Abstract
Deamination of cytosine (C), 5-methylcytosine (mC) and 5-hydroxymethylcytosine (hmC) occurs spontaneously in mammalian DNA with several hundred deaminations occurring in each cell every day. The resulting potentially mutagenic mispairs of uracil (U), thymine (T) or 5-hydroxymethyluracil (hmU) with guanine (G) are substrates for repair by various DNA glycosylases. Here,weshowthat targetedinactivation of the mouse Smug1 DNA glycosylase gene is sufficient to ablate nearly all hmU-DNA excision activity as judged by assay of tissue extracts from knockout mice as well as by the resistance of their embryo fibroblasts to 5-hydroxymethyldeoxyuridine toxicity. Inactivation of Smug1 when combined with inactivation of the Ung uracil-DNA glycosylase gene leads to a loss of nearly all detectable uracil excision activity. Thus, SMUG1 is the dominant glycosylase responsible for hmU-excision in mice as well as the major UNG-backup for U-excision. Both Smug1-knockout and Smug1/Ung-double knockout mice breed normally and remain apparently healthy beyond 1 year of age. However, combined deficiency in SMUG1 and UNG exacerbates the cancer predisposition of Msh2−/− mice suggesting that when both base excision and mismatch repair pathways are defective, the mutagenic effects of spontaneous cytosine deamination are sufficient to increase cancer incidence but do not preclude mouse development.
INTRODUCTION
Uracil (U) can arise in DNA through either misincorporation of dUTP during DNA synthesis (giving rise to U:A pairs) or through deamination of cytosine (C) within the DNA duplex (resulting in U:G mispairs). In addition to C, mammalian cells also contain 5-methylcytosine (mC) and 5-hydroxymethylcytosine (hmC) (1). All three forms of cytosine undergo spontaneous deamination with an estimated 100–500 genomic C bases undergoing deamination per cell every day (2). Thus, if left unrepaired, cytosine deamination would be a major source of spontaneous mutation.
Several DNA glycosylases (UNG, SMUG1, TDG and MBD4) have been described in mammals to remove U or its derivatives from DNA (3). The UNG uracil-DNA glycosylase [whose ancestral counterpart in Escherichia coli was the first to be described (4)] is abundant, widely distributed and can act on U in single or double-stranded DNA. SMUG1, while originally named on the basis of its ability to excise U from single-stranded DNA [‘single-strand-selective monofunctional uracil-DNA glycosylase’; (5)] is in fact much more active on U in double-stranded DNA, especially if mispaired with G (6–10). Thymine-DNA glycosylase (TDG) was named for its ability to excise thymine (T) from T:G mispairs (which arise through spontaneous deamination of mC residues that naturally occur in the context of mCpG dinucleotides), but can also act effectively on U when encountered as a U:G mispair as judged by assays of the recombinant enzyme (11). MBD4 contains both methyl-binding and glycosylase domains and, in in vitro assays, shows a similar target specificity (though lower specific activity) to TDG (12).
Although C deamination has received much attention as a source of spontaneous DNA damage, it has more recently become evident that purposeful enzyme-catalysed deamination of C is used to trigger gene diversification in the immune system. The U:G mismatches generated in the immunoglobulin loci of activated B lymphocytes through the action of the enzyme activation-induced cytidine deaminase (AID) are recognized by the UNG DNA-glycosylase as well as by the MSH2/MSH6 mismatch recognition protein in order to achieve somatic hypermutation of the immunoglobulin variable region and/or class switch recombination of the constant regions (13). The mammalian APOBEC3 enzymes are host restriction factors active against retroviruses which generate U in the DNA of retroviral replication intermediates through C deamination, although the way in which uracilation contributes to retroviral restriction is not fully resolved (14,15).
There have been several suggestions that the DNA glycosylases active on U may also play a role in the erasure of mC from the genome that occurs during mammalian development. An early suggestion was that TDG or MBD4 might directly excise mC from the DNA, since both enzymes exhibit an (albeit very weak) excision activity on mC:G base pairs (16–18). More recently, it has been proposed that mC erasure might involve either deamination of mC to T (catalyzed by a member of the AID/APOBEC family) with subsequent TDG-dependent repair of the resulting T:G mismatch (19–22) or alternatively that oxidation of mC to hmC by one of the TET family dioxygenases is followed by AID/APOBEC-catalysed deamination to hmU and SMUG1-mediated excision of the hmU (23,24). However, definitive evidence demonstrating a role for a DNA glycosylase in physiological pathways of mC demethylation in animal cells is lacking.
Although most of our knowledge about the different DNA glycosylases comes from studies of their substrate specificities and activities as conducted by biochemical assay of the various purified or recombinant proteins or as analysed in cell extracts, genetic studies can obviously give major insights into the functions of the different glycosylases in vivo. Ung−/− mice are largely healthy although they show a deficiency in antibody diversification and an increased propensity to tumour development after 1 year of age (25–27). MBD4−/− mice show increased intestinal tumourigenesis in a cancer-susceptible background (28,29). TDG-deficiency results in embryonic lethality (30), an observation that has especially fuelled interest in possible TDG-dependent mechanisms for genome demethylation. SMUG1-deficient mice have not so far been described although recent work with cell-lines suggests that SMUG1 might play a more major role in excision of U from U:G mismatches in mouse as opposed to human cells (31).
We were especially interested in the effects of SMUG1 deficiency since enforced expression of SMUG1 is able to partially reverse the mutator phenotype of Ung− yeast (32) as well as rescue the antibody gene diversification defect of UNG-deficient B lymphocytes (33,34). This suggested that SMUG1 might act as a backup for UNG, possibly accounting for the relatively mild phenotype of UNG-deficient animals.
MATERIALS AND METHODS
Generation and breeding of Smug1−/− mice
Smug1−/− mice were generated from targeted C57BL/6 embryonic stem (ES) cells that were obtained from the European Conditional Mouse Mutagenesis Consortium (EUCOMM, project ID 23057). These ES cells had been targeted with a construct (35) which, on homologous targeting, will result in the insertion of a β-galactosidase/neoR gene-trap expression cassette into the intron separating the two coding exons of Smug1 (Figure 1A). Transcription of the targeted Smug1 allele is expected to result in the production of a severely truncated SMUG1 protein comprising only the N-terminal 97 amino acids encoded in Smug1 exon 1 followed by 40 amino acids produced by a frameshifted reading of the mouse engrailed 2 gene encoded by the inserted gene trap.
