Abstract
DNA double-strand breaks (DSBs) can be generated not only by reactive agents but also as a result of replication fork collapse at unrepaired DNA lesions. Whereas ubiquitylation of proliferating cell nuclear antigen (PCNA) facilitates damage bypass, modification of yeast PCNA by small ubiquitin-like modifier (SUMO) controls recombination by providing access for the Srs2 helicase to disrupt Rad51 nucleoprotein filaments. However, in human cells, the roles of PCNA SUMOylation have not been explored. Here, we characterize the modification of human PCNA by SUMO in vivo as well as in vitro. We establish that human PCNA can be SUMOylated at multiple sites including its highly conserved K164 residue and that SUMO modification is facilitated by replication factor C (RFC). We also show that expression of SUMOylation site PCNA mutants leads to increased DSB formation in the Rad18−/− cell line where the effect of Rad18-dependent K164 PCNA ubiquitylation can be ruled out. Moreover, expression of PCNA-SUMO1 fusion prevents DSB formation as well as inhibits recombination if replication stalls at DNA lesions. These findings suggest the importance of SUMO modification of human PCNA in preventing replication fork collapse to DSB and providing genome stability.
INTRODUCTION
Cellular DNA is continually damaged by a plethora of extrinsic and intrinsic sources and repaired by a variety of repair processes (1). However, DNA damages can also escape repair and block the replication machinery of which prolonged stalling can result in the collapse of replication fork into DNA double-strand breaks (DSBs) (2–4). Rescue of cells from replication collapse is important since DSB formation can lead to deleterious chromosomal rearrangements and cancer. To sustain the continuity of replication, other defence systems, the so-called DNA damage tolerance pathways are activated which do not repair DNA damage, but their action results in the completion of replication of damaged DNA (5,6). The Rad6–Rad18 ubiquitin-conjugating complex-dependent defence system governs translesion synthesis by specialized DNA polymerases as well as a template switch-dependent post-replicative repair pathway (7–10) Homologous recombination-dependent mechanisms can also rescue the stalled replication fork providing, thereby, alternative means for DNA damage tolerance (2,11–13)
Although replication through damaged DNA as well as homologous recombination can be error-free, unregulated damage bypass and recombination can lead to genome instability (7,14). To minimize the danger, these DNA damage tolerance pathways are thought to be kept under tight control such as by various post-translational modifications of proliferating cell nuclear antigen (PCNA), the processivity factor of replication, which serves also as a platform for giving access for repair players to DNA (15–17). Yeast genetic studies have been instrumental in providing evidence that PCNA monoubiquitylation is required for translesion synthesis, PCNA K63-linked polyubiquitylation governs template switch-dependent replication through DNA lesions, whereas modification of PCNA by small ubiquitin-like modifier (SUMO) prevents recombination and also regulates template switch (12,18–22). It has been suggested that PCNA–SUMO can provide access for the yeast Srs2 helicase to disrupt Rad51 nucleoprotein filaments thereby keeping recombination under control (12,14,23). These studies suggest that reviving replication forks by recombination could be disadvantageous for cells, probably because it can lead to gross chromosomal rearrangements. PCNA–SUMO functions as a guardian during replication of damaged DNA by preventing recombination and facilitating the use of the Rad6–Rad18-dependent damage tolerance pathway (17,24). However, the structure of the DNA substrates that arise from the stalled replication fork in the absence of PCNA–SUMO, the mechanism of recombination and the exact role of PCNA–SUMO at the stalled replication fork have remained elusive.
In mammalian cells, the significance of mono- and poly-ubiquitylation of PCNA has been demonstrated. Although PCNA SUMOylation has been reported in higher eucaryotes such as in Xenopus and chicken cells (25,26), the SUMO-dependent regulation of recombination has been assumed to be important only in particular eukaryotic cells with a naturally high rate of recombination such as yeast (20). Very recently, SUMOylation of human PCNA has also been found and shown that it preferentially interacts with a PCNA interacting protein (PARI) (27). PARI has been suggested to suppress inappropriate recombination events at the replication fork; however, the direct role of SUMO modification of human PCNA has not been studied.
Here, we characterize the modification of human PCNA by SUMO. Notably, we suggest that the presence of the SUMO moiety on human PCNA can prevent DSB formation as well as inappropriate recombination if replication stalls at DNA lesions. We discuss the possibility that in the absence of PCNA-SUMO stalled replication forks collapse more frequently to DSBs, which then become substrates for DSB repair-associated recombination-dependent DNA damage tolerance mechanism.
MATERIALS AND METHODS
Detailed information about materials and methods not provided here can be found in the Supplementary information.
Proteins, cell culture and antibodies
Plasmids for protein expressions in human cells and for protein purifications in yeasts and in Escherichia coli, purified proteins (PCNA and lysine to arginine mutants of PCNA, SUMO1, SUMO2, SUMO3, Pias1, Pias2, Pias3, Pias4, hUBC9, SAE1/SAE2, PCNA-SUMO1 fusion and RFC) and cell cultures used for in vivo/in vitro human PCNA SUMOylation studies are explained in the Supplementary Data. For immunostainings an anti-FLAG mAb 1:400 M2 (Sigma F3165), anti-FLAG 1:400 (Sigma F7425), anti-BrdU 1:500 (Ab-direct Serotech), anti- γH2Ax 1:5000 (Upstate 05-636), anti-mouse Cy3 1:1000 (Sigma C2181), AlexaFluor 488-labelled goat anti-rat antibody 1:1000 (Molecular Probes, Inc.) and anti-rabbit FITC 1:1000 (Sigma F0382) antibodies were used.
