Abstract
Mitochondrial translation is essentially bacteria-like, reflecting the bacterial endosymbiotic ancestry of the eukaryotic organelle. However, unlike the translation system of its bacterial ancestors, mitochondrial translation is limited to just a few mRNAs, mainly coding for components of the respiratory complex. The classical bacterial initiation factors (IFs) IF1, IF2 and IF3 are universal in bacteria, but only IF2 is universal in mitochondria (mIF2). We analyse the distribution of mitochondrial translation initiation factors and their sequence features, given two well-propagated claims: first, a sequence insertion in mitochondrial IF2 (mIF2) compensates for the universal lack of IF1 in mitochondria, and secondly, no homologue of mitochondrial IF3 (mIF3) is identifiable in Saccharomyces cerevisiae. Our comparative sequence analysis shows that, in fact, the mIF2 insertion is highly variable and restricted in length and primary sequence conservation to vertebrates, while phylogenetic and in vivo complementation analyses reveal that an uncharacterized S. cerevisiae mitochondrial protein currently named Aim23p is a bona fide evolutionary and functional orthologue of mIF3. Our results highlight the lineage-specific nature of mitochondrial translation and emphasise that comparative analyses among diverse taxa are essential for understanding whether generalizations from model organisms can be made across eukaryotes.
INTRODUCTION
Mitochondria are multifunctional organelles of virtually all eukaryotic cells and were probably present in the last common ancestor of all extant eukaryotes (1). They take part in production of energy, fatty acid metabolism, apoptosis and many other cellular processes. According to the endosymbiotic hypothesis, mitochondria are of bacterial origin (2), which explains why they contain their own genome and are competent in transcription and translation of their genetic material.
Translation initiation in bacteria is facilitated by three universal and essential initiation factors (IFs), IF1, IF2 and IF3. IF2 in the GTP-bound form promotes binding of aminoacylated and formylated initiator tRNA (fMet-tRNAi) to the small ribosomal subunit and subsequent docking of the large subunit to this pre-initiation complex (3–5). IF1 and IF3 together contribute to selection of the initiator codon and fMet-tRNAi through a delicate kinetic mechanism (6–9), and have equally important roles in ribosomal recycling (10–12). In addition to its role in the IF1/IF3-specific recycling pathway (12), IF3 is required for stable subunit dissociation in the EF-G/RRF-mediated recycling pathway (11).
One important difference between the mitochondrial and bacterial translational systems is that the former deals with a very limited set of different mRNAs coding for a handful of proteins, mostly components of the respiratory complex. Most of what we know about translational control in mitochondria comes from the model organism Saccharomyces cerevisiae. In this yeast, the translational machinery is extremely specialized for translating these mRNAs (13). Sequence-specific translational activators that interact with specific elements in the mRNAs link translation of the specific mRNA to their localization, as well as activate translation (14,15). Thus, the classical IF set in mitochondria in this yeast is less involved in initiation codon and initiator tRNA selection: selection of the initiation region is performed by activators, and the need for fMet-tRNAi is relaxed, allowing initiation with elongator Met-tRNA (16).
The mitochondrial translation machinery has a modified set of classical IFs in comparison to bacteria. IF1 is absent altogether, and mitochondrial IF2 (mIF2), which is universal in mitochondriate eukaryotes, has been suggested to be the functional equivalent of both bacterial IF1 and IF2. A short insertion between domains V and VI has been identified as the element responsible for IF1-like function using complementation assays in Escherichia coli (17). Indeed, cryo-electron microscopy of mIF2 in complex with initiator tRNA and the bacterial ribosome suggests the insertion occupies the same binding site on the ribosome that would be occupied by IF1 (18). However, no detailed comparative sequence analysis of the insertion across a broad distribution of eukaryotes has been carried out to confirm or reject a relationship with IF1 loss.
Orthologues of IF3 (mIF3) are present in a number of eukaryotes including the fission yeast Schizosaccharomyces pombe, but a clear homologue of mIF3 has not been identified in budding yeast S. cerevisiae (19,20). This raises questions regarding the mechanism of translation initiation and recycling in this organism. Recently, S. cerevisiae mitochondrial translational activators Aep3p and Rsm28p have been shown to interact genetically and physically with mIF2 and initiator tRNAi, thus being directly involved in selection of formation of the pre-initiation complex (21–23). These observations raise the possibility that Aep3p and Rsm28p may perform analogous functions of mIF1 and/or mIF3 in this organism (21–23). Again, however, the distributions of Aep3p, Rsm28p and other translational activators and their potential relationship to mIF3 distribution have not previously been addressed.