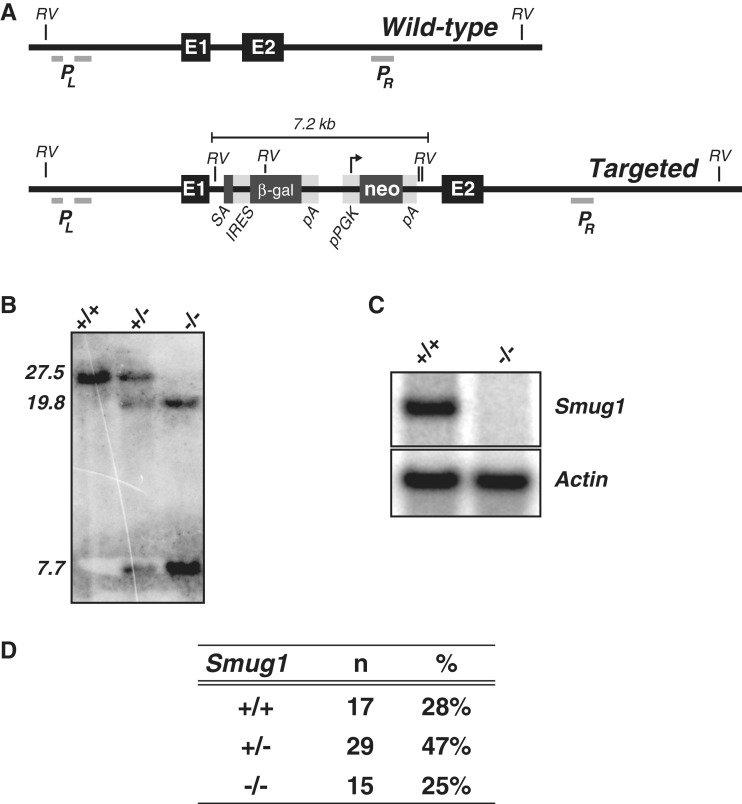
Figure 1.
Targeted inactivation of the Smug1 gene. (A) Map of the wild-type and targeted Smug1 locus. The wild-type Smug1 locus comprises two coding exons (E1 and E2) that, respectively, encode the first 97 and remaining 183 amino acids of the protein, with the latter being essential for catalytic activity (37). The targeted allele is predicted to contain a 7.2-kb cassette insertion into the intron separating these coding exons. This gene-trap cassette comprises a splice acceptor site [SA] (derived from the mouse engrailed 2 gene) followed by: (i) an IRES-driven β-galactosidase [β-gal] reporter and polyadenylation signal [pA] and (ii) an aminoglycoside phosphotransferase cDNA [conferring neomycin resistance (neo)] whose expression is driven from a phosphoglycerate kinase promoter [pPGK]. The locations of the EcoRV restrictions sites [RV] and of the probes PL and PR that were used for Southern-blot analysis (and that lie to the left and right of the DNA used for gene targeting) are indicated. (B) Southern-blot analysis of splenocyte DNA from wild-type, homozygous targeted and heterozygous mice digested with EcoRV and hybridized with a mixture of probes PL and PR . Whereas the wild-type locus is predicted to give a single 27.5-kb EcoRV fragment that hybridizes with both probes, the targeted allele is anticipated to give a 7.7-kb band hybridizing with PL and a 19.8-kb band hybridizing with PR. (C) Northern-blot analysis of liver RNA from wild-type and homozygous targeted mice hybridized with a probe internal to Smug1 exon 2; β-actin served as a loading control. (D) Genotypes of progeny of inter-heterozygote crosses as determined from tissue biopsies taken at the time of weaning.
Cells of the EUCOMM targeted ES cell clone smug1-C05 were injected into blastocysts from albino C57BL/6-Tyr mice to generate chimeras, which were then crossed with C57BL/6 mice to generate heterozygous and ultimately homozygous Smug1−/− mice. A second line of Smug1−/− mice was established from EUCOMM clone smug1-B07 but showed no differences from animals established using clone smug1-C05 in respect of mouse health and deficiency in glycosylase activity. The Smug1−/− mice were interbred with Ung−/− mice (25) that were kindly provided by Deborah Barnes and Tomas Lindahl (London Research Institute, CRUK, UK) as well as with Msh2−/− mice (36) that were kindly provided by Dr H. te Reile through Dr K. Brown (CRUK Beatson Laboratories, University of Glasgow). Animal studies were approved by the Laboratory of Molecular Biology Ethical Review Committee and performed under United Kingdom Home Office Project License PPL80/2226.
Embryo fibroblast cultures
Spontaneously, immortalized cell lines were obtained through extended cultivation (>20 passages) in DMEM/10% FBS of primary fibroblasts from day 13.5 male embryos of the desired genotypes. Cells were passaged at ∼70% confluence during culture and kept subconfluent for all experiments. For toxicity studies, 3000 cells per well were seeded in a 96-well plate, incubated in medium overnight and then exposed to the compound of interest for 48 h. Cell viability was determined immediately after medium change using Cell Counting Kit 8 (Dojindo Molecular Technologies).
Staining of mouse tissues
Freshly isolated mouse tissues were rinsed with cold PBS, then fixed in 4% paraformaldehyde at 4°C for 1 h. After 3 × 10 min rinses with cold PBS, tissues were stained at 37°C in a solution made by mixing nine parts 0.4 mg/ml 5-bromo-4-chloro-indolyl-β-d-galactopyranoside (X-gal), 1 mM MgCl2 in PBS with one part 0.5 M K3[Fe(CN)6]/0.5 M K4[Fe(CN)6].
Analysis of DNA and RNA
For Southern-blot analysis, DNA was extracted from mouse tail biopsies, digested with EcoRV and subjected to electrophoresis on an 0.8% agarose gel prior to transfer onto Hybond-XL nylon membranes (GE Healthcare). Membranes were then hybridized with PCR-generated probes specific for regions of the Smug1 locus located either up- or downstream of the homology arms used in the targeting construct (Figure 1A). Primers for the downstream probe were 5′-CCCTTTACCTTGAGCCCTTC and 5′-TCTTGGACGTCCTTGTTTCC. Two PCR amplifications yielding products that flank a region of repetitive sequence were used for the upstream probe: 5′-TCAGTGCTGAGGGGCTAGTT/5′-ACATCTGCCAGCCTGTATCC and 5′-GTAAACAGTTAATGGCCCTTGG/5′-GCCTTAAGAAACATGTCACACA.
