Abstract
DNA polymerase IV (Pol IV) is one of three translesion polymerases in Escherichia coli. A mass spectrometry study revealed that single-stranded DNA-binding protein (SSB) in lysates prepared from exponentially-growing cells has a strong affinity for column-immobilized Pol IV. We found that purified SSB binds directly to Pol IV in a pull-down assay, whereas SSBΔC8, a mutant protein lacking the C-terminal tail, failed to interact with Pol IV. These results show that the interaction between Pol IV and SSB is mediated by the C-terminal tail of SSB. When polymerase activity was tested on an SSBΔC8-coated template, we observed a strong inhibition of Pol IV activity. Competition experiments using a synthetic peptide containing the amino acid sequence of SSB tail revealed that the chain-elongating capacity of Pol IV was greatly impaired when the interaction between Pol IV and SSB tail was inhibited. These results demonstrate that Pol IV requires the interaction with the C-terminal tail of SSB to replicate DNA efficiently when the template ssDNA is covered with SSB. We speculate that at the primer/template junction, Pol IV interacts with the tail of the nearest SSB tetramer on the template, and that this interaction allows the polymerase to travel along the template while disassembling SSB.
INTRODUCTION
During processes of genome maintenance such as DNA replication, repair and recombination, double-stranded DNA transiently becomes single-stranded. Single-stranded DNA-(ssDNA) binding protein (SSB) is essential for cell survival in all organisms. It coats ssDNA to prevent the formation of secondary structure on ssDNA, thereby allowing DNA processing enzymes to access their substrate (1–3). In addition to its intrinsic ability to bind ssDNA, SSB has an important role in recruiting genome maintenance proteins to their target ssDNA through physical interaction. To date, 14 such proteins have been reported to interact with SSB in Escherichia coli (3).
Five DNA polymerases have been identified in E. coli (4,5). DNA polymerase III (Pol III) replicates chromosomal DNA with high fidelity (6–8). Pol I functions in processing Okazaki fragments during lagging strand synthesis and also in the nuclear excision repair pathway. The other three polymerases, Pol II, Pol IV and Pol V, have been identified as specialized DNA polymerases and are upregulated by the SOS response. These low-fidelity enzymes are thought to act as lesion-bypass polymerases.
Among these five DNA polymerases, three have been reported to be associated with SSB (3). Pol II can bind to SSB and their interaction stimulates Pol II processivity (9). In the presence of SSB on a template ssDNA, Pol II can replicate an abasic lesion together with the β clamp (10). It is also reported that Pol III holoenzyme (Pol III HE), a multi-subunit complex composed of 17 proteins, binds directly to SSB (11,12). The main interaction between Pol III and SSB is mediated by the χ subunit in the clamp-loading complex of Pol III HE. This interaction has been proposed to be important for Pol III to load the β clamp onto SSB-coated ssDNA and for synthesis of Okazaki fragments on the lagging strand during DNA replication (11,12). A recent report suggested a discrete role for the interaction between Pol III and the χ subunit in the replisome establishment and maintenance (13). In addition, the interaction between Pol III and SSB is necessary for the strand displacement reaction and to stimulate initiation complex formation by Pol III on an SSB-coated template (14,15). Finally, Pol V, the main polymerase for the DNA damage tolerance mechanism, interacts physically with SSB, which stimulates the translesion synthesis reaction of Pol V by recruiting Pol V to the 3′-primer terminus on ssDNA coated with RecA (16).
Here, we identified a new interaction between SSB and Pol IV. Pol IV, encoded by dinB, is conserved among diverse organisms including human (17), and can both replicate undamaged DNA and bypass various lesions in vitro (18,19). We found that Pol IV binds to the C-terminus of SSB and, when it does so, elongates a primer 3′-terminus more rapidly and stably on SSB-coated ssDNA. Our results suggest that this interaction enables Pol IV to dislodge or translocate SSB protein to facilitate the replication of SSB-coated ssDNA.
MATERIALS AND METHODS
Nucleic acids and peptides
M13mp18 ssDNA primed with the 32P-labeled or unlabeled 25-mer primer uni25 was prepared as described previously (20). A 35-mer DNA, hook10 (5′-tttgttcttttggcaccaactatatgttggtgcca-3′), was synthesized to produce a hook-like structure with a single-stranded 5′ tail of 10 nt. The peptide SSB-Ct (Trp–Met–Asp–Phe–Asp–Asp–Asp–Ile–Pro–Phe) was synthesized and dissolved in 100% dimethyl sulfoxide (DMSO), and quantified spectrophotometrically (21,22).
Proteins
His-tagged, wild-type Pol IV and mutant Pol IV proteins were purified as described (20). Wild-type SSB and the β clamp were purified as described (23). The γ complex was a generous gift of Dr Tsutomu Katayama (Kyushu University). An overexpression plasmid encoding SSBΔC8 was a generous gift from Dr Michael M. Cox (University of Wisconsin, Madison). SSBΔC8 protein was expressed as described previously (24) and purified from lysed cells by HiTrap heparin and Sephacryl HR S-200 (GE Healthcare) column chromatography.
Pol IV-affinity column chromatography
Purified His-Pol IV was dialyzed against 100 mM HEPES–NaOH pH 7.4 and 200 mM NaCl at 4°C. The coupling reaction was performed by addition of 4.7 mg of His-Pol IV to 0.5 ml AffiGel 10 (Bio-Rad) and mixing for 4 h, followed by a blocking reaction in 20 mM ethanolamine-HCl for 1 h. Pol IV-crosslinked beads were packed into a column (0.5 ml) and were equilibrated in Buffer A (50 mM HEPES–NaOH pH 7.4, 100 mM KCl, 5% glycerol and 1 mM dithiothreitol) as described (25). Escherichia coli ΔdinB strain MK7003 (MG1655 rpsL(Smr) ΔdinB, laboratory stock) cells were grown at 37°C in LB medium until the OD600 reached ∼0.8 and were then harvested, and 3 g of the cells were lysed by sonication in 90 ml Buffer A supplemented with 15 mM MgCl2 in the presence of 0.2 mM phenylmethylsulfonyl fluoride, 0.5 mg/ml lysozyme and 2.3 U/ml benzonase (Novagen). The lysate was centrifuged at 17 000 g for 40 min and the supernatant was applied to the Pol IV-affinity column. The column was washed with 25 ml of Buffer A and 5 ml of 0.1 M glycine–HCl pH 2.5. Eluted proteins in the wash fractions were quantified and separated on a NuPAGE 4–12% Bis–Tris gel (Invitrogen), and were identified by silver staining and mass spectrometry. Proteins retained in the column were eluted by the addition of NuPAGE LDS-loading buffer (Invitrogen) and heated at 70°C for 10 min. Eluted proteins were then separated by NuPAGE 4–12% Bis–Tris gel, stained by silver staining and identified by mass spectrometry. For control Lysozyme column, 4.7 mg of Lysozyme (Sigma) was coupled with 0.5 ml AffiGel 10, and the recovery of Lysozyme-interacting factors were performed identically to Pol IV column chromatography using the same lysate. All operations for protein coupling and chromatography were performed at 4°C.
Pull-down assay
Pull-down assays with Ni–NTA magnetic beads (QIAGEN) were performed using purified His-Pol IV, SSB and SSBΔC8. Proteins were mixed at the indicated concentrations (His-Pol IV 0.42 µM; SSB or SSBΔC8 0, 0.25, 0.5 and 1 µM as a tetramer) and incubated at 4°C for 2 h in 25 µl of Buffer B (50 mM HEPES–NaOH pH 7.4, 100 mM KCl, 20 mM imidazole, 5% glycerol, 1 mM dithiothreitol and 0.005% Tween-20). Magnetic beads were added to each reaction and mixed at 4°C for 1 h. The beads were separated and washed three times with 0.5 ml of Buffer B, and bound proteins were eluted in 25 µl of Buffer B supplemented with 100 mM EDTA. Eluted proteins were analyzed by NuPAGE 4–12% Bis–Tris gel followed by silver staining. In Figure 4A, the indicated concentrations of SSB-Ct peptide and His-Pol IV were pre-incubated at 4°C for 5 min before mixing with SSB.