In vitro assays for PCNA SUMOylation and polymerase stimulation
In vitro SUMOylation reaction of PCNA contained 40 nM PCNA, 10 nM SAE1/2, 100 nM Ubc9, 500 nM SUMO1, 10 nM RFC and 2 nM nicked PUC19 plasmid DNA. Reactions were incubated at 37°C for 60 min and the products were separated on 10% denaturing polyacrylamide gel and visualized by western blot using anti-PCNA antibody (Santa Cruz PC10).
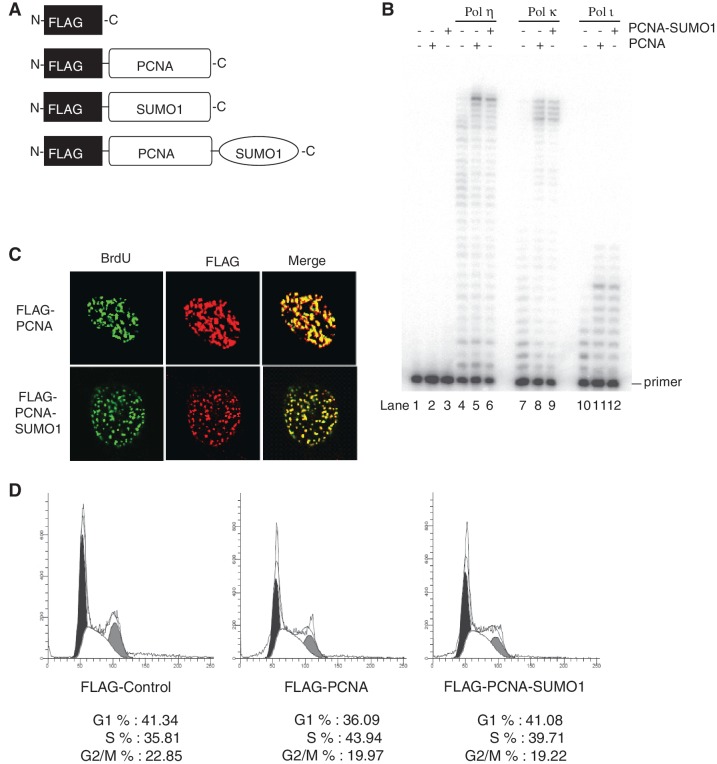
For DNA polymerase stimulation assay (Figure 3B) Polη, Polκ or Polι (2 nM each) was incubated with a 75/27-nt partial heteroduplex oligonucleotide DNA substrate (10 nM) containing biotin–streptavidine at both ends in the presence of RFC (5 nM), and either PCNA (10 nM) or PCNA-SUMO1 fusion protein (10 nM) at 37°C for 10 min.
Figure 3.
Effect of PCNA-SUMO1 fusion protein on translesion synthesis polymerases and qualitative analysis of PCNA-SUMO1 fusion protein. (A) Schematic representation of the fusion of SUMO1 at the C-terminus of a FLAG-tagged PCNA and the control vectors expressing only FLAG, FLAG-SUMO1 or FLAG-PCNA. (B) DNA polymerase reactions were carried out using various TLS DNA polymerases on DNA substrate containing biotin–streptavidin at both ends generated by annealing a 75-nt long oligonucleotide template to a 5′ labelled 27-nt primer DNA in the presence of RFC and either PCNA or PCNA-SUMO1. The reaction products were analysed on 10% polyacrylamide gels containing 8 M urea, and the DNA bands were visualized by autoradiography. (C) HeLa cells stably expressing FLAG-PCNA and FLAG-PCNA-SUMO1 were pulse labelled with 10 µM BrdU for 1 h and immunostained with antibodies against FLAG (red) and BrdU (green). (D) Stable cell lines expressing FLAG-control, FLAG-PCNA and FLAG-PCNA-SUMO1 were subjected to flow cytometric analysis. Cell number is plotted on the y axis; DNA content on the x axis. Black, G1 peak; grey, G2/M peak; white, S phase fraction.
In vivo SUMOylation of PCNA and analysis of recombination and cell survival
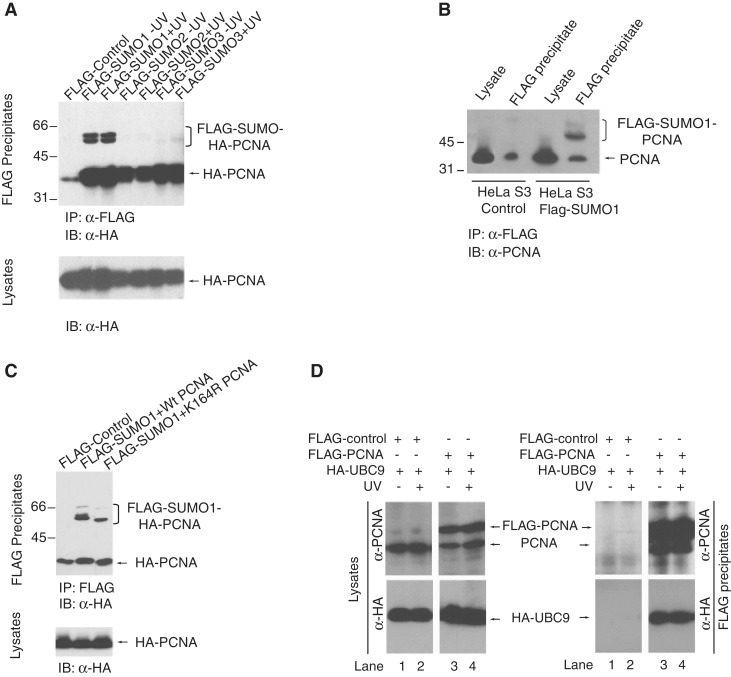
To detect SUMOylation of endogenous human PCNA in vivo cells expressing hemagglutinin epitope tagged (HA)-PCNA together with either FLAG epitope-tagged (FLAG)-SUMO1, or FLAG-SUMO2, or FLAG-SUMO3 (Figure 1A and C) or stably expressing FLAG-SUMO1 (Figure 1B) were lysed and total cell extracts in a buffer containing 50 mM Tris–HCl (pH7.5), 200 mM NaCl, 1% NP-40, 0.1% SDS, 5 mM EDTA, 10% glycerol and 1 mM PMSF were used for immunoprecipitation on FLAG beads (Sigma) followed by immunoblot detection with anti-HA (Roche 3F10) or anti-PCNA (Santa Cruz PC-10) antibodies.