Here, present a systematic in silico analysis of mitochondrial IF2, IF3 and translational activators. We identify S. cerevisiae Aim23p as the hitherto unidentified mIF3 in Saccharomycetales. By means of in vivo complementation assays, we show that S. pombe mIF3 effectively complements a genomic disruption of S. cerevisiae AIM23, verifying that Aim23p is a bona fide mitochondrial IF3.
MATERIALS AND METHODS
Sequence retrieval and phylogenetic analysis
Sequences homologous to mIF2, mIF3 and 17 S. cerevisiae translational activators were retrieved by BlastP and PSI-Blast searches at the NCBI. Sequences were aligned using MAFFT (24), and maximum likelihood (ML) and Bayesian inference (BI) phylogenetic analyses were carried out using RAxML v7.0.4 (25) and MrBayes v3.1.2 (26). Full methods for sequence analysis are presented in Supplementary Text S1: SI Methods.
Amino acid composition, subcellular targeting and conservation analyses
The amino acid composition of peptides was calculated using the Expasy ProtParam tool (27). Mitochondrial and plastid targeting peptides were predicted using TargetP (28), MitoProt (II) (29), PATS (30) and Plasmit (31). Consensus sequences were calculated using the Consensus Finder Python script (32).
Aim23p in vivo complementation experiments
To investigate whether Aim23p is a functional orthologue of mIF3, a strain of S. cerevisiae lacking Aim23p was first obtained. The heterozygous AIM23 knockout diploid strain Y21294, which carries the chromosomal AIM23 gene disrupted by a geneticin (G418) resistance cassette was purchased from EUROSCARF, and sporulation was induced to obtain the haploid AIM23::kanMX4 strain (referred to here as AIM23Δ). This was complemented by a plasmid expressing the gene for S. pombe mIF3 (gene name SPBC18E5.13, henceforth referred to as S.p.MIF3), and growth was assayed in fermentable and non-fermentable media. Mitochondrial oxygen uptake was measured by Clark electrode and mitochondrial membrane potential was assayed by Rhodamine 123 and DiOC6 labelling. Full methods for complementation analyses are presented in Supplementary Text S1: SI Methods, and the full list of strains and plasmids used in this study is found in Supplementary Table S1.
RESULTS
The conserved insertion within mIF2 is only found in vertebrates
IF2 is a translational GTPase (trGTPase) and is the only IF universal in bacteria, archaea, eukaryotes and both the eukaryotic organelles capable of translation: plastids (chloroplasts and apicoplasts derived from secondary endosymbiosis) and mitochondria. We therefore use IF2 as a framework for mapping the presence and absence of other IFs. Phylogenetic analysis of the ‘full IF2 data set’, that is IF2 homologues across bacteria, eukaryotes and archaea, rooted with archaeal and eukaryotic orthologues a/eIF5B shows a lack of resolution in the backbone of the bacterial and organellar IF2 part of the tree (Supplementary Figure S1). This is not surprising given the vast evolutionary distances covered in the whole tree (dating back to the last common ancestor of all life) and the differences in evolutionary rate among organellar sequences (as seen by the long branches for organellar IF2, Supplementary Figure S1). The longest branched protist organellar IF2 sequences are mostly found at the base of the IF2 part of the tree, but relationships among groups have no statistical support in this part of the tree, and possibly represent long-branch attraction (LBA) to the out-group. Scans for organellar transit peptides enable grouping of sequences into putative organelle-specific groups (Supplementary Figure S1). A surprising feature of the tree is a long-branched clade referred to here as mIF2-2, which is a second copy of mIF2 in members of alveolates (dinoflagellates and apicomplexa) and kinetoplastida. mIF2-2 forms a strong clade (83%) but there is no support for its placement within the IF2 tree. Its identity as an IF2-like homologue rather than another member of the trGTPase superfamily is supported by a more significant hit to the IF2 family hidden Markov model in the PFam database than to other trGTPase subfamilies (33). Apicomplexa contain two translationally competent organelles: apicoplasts and mitochondria, both of which require IF2. However, Plasmit (31), PATS (28) and TransitP (28) predict that mIF2-2 are mitochondria- rather than apicoplast-targeted proteins, while the predicted apicoplast IF2s are nested without statistical support in the bacterial part of the tree (Supplementary Figure S1).
