chromosomal rearrangements (20,41) and are likely to have lost much of their expressional fine tuning. Clearly, tumour cells can survive catastrophic rearrangements, including the shattering of chromosomes into hundreds of fragments (41). In the present study we asked if (and if so how) several aneuploid regions affect each other. In Drosophila monosomy of up to 1% of the genome can be tolerated (3). We can hypothesize that buffering is a limited resource and that increasing degrees of monosomy are likely to result in decreased buffering. However, we can also hypothesize that buffering is an induced response and increased monosomy may in fact lead to increased buffering. Based on our results on the pairwise analysis we find no evidence for a directional effect, i.e. increases in monosomic content will not increase buffering, nor will a Df show a directional effect in combination. Buffering seems to be a general effect that varies slightly between different regions, but it is not influenced by combinations.
Gene length is the primary determinant of buffering
We asked which factors influence buffering at both Df region and individual gene levels. We noted that shorter Df regions are slightly better buffered than longer Df regions. Although this effect is minor it conflicts with expectations based on the inverse regulatory model. The longer the region, the more likely it is to include both target genes and the linked negative regulators predicted in the model. At the gene level, buffering is primarily determined by gene length and longer genes are better buffered. In a previous study we showed that the expression pattern is important, i.e. differentially expressed genes are better buffered than ubiquitously expressed genes (19). Results presented here confirm this, but also show that the effect of differential expression is subordinate to gene length. However, among the genes classified as short genes only those differentially expressed are buffered. Long genes are buffered independently of their expression levels, while short genes are not buffered if highly expressed. The different gene length criteria have correlated associations with buffering patterns, which makes it difficult to determine whether the total exon length or total length of the gene is the primary determinant for buffering. However, the finding that gene length correlates with buffering makes it tempting to speculate that the buffering mechanism involves transcription elongation. In Drosophila, dosage compensation for the monosomy of the male X-chromosome is achieved by a 2-fold increase in the expression of genes on the chromosome (32,34,42,43), putatively arising from the combined effect of the general buffering effect as analysed here and the male X-chromosome specific enhancement mediated by the MSL-complex (18,20,32). Interestingly, dosage compensation in Drosophila has been proposed to be mediated by enhanced transcription elongation (44) and this has also recently gained experimental support (45).Whether enhanced transcription elongation is the mechanism mediating the buffering effects observed here remains to be determined.
The observed buffering patterns suggest a general effect
In principle, buffering could be envisioned as a general effect acting on many genes or the sum of feed-back regulation of a smaller number of genes. The expression ratio of genes within monosomic regions shows an approximately normal distribution. This distribution is indicative of a general effect, however it could still be the sum of feed-back regulation of individual genes followed by local spreading of this positive effect. Spreading is a well-documented effect in dosage compensation mechanisms and there is evidently local spreading of the dosage compensation complex in Drosophila (46–50). However, we have not detected any signs of spreading within the deficiency regions nor spreading over the diploid breakpoints. Taken together, our results are consistent with the hypothesis that buffering is a general effect acting on monosomic regions and preferentially on long genes. The observed buffering could be the result of an induced increase in transcription output when a gene is in monosomic condition, which could be caused by a passive mechanism, e.g. a monosomic region could have an increased tendency to loop out from its normal nuclear position into more permissive environments since it is unpaired. Alternatively, unknown targeting mechanisms may recognize monosomic regions and stimulate their transcription output. We have previously shown that the fourth chromosome in D. melanogaster is compensated when in monosomic condition (19,22). This compensation of a monosomic fourth condition is slightly more efficient than the buffering of segmental monosomic regions analysed here. In the case of the fourth chromosome a specific targeting and stimulating mechanism is provided by the protein POF (19). However, POF targeting is chromosome-specific, and both the targeting and stimulation occur irrespectively of the copy number of the fourth chromosome (22).