Figure 1.
In vivo SUMO modification of human PCNA. (A) HEK293T cells were co-transfected with HA-PCNA, His-UBC9 and either FLAG-SUMO1, or FLAG-SUMO2, or FLAG-SUMO3. In 48 h, post-transfection cells were UV-treated (30 J/m2) or mock-irradiated and, after 3 h lysed and immunoprecipitated on FLAG-beads. FLAG-SUMO precipitates were immunoblotted with anti-HA antibody to detect PCNA and the SUMO-modified forms of PCNA. The lower panel shows the anti-HA western blot of the lysates. (B) Cell lysates and FLAG immunoprecipitates from control HeLa S3 cells and HeLa S3 cells stably expressing FLAG-SUMO1 were immunoblotted with anti-PCNA antibody to detect SUMOylated forms of endogenous PCNA. (C) HEK293T cells were co-transfected with FLAG-SUMO1, His-UBC9 and either HA-PCNA or K164R PCNA and 48-h post transfection cells were lysed and immunoprecipitated on FLAG-beads. FLAG-SUMO1 precipitates were immunoblotted with anti-HA antibody to detect the effect of the K164R mutation on the SUMOylation of PCNA. (D) Interaction of PCNA with UBC9 was tested by co-expressing FLAG-PCNA and HA-UBC9 in HEK293 cells followed by FLAG immunoprecipitation and then by western blot analysis with anti-HA and anti-FLAG antibodies for UBC9 and PCNA, respectively.
Frequency of I-SceI induced and spontaneous recombination were measured by a modified GFP-based reporter system as described in the Supplementary Methods (28).
Cell survival was determined using Vialight Plus cell proliferation/cytotoxicity Kit assay (Lonza) according to the manufacturer’s instructions.
Monitoring DSB formation by detection of γH2AX foci and neutral comet assay
For the detection of γH2AX foci cells stably expressing FLAG, FLAG-SUMO1, FLAG-PCNA or FLAG PCNA-SUMO1 were grown on coverslips and mock- or MMS (0.01%)-treated for 1 h before immunostaining with the anti-γH2AX antibody (Upstate 05-636) and analysis by confocal laser scanning microscope.
For neutral comet assays (29), cells stably expressing FLAG, FLAG-PCNA or FLAG-PCNA-SUMO1 were treated with 0.01% MMS or mock for 1 h. After various time points, cells were embedded in agarose blocks and analysed by non-denaturing electrophoresis conducted at 4°C for 20 min at the electric field strength of 25 V and 15 mA as described in the Supplementary Methods. The data were analysed for statistical significance using the Student’s t-test. A value of P < 0.001 was considered to be statistically significant for comparison between data sets.
RESULTS
In vivo modification of human PCNA by SUMO
In our previous studies on polyubiquitin modification of human PCNA, we occasionally detected higher mobility PCNA shifts that raised the possibility that covalent attachment of SUMO to human PCNA might occur (30,31). To test directly for SUMOylation of human PCNA, we transiently co-expressed HA-tagged PCNA with FLAG-tagged SUMO1, SUMO2 or SUMO3 in human cells, and then performed FLAG immunoprecipitations. Western blotting of the resulting samples with anti-HA antibody established that PCNA was indeed SUMOylated (Figure 1A). SUMOylation of human PCNA was further confirmed by HA immunoprecipitations of the samples from the same transfections and western blotting with anti-FLAG antibody (Supplementary Figure S1). Notably, we detected two higher mobility shifts suggesting at least two potential SUMOylation sites in human PCNA, and although SUMO1 predominantly conjugated to PCNA weak attachment of SUMO2 and SUMO3 was also detectable. Furthermore, the SUMOylation of PCNA did not require exogenous DNA damage (Figure 1A). Next, we also evaluated the endogenous PCNA SUMOylation in cells stably expressing FLAG-SUMO1 by FLAG immunoprecipitation followed by probing with anti-PCNA antibody (Figure 1B). Also, we tested for the SUMO1 acceptor residues in human PCNA by mutating the highly conserved lysine 164 residue the main SUMO acceptor residue of yeast PCNA, to arginine, which indeed eliminated one of the main SUMOylated PCNA species (Figure 1C). Nevertheless, similar experiments using all 16 single lysine mutant PCNAs did not help to unambiguously define the other SUMO attachment sites in PCNA (data not shown), presumably because SUMOylation of its different acceptor lysine residues can give rise to similar mobility PCNA shifts. In line with PCNA SUMOylation, PCNA could be co-immunoprecipitated with UBC9, the SUMO conjugating enzyme (Figure 1D).