Figure 4.
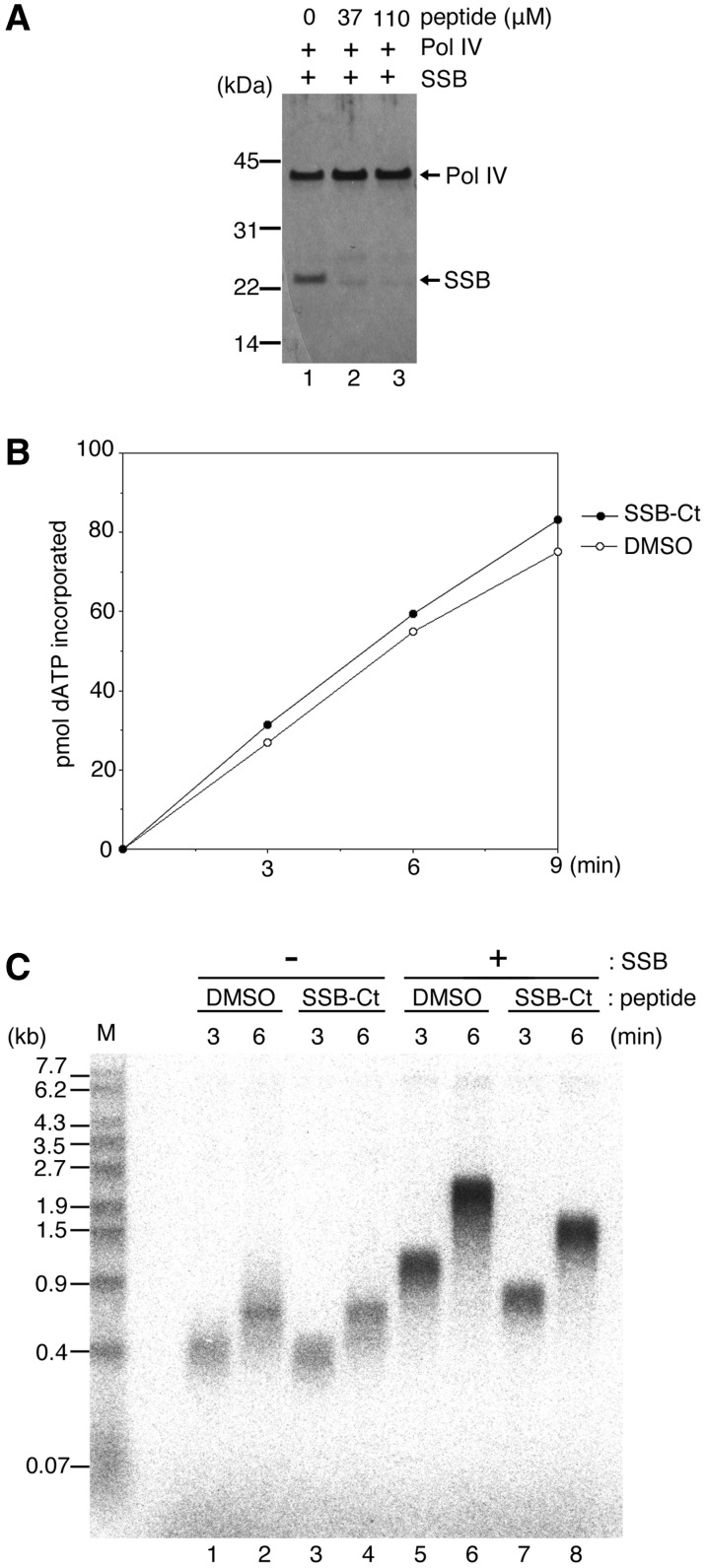
Effects of SSB-Ct peptide on Pol IV DNA synthesis on SSB-coated ssDNA. (A) Pull-down assay using Pol IV (final concentration of 0.42 µM) and SSB (final concentration of 1 µM as a tetramer) in the presence of SSB-Ct peptide (final concentration of 0, 37 and 110 µM), as in Figure 2A. SSB-Ct peptide was pre-incubated with Pol IV for 5 min before the addition of SSB. (B) Polymerase activity of Pol IV on hook DNA in the absence or presence of SSB-Ct peptide. Pol IV was incubated with a 35-mer hook DNA in the presence (110 µM, filled circles) or absence (0 µM, open circles) of SSB-Ct peptide. In the latter reaction, the same volume of DMSO as the volume of SSB-Ct peptide used was added to the reaction. After 3 -, 6 - and 9-min incubations, the amount of incorporated nucleotides in the reactions was quantified. See ‘Materials and Methods’ section for details. (C) The effect of SSB-Ct peptide on Pol IV DNA synthesis on primed M13mp18 circular ssDNA coated with SSB. M13mp18 ssDNA (final concentration of 1 nM) annealed with a non-labeled primer was pre-incubated for 3 min in a reaction containing the γ complex and the β clamp, without SSB (lanes 1–4) or with SSB (lanes 5–8). After the clamp–template DNA complex had formed, Pol IV (final concentration of 22 nM) together with SSB-Ct peptide (final concentration of 0 µM in lanes 1, 2, 5 and 6 or 110 µM in lanes 3, 4, 7 and 8) was added to the reaction at the start of DNA synthesis. After 3 - or 6-min incubations, replication products labeled by 32P-dATP were analyzed by alkaline agarose gel electrophoresis. Size markers are as in Figure 3B. See ‘Materials and Methods’ section for details.
Assay for Pol IV DNA polymerase activity
Polymerase activity of Pol IV was measured as the amount of nucleotide incorporated into DNA. Template DNA hook10 (200 pmol) and His-Pol IV (22 nM) were incubated at 30°C for 3, 6 and 9 min in 20 µl of EDBG (20 mM Tris–HCl pH 7.5, 4% glycerol, 8 mM dithiothreitol, 80 µg/ml bovine serum albumin, 1 mM ATP, 8 mM MgCl2) containing 100 µM each of dCTP, dGTP and [α-32P]dATP, in the presence or absence of SSB-Ct peptide (110 µM). After incubation, reactions were stopped by addition of an equal volume of 50 mM EDTA pH 8.0, and aliquots were spotted onto DE81 paper (Whatman), which was dried, washed by 0.5 M Na2HPO4, exposed to an imaging plate and analyzed by BAS 2500 (Fuji Film). Protein and peptide concentrations are indicated as their final concentrations in the reaction.