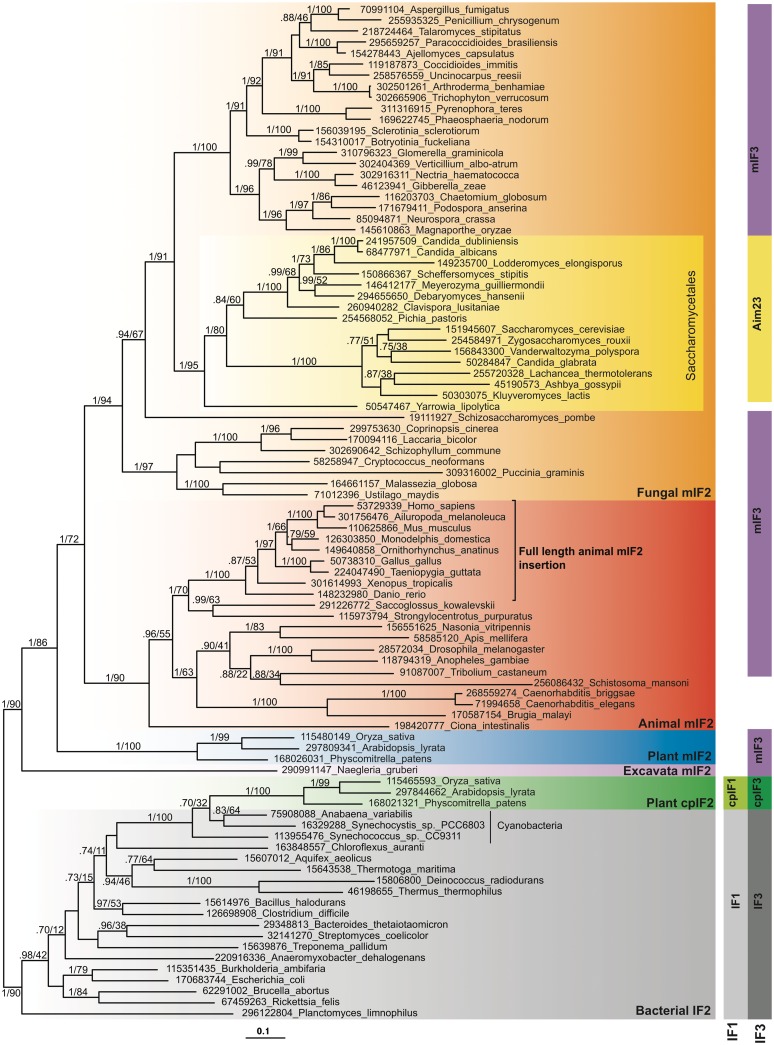
The full IF2 data set comprises alignment columns that are universally alignable across the IF2 family of bacteria, archaea and eukaryotes. Reducing the data set to the bacterial and organellar IF2 group alone and removing the long-branched protist sequences means more positions can be used, and LBA artefacts minimized. Phylogenetic analysis of this cut-down IF2 data set shows much more resolution of the organellar sequences (Figure 1). There is strong support (1.0 BIPP, 90% MLBP) for the monophyly of mIF2 and also full support (1.0 BIPP, 100% MLBP) for the grouping of cpIF2 with cyanobacteria.
Figure 1.
Phylogenetic tree of mitochondrial IF2 (mIF2), bacterial IF2 and chloroplast IF2 (cpIF2). The tree is a MrBayes consensus tree, generated from 405 aligned amino acids. The standard deviation of split frequencies at the end of the MrBayes run was 0.018. Bayesian inference posterior probability (BIPP) and maximum likelihood bootstrap percentage (MLBP) support are indicated on branches. Only branches with >0.70 BIPP support are labelled with BIPP and MLPP values. The scale bar below the tree shows the evolutionary distance expressed as substitutions per site. Numbers in taxon names are NCBI GI numbers. Vertical blocks show the distribution of IF1, mitochondrial IF1 (mIF1), chloroplast IF1 (cpIF1), IF3, mitochondrial IF3 (mIF3), Aim23p and chloroplast IF3 (cpIF3). Blocks in the same column indicate orthologous proteins. The black bracket shows the taxonomic boundary of the full-length conserved animal IF2 insertion.
The mIF2 sequence alignment (Figure 2) shows the distribution and extent of conservation of the insertion in mtIF2 in the region between domains V and VI (Figure 2), which on the basis of complementation assays between E. coli IF1 and bovine mIF2 was suggested to replace the universally lost mitochondrial IF1 (17). The insertion was first noted in bovine mIF2 (34) and alignment of several available sequences indicated the absence of universal conservation, with conservation limited to mammals (35,36). Our alignment samples more broadly across the eukaryotic tree and shows that the well-conserved, full-length animal insertion is limited to vertebrates (Figure 2). PSI-Blast searches for IF1 in eukaryotes identified cpIF1 and cytoplasmic eIF1A, but failed to identify IF1 mitochondrial homologues (Figure 1 and Supplementary Table S2). Thus, mitochondrial IF1 loss appears to be universal for all eukaryotes, most likely occurring millions of years earlier than the IF2 insertion evolved in animals. The insertion has a strongly biased amino acid composition; considering the whole region that cannot be confidently aligned with bacterial IF2 (63 amino acids, 15–111 in Figure 2), glutamate and lysine are particularly over-represented (20.3 and 21.9%, respectively, for human IF2; Supplementary Table S3). Although the IF2 insertion is proposed to be carrying out the function of IF1 in mitochondria, such dramatic bias is not seen in E. coli IF1, which is only slightly over-represented in glutamate and lysine (by 1.54 and 1.05%, respectively; Supplementary Table S3). Due to ambiguous homology, the boundaries of the insertion are impossible to define with confidence. There appear to have been many independent insertions and deletions, as the region is variable in length across all taxa, including bacteria and cpIF2, both of which carry IF1 (Figure 1). There has been a particularly large insertion in Ustilago maydis mIF2 (Figure 2). This suggests that this region is able to accommodate quite large variations in size and sequence seemingly without perturbing protein function.