SUMOylation of human PCNA in a reconstituted system using purified proteins
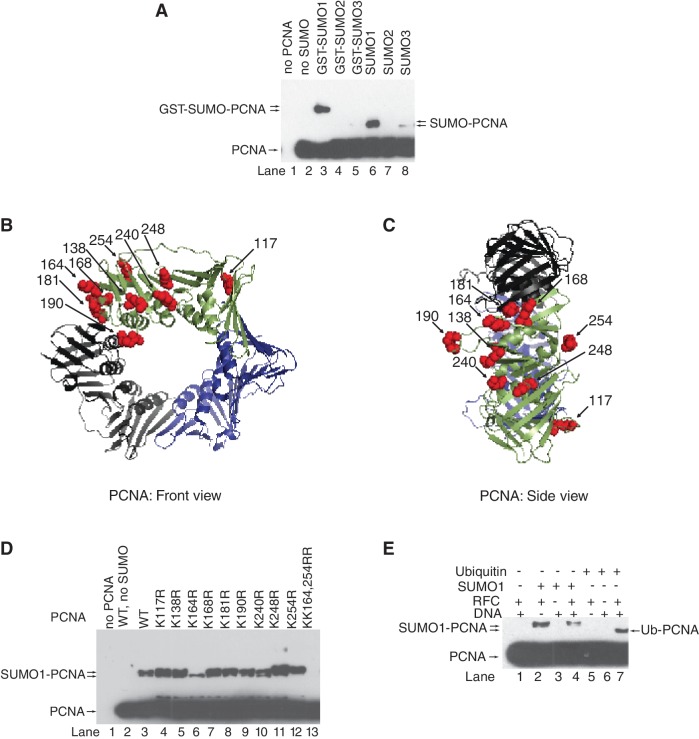
To further characterize, we set up an in vitro PCNA SUMOylation system using purified E1 SUMO-activating enzyme (SAE1/SAE2), E2 SUMO-conjugating enzyme (Ubc9), various E3 SUMO-ligases (Pias1, Pias2, Pias3 and Pias4) and all three SUMO isoforms (SUMO1, SUMO2 and SUMO3; Figure 2 and Supplementary Figure S2). Consistently with the in vivo findings, we established that out of the three SUMO isoforms, only SUMO1 could be efficiently conjugated to PCNA (Figure 2A) and that the lysine 164 residue of PCNA was one of the main SUMO attachment site (Figure 2D). In addition, using surface lysine mutant PCNAs, we managed to identify the K254 residue as a second SUMOylation site in PCNA that locates in a consensus ΨKxE SUMO attachment site, where Ψ is an aliphatic residue (Figures 2D). Consistently with alternate lysine modifications that we concluded from our in vivo experiments, at higher enzyme concentrations, new SUMO-PCNA shifts also became apparent, particularly for certain lysine mutants (Supplementary Figure S2A). Strikingly, PCNA SUMOylation was dependent on replication factor C (Figure 2E), but did not require any of the four PIAS E3 ligases (Supplementary Figure S2B). Moreover, comparing PCNA SUMOylation with ubiquitylation in the presence of RFC, nicked plasmid DNA or their combination revealed that in contrast to ubiquitylation, which requires RFC-dependent loading of PCNA onto DNA, PCNA SUMOylation was dependent on RFC but not DNA (Figures 2E and Supplementary Figure S2C). Thus, interaction between PCNA and RFC is a prerequisite for PCNA SUMOylation presumably by exposing residues in PCNA or giving access for Ubc9.
Figure 2.
In vitro SUMO modification of human PCNA. (A) in vitro SUMOylation reaction of human PCNA (40 nM) was carried out in the presence of purified SAE1/2 (10 nM), Ubc9 (100 nM), RFC (10 nM) nicked PUC19 plasmid DNA (2 nM) and either GST-SUMO1, or GST-SUMO2, or GST-SUMO3, or SUMO1, or SUMO2, or SUMO3 (500 nM) at 37°C for 60 min. Samples containing unmodified and SUMOylated PCNA were separated on 10% denaturing polyacrylamide gel and visualized by western blot using anti-PCNA antibody. Structure of human PCNA from the front (B) and side (C) views; surface lysine residues are represented by red spheres (K117, K138, K164, K168, K181, K190, K240, K248 and K254). PCNA structures showing the surface lysine residues were generated using the PyMOL version 0.96 by DeLano scientific (http.//www.pymolsourceforge.net). (D) Wild-type and lysine point-mutant PCNA samples were subjected to in vitro SUMOylation reaction as described above. (E) In vitro SUMOylation and ubiquitylation reactions of PCNA were compared in the absence or presence of combinations of RFC and nicked plasmid DNA as indicated.
Characterization of PCNA-SUMO fusion protein
To directly investigate the role of SUMO modification of human PCNA, we sought to generate a C-terminal fusion of SUMO1 to PCNA resulting in a PCNA-SUMO1 conjugate, a known strategy (32–36) used to study the effect of SUMOylation of target proteins (Figure 3A). Purified PCNA and PCNA-SUMO1 chimera (Supplementary Figure S3A) stimulated the DNA synthetic activity of translesion polymerases η, κ and ι equally well indicating the proficiency of loading PCNA-SUMO1 by RFC and suggesting that the SUMO-moiety on PCNA does not affect the interaction between PCNA and translesion synthesis polymerases (Figure 3B). Also, PCNA-SUMO1 was proficient in ubiquitylation reaction and localized to replication foci similarly to PCNA, further corroborating the functionality of the PCNA-SUMO1 chimera (Supplementary Figure S3B and Figure 3C). In addition, comparing flow cytometric profiles of stable cell lines expressing PCNA or PCNA-SUMO1 confirmed that PCNA-SUMO1 did not influence the cell cycle (Figure 3D and Supplementary Figure S4B).