For analysis of RNA from mouse liver, polyA+ RNA was purified using RNeasy and Oligotex kits (Qiagen) according to the manufacturer’s instructions and aliquots (4 µg) subjected to electrophoresis on 1.2% agarose/MOPS-acetate gels, transferred onto Hybond-XL membranes and hybridized with a 317-nt probe for Smug1 exon 2 generated by PCR using primers 5′-GTTGGGGGCCCTGTGCTGAC and 5′-CCGCCCCACTCCCACTACCA.
DNA-glycosylase assays
Nuclear lysates were prepared according to a modified version of the procedure described in (6). Snap-frozen tissues were thawed on ice and finely minced with a razorblade before passage through a 70 µm nylon mesh cell strainer (BD Falcon). Fragments were washed twice in ice-cold PBS in the presence of protease inhibitors (Roche) and then resuspended in twice the packed cell mass of hypotonic buffer A (10 mM HEPES/KOH pH 7.7, 0.5 mM MgCl2, 10 mM KCl, 1 mM DTT plus protease inhibitors). After 15-min incubation to lyse cell membranes, nuclei were pelleted by centrifugation (2000 g, 10 min) and incubated in two volumes of nuclear lysis buffer (20 mM HEPES/KOH pH 7.7, 0.5 mM MgCl2, 0.42 M NaCl, 0.2 mM EDTA, 1 mM DTT, 25% glycerol plus protease inhibitors). After 20-min incubation on ice, particulates were removed by centrifugation (14 000 g, 10 min) and the supernatants dialysed for 6 h against 25 mM HEPES/KOH pH 7.7, 50 mM KCl, 2 mM DTT through a 10-kD molecular weight cut-off membrane, aliquoted and snap-frozen for storage. Protein content was determined using a BioRad Bradford kit according to the manufacturer’s instructions.
Excision activities in nuclear extracts were determined by mixing 5 µg protein with 25 fmol oligonucleotide substrate and 2.5 U of APE1 (NEB) in 20 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 100 µg/ml BSA (final volume 10 µl) for 1 h at 37°C. After addition of an equal volume of formamide, reactions were boiled for 20 min at 100°C (to enhance cleavage at abasic sites in single-stranded DNA substrates which are poor substrates for APE1) and run on 10% denaturing polyacrylamide/TBE-Urea gels (Invitrogen). The oligonucleotide substrate and cleaved product were visualized using a Typhoon scanner and band intensities quantified using ImageJ. The single-stranded oligonucleotide substrates (3′-fluoresceinated 5′-CATAAAGTGxAAAGCCTGGA or 5′-CATAAAGTGxAAGCCTGGA, where x = U, hmU or FU), and their complements with a centrally placed A or G were obtained HPLC-purified from Integrated DNA Technologies or the Midland Certified Reagent Company.
RESULTS
Mice carrying a targeted β-galactosidase insertion in the Smug1 locus
Embryonic stem cells of C57BL/6 origin were obtained that carried a targeted insertion between the two coding exons of Smug1. The inserted cassette comprises (from 5′- to 3′-ends): an RNA splice acceptor site, an internal ribosomal entry site (IRES), a promoterless β-galactosidase gene and a phosphoglycerate kinase (PGK) promoter-driven aminoglycoside phosphotransferase gene which confers neomycin resistance (Figure 1A). It is thus anticipated that the β-galactosidase gene will be expressed under control of the endogenous Smug1 transcription regulatory signals but that the second exon of Smug1 (which contains most of the SMUG1 protein-coding sequence) will not be transcribed into mRNA. These Smug1:βgal/neo ES cells were injected into blastocysts isolated from albino C57BL/6-Tyr mice and the resultant chimaeras bred to obtain heterozygous mice carrying the Smug1:βgal/neo insertion on one allele in their germline.
Southern-blot analysis of tail DNA from Smug1:βgal/neo heterozygous mice revealed the expected change in Smug1-hybridizing restriction fragments (Figure 1B). Interbreeding of the heterozygotes yielded Smug1:βgal/neo homozygotes at the expected Mendelian frequency with northern-blot analysis revealing that these homozygotes had indeed lost Smug1 mRNA as judged by northern blotting with an exon 2 probe (Figure 1C and D). As indicated under ‘Materials and Methods’ section, the nature of the gene targeting might allow the production of a polypeptide including the first 97 amino acids of SMUG1. However, not only would such a polypeptide lack essential catalytic residues of SMUG1, experiments designed to detect a possible dominant inhibitory effect of liver nuclear extracts from homozygous Smug1:βgal/neo mice in base excision repair assays proved negative (Supplementary Figure 1). Thus, Smug1:βgal/neo homozygous mice are viable, show no obvious impairment in health or fertility with any phenotype likely attributable to the loss of Smug1 gene expression. We designate these animals Smug1−/− mice.
Targeted Smug1 inactivation results in loss of hmU-excision activity
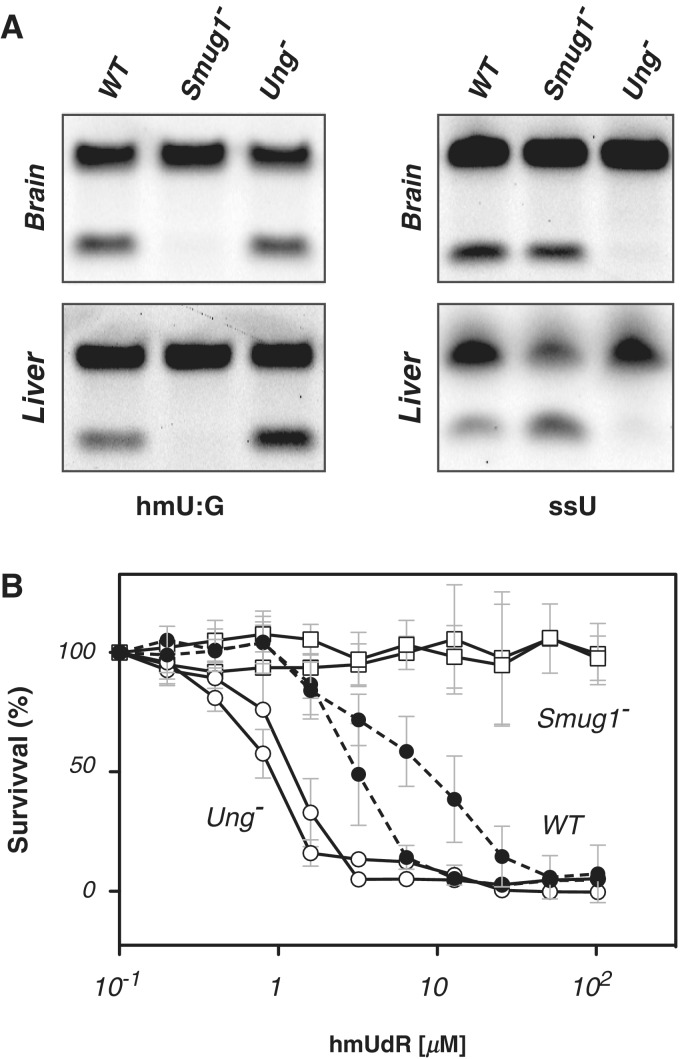
Recombinant SMUG1 has been shown not only to be active in excising U from DNA, but also active on several pyrimidines that carry substitutions at the C5 position such as hmU, 5-formyluracil, 5-hydroxyuracil and 5-fluorouracil (8,37,38). Analysis of both brain and liver cell extracts revealed that Smug1 inactivation results in loss of nearly all detectable hmU-excision activity (without notably impairing the total U-excision activity in the extracts). A similar ablation of hmU-excision activity was noted in all other tissues analysed (data not shown). The results indicate that SMUG1 is the major if not sole enzyme responsible for hmU excision in the mouse (Figure 2A).