Figure 2.
Sequence alignment of the mIF2 insertion region. Example sequences from across the IF2 tree are aligned, with their major taxonomic groupings indicated. The domain structure of human mIF2 is shown above the alignment, with dotted lines indicating the location of the insertion.
Aim23p is the orthologue of mitochondrial initiation factor mIF3
BlastP searches alone failed to identify mIF3 homologues from distantly related eukaryotes. However homologues from across the eukaryotic tree of life were identified with the more sensitive PSI-Blast (Supplementary Table S2). The PSI-Blast hits included previously identified mIF3s from human and S. pombe, chloroplast IF3s (cpIF3s), and a Saccharomycetale protein called Aim23p/YJL131C in S. cerevisiae (henceforth referred to as Aim23p). Aim23p has been identified in the S. cerevisiae mitochondrial proteome, and mutation of Aim23p results severe respiratory defect suggesting its importance for mitochondrial functionality (37–39). However, no specific function has previously been assigned or predicted for Aim23p.
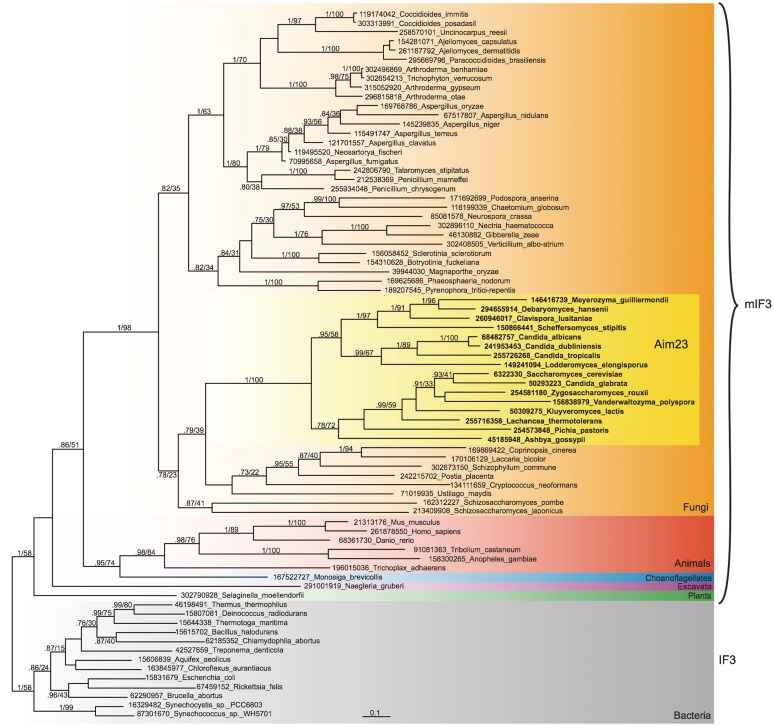
Phylogenetic analysis of the full data set comprising IF3/mIF3/cpIF3 and Aim23p shows that the Aim23p sequences group with other fungi (Supplementary Figure S2). Although this has only weak support in the full IF3 tree (51% MLBP), phylogenetic analysis of a cut-down IF3 data set with the longest branched protist IF3 sequences removed and more sites included has very strong support for the monophyly of Aim23p and fungal mIF3s (1.0 BIPP, 98% MLBP; Figure 3). Thus, although the position of Aim23p within fungi has no significant support, it clearly groups with fungi, and the most parsimonious explanation is that Aim23p is in fact the previously unidentified mIF3 orthologue. The inability to find significant mIF3 hits across fungi using BlastP alone is probably due to the short length of IF3 and its highly biased amino acid composition. IF3 is enriched in charged amino acids, particularly lysine; E. coli IF3, human mIF3 and S. cerevisiae Aim23p contain 11.7, 10.4 and 14.9% lysine, respectively (Supplementary Table S3). Schizosaccharomyces pombe mIF3 is also surprisingly enriched in serine (12.4%). The bias in lysine composition in IF3 and the IF2 insertions may reflect the role of these proteins in binding negatively charged RNA; enrichment of lysine and other charged amino acids is a feature of ribosomal proteins (40).