Segmental monosomic conditions induce proteolysis gene networks
Since our analysis included a large number of samples with segmental monosomywe were interested to test if their monosomic condition induced any general expression response outside the aneuploid regions. The results indicate that induction of genes involved in proteolysis is a universal response to the segmental monosomic condition. Interestingly, in disomic yeast (with an extra chromosome copy) increased sensitivity to proteotoxic stress has been observed (31,51) and proteotoxic stress has been suggested to be a key source of the antiproliferative effects of aneuploidy (52). Trisomic mouse cells show similar signs of proteotoxic stress (9,53) and proteotoxic stress is sometimes considered a hallmark of cancer (54). It is important to note that in the cited cases the proteotoxic stress is induced by addition of extra chromosome copies. In yeast it has also been shown that mutations in Ubp6 enhance both proteolysis and aneuploidy tolerance (31). Our results show that genes involved in proteolysis are in fact induced by aneuploidies and the Ubp6 D. melanogaster ortholog is down-regulated. Furthermore, proteolysis induction occurs not only as a response to extra chromosome copies but also in response to a segmental monosomic condition. Thus, taken together the results suggest that induction of proteolysis is a general response to the genomic imbalance resulting from aneuploidy. Notably, although the Df/+ flies that we analysed were created by outcrosses of flies carrying the Df to wild-type (or pairwise Df1/+ × Df2/+ crosses), the deficiencies are also heterozygous (Df/Balancer) in the parental strains. This means that these strains have adjusted to their aneuploidy condition for years, but still show increased expression of genes involved in proteolysis. Furthermore, the degree of this increased proteolysis negatively correlates with the degree of buffering. We speculate that in segmental monosomic conditions genes are buffered to restore the imbalances. However, since not all genes are buffered and the buffering is not complete the cells harbouring segmental deficiencies will be subject to a stoichiometric imbalance that will induce proteolysis.
The majority of tumours in humans are aneuploid, and although the role of aneuploidy in tumorigenesis is controversial it provides a hallmark for human cancer (8,55). Identification of pathways induced by aneuploidy could provide insights into how cancer cells evolve and how they escape the negative effects associated with aneuploidy (31,52,56). Interestingly, drugs that increased proteotoxic stress have recently been suggested as promising cancer therapy candidates (52). Previous studies in yeast have suggested that trisomy causes proteotoxic stress and have highlighted the importance of ubiquitin-proteasomal degradation in suppressing the adverse effects of aneuploidy (31). Our study focused on segmental monosomic states in Drosophila identifies proteolysis as a main induced pathway in aneuploid individuals. This finding has two important implications. First, aneuploidy-induced proteotoxic stress appears to be conserved in evolution. Second, increased proteolysis appears to be induced by aneuploidy per se; not only aneuploidies that increase the copy number of certain genomic regions but also, as shown here, reductions of some genomic regions.
ACCESSION NUMBERS
The microarray data reported in this article have been deposited at http://www.ncbi.nlm.nih.gov/geo/ (Accession: GSE34400).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1.
FUNDING
Kempe Foundation (to L.L.); Carl Tryggers, Åke Wibergs and Erik Philip-Sörensens Foundations (to P.S.); Swedish Research Council (to J.L.). Funding for open access charge: the Swedish Research Council (to J.L.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Karin Ekström for technical assistance, Philge Philip for help with annotations and members of the Larsson and Stenberg groups for stimulating discussions.
REFERENCES
- 1.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassold TJ, Jacobs PA. Trisomy in man. Annu. Rev. Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- 3.Lindsley DL, Sandler L, Baker BS, Carpenter AT, Denell RE, Hall JC, Jacobs PA, Miklos GL, Davis BK, Gethmann RC, et al. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown S. Miscarriage and its associations. Semin. Reprod. Med. 2008;26:391–400. doi: 10.1055/s-0028-1087105. [DOI] [PubMed] [Google Scholar]
- 5.McClintock B. A cytological and genetical study of triploid maize. Genetics. 1929;14:180–222. doi: 10.1093/genetics/14.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigurdson DC, Herman RK, Horton CA, Kari CK, Pratt SE. An X-autosome fusion chromosome of Caenorhabditis elegans. Mol. Gen. Genet. 1986;202:212–218. doi: 10.1007/BF00331639. [DOI] [PubMed] [Google Scholar]
- 8.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devlin RH, Holm DG, Grigliatti TA. Autosomal dosage compensation Drosophila melanogaster strains trisomic for the left arm of chromosome 2. Proc. Natl Acad. Sci. USA. 1982;79:1200–1204. doi: 10.1073/pnas.79.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devlin RH, Holm DG, Grigliatti TA. The influence of whole-arm trisomy on gene expression in Drosophila. Genetics. 1988;118:87–101. doi: 10.1093/genetics/118.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birchler JA, Newton KJ. Modulation of protein levels in chromosomal dosage series of maize: the biochemical basis of aneuploid syndromes. Genetics. 1981;99:247–266. doi: 10.1093/genetics/99.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birchler JA, Hiebert JC, Paigen K. Analysis of autosomal dosage compensation involving the alcohol dehydrogenase locus in Drosophila melanogaster. Genetics. 1990;124:679–686. [PMC free article] [PubMed] [Google Scholar]