Figure 2.
Smug1 inactivation ablates hmU excision activity and gives resistance to hmUdR. (A) Analysis of base excision activity in brain and liver nuclear extracts from wild-type (WT), Ung−/− and Smug1−/− mice assayed on a 3′-fluorescently labelled double-stranded oligodeoxyribonucleotide substrate containing a centrally placed hmU:G mispair (left), as well as on a single-stranded 3′-fluorescently labelled substrate containing a single centrally placed U as an UNG activity control. Excision creates an abasic site that is subject to cleavage by APE1; cleavage products of the 3′-fluorescently labelled oligonucleotides were visualized after polyacrylamide gel electrophoresis. Similar results regarding a loss of hmU-excision activity were obtained on assaying multiple different tissues from Smug1−/− mice of different ages. (B) Effects of a 48-h culture in the presence of various concentrations of hmUdR on the viability of embryo fibroblast cell-lines derived from mice of different genotypes. Survival is expressed relative to that obtained in the absence of hmUdR. The results are shown for triplicate assays performed on two independently derived cell-lines for each indicated genotype.
Smug1−/− embryo fibroblasts are resistant to 5-hydroxymethyldeoxyuridine toxicity
HmU can arise in DNA not only as a result of modification of one of the bases naturally present in DNA but also from misincorporation of 5-hydroxymethyldeoxyuridine (hmUdR) during DNA synthesis. Indeed, hmUdR is toxic to cells (39) with studies in CHO cells having revealed that this toxicity is likely due to excessive excision of misincorporated hmUdR (40,41). To test whether SMUG1 is responsible for hmUdR toxicity in mouse cells, we established embryo fibroblast lines from Smug1−/− as well as SMUG1-proficient control embryos and tested their sensitivity to culture with varying concentrations of hmUdR. The results (Figure 2B) show that deficiency in SMUG1 is indeed sufficient to overcome sensitivity to hmUdR, consistent with SMUG1 being the dominant if not sole glycosylase involved in hmUdR excision from genomic DNA in mice. Curiously, the UNG-deficient cells appear slightly more sensitive to hmUdR than the wild-type controls. We do not know the basis for this increased sensitivity. It does not seem to be due to a compensatory increase in Smug1 expression since RT-PCR analysis reveals no major perturbation in Smug1 RNA levels (data not shown) but it might conceivably reflect an increased efficacy with which SMUG1 itself acts on the hmU in DNA or with which the abasic site is processed in the absence of UNG.
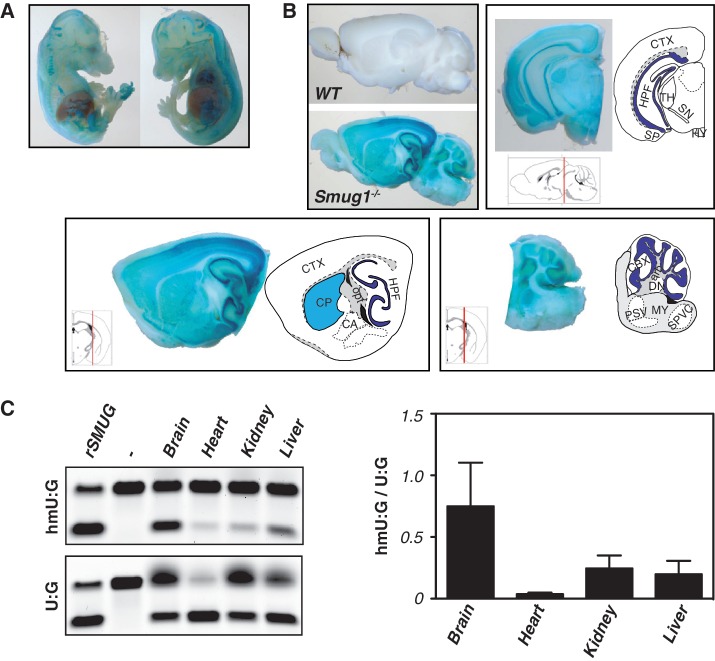
Although SMUG1 is therefore responsible for making mouse cells sensitive to being cultured in the presence of hmUdR, a more likely physiological function of SMUG1 is to mediate the repair of hmU that arises in DNA through either the oxidation of T or the spontaneous deamination of hmC. Indeed, recent data have focused on hmC as a component of mammalian genomic DNA with indications that it is especially abundant in brain (42–44). However, SMUG1-deficiency does not lead to any gross abnormalities in the brain as judged by crude histological examination (Figure 3A and B), even though in keeping with earlier studies (45) we find that brain shows a significantly higher ratio of hmU to U excision activity as compared to extracts from other tissues tested (Figure 3C).
Figure 3.
Activity of the Smug1 locus in different tissues as judged by β-gal reporter and hmU-excision activity. (A and B) X-gal staining of day 13.5 embryos and of sagittal and coronal sections of adult mouse brains from wild-type and Smug1−/− mice (which carry the β-gal/neo gene-trap expression cassette in the Smug1 locus, driven by the endogenous Smug1 promoter) as well as from wild-type (WT) controls. arb, arbor vitae; CA, central amygdalar nuclei; CBX, cerebellar cortex; CP, caudoputamen; CTX, cerebral cortex; DN, dentate nucleus; HPF, hippocampus; HY, hypothalamus; MY, myelencephalon; opt, optical tract; PSV, principal sensory nucleus of the trigeminal; SN, substantia nigra; SP, pyramidal layer; SPVC, spinal nucleus of the trigeminal (caudal part); TH, thalamus. (C) Comparison of the activity of nuclear extracts from various tissues on double-stranded oligodeoxyribonucleotide substrates containing central hmU:G and U:G mismatches. Recombinant SMUG1 (rSMUG; New England Biolabs) served as control. On the right hand side, the excision activity on hmU:G relative to that on U:G is shown for different tissues. Activities were determined as the product band intensity divided by the integrated total band intensity in each assay and is based on the analysis of tissues from three mice.