Figure 3.
Phylogenetic tree of mitochondrial IF3 (mIF3), Aim23p and bacterial IF3. The tree is a MrBayes consensus tree, generated from 156 aligned amino acids. The standard deviation of split frequencies at the end of the MrBayes run was 0.015. Branch support, GI numbers and substitutions per site are indicated as per Figure 1. The Aim23p clade is indicated with a dashed box.
Although we have identified the missing mIF3 orthologue in S. cerevisiae, and we find mIF3 is represented in all major groups of mitochondriate eukaryotes, we could not identify mIF3 in a handful of species: alveolate Toxoplasma gondii, fungus Yarrowia lipolytica, platyhelminth Schistosoma mansoni and chromodorean nematodes Caenorhabditis briggsae, Caenorhabditis elegans and Brugia malayi (although a homologue was found in Enoplean nematode Trichinella spiralis; Figure 1). Kinetoplastida, on the other hand, have experienced a duplication of mIF3 (NCBI GI numbers in Supplementary Table S2). These paralogues are more similar to each other than to any other mIF3 orthologue, indicating that they are in-paralogues, i.e. the duplication occurred in the lineage to kineoplastida. The PSI-Blast results suggest kinetoplastida mIF3-1 and mIF3-2 are genuine mIF3 homologues, and TargetP predicts mitochondrial transit peptides. However, the sequences are too divergent to be reliably aligned and included in either the full or reduced mIF3 phylogenies.
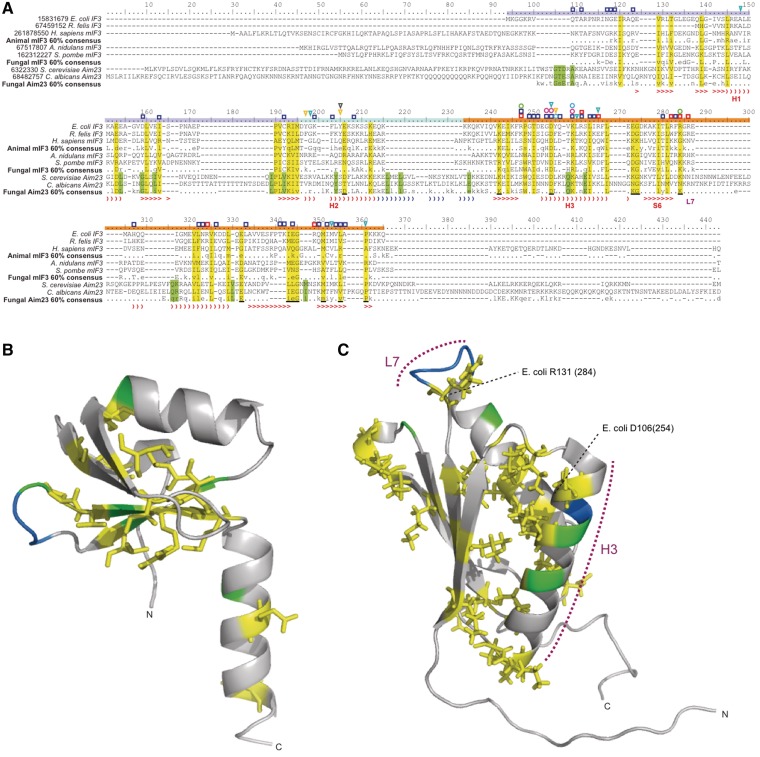
IF3 is comprised of two domains joined by a flexible linker (41). NMR has shown that the ribosome interacting sites of bacterial IF3 are mainly found in the C-terminal domain (CTD), with fewer interacting sites in the N-terminal domain (NTD) (42). Sequence alignment shows that Aim23p, mIF3 and bacterial IF3 can be aligned and contain conserved positions across both the NTD and CTD (yellow columns, Figure 4A–C). A striking clustering of functionally important sites is found in and around helix H3 and sheet S6 of the CTD, regions shown with NMR and X-ray crystallography to interact with the ribosome (42,43). Several sites are conserved in Aim23p and other mIF3s in these regions (Figure 4A). The linker between the N and C domains is extended by an insertion of up to 19 amino acids in Aim23p (alignment positions 216–234; Figure 4A), and secondary structure prediction of S. cerevisiae Aim23p with PSIPred (44) suggests the insertion in this region extends the H2 helical region of the linker (Figure 4A). There are low complexity regions at both the extreme N and C termini of mIF3; N and C-terminal extensions have been previously reported in human mtIF3, relative to bacterial IF3 (45), but these are longer in Aim23p (Figure 4A). The extensions are poorly conserved in primary sequence, but are rich in charged amino acids, particularly glutamate, glutamine, asparagine and lysine, similarly to the insertion in mIF2 (Figure 4A).