Assay for Pol IV DNA synthesis on SSB-coated ssDNA
In Figure 3B and C, primed M13mp18 (32P-labeled, 1 nM as a template molecule and 7.25 µM as nucleotide residues), the γ complex (5 nM), the β clamp (0.12 µg) and SSB or SSBΔC8 (230 nM as a tetramer) were pre-incubated at 30°C for 3 min in Buffer 1 (7.5 µl, EDBG containing 100 µM each of dATP, dCTP and dGTP) to load the β clamp onto DNA. DNA chain elongation was started by adding Pol IV (44 nM) with pre-warmed Buffer 2 (22.5 µl, EDBG containing 100 µM each of dATP, dCTP and dGTP and 133 µM dTTP) to the template. After a 1- or 3-min incubation at 30°C, the reaction was stopped by adding an equal volume of stop buffer (50 mM EDTA, 0.15% SDS, pH 8.0), and the replication products were then analyzed by alkaline agarose gel electrophoresis as described (20) or by 8 M urea-denaturing 8% polyacrylamide-sequencing gel electrophoresis. In Figure 6, after a 3-min pre-incubation of labeled DNA, SSB, the γ complex and the β clamp, Pol IV was added to Buffer 1 to a final concentration of 0.3 nM and the reaction mixture was further pre-incubated for 1 min, prior to the addition of Buffer 2 containing SSB-Ct peptide (0 or 110 µM). The reaction was incubated for the indicated time at 30°C after addition of Buffer 2, and the products were then analyzed as described above. In Figure 4C, primed M13mp18 (unlabeled, 1 nM), the γ complex (5 nM), the β clamp (0.12 µg) and SSB (0.23 µM as a tetramer) were pre-incubated at 30°C for 3 min in Buffer 1 (7.5 µl, EDBG containing 100 µM each of dATP, dCTP and dGTP). DNA chain elongation was started at 30°C by adding Pol IV (22 nM) in pre-warmed Buffer 2 (22.5 µl, EDBG containing 100 µM each of [α-32P]dATP, dCTP and dGTP and 133 µM dTTP) with or without SSB-Ct peptide (110 µM). The reaction was stopped at each time point and analyzed as describe above. In Figure 5, after a 3-min pre-incubation of primed M13mp18 (unlabeled), SSB, the γ complex and the β clamp, Pol IV (22 nM) was added to Buffer 1. The reaction mixture was further pre-incubated for 1 min prior to the addition of Buffer 2 containing the indicated amount of SSB-Ct peptide (0, 37, 110, 220 or 330 µM). The reaction was incubated for 3 min at 30°C after addition of Buffer 2, and the products were then analyzed as describe above. In control 0 µM SSB-Ct peptide reactions, the same volume of DMSO as that of SSB-Ct peptide was added to the reaction in each experiment.
Figure 3.
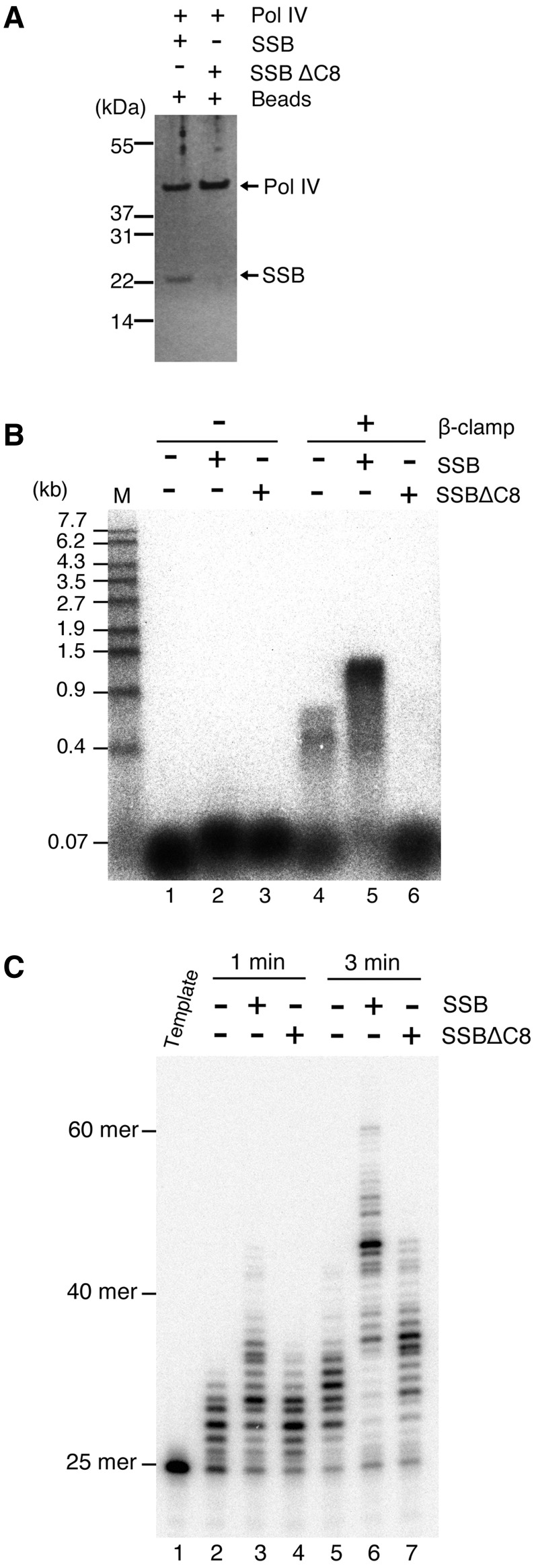
SSBΔC8 fails to interact with Pol IV and abolishes DNA synthesis by Pol IV on ssDNA. (A) Pull-down assay using Pol IV (final concentration of 0.42 µM) and SSB or SSBΔC8 (final concentration of 1 µM as a tetramer), as in Figure 2A. (B) Assays for Pol IV DNA synthesis on SSB-coated ssDNA were carried out as described under ‘Materials and Methods’ section. Briefly, a 7.25-kb M13mp18 circular ssDNA (final concentration of 1 nM) annealed with a 5′-32P-labeled 25-mer primer was pre-incubated for 3 min in a reaction containing the γ complex, without SSB (lanes 1 and 4) or with either SSB (lanes 2 and 5) or SSBΔC8 (lanes 3 and 6), in the presence or absence of the β clamp (as indicated at the top of the figure). After the clamp–template DNA complex had formed, Pol IV (final concentration of 44 nM) was added at the start of DNA synthesis, and replication products formed after a 3-min incubation were analyzed by alkaline agarose gel electrophoresis. Size markers (M; 5′-32P-labeled λ/EcoT14I) are indicated on the left of the figure. (C) Replication products at 1 and 3 min without SSB (lanes 2 and 5) or with either SSB (lanes 3 and 6) or SSBΔC8 (lanes 4 and 7) in the absence of the β clamp were analyzed on an 8% polyacrylamide sequencing gel. A 5′-32P-labeled template DNA was included as a size marker (lane 1).
Figure 6.
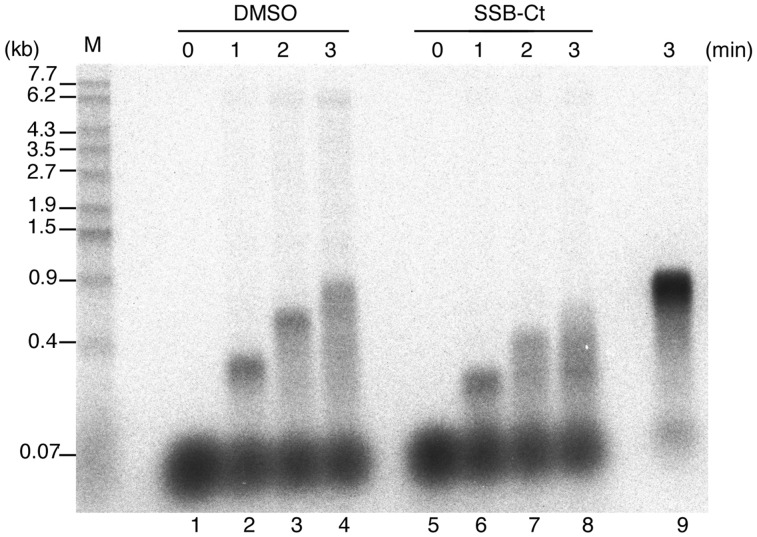
Interaction with SSB C-terminal tail is required by Pol IV for rapid and stable elongation on SSB-coated ssDNA. Assays for Pol IV DNA synthesis on SSB-coated ssDNA were carried out at a final concentration of 0.3 nM Pol IV, as in Figure 5. M13 annealed with 5′-32P-labeled primer (final concentration of 1 nM) was used as a template DNA, and Pol IV was added to the reaction 1 min before the start of DNA synthesis. Replication products in the presence of SSB-Ct peptide (0 or 110 µM) after the indicated times are shown. A control reaction with 22 nM Pol IV was carried out (lane 9). Size markers are as in Figure 3B. See ‘Materials and Methods’ section for details.