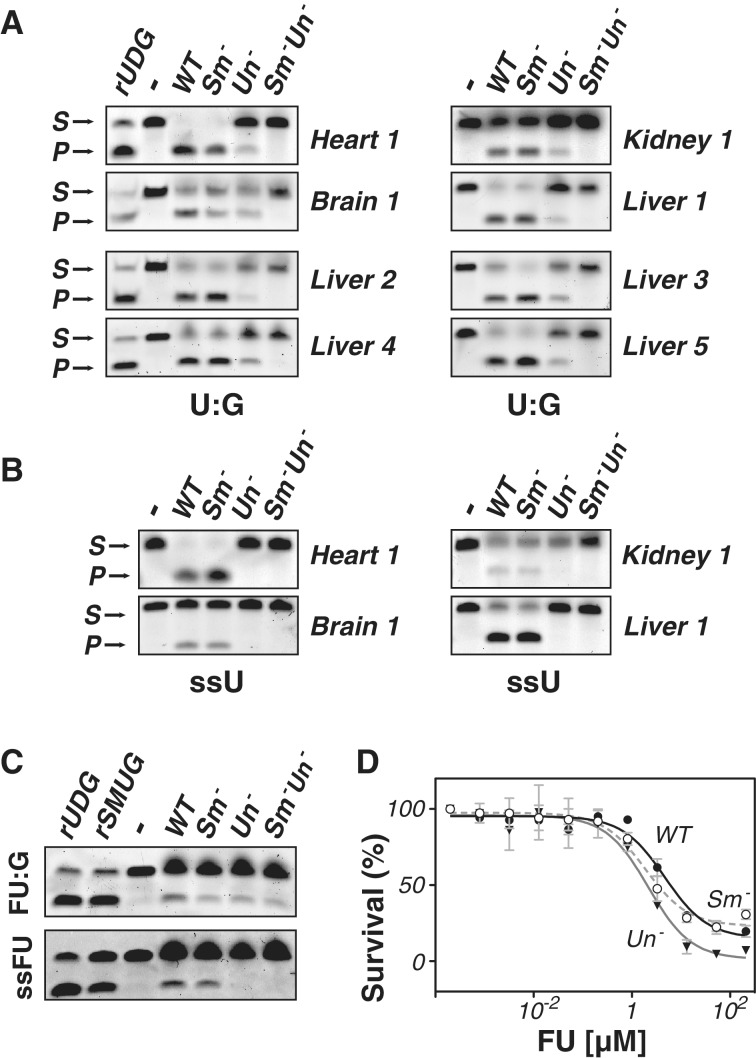
Lack of detectable U-excision activity in Ung−/− Smug1−/− double knockouts
Mice deficient in the UNG uracil-DNA glycosylase retain residual U excision activity as judged by biochemical assay of tissue extracts. Antibody inhibition experiments as well as analysis of fibroblast lines carrying a Smug1 knockdown expression construct have indicated that SMUG1 is responsible for much of this backup activity (6,25,38,46). Comparison of tissue extracts from mice carrying homozygous disruptions of either Ung alone, Smug1 alone or both Ung and Smug1 reveals that residual U-excision activity as judged on a double-stranded oligodeoxyribonucleotide substrate containing a U:G mismatch is still evident in tissues from Ung−/− mice but that this residual activity is essentially absent in extracts from Ung−/− Smug1−/− double knockouts (Figure 4A). Thus, in all the tissues examined, SMUG1 is the major UNG backup for excising U from U:G mismatches—indeed, the sole detectable backup within the sensitivity of these assays. Under the conditions in which these assays were performed, there is little evidence of any UNG-backup activity when a single-stranded U-containing oligonucleotide was used as substrate (Figure 4B), consistent with previous observations (6) that SMUG1 exhibits much less activity on single- as opposed to double-stranded DNA substrates.
Figure 4.
SMUG1 is the major UNG-backup for U excision from U:G-containing double-stranded oligodeoxyribonucleotides. (A) Assay of nuclear extracts of tissues from wild-type (WT), Smug1−/− (Sm−), Ung−/− (Un−) and Smug1−/−Ung−/− (Sm− Un−) mice for excision activity on a double-stranded oligodeoxyribonucleotide substrate (S) containing a centrally placed U:G mispair. P denotes the product following cleavage. Recombinant UDG (rUDG; NEB) served as a positive control. Representative results are shown for different tissues from one set of mice as well as from livers of four other sets. (B) Assay of nuclear extracts for excision activity on a single-stranded oligodeoxyribonucleotide containing a single centrally placed U. (Similar results were obtained if samples were boiled under alkaline conditions confirming that the weak signals obtained on single-stranded DNA substrates do not reflect incomplete strand cleavage at abasic sites.) (C) Assay of liver nuclear extracts for excision activity on a double-stranded oligodeoxyribonucleotide containing a centrally placed FU:G mispair. The result presented is representative of three such analyses. (D) Effects of a 48-h culture in the presence of various concentrations of FU on the viability of embryo fibroblast cell-lines obtained from mice of different genotypes (WT, filled circles; Smug1−/−, open circles; Ung−/−, filled triangles). Survival is expressed relative to that obtained in the absence of FU. The results are shown as the average of triplicate assays performed on embryonic fibroblast cell-lines derived from mice of each indicated genotype.
Fluorouracil excision activity in Smug1−/− and Ung−/− Smug1−/− mice
5-Fluorouracil (FU) is a base analogue that has found widespread use in cancer treatment and can be incorporated into both RNA and DNA. Both UNG and SMUG1 are known to be able to excise FU from double-stranded oligonucleotides (46–48) and assays of liver nuclear extracts from knockout mice indicate that inactivation of either UNG or SMUG1 is sufficient to reduce the overall excision activity detected on an FU:G substrate (Figure 4C). However, inactivation of both Ung and Smug1 does not give an additive reduction in excision activity with residual FU:G excision activity still being detected in Smug1−/−Ung−/− extracts. The results therefore indicate a contribution from at least one other glycosylase, with TDG being a possible candidate (48,49). When assayed on a single-stranded substrate, UNG appears to be the dominant enzyme responsible for FU excision, consistent with observations made previously by Pettersen et al. (50). It is nevertheless notable that deficiency in either UNG or SMUG1 has little effect on the sensitivity of cells to FU (Figure 4D), consistent with the proposal that FU toxicity is largely due to its incorporation into RNA, rather than DNA (50).