Figure 4.
IF3 NTD and CTD structures showing patterns of sequences conservation. IF3 subgroup consensus and example sequences are aligned in (A), with consensus sequences calculated at the 60% level using the Python script Consensus Finder (32). Yellow columns indicate those sites that are conserved in Aim23p, mIF3 and/or bacterial IF3, exposed sites of which are underlined in black. Green columns are those sites limited in conservation to Aim23p. The secondary structure of E. coli IF3 (41) is shown in red below the alignment, with ‘closing parenthesis’ representing helices and ‘greater than’ symbol representing sheets. Secondary structure in blue shows the extension of the linker helical region in S. cerevisiae Aim23p, as predicted with PSI-Pred symbols above the alignment show sites predicted to affect IF3 function. Dark blue squares: (42); red squares: (49); yellow triangles: (51); turquoise triangles: (48); green circles: (50); light blue circles: (47); pink circles: (46). The structure of (B) Bacillus stearothermophilus IF3 NTD [PDB entry 1TIF (41)], and (C) Mus musculus mIF3 CTD (NMR structure, PDB entry 2CRQ). Helix H3 is indicated, along with the universally conserved D106 (E. coli numbering), which is found at alignment position 254 in (A). As in (A), yellow residues indicate those sites that are conserved in Aim23p, mIF3 and/or bacterial IF3. The side-chains of these residues are also shown. Green residues are those limited in conservation to Aim23p. Blue residues show the location of insertions in Aim23p.
Plotting the sites that are conserved in Aim23p and mIF3 or bacterial IF3 onto the 3D structures of the NTDs and CTDs shows that these sites are largely buried, with the side chains oriented into the center of each domain (Figure 4B and C). This is particularly apparent in the CTD, where the conserved residues may be involved in intramolecular interactions to maintain the bundle shape.
Residues H170, D171 and K175 of human mIF3 (positions 253, 254 and 259, respectively in Figure 4A) have been found to be particularly important for the dissociation of mitochondrial 55S ribosomes (46). D254 is universally conserved in Aim23p and IF3, H253 is unconserved in Aim23p, and K259 is universally conserved as K in Aim23, with a conservative substitution to R in some mIF3s (Figure 4A). The positions D254 (D106 in E. coli) and K259 (K110 in E. coli) have also been implicated in ribosome interactions in bacteria (42,47,48). The functional importance of all arginine residues in the E. coli IF3 CTD has been determined (49). However, none of these arginines are conserved outside of bacteria, with the possible of exceptions of R112 (alignment position 262), which has the chemically similar lysine in this position in mIF3 (but not Aim23p), and R131, which has lysine at this position in both mIF3 and Aim23p (Figure 4A). Mutation of R112 on the surface of helix H3 severely disrupts ribosome binding and subunit dissociation in E. coli (49). R131 is part of an exposed loop (Figure 4A and C) and is proposed to contact the mRNA (49), as is supported by its involvement in start codon discrimination (50). Another critical residue of E. coli IF3, Y75 (51) is unconserved outside of bacteria (Figure 4A).
Validation of Aim23p as a bona fide mitochondrial IF3
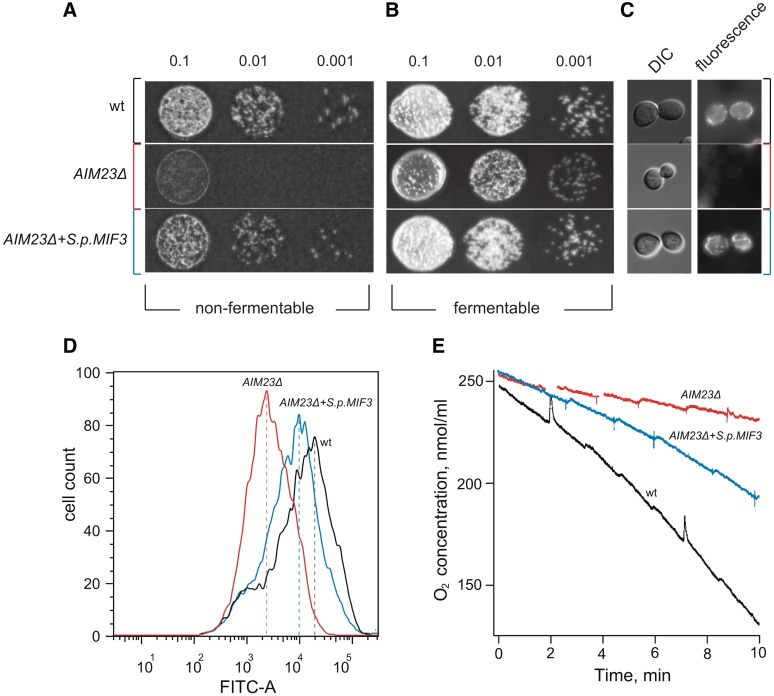
To test whether Aim23p is a functional mIF3 in S. cerevisiae, we employed an in vivo complementation strategy. We obtained a haploid S. cerevisiae strain with the chromosomal AIM23 gene disrupted by a geneticin (G418) resistance cassette by inducing sporulation in a heterozygous AIM23 knockout diploid yeast strain Y21294 (EUROSCARF). By assessing growth of the wild-type and AIM23Δ strains on fermentable (i.e. not requiring mitochondrial functionality) and non-fermentable (i.e. requiring mitochondrial functionality) media, we confirm the AIM23Δ respiration deficient phenotype reported in (37–39) (Figure 5A and B). This defect in growth on non-fermentable media is reversed by complementation with a plasmid encoding the gene for S. pombe mIF3 (S.p.MIF3) with S. cerevisiae 5′ and 3′ flanking regions driving its expression (Figure 5A and B).