Figure 5.
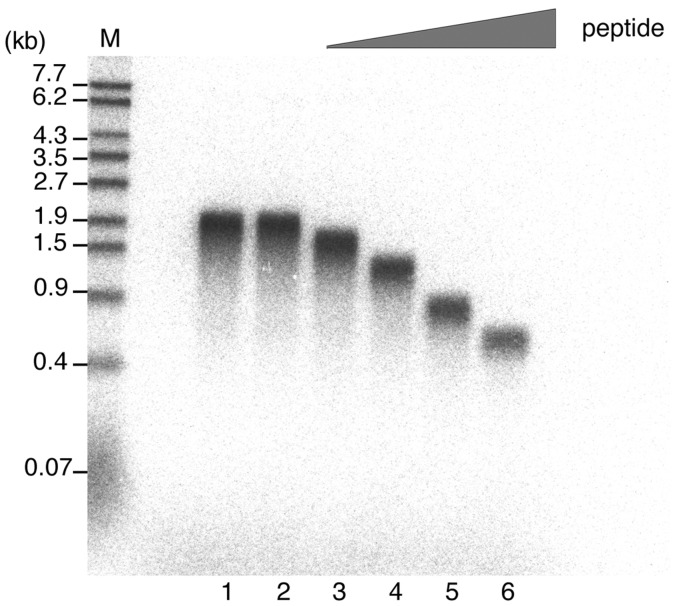
Effect of SSB-Ct peptide on Pol IV DNA synthesis is peptide concentration-dependent. After the clamp–template DNA (M13 annealed with non-labeled primer) complex had formed, Pol IV was added 1 min before the start of DNA synthesis to a final concentration of 22 nM. Increasing amounts of SSB-Ct peptide were then added to the Pol IV DNA synthesis reaction (final concentrations of 0, 37, 110, 220 or 330 µM; lanes 2–6, respectively) at the start time of DNA synthesis, and, after a 3-min incubation, replication products labeled with 32P-dATP were analyzed by alkaline agarose gel electrophoresis. A reaction with neither DMSO nor SSB-Ct peptide was carried out as a control (lane 1). Size markers are as in Figure 3B. See ‘Materials and Methods’ section for details.
RESULTS
The β clamp and SSB bind to a Pol IV affinity column
Pol IV is the most abundant DNA polymerase in E. coli (250 molecules in a normal cell and 2500 molecules in an SOS-induced cell) (26). To elucidate the regulatory mechanism of Pol IV, Godoy et al. (25) identified Pol IV-interacting proteins from lysates of constitutively SOS-induced E. coli. They reported that UmuD, UmuD′ and RecA physically bind to Pol IV and regulate its activity both in vivo and in vitro. However, since the cellular levels of all these proteins increase at least 10-fold in the SOS response, we hypothesized that there might be a different binding partner(s) and regulatory mechanism at the lower protein concentrations that pertain in a normally growing cell. To test this idea, we took the same approach as Godoy et al. (25). We recovered Pol IV-interacting proteins in lysates prepared from SOS-uninduced, exponentially growing cells, using column-immobilized recombinant His-tagged Pol IV. The ΔdinB strain was used in an effort to isolate any low-abundance factors that might not be available to column-bound His-Pol IV if they interact with endogenous Pol IV in wild-type cells.
When we loaded the cell lysate onto the Pol IV column and tried to elute binding proteins with a low-pH buffer, >90% of the protein remained bound in the column (data not shown). We then retrieved the resin from the column and heated it with SDS–PAGE sample buffer to recover all proteins in the column. Pol IV column-interacting proteins were then compared by SDS–PAGE with proteins recovered similarly from the lysozyme-immobilized control column (Figure 1, compare lane 1 with lane 2). In the Pol IV column eluate, specific bands of 45, 42 and 22 kDa were detected and were identified by mass spectrometry as the β clamp, Pol IV and SSB, respectively. The Pol IV must come from the column, because there can be no endogenous Pol IV in the lysate of the ΔdinB strain used. Other proteins observed here should be Pol IV column-interacting proteins, but they were different from those observed in Godoy et al.’s (25) report. In that study, GroEL, RecA and UmuD′ were detected as the major Pol IV-interacting proteins, whereas the β clamp and SSB were not recovered. It is possible that the former proteins were washed out by the low-pH buffer in our experiments, although we could not detect any of them in low-pH fractions by mass spectrometry (data not shown). Their interactions with Pol IV may be weak, and/or the proteins may bind to Pol IV only when the SOS is induced.
Figure 1.
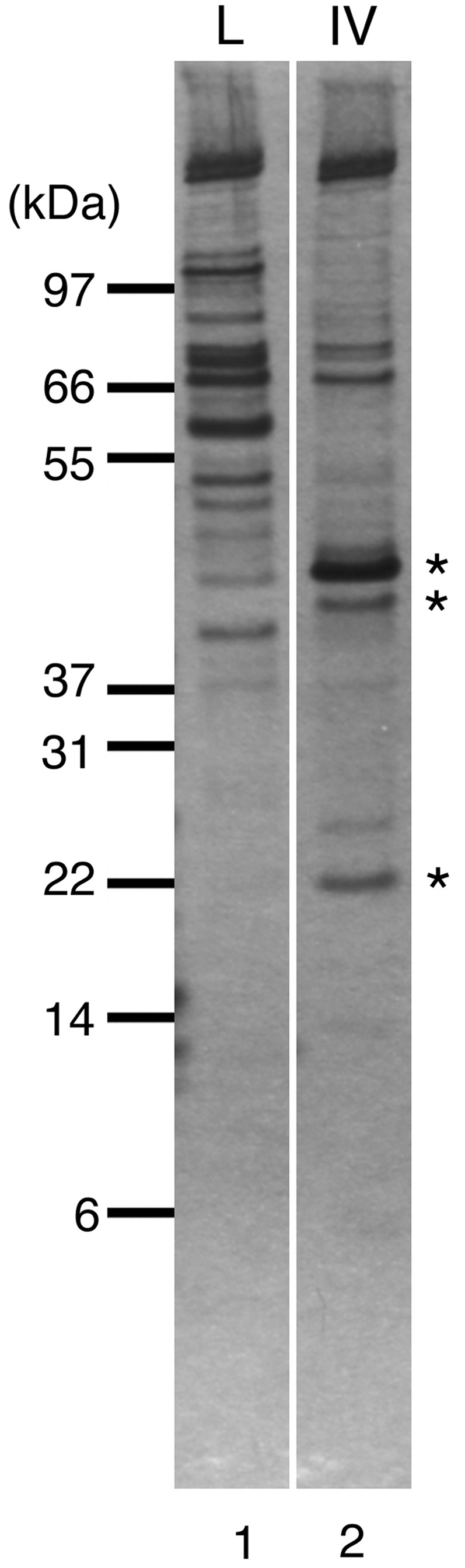
Proteins bound to Pol IV and lysozyme affinity column. Fractions eluted by heating in NuPAGE-loading buffer from the lysozyme control column (L) and the Pol IV column (IV) were separated on NuPAGE Bis–Tris gel and visualized by silver staining. Asterisks indicate protein bands specific to the Pol IV column that were analyzed by mass spectrometry.