PCNA-SUMO1 fusion inhibits spontaneous as well as DSB-induced homologous recombination
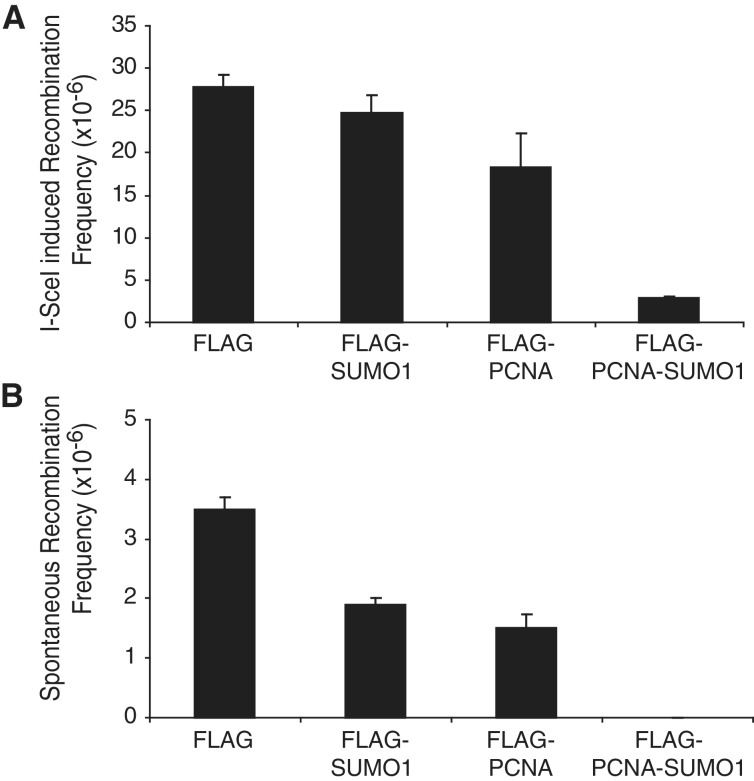
To measure the effect of the SUMO modification of PCNA on the frequency of homologous recombination, we employed a chromosomally integrated reporter system, which measures the reconstituted expression of green fluorescent protein (GFP) as events of homologous recombination. Supporting the findings in yeast studies, the expression of PCNA-SUMO1 conjugate in human cells significantly lowered the rate of I-SceI-generated DSB-induced homologous recombination (Figures 4A and Supplementary Figure S4A), which indicated that SUMO modification of human PCNA can control recombination. Strikingly, the presence of PCNA-SUMO1 in human cells had even stronger inhibitory effect on spontaneous recombination, which was almost completely eliminated (Figure 4B) indicating that it might prevent formation of DSBs. These experiments suggested that PCNA-SUMO1 fusion could have two seemingly independent functions in preventing recombination; on one hand, it directly inhibits DSB-initiated homologous recombination, and on the other hand, it could prevent DSB formation, which indirectly inhibits recombination.
Figure 4.
Effect of SUMO modification of PCNA on homologous recombination. (A) I-SceI-induced recombinations and (B) spontaneous recombinations were measured as GFP positive cells after expressing control FLAG, FLAG-SUMO1, FLAG-PCNA or FLAG-PCNA-SUMO1. Error bars show standard deviation of the data obtained from three independent experiments.
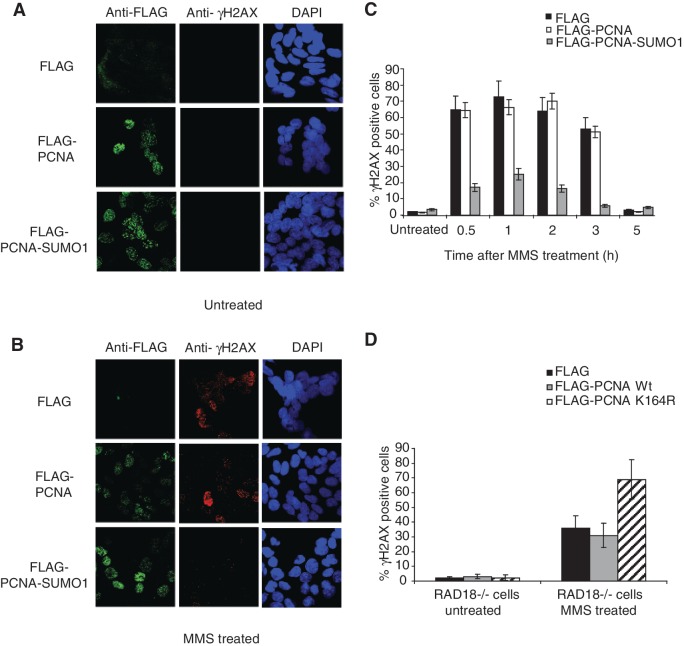
PCNA-SUMO1 reduces the accumulation of γ-H2AX foci
To test for inhibition of DSB formation by PCNA-SUMO1, we treated HeLa cells stably expressing PCNA-SUMO1 with methyl methanesulphonate (MMS), which does not generate DSBs directly, but induces replication fork stalling, which can then be converted into DSBs (3). The occurrence of MMS-induced DSBs was measured by a sensitive indicator of DNA DSBs in cells, histone H2AX phosphorylation (γ-H2AX). Notably, cells expressing PCNA-SUMO1 showed much fewer MMS-induced γ-H2AX foci than control cells as revealed by anti-γ-H2AX immunostaining (Figure 5A and B). Moreover, kinetic analysis of γ-H2AX foci formation and disappearance further confirmed this finding and revealed no difference in the kinetics of DSB repair indicating that the effect of PCNA-SUMO1 indeed can stem from the inhibition of DSB formation (Figure 5C and Supplementary Figure S4C). These data were further confirmed in HCT116 and RAD18−/− cell lines transiently transfected with PCNA-SUMO1 and, in parallel, using additional control transfections with SUMO1 and PCNA-ubiquitin conjugate apart from PCNA alone and control cell lines. We found PCNA-SUMO1 specific inhibition of DSB formation in HCT116 cell lines, as well (Supplementary Figure S5A and B).
Figure 5.