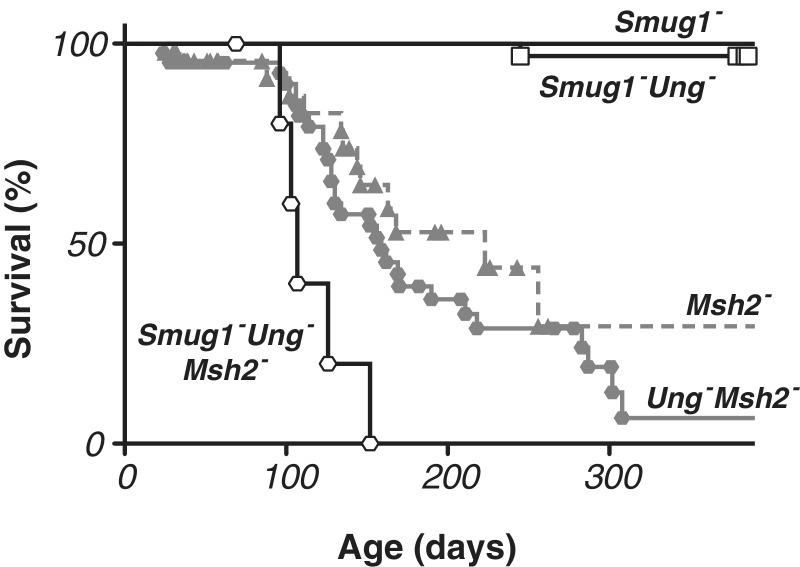
Longevity of Smug1−/−, Ung−/− Smug1−/− and Ung−/− Smug1−/− Msh2−/− mice
Not only were Smug1−/− mice born at the expected Mendelian ratio from Smug1+/− intercrosses, we have also not detected any effects of SMUG1-deficiency on mouse health or survival as monitored out beyond 1 year of age (Figure 5). Strikingly, despite the lack of detectable uracil-excision activity in their tissues, Ung−/− Smug1−/− double knockout mice were also born at the expected frequency from inter-heterozygote crosses and appear healthy. Out of 32 Ung−/− Smug1−/− mice followed for >1 year, only one has died so far, with the cause of death likely being unrelated to the genetic defect (fight wounds). This is consistent with the observations of Nilsen et al. (27) who found that a modest increase in morbidity of Ung−/− mice only became manifest at >18 months of age. Thus, it is evident that mouse health and survival up to 1 year of age is not severely compromised by a lack of detectable uracil excision activity.
Figure 5.
Effects of deficiency in SMUG1, UNG and MSH2 on mouse longevity. Kaplan–Meyer survival curves for mice of different genotypes as monitored up to >1 year of age. Data for Smug1−/− (n = 39), Smug1−/− Ung−/− (n = 32) and Smug1−/− Ung−/− Msh2−/− (n = 6) mice is based on a prospective study of cohorts of mice that were left to age; data for Msh2−/− (n = 47) and Ung−/− Msh2−/− (n = 42) is derived from retrospective analysis of all mice in the colony. In the prospective cohort studies, each symbol indicates a death whereas in the retrospective analyses each symbol indicates the end of observations for one animal with the survival line only coming down if this observation endpoint was due to a disease-related death rather than caused by a healthy animal being removed from the colony (e.g. for use in experimentation). The median life expectancy, determined from retrospective analysis, was >420 days for Smug1−/− and >510 days for Smug1−/− Ung−/− mice (the cohorts having so far only been followed for those numbers of days with too few mice having died to give a median life expectancy), 223 days for Msh2−/−, 159 days for Ung−/− Msh2−/− and 107 days for Smug1−/− Ung−/− Msh2−/− mice.
We wondered if the apparent health of the double knockout animals could reflect the fact that the U:G mispairs generated through spontaneous C deamination might, in the absence of UNG and SMUG1, be corrected by mismatch repair. We therefore asked whether it was possible to generate mice that simultaneously lack UNG, SMUG1 and MSH2 (with MSH2 being essential for mismatch repair). We found that such mice could be generated although the reduced fertility of Msh2−/− mice and the complexity of the breedings mean that we have so far generated only six triple knockout mice. The triple knockout animals were obtained at somewhat below the expected frequency since, out of the 159 animals screened (which had been weaned from 55 litters arising from 21 different breeding pairs), we would have anticipated obtaining 10.8 triple knockouts on a Mendelian basis given the nature of the genotypes used for the various crosses.
Msh2−/− mice are known to exhibit greatly reduced survival with a mean longevity of ∼6 months (51). Although we have only so far obtained six Ung−/− Smug1−/− Msh2−/− triple knockout mice, they show a greatly reduced longevity with a mean life expectancy of 107 days (as compared to 159 days for the Ung−/− Msh2−/−, 238 days for the Smug1−/− Msh2−/− mice and 223 days for the Msh2−/− mice). It therefore appears that combined deficiency in uracil excision and mismatch repair results in a more severe phenotype than mismatch repair deficiency alone (P < 0.01). Indeed, not only is the reduced mean life expectancy of the triple knockout mice statistically highly significant, it is also most striking that all deaths in the triple knockout mice occurred before 152 days of age whereas the mean life expectancy of all the other single/double knockout strains analysed is ≥157 days. Similar to what has been noted with Msh2−/− mice (36,52), the triple knockout mice that have died so far have all died from tumours, which were lymphoid tumours in four of the five cases.
The results therefore show that it is possible to generate mice that are deficient in both uracil-excision repair and mismatch repair but that such mice show severely compromised survival that appears to be largely due to tumours and that the compromised survival of these animals seems somewhat more severe than is attributable to MSH2-deficiency alone.
DISCUSSION
The results described here reveal that ablation of the Smug1 gene in mice removes essentially all detectable hmU-excision activity as well as the residual U-excision activity that is observed in tissue extracts of Ung−/− mice.