Instead of RecA and UmuD′, we found that the β clamp binds to the Pol IV column. The β clamp, a ring-shaped processivity factor for DNA replication, has been reported previously to interact with Pol IV and stimulate its polymerase activity (27,28). Unexpectedly, we found that SSB was one of the major protein recovered from Pol IV-column. This suggests that SSB interacts strongly with Pol IV in the normal E. coli cell, but no evidence of any physical interaction between SSB and Pol IV has previously been reported. We therefore investigated whether SSB indeed binds to Pol IV and how such an interaction might affect the activity of Pol IV in vitro.
SSB directly interacts with Pol IV in vitro
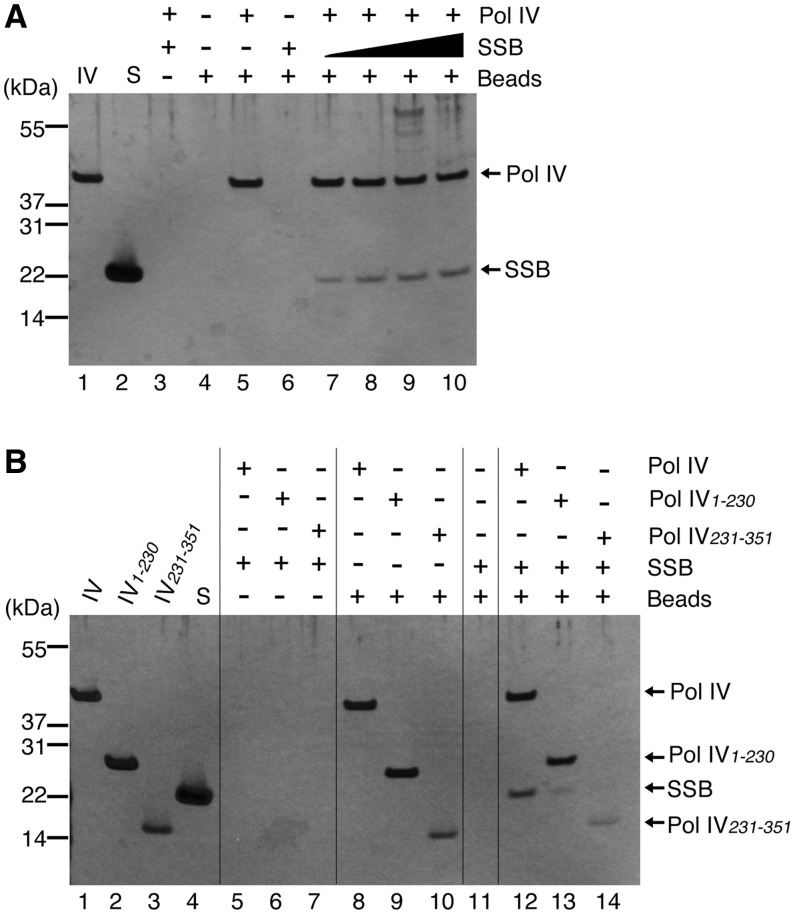
Pull-down assays using recombinant His-tagged Pol IV and SSB demonstrated that SSB binds directly to Pol IV (Figure 2A). We used Pol IV at its concentration in the normal cell (0.42 µM) and varied the concentration of SSB as a tetramer from 0.25 to 1 µM. Increasing the concentration of SSB resulted in an increased amount of SSB co-precipitating with Pol IV. Since the concentration of SSB tetramer in vivo is ∼1.7– 3.4 µM (29), we conclude that SSB is able to interact with Pol IV at concentrations that approximate to those found in normal cells.
Figure 2.
Pol IV and SSB interact directly in vitro. (A) Pull-down assay using Pol IV (final concentration of 0.42 µM) and SSB (final concentrations as a tetramer of 1 µM in lanes 3 and 6, and 0.25, 0.5, 0.75 and 1 µM in lanes 7–10, respectively) as described under ‘Materials and Methods’ section. Precipitated proteins were separated on a NuPAGE Bis–Tris gel and detected by silver staining. Proteins and magnetic beads added to reactions are indicated at the top of the figure. Purified Pol IV and SSB (100 ng each) were loaded in lanes 1 and 2, respectively as markers. (B) Pull-down assay using Pol IV, mutant Pol IV (final concentrations of 0.42 µM) and SSB (final concentration as a tetramer of 1 µM), as in (A). Proteins and magnetic beads added to reactions are indicated at the top of the figure. Purified Pol IV (labeled IV; 100 ng), Pol IV1–230 (IV1–230; 63 ng), Pol IV231–351 (IV231–351; 35 ng) and SSB (S; 53 ng) (2.5 pmol each) were loaded in lanes 1–4 as markers.
Pol IV binds to the β clamp via conserved motifs in the little finger domain located in the C-terminal region of Pol IV (30). Next, we investigated whether the interaction domain between Pol IV and SSB also resides in the C-terminal region of Pol IV, by a pull-down assay using His-tagged mutant Pol IVs. The results showed that neither Pol IV1–230 (the N-terminal two-thirds of Pol IV) nor Pol IV231–351 (the C-terminal one-third of Pol IV including little finger domain) could co-precipitate an amount of SSB comparable to that observed using full-length Pol IV, although small amounts of SSB did co-precipitate with these truncated polypeptides (Figure 2B, compare lane 12 with lanes 13 and 14). These findings indicate that both the N- and C-terminal regions of Pol IV are required for tight binding with SSB. Pol IV may have a binding site for SSB that is composed of residues from both the N- and C-terminal regions, or there may be multiple weak interaction sites for SSB within full-length Pol IV.
The eight C-terminal amino acids of SSB are essential for the interaction with Pol IV
Previous studies have shown that SSB is composed of two characteristic structures: an N-terminal, tightly folded region containing the oligonucleotide/oligosaccharide-binding (OB) domain for ssDNA binding and a C-terminal, dynamic tail (2,31). SSB interacts with various partner proteins through this tail (3). The eight amino acid sequence at the end of the tail is highly conserved among eubacteria, and mutant SSB lacking this sequence fails to interact with many partner proteins (3,11,14,32). To test whether this C-terminal conserved region is also essential for the interaction between SSB and Pol IV, we used SSBΔC8, a mutant SSB which lacks the C-terminal eight amino acids, in a pull-down assay. The amount of SSBΔC8 precipitated with Pol IV was almost undetectable (Figure 3A), showing that Pol IV cannot bind to SSBΔC8. This result demonstrates that the C-terminal tail of SSB is required for the interaction between Pol IV and SSB, suggesting that Pol IV binds to the conserved motif in the SSB C-terminal tail, as other proteins do.
SSBΔC8 inhibits processive elongation by Pol IV on SSB-coated ssDNA
We next tested whether the physical interaction between Pol IV and SSB affects the DNA-synthesizing ability of Pol IV on SSB-coated ssDNA, using an assay for synchronized DNA synthesis by Pol IV on a singly primed M13mp18 circular ssDNA. Template DNA was pre-incubated with SSB or SSBΔC8, the γ complex and the β clamp for 3 min in the presence of ATP. During pre-incubation, SSB binds to ssDNA and recruits the γ complex to the primer/template junction through a physical interaction, and the γ complex loads the β clamp onto the primer. Pol IV was then added to the reaction together with dNTP to start DNA synthesis, and the reaction was incubated for another 3 min. The replication products were then analyzed by alkaline agarose gel electrophoresis (Figure 3B).