SUMO modification of PCNA reduces the accumulation of γH2AX foci. (A) HeLa cells stably expressing FLAG, FLAG-PCNA or FLAG-PCNA-SUMO1 were fixed 3 h after mock or (B) MMS treatments and stained with anti-FLAG (green), anti-γH2AX (red) and DAPI (blue). (C) Percentage of cell populations that showed more than two foci for γH2AX at 0.5, 1, 2, 3 and 5 h after MMS treatment was calculated from three independent experiments; error bars show standard deviations. (D) Effect of the expression of K164R PCNA in RAD18−/− HCT116 cells was revealed by γH2AX staining 3 h after MMS treatment as described above.
PCNA SUMOylation site mutants elevate the accumulation of γ-H2AX foci
To validate that SUMOylation of endogenous PCNA has similar consequences, we over-expressed K164R PCNA, which can compete against K164 SUMOylation of endogenous PCNA, in the Rad18−/− cell line where the effect of Rad18-dependent K164 PCNA ubiquitylation can be ruled out (Supplementary Figure S4D). In line with the assumption that PCNA-SUMO1 inhibits DSB formation, expression of K164R PCNA caused a significant increase in the number of γ-H2AX foci after MMS treatment (Figure 5D). We also tested the consequences of the overexpression of the alternative SUMOylation site mutant K254R PCNA and found elevated γ-H2AX foci level. Moreover, expression of the KK164, 254RR double mutant showed some additive effect (Supplementary Figure S5C and S5D). Taken together that mutations in SUMOylation sites of PCNA facilitate DSB formation with the results of overexpression of inhibiting DSB formation by PCNA-SUMO1 fusion suggest a role for SUMOylation of PCNA in preventing DSB formation.
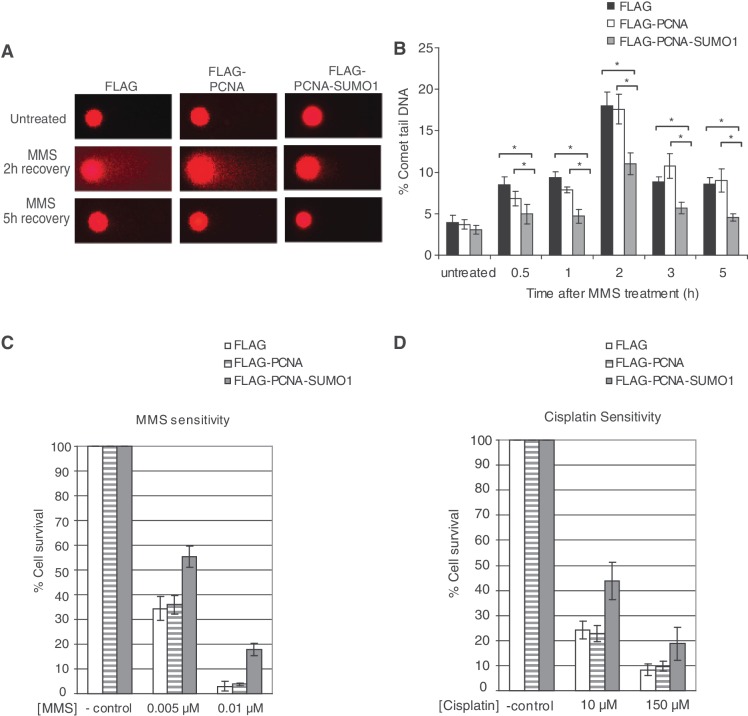
PCNA-SUMO1 suppresses DNA DSB formation
To examine directly whether PCNA-SUMO1 prevents DSB formation, we adopted the comet assay under neutral conditions, which allows the detection of DSBs at single cell level (37,38). Remarkably, cells expressing PCNA-SUMO1 exhibited a lower amount of fragmented double-stranded DNA upon replication-blocking MMS treatment than did control cell lines expressing PCNA (Figure 6A). For example, 2 h after MMS treatment the comet tails were 40% shorter in the presence of PCNA-SUMO1 fusion than in the control sample expressing PCNA. Furthermore, kinetic analysis by measuring the percentage of double-stranded DNA present in the tail as compared to the intact head DNA proved that PCNA-SUMO1 expressing cells are less prone to DSB formation (Figure 6B), which is in accordance with our data obtained by γ-H2AX staining. As can be expected from these findings, the PCNA-SUMO1 conjugate provided enhanced cell survival against treatment with DNA damaging agents such as MMS and cisplatin but not against UV suggesting that the rescue mechanisms could depend on the particular type of DNA damage at stalled replication forks (Figure 6C and D and Supplementary Figure S6). Thus, it is possible that UV lesions are more efficiently bypassed, perhaps due to Poleta specificity to this damage, than other lesions irrespective of the presence of PCNA-SUMO1. Nevertheless, these data collectively suggest that SUMOylated PCNA can provide protection against DSB formation, if replication stalls at an unrepaired lesion.
Figure 6.
SUMO modification of PCNA limits MMS-induced DNA DSB formation and effects DNA damage sensitivity. (A) Representative images of HeLa cells stably expressing FLAG, FLAG-PCNA or FLAG-PCNA-SUMO1 either untreated or 2 and 5 h after MMS treatment subjected to comet assay under neutral conditions. (B) Percentage of comet tail DNA, a measure of DNA DSBs, at 0.5, 1, 2, 3 and 5 h after MMS treatment were calculated from three independent experiments using three independent stable cell lines; error bars show standard deviations. Significant difference was detected between the levels of double stand breaks of FLAG FLAG-PCNA-SUMO1, FLAG-PCNA and FLAG-PCNA-SUMO1 expressing cell lines *P < 0.001). HeLa cells stably expressing FLAG, FLAG-PCNA and FLAG-PCNA-SUMO1 were assayed for cell survival after treatment with 0.005 and 0.01 µM MMS (C) or 10 and 150 µM Cisplatin (D) as compared to the untreated cells. Error bars show standard deviation from the results of three independent experiments.