That SMUG1 accounts for the residual uracil-excision observed in tissue extracts from Ung−/− mice when assayed on oligodeoxyribonucleotide substrates containing a U:G mismatch is consistent with previous depletion and inhibition studies (6,25,38,46). It is, however, striking that Smug1−/− Ung−/− double knockout mice in which all such readily detectable uracil-excision activity has been ablated can nevertheless be easily generated and are largely healthy up till at least 1 year of age. This indicates that the lesions generated by spontaneous C and hmC deamination in these animals must either be dealt with by another pathway or that these do not occur at a frequency that jeopardizes mouse development and viability.
It is conceivable that the U or hmU residues generated by C or mC deamination, respectively, could be repaired by TDG, since in vitro studies have shown that recombinant TDG can act on both U:G and hmU:G mismatches (53). It is difficult to test whether TDG actually acts as an essential backup in repairing U:G lesions in vivo since deficiency in TDG alone is embryonically lethal (30). However, it is notable that not only do we not readily detect any back up uracil excision activity in extracts from Smug1−/− Ung−/− mice (which could obviously reflect the sensitivity of our in vitro detection), but we have previously observed that enforced overexpression of TDG in mouse B cells is not sufficient to process AID-generated U:G lesions into class-switch recombination, whereas both UNG and SMUG1 function effectively in such assays (34). Thus, although TDG may well act on T:G mismatches that result from deamination at mCpG dinucleotides in vivo, there must be some question as to whether TDG plays a major role in the general in vivo repair of genomic U:G lesions.
A likely backup pathway for the repair of U:G mismatches is provided by mismatch repair. Indeed, studies of antibody diversification provide good evidence that MSH2/MSH6 does indeed recognize and process U:G mismatches in vivo (13). We find in this work that Smug1−/− Ung−/− Msh2−/− triple knockout mice are viable though, like Msh2−/− single knockouts, they are cancer-prone. There is a clear indication that the cancer predisposition of Msh2−/− mice is exacerbated if the animals are also rendered deficient in UNG and SMUG1. These results suggest that when both base excision and mismatch repair pathways are blocked, spontaneous cytosine deamination occurs at a frequency sufficient to increase cancer incidence but not to preclude mouse development.
The presence of an hmU-excision activity in mammalian cell extracts was noted >20 years ago (45,54) with this activity being found to be especially effective on an hmU:G mismatch (55). Subsequent purification of the predominant hmU-excision activity attributed it to SMUG1 (56) although recombinant TDG has also been shown to be able to excise hmU from DNA in vitro (24,53). Our results with Smug1−/− mice not only confirm that SMUG1 accounts for essentially all the hmU-excision activity detectable in mouse tissue extracts as judged by biochemical assay on an hmU:G double-stranded oligonucleotide substrate, they also show that SMUG1 is responsible for the sensitivity of mouse cells to culturing with hmUdR—analogously to what has been observed with a mutant Syrian hamster cell line (40).
It was speculated several years ago on the basis of phylogenetic evidence that hmU-DNA glycosylase might function in the maintenance of 5-methylcytosine in genomic DNA (57). Interest in this possibility has recently gained a considerable boost in light of findings concerning hmC as a natural component of genomic DNA in mammals (42,43,58) as well as from suggestions that oxidation of mC → hmC followed by deamination of hmC → hmU and subsequent hmU excision might constitute physiological steps in the demethylation of mC (23,24). Although the viability, fertility and apparent health of SMUG1-deficient mice do not wholly exclude a role for hmU excision in the physiological demethylation of mC, the results do suggest that hmU excision is not essential for the process. Given the presence of hmC as a natural component of mammalian DNA, it may simply be that SMUG1 predominantly evolved to take care of the hmU arising out of spontaneous hmC deamination, as well as from thymine oxidation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
Medical Research Council [U105178806]; German Academic Exchange Service (DAAD) (stipends to K.K.); and Cambridge European Trust (stipends to K.K.). Funding for open access charge: Medical Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Andreas Hörlein from the EUCOMM consortium for ES cell lines, Shona Butler and her team for animal husbandry, Richard Pannell for assistance with ES cell culture and generation of chimaeric mice, Alison Lane and Leslie Drynan for help with genotyping and Neil Grant for photographic images and assistance with illustrations.
REFERENCES
- 1.Dahl C, Grønbæk K, Guldberg P. Advances in DNA methylation: 5-hydroxymethylcytosine revisited. Clin. Chim. Acta. 2011;412:831–836. doi: 10.1016/j.cca.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Krokan HE, Drabløs F, Slupphaug G. Uracil in DNA–occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. U. S. A. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haushalter KA, Stukenberg PT, Kirschner MW, Verdine GL. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol. 1999;9:174–185. doi: 10.1016/s0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- 6.Nilsen H, Haushalter KA, Robins P, Barnes DE, Verdine GL, Lindahl T. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 2001;20:4278–4286. doi: 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masaoka A, Matsubara M, Hasegawa R, Tanaka T, Kurisu S, Terato H, Ohyama Y, Karino N, Matsuda A, Ide H. Mammalian 5-formyluracil-DNA glycosylase. 2. Role of SMUG1 uracil-DNA glycosylase in repair of 5-formyluracil and other oxidized and deaminated base lesions. Biochemistry. 2003;42:5003–5012. doi: 10.1021/bi0273213. [DOI] [PubMed] [Google Scholar]
- 8.Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol. Cell. 2003;11:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 9.Pettersen HS, Sundheim O, Gilljam KM, Slupphaug G, Krokan HE, Kavli B. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35:3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darwanto A, Theruvathu JA, Sowers JL, Rogstad DK, Pascal T, Goddard W, Sowers LC. Mechanisms of base selection by human single-stranded selective monofunctional uracil-DNA glycosylase. J. Biol. Chem. 2009;284:15835–15846. doi: 10.1074/jbc.M807846200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neddermann P, Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc. Natl Acad. Sci. USA. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 13.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 14.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 15.Sire J, Quérat G, Esnault C, Priet S. Uracil within DNA: an actor of antiviral immunity. Retrovirology. 2008;5:45. doi: 10.1186/1742-4690-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, Thiry S, Jost JP. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc. Natl Acad. Sci. USA. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu B, Zheng Y, Angliker H, Schwarz S, Thiry S, Siegmann M, Jost JP. 5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence. Nucleic Acids Res. 2000;28:4157–4165. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jost JP, Siegmann M, Sun L, Leung R. Mechanisms of DNA demethylation in chicken embryos. Purification and properties of a 5-methylcytosine-DNA glycosylase. J. Biol. Chem. 1995;270:9734–9739. doi: 10.1074/jbc.270.17.9734. [DOI] [PubMed] [Google Scholar]