Figure 5.
S. pombe mIF3 rescues a respiratory defect in an S. cerevisiae strain lacking the genomic copy of AIM23. (A and B) Ten-fold serial dilutions (made on the basis of measurement of OD600 of wild-type (‘wt’, Y21294a), AIM23Δ and AIM23Δ + S.p.MIF3 strains were spotted on YPGly (A, non-fermentable) or YPD (B, fermentable) plates and incubated at 30°C for 4 and 1 days, respectively. (C) Differential interference contrast (DIC, left column) and fluorescence (right column) images of the yeast cells stained with mitochondrial membrane potential marker Rhodamine 123. (D) Flow cytometry analysis of DiOC6-stained wt (black trace), AIM23Δ (red trace) and AIM23Δ + S.p.MIF3 (blue trace) yeast cells with the FITC-A detection channel. (E) Measurements of oxygen consumption using a Clark-type oxygen electrode. Oxygen consumption values (in pmoles O2 per minute per 10Е6 cells) prior to FCCP treatment /after FCCP treatment/after treatment with 0.5 mM KCN are 49/92/2 for wt (black trace), 26/29/18 for AIM23Δ (red trace) and 29/58/2 for AIM23Δ + S.p.MIF3 (blue trace).
To demonstrate that the AIM23 knockout indeed specifically affects mitochondrial functionality and that these effects are rescued by S. pombe mIF3, we analysed mitochondrial membrane potential of live yeast cells both by Rhodamine 123 staining followed by fluorescence microscopy and by DiOC6 staining followed by flow cytometry. In the AIM23Δ strain, unlike wild-type, Rhodamine 123 fluorescence signal is almost indetectable, indicating failure in the electron transport chain and resulting lack of membrane potential. Fluorescence signal is restored in the mIF3 complementation strain (Figure 5C). Similarly, mean DiOC6 fluorescence decreases in the AIM23Δ strain by around an order of magnitude in comparison to wild-type and over half an order of magnitude in comparison to the S.p.MIF3 complementation strain (Figure 5D). To assess effective oxygen uptake by all strains, oxygen consumption was followed using a Clark-type oxygen electrode. FCCP-induced oxygen consumption is severely affected in the AIM23Δ strain as compared to wild-type, and, again, this defect is largely (although not entirely) rescued by complementation with S.p.MIF3 (Figure 5E). Importantly, oxygen uptake by the knockout strain was insensitive to inhibition of cytochrome oxidase by 0.5 mM potassium cyanide, unlike the wild-type and complemented strains (data not shown), suggesting that only the latter two display bona fide mitochondrial respiration. Finally, we tested binding of Aim23p in vitro using E. coli ribosomes and showed that, indeed, purified Aim23p forms a tight complex with the E. coli 30S ribosome, suggestive of at least partial function in this heterologous system (Supplementary Figure S3A). It was, however, not able to split the E. coli 70S ribosome (Supplementary Figure S3B).
Distribution of yeast mitochondrial translational activators
The S. cerevisiae translational activator Aep3p has been observed to interact with mIF2 genetically and physically, suggesting it is a mitochondrial translation initiation accessory factor (21). Similarly, genetic screens suggest the Rsm28p protein of the same organism may have overlapping roles with mIF2, and is possibly performing IF1 or IF3-like functions in translation initiation (22,23). Therefore, the distributions of Aep3p and Rsm28p are of interest in relation to the distribution of mitochondrial initiation factors. In fact, these proteins are just two of many initiation-associated activators that have been discovered using S. cerevisiae as a model system [Aep1p (52), Aep2p (53), Atp22p (54,55) Cbp1p (56), Cbp6p, Cbs1p, Cbs2p (57), Cbt1p (58,59), Mss51p (60), Mtf2p (61), Nca2p (62), Nca3p (63), Pet111p (64), Pet54p, Pet112p and Pet494p (65), Pet122p (66), Pet309p (67) and Rmd9p (23)]. As the distributions of these activators across eukaryotes have not been systematically addressed, we searched with PSI-Blast for these 21 activators to uncover their patterns of presence and absence (Supplementary Table S2).