- 14.Guo M, Birchler JA. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science. 1994;266:1999–2002. doi: 10.1126/science.266.5193.1999. [DOI] [PubMed] [Google Scholar]
- 15.Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B. Global analysis of X-chromosome dosage compensation. J. Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarevitch I, Phillips RL, Springer NM. Profiling expression changes caused by a segmental aneuploid in maize. BMC Genomics. 2008;9:7. doi: 10.1186/1471-2164-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.FitzPatrick DR, Ramsay J, McGill NI, Shade M, Carothers AD, Hastie ND. Transcriptome analysis of human autosomal trisomy. Hum. Mol. Genet. 2002;11:3249–3256. doi: 10.1093/hmg/11.26.3249. [DOI] [PubMed] [Google Scholar]
- 18.Stenberg P, Larsson J. Buffering and the evolution of chromosome-wide gene regulation. Chromosoma. 2011;120:213–225. doi: 10.1007/s00412-011-0319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenberg P, Lundberg LE, Johansson AM, Rydén P, Svensson MJ, Larsson J. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet. 2009;5:E1000465. doi: 10.1371/journal.pgen.1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, Macalpine DM, Oliver B. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McAnally AA, Yampolsky LY. Widespread transcriptional autosomal dosage compensation in Drosophila correlates with gene expression level. Genome Biol. Evol. 2010;2:44–52. doi: 10.1093/gbe/evp054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson AM, Stenberg P, Bernhardsson C, Larsson J. Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. EMBO J. 2007;26:2307–2316. doi: 10.1038/sj.emboj.7601604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson J, Chen JD, Rasheva V, Rasmuson-Lestander A, Pirrotta V. Painting of fourth, a chromosome-specific protein in Drosophila. Proc. Natl Acad. Sci. USA. 2001;98:6273–6278. doi: 10.1073/pnas.111581298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson J, Svensson MJ, Stenberg P, Mäkitalo M. Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc. Natl Acad. Sci. USA. 2004;101:9728–9733. doi: 10.1073/pnas.0400978101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, Webster J, Gubb D, Gunton N, Johnson G, et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G, Morley T, Chan YS, Blows F, Coulson D, et al. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics. 2007;177:615–629. doi: 10.1534/genetics.107.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prestel M, Feller C, Becker PB. Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol. 2010;11:216. doi: 10.1186/gb-2010-11-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu. Rev. Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 34.Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliver B. Sex, dose, and equality. PLoS Biol. 2007;5:e340. doi: 10.1371/journal.pbio.0050340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aït Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, Rossier J, Personnaz L, Creau N, Bléhaut H, et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am. J. Hum. Genet. 2007;81:475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birchler JA, Pal-Bhadra M, Bhadra U. Dosage dependent gene regulation and the compensation of the X chromosome in Drosophila males. Genetica. 2003;117:179–190. doi: 10.1023/a:1022935927763. [DOI] [PubMed] [Google Scholar]
- 38.Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 2011;43:1179–1185. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong Y, Chen X, Chen Z, Wang X, Shi S, Zhang J, He X. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat. Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- 41.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hallacli E, Akhtar A. X chromosomal regulation in flies: when less is more. Chromosome Res. 2009;17:603–619. doi: 10.1007/s10577-009-9054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsson J, Meller VH. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 2006;14:417–431. doi: 10.1007/s10577-006-1064-3. [DOI] [PubMed] [Google Scholar]
- 44.Lucchesi JC. Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr. Opin. Genet. Dev. 1998;8:179–184. doi: 10.1016/s0959-437x(98)80139-1. [DOI] [PubMed] [Google Scholar]
- 45.Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- 47.Sass GL, Pannuti A, Lucchesi JC. Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc. Natl Acad. Sci. USA. 2003;100:8287–8291. doi: 10.1073/pnas.1332749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorchakov AA, Alekseyenko AA, Kharchenko P, Park PJ, Kuroda MI. Long-range spreading of dosage compensation in Drosophila captures transcribed autosomal genes inserted on X. Genes Dev. 2009;23:2266–2271. doi: 10.1101/gad.1840409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh H, Park Y, Kuroda MI. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev. 2003;17:1334–1339. doi: 10.1101/gad.1082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 52.Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.