DISCUSSION
Both in vitro and in vivo experiments in this study demonstrate the SUMOylation of human PCNA with all three SUMO paralogues, with SUMO1 being predominant. We revealed that PCNA SUMOylation depends on RFC, suggesting the importance of the presence of PCNA in a replication ensemble for its SUMOylation to occur. Remarkably, we observed that cells encountered less DSBs in the presence of PCNA-SUMO1 fusion protein as measured by phosphohistone foci and neutral comets. In line with these data, linear attachment of SUMO1 to PCNA not only lowered I-SceI-generated DSB-induced recombination frequencies but also dramatically reduced spontaneous recombination and conferred damage resistance against replication fork blocking lesions. Although previous studies have used successfully linear PCNA-SUMO fusion to obtain insight into the function of SUMO modification of PCNA, some concern remained that what degree the PCNA-SUMO1 fusion protein mirrors the endogenously conjugated PCNA function. Thus, to provide further evidence for the action of SUMO modification of PCNA in limiting DSB formation, we overexpressed SUMOylation-site mutant PCNA proteins, the K164R, K254R and KK164,254RR PCNA proteins in the Rad18−/− cell line that is defective in Rad18-dependent K164 PCNA ubiquitylation and found an increase in phosphohistone foci. Thus, impairment of SUMOylation of human PCNA facilitates DSB formation as predicted from the results with PCNA-SUMO1 fusion.
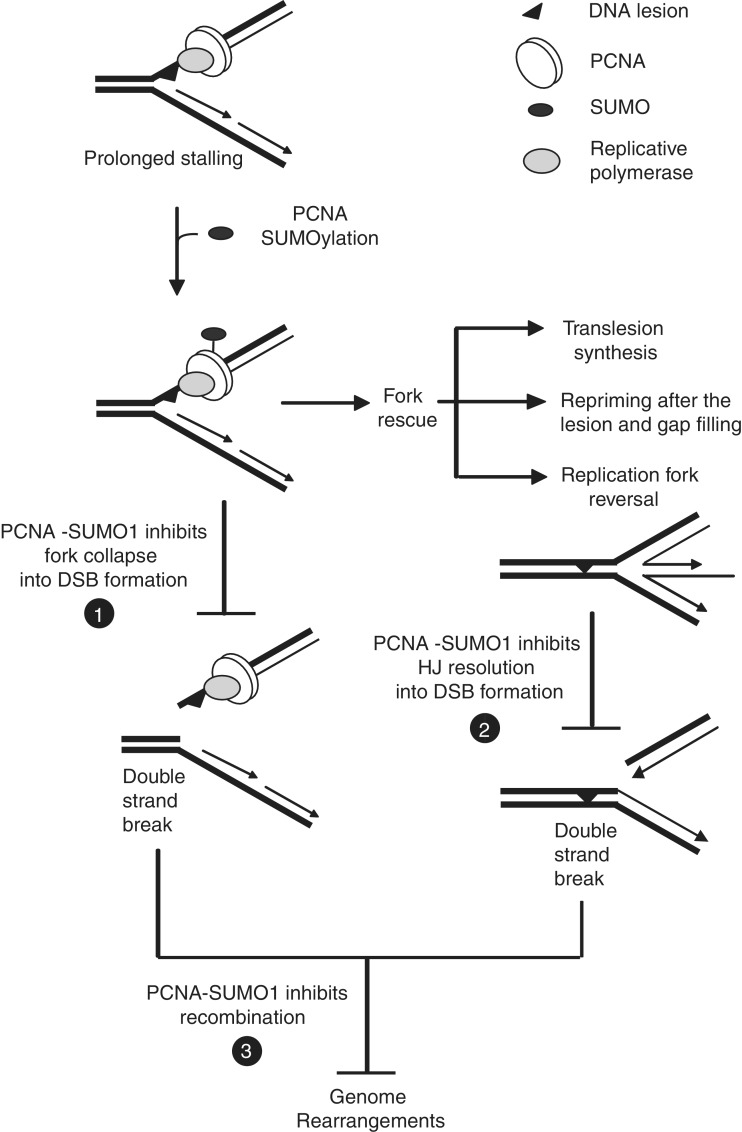
For the first time, our results reveal the importance of the SUMO-PCNA-dependent mechanisms in human cells, and we show that PCNA SUMOylation-dependent regulatory functions are not restricted to organisms with a naturally high rate of recombination such as yeast. Importantly, this study also provides a previously unknown role for the SUMO modification of PCNA in limiting DSB formation, and suggests that in the presence of SUMO-modified PCNA replication forks encountering DNA lesions are less likely to be converted to DSBs (Figure 7). We envision various scenarios for the mechanism by which PCNA-SUMO could prevent DSB formation, such as providing a higher stability for the replication ensemble, inhibiting the access of particular fork remodelling enzymes or nucleases and promoting damage bypass by giving access to players of translesion synthesis or template switch-dependent replication. Moreover, transient fork reversal could provide an error-free mechanism for damage bypass whereas stabilized reversed forks could be accessed by Holliday-junction-specific nucleases leading to DSB. Therefore, we hypothesize that in the presence of PCNA-SUMO fork reversal remains transient and productive (Figure 7). Although these scenarios are not mutually exclusive, supportive yeast studies have indicated that PCNA-SUMO can facilitate the Rad6–Rad18-dependent damage tolerance pathways (19,22).
Figure 7.