The β clamp, loaded on the primer, tethers Pol IV to the template DNA for processive DNA synthesis (27). As expected, when the β clamp was omitted from the reaction, Pol IV extended the 5′-32P-labeled primer very slowly (Figure 3B, lanes 1–3). Even after a 3-min incubation, replication products were shorter than ∼100 bases on an alkaline agarose gel. However, we observed small differences in the mobility of these products in the presence or absence of SSB or SSBΔC8 (Figure 3B, compare lane 1 with lanes 2 and 3). To confirm this, we analyzed the replication products at 1 and 3 min in the absence of the β clamp using a denaturing sequencing gel (Figure 3C). In the absence of SSB, Pol IV extended the 25-mer primer by <10 bases within 1 min and <20 bases within 3 min (Figure 3C, compare lane 1 with lanes 2 and 5). Addition of SSB and SSBΔC8 enhanced the primer extension by Pol IV (Figure 3C, compare lane 5 with lanes 6 and 7). Since SSB binding eliminates the secondary structure on ssDNA (1–3), both SSB and SSBΔC8 may stimulate Pol IV activity by preventing the formation of a blocking structure at the primer end. However, the longest products with SSB were ∼60 bases after a 3-min incubation, but were ∼45 bases in the presence of SSBΔC8, showing that the stimulation by SSBΔC8 is weaker than that by wild-type SSB (Figure 3C, compare lanes 5, 6 and 7). In these reactions without the β clamp, Pol IV extended the primer distributively. Therefore, it is likely that the enhancement of Pol IV extension is caused by the frequent action of Pol IV at the primer end. This suggests that the C-terminal tail of SSB has some role in the recruitment or stabilization of Pol IV on the primer end in the absence of the β clamp.
Together with the β clamp, Pol IV could extend the primer to products >0.4 kb in the absence of SSB (Figure 3B, lane 4). However, the length of the elongated products was <1 kb and paused products were visible. The number of primers elongated was also low, showing that the γ complex cannot properly load the β clamp on the primer without SSB. The addition of SSB allowed Pol IV to elongate most of the primer to products of 1 kb within 3 min, at a rate of ∼6 nt/s, as reported previously (27) (Figure 3B, lane 5). This shows that SSB can effectively assist Pol IV in elongating a primer with the β clamp on a long ssDNA template.
On the other hand, the long elongation products were not observed when SSBΔC8 was added instead of SSB (Figure 3B, lane 6). Pol IV thus cannot processively synthesize DNA together with the β clamp in the presence of SSBΔC8, whereas it can do so without SSB if the β clamp is loaded on the primer (Figure 3B, compare lane 4 with lane 6). This is surprising because we observed some stimulation of Pol IV’s primer extension activity by SSBΔC8, which probably prevented the formation of secondary structure, in Figure 3C. These data suggest that the C-terminal tail of SSB is essential for processive elongation by Pol IV, not only for removing the secondary structure or recruiting Pol IV on the primer end but also for facilitating the action of Pol IV itself on SSB-coated ssDNA. Pol IV may thus need to interact with the SSB tail for rapid and processive elongation when the template ssDNA is coated by SSB.
SSB-Ct peptide inhibits the interaction between Pol IV and SSB
In the above experiments, we could not rule out the possibility that the intrinsic ssDNA-binding ability of SSB is changed by removal of the C-terminal tail in such a way that SSBΔC8 might gain an extra inhibitory activity that prevents the movement of Pol IV. Another possibility is that the γ complex failed to load the β clamp onto DNA covered with SSBΔC8, because the γ complex itself also binds to the C-terminal tail of SSB (11,12). To exclude these possibilities, we next inhibited the interaction between Pol IV and SSB by the addition of a synthetic peptide, SSB-Ct (Trp–Met–Asp–Phe–Asp–Asp–Asp–Ile–Pro–Phe), containing the conserved C-terminal eight amino acids of the SSB tail. Lu et al. (21) have reported that this peptide effectively and competitively inhibited the interaction between SSB and Exo I. Addition of the peptide after the β clamp-loading step should enable us to eliminate the effect of inhibition on the γ complex.
We first used a pull-down assay to test whether the SSB-Ct peptide competes with SSB for binding to Pol IV. SSB-Ct (at the final concentration of 37 or 110 µM) was incubated with Pol IV for 5 min prior to the addition of SSB, and the pull-down assay was carried out. Pol IV failed to co-precipitate SSB when SSB-Ct peptide was added to the reaction (Figure 4A, lanes 2 and 3), showing that SSB-Ct peptide can inhibit the interaction between Pol IV and SSB. This result also indicates that the binding site of Pol IV is the conserved motif in the SSB C-terminal tail, as suggested by earlier experiments (Figure 3A).
SSB-Ct peptide inhibits DNA synthesis by Pol IV on SSB-coated ssDNA
The effects of SSB-Ct peptide on Pol IV DNA synthesis were then tested. First, we confirmed that 110 µM SSB-Ct peptide had no inhibitory effect on Pol IV’s polymerase activity itself, using a 35-nt hook DNA as a substrate (Figure 4B). In this reaction, hook DNA and Pol IV were incubated with 32P-dATP, in the absence of SSB. The amount of the dATP incorporated into hook DNA in the presence of SSB-Ct peptide increased at almost the same rate as it did in the absence of SSB-Ct peptide.
Next, we tested the effect of the peptide on Pol IV activity on a long, SSB-coated ssDNA. Primed M13mp18 circular ssDNA template was pre-incubated with SSB, the γ complex and the β clamp for 3 min to load the β clamp. Pol IV was then added to the reaction, together with SSB-Ct peptide at a final concentration of 110 µM, and the DNA synthesis reaction was started. After 3- or 6-min incubations with dNTPs and 32P-dATP, labeled, newly synthesized products were analyzed.
In the absence of SSB, the lengths of the observed products were indistinguishable with or without SSB-Ct peptide (Figure 4C, compare lanes 1 and 2 with 3 and 4); this demonstrates that SSB-Ct peptide has no effect on Pol IV activity, consistent with the observations in Figure 4B. On the other hand, when SSB-Ct peptide was added to the reaction in the presence of SSB, the product lengths at each time point were shorter than those in reactions lacking the peptide (Figure 4C, compare lanes 5 and 6 with lanes 7 and 8). The amount of primer elongated by Pol IV appeared to be similar in both reactions, indicating that the amount of β clamp loaded on the primer was unaffected by the addition of the SSB-Ct peptide. The elongation rate was reduced to ∼75% of the normal rate by the addition of SSB-Ct peptide: in the absence of SSB-Ct peptide, the elongation rate was ∼6 nt/s, but the rate was ∼4.5 nt/s at 3 and 6 min in its presence (Figure 4C, compare lanes 5 and 6 with lanes 7 and 8). These data clearly indicate that SSB-Ct peptide has an inhibitory effect on DNA synthesis by Pol IV, but only when the template is covered with SSB.
When different amounts of SSB-Ct peptide were added to the reaction, an increasing concentration of SSB-Ct peptide resulted in progressively shorter products (Figure 5), the elongation rate at 330 µM peptide being approximately one-third of the normal rate (Figure 5, compare lane 2 with lane 6). Higher concentrations of peptide cause stronger inhibition of the interaction between SSB and Pol IV, as the peptide competes with the SSB C-terminal tail for binding to Pol IV. Therefore, we conclude that the elongation rate of Pol IV decreased as a result of the inhibition of Pol IV binding to the C-terminal tail of SSB on the ssDNA template.