- 19.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 23.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, Barnes DE. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 26.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 27.Nilsen H, Stamp G, Andersen S, Hrivnak G, Krokan HE, Lindahl T, Barnes DE. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 2003;22:5381–5386. doi: 10.1038/sj.onc.1206860. [DOI] [PubMed] [Google Scholar]
- 28.Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 29.Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, Fazzari M, Jin B, Brown AM, Lipkin M, et al. Mbd4 inactivation increases C → T transition mutations and promotes gastrointestinal tumor formation. Proc. Natl Acad. Sci. USA. 2002;99:14937–14942. doi: 10.1073/pnas.232579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 31.Doseth B, Visnes T, Wallenius A, Ericsson I, Sarno A, Pettersen HS, Flatberg A, Catterall T, Slupphaug G, Krokan HE, et al. Uracil-DNA glycosylase in base excision repair and adaptive immunity: species differences between man and mouse. J. Biol. Chem. 2011;286:16669–16680. doi: 10.1074/jbc.M111.230052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elateri I, Tinkelenberg BA, Hansbury M, Caradonna S, Muller-Weeks S, Ladner RD. hSMUG1 can functionally compensate for Ung1 in the yeast Saccharomyces cerevisiae. DNA Repair. 2003;2:315–323. doi: 10.1016/s1568-7864(02)00221-5. [DOI] [PubMed] [Google Scholar]
- 33.Di Noia JM, Rada C, Neuberger MS. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J. 2006;25:585–595. doi: 10.1038/sj.emboj.7600939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Noia JM, Williams GT, Chan DT, Buerstedde JM, Baldwin GS, Neuberger MS. Dependence of antibody gene diversification on uracil excision. J. Exp. Med. 2007;204:3209–3219. doi: 10.1084/jem.20071768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 37.Matsubara M, Tanaka T, Terato H, Ohmae E, Izumi S, Katayanagi K, Ide H. Mutational analysis of the damage-recognition and catalytic mechanism of human SMUG1 DNA glycosylase. Nucleic Acids Res. 2004;32:5291–5302. doi: 10.1093/nar/gkh859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An Q, Robins P, Lindahl T, Barnes DE. C –> T mutagenesis and gamma-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 2005;24:2205–2213. doi: 10.1038/sj.emboj.7600689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waschke S, Reefschläger J, Bärwolff D, Langen P. 5-hydroxymethyl-2′-deoxyuridine, a normal DNA constituent in certain Bacillus subtilis phages is cytostatic for mammalian cells. Nature. 1975;255:629–630. doi: 10.1038/255629a0. [DOI] [PubMed] [Google Scholar]
- 40.Boorstein RJ, Chiu LN, Teebor GW. A mammalian cell line deficient in activity of the DNA repair enzyme 5-hydroxymethyluracil-DNA glycosylase is resistant to the toxic effects of the thymidine analog 5-hydroxymethyl-2′-deoxyuridine. Mol. Cell. Biol. 1992;12:5536–5540. doi: 10.1128/mcb.12.12.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaung W, Boorstein RJ. Molecular spectrum of mutations induced by 5-hydroxymethyl-2′-deoxyuridine in (CHO)-PL61 cells. Mutat. Res. 1997;373:125–137. doi: 10.1016/s0027-5107(96)00197-2. [DOI] [PubMed] [Google Scholar]
- 42.Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan R. Tissue-type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012;22:467–477. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boorstein RJ, Levy DD, Teebor GW. 5-Hydroxymethyluracil-DNA glycosylase activity may be a differentiated mammalian function. Mutat. Res. 1987;183:257–263. doi: 10.1016/0167-8817(87)90008-3. [DOI] [PubMed] [Google Scholar]
- 46.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 47.Baker D, Liu P, Burdzy A, Sowers LC. Characterization of the substrate specificity of a human 5-hydroxymethyluracil glycosylase activity. Chem. Res. Toxicol. 2002;15:33–39. doi: 10.1021/tx010113b. [DOI] [PubMed] [Google Scholar]
- 48.Liu P, Burdzy A, Sowers LC. Substrate recognition by a family of uracil-DNA glycosylases: UNG, MUG, and TDG. Chem. Res. Toxicol. 2002;15:1001–1009. doi: 10.1021/tx020030a. [DOI] [PubMed] [Google Scholar]
- 49.Kunz C, Focke F, Saito Y, Schuermann D, Lettieri T, Selfridge J, Schär P. Base excision by thymine DNA glycosylase mediates DNA-directed cytotoxicity of 5-fluorouracil. PLoS Biol. 2009;7:e91. doi: 10.1371/journal.pbio.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen HS, Visnes T, Vågbø CB, Svaasand EK, Doseth B, Slupphaug G, Kavli B, Krokan HE. UNG-initiated base excision repair is the major repair route for 5-fluorouracil in DNA, but 5-fluorouracil cytotoxicity depends mainly on RNA incorporation. Nucleic Acids Res. 2011;39:8430–8444. doi: 10.1093/nar/gkr563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reitmair AH, Redston M, Cai JC, Chuang TC, Bjerknes M, Cheng H, Hay K, Gallinger S, Bapat B, Mak TW. Spontaneous intestinal carcinomas and skin neoplasms in Msh2-deficient mice. Cancer Res. 1996;56:3842–3849. [PubMed] [Google Scholar]
- 52.Reitmair AH, Schmits R, Ewel A, Bapat B, Redston M, Mitri A, Waterhouse P, Mittrücker HW, Wakeham A, Liu B. MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat. Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- 53.Hardeland U, Bentele M, Jiricny J, Schär P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hollstein MC, Brooks P, Linn S, Ames BN. Hydroxymethyluracil DNA glycosylase in mammalian cells. Proc. Natl Acad. Sci. USA. 1984;81:4003–4007. doi: 10.1073/pnas.81.13.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rusmintratip V, Sowers LC. An unexpectedly high excision capacity for mispaired 5-hydroxymethyluracil in human cell extracts. Proc. Natl Acad. Sci. USA. 2000;97:14183–14187. doi: 10.1073/pnas.97.26.14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boorstein RJ, Cummings A, Marenstein DR, Chan MK, Ma Y, Neubert TA, Brown SM, Teebor GW. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 57.Boorstein RJ, Chiu LN, Teebor GW. Phylogenetic evidence of a role for 5-hydroxymethyluracil-DNA glycosylase in the maintenance of 5-methylcytosine in DNA. Nucleic Acids Res. 1989;17:7653–7661. doi: 10.1093/nar/17.19.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.