Schematic representation of the suggested multiple roles of SUMOylated PCNA at stalled replication forks. We recommend a novel function for PCNA-SUMO in preventing DSB formation if replication stalls at DNA lesions (number one in filled circle) or in preventing the conversion of reversed replication forks to one-ended DSB (number two in filled circle). We also suggest that PCNA-SUMO prevents DSB-induced recombination and genome rearrangements (number three in filled circle).
Further support for our model on human PCNA-SUMO function is provided by the recent characterization of human PARI that has been carried out in parallel with our study (27). PARI exhibits a PCNA-, a Rad51- and a SUMO-interacting motif, and it binds preferentially to PCNA-SUMO1 fusion protein over PCNA and can disrupt Rad51 nucleofilaments. Since PARI also suppresses homologous recombination, it was proposed that PARI resembles yeast Srs2, which is recruited to stalled fork by PCNA-SUMO and abolishes the access of Rad51 (12,14). While the role of PARI was described in great detail, no functional assay for the cellular function of PCNA-SUMO has been carried out. Taken together, the findings on PARI with our functional assay on PCNA-SUMO strongly support their functional interaction. Human FBH1 and RTEL1 have also been implicated as functional homologues of yeast Srs2, and it would be interesting to examine whether these proteins can also interact with and compete for PCNA-SUMO, which could provide an additional level of recombination control in human cells.
Better understanding the role of SUMOylation of human PCNA and its interplay with its interacting partners can give more insight into how human cells suppress aberrant recombination events and cancer development. In summary, our study reveals the importance of SUMO modification of human PCNA in preventing DSB formation at stalled replication fork and suggests that PCNA SUMOylation has a more complex role in safeguarding genome integrity than previously anticipated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Figures 1–6 and Supplementary References [39–44].
FUNDING
Howard Hughes Medical Institute [55005612]; Hungarian Science Foundation [OTKA 77495]; Hungary-Serbia IPA Cross-border Co-operation Programme [HUSRB/1002/214/126]. Funding for open access charge: Hungarian Science Foundation [OTKA 77495].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ronald T. Hay for providing SUMO-1, SUMO-2, SUMO-3, UBC9 and SAE1/SAE2 plasmids for protein expressions, Barnabas Szakal for sharing his expertise on cell biological assays and carrying out preliminary experiments on this project, and Zsofia Kohajda for her technical assistance.
REFERENCES
- 1.Prakash S, Sung P, Prakash L. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikolova T, Ensminger M, Löbrich M, Kaina B. Homologous recombination protects mammalian cells from replication-associated DNA double-strand breaks arising in response to methyl methanesulfonate. DNA Repair. 2010;9:1050–1063. doi: 10.1016/j.dnarep.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair. 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 6.Unk I, Hajdü I, Blastyák A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair. 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 7.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl Acad. Sci. USA. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 2010;6:e1001205. doi: 10.1371/journal.pgen.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita YM, Okada T, Matsusaka T, Sonoda E, Zhao GY, Araki K, Tateishi S, Yamaizumi M, Takeda S. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 2002;21:5558–5566. doi: 10.1093/emboj/cdf534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 13.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Bozza W, Zhuang Z. Ubiquitination of PCNA and its essential role in eukaryotic translesion synthesis. Cell Biochem. Biophys. 2011;60:47–60. doi: 10.1007/s12013-011-9187-3. [DOI] [PubMed] [Google Scholar]
- 16.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 17.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Hishida T, Kubota Y, Carr AM, Iwasaki H. RAD6-RAD18-RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature. 2009;457:612–615. doi: 10.1038/nature07580. [DOI] [PubMed] [Google Scholar]
- 19.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 20.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 21.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 22.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seong C, Colavito S, Kwon Y, Sung P, Krejci L. Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase. J. Biol. Chem. 2009;284:24363–24371. doi: 10.1074/jbc.M109.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 25.Arakawa H, Moldovan GL, Saribasak H, Saribasak NN, Jentsch S, Buerstedde JM. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS. Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leach CA, Michael WM. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J. Cell. Biol. 2005;171:947–954. doi: 10.1083/jcb.200508100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, D'Andrea AD. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 30.Unk I, Hajdü I, Fátyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl Acad. Sci. USA. 2008;105:3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unk I, Hajdü I, Fátyol K, Szakál B, Blastyák A, Bermudez V, Hurwitz J, Prakash L, Prakash S, Haracska L. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl Acad. Sci. USA. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hishida T, Ohya T, Kubota Y, Kamada Y, Shinagawa H. Functional and physical interaction of yeast Mgs1 with PCNA: impact on RAD6-dependent DNA damage tolerance. Mol. Cell Biol. 2006;26:5509–5517. doi: 10.1128/MCB.00307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long J, Wang G, He D, Liu F. Repression of Smad4 transcriptional activity by SUMO modification. Biochem. J. 2004;379:23–29. doi: 10.1042/BJ20031867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross S, Best JL, Zon LI, Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell. 2002;10:831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- 35.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc. Natl Acad. Sci. USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- 37.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 38.Patro BS, Frøhlich R, Bohr VA, Stevnsner T. WRN helicase regulates the ATR-CHK1-induced S-phase checkpoint pathway in response to topoisomerase-I-DNA covalent complexes. J. Cell Sci. 2011;124:3967–3979. doi: 10.1242/jcs.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol. Cell. Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc. Natl Acad. Sci. USA. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol. Cell. Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiomi N, Mori M, Tsuji H, Imai T, Inoue H, Tateishi S, Yamaizumi M, Shiomi T. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2007;35:e9. doi: 10.1093/nar/gkl979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 44.Wojewódzka M, Buraczewska I, Kruszewski M. A modified neutral comet assay: elimination of lysis at high temperature and validation of the assay with anti-single-stranded DNA antibody. Mutat. Res. 2002;518:9–20. doi: 10.1016/s1383-5718(02)00070-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.