SSB-Ct peptide suppresses both the processivity and the elongation rate of Pol IV
These results demonstrate that Pol IV needs to interact with the SSB tail to elongate a primer on SSB-coated ssDNA, but it is not clear how the interaction contributes to Pol IV function. As the data in Figure 3C suggested, the interaction between Pol IV and SSB tail might recruit Pol IV to the primer/template junction and accelerate the formation of the elongation complex with the β clamp. Another possible explanation is that the interaction stabilizes the elongation complex of Pol IV and promotes processive DNA synthesis; addition of the peptide might cause Pol IV to dissociate from the primer end, resulting in a decrease in the apparent elongation rate. However, it seemed improbable that these explanations would account for such a large decline in the elongation rate of Pol IV. We therefore hypothesized that the actual speed of Pol IV movement might be reduced by the addition of SSB-Ct peptide. This would mean that Pol IV requires the physical interaction with SSB tail while it replicates SSB-coated ssDNA processively.
To test this idea, we analyzed the effect of SSB-Ct peptide on the elongation rate of Pol IV during processive DNA synthesis. In Figure 5, the number of Pol IV molecules was ∼20-fold higher than that of the template DNA, under which conditions Pol IV can easily return to a 3′-primer end from which it has dissociated. On the other hand, in Figure 6, the number of Pol IV molecules is reduced to less than one-third that of the template DNA (0.3 nM Pol IV and 1 nM template DNA); Pol IV is now more likely to bind to an unused primer when it dissociates from a primer end, so that the elongation rate and processivity can be measured accurately.
M13 ssDNA annealed with a 5′-32P-labeled primer was used as a template, and Pol IV was added to the reaction 1 min before the start of DNA synthesis. We confirmed that the β clamp was loaded on all primer/template DNA by adding Pol IV to 22 nM in the control reaction (Figure 6, lane 9). One minute after the start of DNA synthesis, a portion of the primer had been homogeneously extended by ∼300 nt, although most of the primer remained unused (Figure 6, lane 2). After a 3-min incubation, a smearing pattern of replication products was observed, showing that Pol IV had started to dissociate from the primer end (Figure 6, lane 4). The elongation rate was ∼5 nt/s, and the processivity was between 300 and 900 nt, consistent with previous observations of Pol IV with the β clamp (27).
SSB-Ct peptide was next added to the reaction at the same time as the start of DNA synthesis (Figure 6, lanes 5–8). The amount of elongated primer was quite similar to that without SSB-Ct peptide (Figure 6, compare lanes 2 and 6), showing that the amount of Pol IV that binds to the β clamp and the 3′ primer end is the same in the presence or absence of SSB-Ct peptide. At 1 min, Pol IV was synthesizing DNA processively and the length of replication products was homogeneous, but the product length was markedly shorter than the normal product length (Figure 6, compare lanes 2 and 6). This clearly shows that the elongation rate is suppressed by the addition of SSB-Ct peptide, while Pol IV still replicates processively. Pol IV therefore cannot travel smoothly on SSB-coated ssDNA unless it interacts with the SSB tail.
In addition, after a 2-min incubation, the pattern of replication products becomes smeared slightly earlier in the presence than in the absence of peptide (Figure 6, compare lanes 3 and 7), showing that the processivity of Pol IV is also reduced by the addition of the peptide. This indicates that binding to the SSB C-terminal tail contributes to the stability of Pol IV to some extent, not only for the distributive extension by Pol IV without the β clamp (Figure 3C) but also even if the β clamp tethers Pol IV tightly to the primer.
Taken together, these data show that a physical interaction with the SSB C-terminal tail is an important step in DNA synthesis by Pol IV. When Pol IV binds to the β clamp and the 3′-primer end, it probably interacts with the C-terminal tail of the SSB molecule nearest to the primer on the ssDNA template. While this interaction augments the stability of the Pol IV elongation complex, at the same time it is required to enable Pol IV to migrate forward while disassembling the SSB–ssDNA complex. How the interaction mediates the movement of Pol IV on SSB-coated ssDNA is an interesting question. We propose that Pol IV actively dislodges or translocates SSB, which is potentially an obstacle to processive elongation of the polymerase, by interacting with its C-terminal tail.
DISCUSSION
We have found a previously unrecognized factor that may be involved in the function of Pol IV in the cell. SSB was one of the major proteins that bound to a Pol IV affinity column (Figure 1), and a direct interaction was detected between Pol IV and the C-terminal tail of SSB (Figures 2, 3A and 4A). The data suggest that the SSB tail functions in the recruitment or stabilization of Pol IV on the primer end in the absence of the β clamp (Figure 3C). Surprisingly, we found that the C-terminal tail of SSB is essential for processive elongation by Pol IV on SSB-coated ssDNA (Figure 3), and that Pol IV activity on SSB-coated ssDNA was greatly impaired when the interaction between Pol IV and the SSB C-terminal tail was inhibited (Figures 4C, 5 and 6). This clearly indicates that Pol IV needs the physical interaction with the SSB C-terminal tail to replicate SSB-coated ssDNA.
On the other hand, our results showed that the interaction between SSB and Pol IV is not required for the recruitment of Pol IV to the 3′-primer end when the β clamp is loaded on the primer (Figure 4C). Moreover, even when the concentration of Pol IV in the reaction was low, the amount of elongated primer was similar with or without SSB-Ct peptide (Figure 6, compare lanes 2 and 6). This shows that Pol IV does not bind indiscriminately to SSB, which coated the entire ssDNA of the circular M13 template used in our assay. The β clamp on the primer probably provides a sufficiently strong interaction to recruit Pol IV to the primer end, so that Pol IV interacts with the neighboring SSB at the primer junction only when it is bound stably to the β clamp. This speculation is consistent with a report that the presence or absence of SSB has little effect on the formation of a stable initiation complex on 30/90-mer linear DNA, whereas the β clamp greatly stabilizes Pol IV at primer/template junction (27).
However, the processivity of Pol IV decreased slightly when the interaction between Pol IV and SSB C-terminal tail was inhibited (Figure 6). This suggests that the physical interaction contributes, to some extent, to the stability of the Pol IV elongation complex on SSB-coated ssDNA even with the tight anchoring of the β clamp to the template/primer junction.
Unexpectedly, the elongation rate of Pol IV was dramatically reduced in the presence of inhibitor peptide (Figure 5 and 6). We infer that the inhibition of Pol IV DNA synthesis observed in Figures 4C and 5 was attributable mainly to a decrease in the elongation rate, as the effect of SSB-Ct peptide on Pol IV processivity was only modest. It is surprising that in the presence of the inhibitor peptide, Pol IV moves more slowly than its normal elongation rate on SSB-coated ssDNA while it is bound stably to the primer end with the β clamp (Figure 6, compare lanes 2 and 6). Moreover, Pol IV cannot processively synthesize DNA together with the β clamp in the presence of SSBΔC8 (Figure 3B). These results strongly suggest that Pol IV needs to interact physically with the SSB C-terminal tail to ensure a rapid elongation process across bound SSB on ssDNA.
There are two possible explanations for the requirement of an interaction between Pol IV and the SSB tail. One is that binding to the C-terminal tail of SSB enhances the activity of Pol IV strongly enough to dislodge SSB from the template, allowing Pol IV to rapidly elongate the primer. Although we did not observe a strong stimulation of Pol IV activity by the addition of SSB-Ct peptide in the absence of SSB (Figure 4B and C), such stimulation may be detectable only when the template is coated with SSB.
The other possibility is that Pol IV actively promotes SSB’s dissociation from, or translocation along, ssDNA by interacting with the SSB C-terminal tail. It has been reported that the C-terminal tail of SSB inhibits the ssDNA-binding activity of SSB tetramer (33,34), and that removal of the tail stimulates the transition between two major ssDNA-binding modes (35). Although it is still unclear how SSB regulates its binding mode by means of the C-terminal tail during DNA replication, the interaction of Pol IV with the tail may stimulate the switch of SSB binding mode to that which is more conducive to elongation by Pol IV.
As well as Pol IV, three of the five DNA polymerases in E. coli are already known to bind to SSB (3). Does the requirement for an interaction with the SSB tail also apply to other polymerases? Similar to the result observed in Figure 3B, SSBΔC8 greatly inhibits DNA synthesis and strand displacement by Pol III (14,15). In addition, Pol III HE requires the χ/ψ subunit, which is necessary for SSB binding, to overcome an inhibition of DNA synthesis by SSB on an SSB-coated linear ssDNA template (12). We have also observed some inhibition of burst DNA synthesis by Pol III on an SSB-covered M13 template when the SSB-Ct peptide is present in the reaction (data not shown). These results imply that Pol III, too, needs to interact with the SSB tail to replicate SSB-coated ssDNA efficiently. In vivo, exposed ssDNA is rapidly covered with SSB, so that any polymerase must overcome SSB to replicate ssDNA. We suggest that all polymerases may possess a common mechanism to replicate SSB-coated ssDNA that is mediated by an interaction between the polymerase and the SSB C-terminal tail. It will be interesting to ascertain whether the requirement for the SSB tail that we describe here for E. coli Pol IV is a fundamental characteristic of polymerases in other organisms.
FUNDING
Ministry of Education, Culture, Sports, Science and Technology, Japan, Grant-in-Aid for Young Scientists (Start-up) (21870023 to A.F.); and Grant-in-Aid for Scientific Research on Innovative Areas (23131518 to H. M.). Funding for open access charge: Intramural funding of Nara Institute of Science and Technology.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Daichi Ogawara for ΔdinB strain construction, Drs Yoichiro Fukao and Masayuki Fujiwara (Plant Science Education Unit, NAIST) for mass spectrometry analysis, Dr Tsutomu Katayama (Kyushu University) for the generous gift of the γ complex, Drs Akiko Sakai and Michael M. Cox (University of Wisconsin-Madison) for their generous gift of SSBΔC8 overexpression plasmid and for help in purification of SSBΔC8. The authors thank Dr Hisashi Tatebe for useful discussions. The authors are grateful to Dr Satoko Maki for helpful discussions and generous support, and to Dr Ian Smith for helpful discussions and comments on the manuscript.
REFERENCES
- 1.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 3.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kornberg A, Baker TA. DNA Replication. 2nd edn. New York, NY: Freeman; 1992. [Google Scholar]
- 5.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and Mutagenesis. 2nd edn. Washington, DC: ASM Press; 2006. [Google Scholar]
- 6.McHenry CS. DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- 7.McHenry CS. Chromosomal replicases as asymmetric dimers: studies of subunit arrangement and functional consequences. Mol. Microbiol. 2003;49:1157–1165. doi: 10.1046/j.1365-2958.2003.03645.x. [DOI] [PubMed] [Google Scholar]
- 8.Pomerantz RT, O'Donnell M. Replisome mechanics: Insights into a twin DNA polymerase machine. Trends Microbiol. 2007;15:156–164. doi: 10.1016/j.tim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Molineux IJ, Gefter ML. Properties of the Escherichia coli in DNA binding (unwinding) protein: interaction with DNA polymerase and DNA. Proc. Natl Acad. Sci. USA. 1974;71:3858–3862. doi: 10.1073/pnas.71.10.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonner CA, Stukenberg PT, Rajagopalan M, Eritja R, O'Donnell M, McEntee K, Echols H, Goodman MF. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J. Biol. Chem. 1992;267:11431–11438. [PubMed] [Google Scholar]
- 11.Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glover BP, McHenry CS. The χ ψ subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 13.Marceau A, Bahng S, Massoni S, George N, Sandler S, Marians K, Keck J. Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO J. 2011;30:4236–4247. doi: 10.1038/emboj.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Q, McHenry CS. Strand displacement by DNA polymerase III occurs through a τ-ψ-χ link to single-stranded DNA-binding protein coating the lagging strand template. J. Biol. Chem. 2009;284:31672–31679. doi: 10.1074/jbc.M109.050740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Downey CD, McHenry CS. Chaperoning of a replicative polymerase onto a newly assembled DNA-bound sliding clamp by the clamp loader. Mol. Cell. 2010;37:481–491. doi: 10.1016/j.molcel.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arad G, Hendel A, Urbanke C, Curth U, Livneh Z. Single-stranded DNA-binding protein recruits DNA polymerase V to primer termini on RecA-coated DNA. J. Biol. Chem. 2008;283:8274–8282. doi: 10.1074/jbc.M710290200. [DOI] [PubMed] [Google Scholar]
- 17.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 18.Tippin B, Pham P, Goodman MF. Error-prone replication for better or worse. Trends Microbiol. 2004;12:288–295. doi: 10.1016/j.tim.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Jarosz DF, Beuning PJ, Cohen SE, Walker GC. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15:70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Furukohri A, Goodman MF, Maki H. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J. Biol. Chem. 2008;283:11260–11269. doi: 10.1074/jbc.M709689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu D, Windsor MA, Gellman SH, Keck JL. Peptide inhibitors identify roles for SSB C-terminal residues in SSB/exonuclease I complex formation. Biochemistry. 2009;48:6764–6771. doi: 10.1021/bi900361r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pace C, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugaya Y, Ihara K, Masuda Y, Ohtsubo E, Maki H. Hyper-processive and slower DNA chain elongation catalysed by DNA polymerase III holoenzyme purified from the dnaE173 mutator mutant of Escherichia coli. Genes Cells. 2002;7:385–399. doi: 10.1046/j.1365-2443.2002.00527.x. [DOI] [PubMed] [Google Scholar]
- 24.Hobbs MD, Sakai A, Cox MM. SSB protein limits RecOR binding onto single-stranded DNA. J. Biol. Chem. 2007;282:11058–11067. doi: 10.1074/jbc.M611007200. [DOI] [PubMed] [Google Scholar]
- 25.Godoy VG, Jarosz DF, Simon SM, Abyzov A, Ilyin V, Walker GC. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell. 2007;28:1058–1070. doi: 10.1016/j.molcel.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics. 2001;266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- 27.Wagner J, Fujii S, Gruz P, Nohmi T, Fuchs RP. The β clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 2000;1:484–488. doi: 10.1093/embo-reports/kvd109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertram JG, Bloom LB, O'Donnell M, Goodman MF. Increased dNTP binding affinity reveals a nonprocessive role for Escherichia coli β clamp with DNA polymerase IV. J. Biol. Chem. 2004;279:33047–33050. doi: 10.1074/jbc.C400265200. [DOI] [PubMed] [Google Scholar]
- 29.Bobst EV, Bobst AM, Perrino FW, Meyer RR, Rein DC. Variability in the nucleic acid binding site size and the amount of single-stranded DNA-binding protein in Escherichia coli. FEBS Lett. 1985;181:133–137. doi: 10.1016/0014-5793(85)81128-5. [DOI] [PubMed] [Google Scholar]
- 30.Lenne-Samuel N, Wagner J, Etienne H, Fuchs RP. The processivity factor β controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 2002;3:45–49. doi: 10.1093/embo-reports/kvf007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savvides SN, Raghunathan S, Fütterer K, Kozlov AG, Lohman TM, Waksman G. The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA. Protein Sci. 2004;13:1942–1947. doi: 10.1110/ps.04661904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witte G, Urbanke C, Curth U. DNA polymerase III χ subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–4440. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marintcheva B, Marintchev A, Wagner G, Richardson CC. Acidic C-terminal tail of the ssDNA-binding protein of bacteriophage T7 and ssDNA compete for the same binding surface. Proc. Natl Acad. Sci. USA. 2008;105:1855–1860. doi: 10.1073/pnas.0711919105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozlov AG, Cox MM, Lohman TM. Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J. Biol. Chem. 2010;285:17246–17252. doi: 10.1074/jbc.M110.118273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J. Mol. Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]