Activators are often members of multi-protein families sharing a common domain (particularly the pentatricopeptide repeat (PPR) domain). Therefore, in some cases, the sequence searching identified distant paralogues in other groups of organisms. However, such paralogues were not included, in order to record only putative functional orthologues that form a distinct clade in their phylogenetic tree. Using these criteria, putative orthologues were only identified in fungi (Supplementary Table S2), but with differences in distributions within fungi in each protein. We find that Rsm28p, Pet309p, Pet494p, Pet122p, Cbs1p, Cbs2p and Cbt1p are limited to the Saccharomycetaceae family. Aep1p, Aep3p, Atp22p, Rmd9p, Pet54p and Pet111p have a wider distribution, being specific to the order Saccharomycetes. Rmd9p has duplicates in Candida glabrata, S. cerevisiae and Vanderwaltozyma polyspora, possibly as a result of the whole genome duplication in Saccharomycete evolution (68). Aep2p and Cbp1p have representatives across the phylum Ascomycota, although Aspergillus Cbp1ps are very different in sequence. Finally, Cbp6p, Nca2p, Mtf2p and Mss51p are found across the fungal kingdom (Supplementary Table S2). Nca3p is also found in many species of fungi, but this gene has experienced multiple duplications in fungi, and therefore was not included in the table.
Pet309, a member of the PPR protein family (67) was earlier identified to have a human homolog [LRPPRC, (69,70)]. However, sequence alignment and phylogenetic analysis suggest that while they are homologous, they are distant relatives, and may be paralogues. As such, the mammalian homologues are not included in Supplementary Table S2. Pet111p also has homologues in animals and may be a highly diverged orthologue of the nucleus-localized human RNA polymerase-associated protein CTR9 homologue. Rsm28p is targeted to mitochondria in Saccharomycetales, while its closest homologue outside of this subset (aromatic amino acid aminotransferase Aro8/YGL202Wp) is seemingly cytoplasmic (38,71).
DISCUSSION
IF2 functional evolution and loss of IF1
Formation of the 70S initiation complex (IC) in bacteria requires the initiation factors IF1, IF2 and IF3. Together with IF3, IF1 inhibits the formation of initiator tRNA-less 70S ribosomes in E. coli by accelerating the binding of initiator tRNA to the 30S subunit and inhibiting joining of 50S subunits with initiator tRNA-less 30S subunits, thereby minimizing the abundance of initiator-tRNA lacking 70S ribosomes (7). IF1 plays an additional role in translation fidelity, increasing the accuracy of initiator tRNA selection over elongator aa-tRNAs (6). In addition to their role in initiation, IF1 and IF3 are also involved in ribosomal recycling (10–12).
Given all its crucial functions, it is unsurprising that IF1 is universal in bacteria, archaea (aIF1A) and the eukaryotic cytoplasm (eIF1A), as well as eukaryotic plastids (Figure 1) (72,73). However, it is not present in mitochondria, where mIF2 and mIF3 are able to function in in vitro IC formation without a mIF1 (Figure 1), (19). Based on in vivo complementation experiments in E. coli and cryoEM reconstructions of the 70S-bound bovine mIF2, it has been hypothesized that an insertion in mitochondrial mIF2 is the sole determinant needed to compensate for loss of IF1 in mitochondria (17,18).
One would expect this insertion to be universally conserved in mIF2 in order to substitute for universal loss of IF1. In fact, we find the insertion in its full form is not conserved across eukaryotes, being limited in length and conservation to vertebrates (Figures 1 and 2), consistent with an earlier report using a much smaller dataset (35). IF1 is a small protein (72 amino acids in E. coli), but the vertebrate insertion is considerably smaller (the maximum difference in length in this region in human versus E. coli IF2 is 37 amino acids). The insertion has no sequence homology to IF1, and the former has a striking bias for glutamate and lysine (∼14% and ∼16% more enriched than the Uniprot database values for the respective residues). Escherichia coli IF1, on the other hand, is only ∼1% more enriched than the Uniprot database values for these residues (Supplementary Table S3). Thus, the weight of evolutionary evidence suggests that the IF2 insertion compensation hypothesis, while it may hold true for a subset of animals, cannot hold true for all eukaryotes, as most simply do not carry a homologous insertion in mIF2. Despite this, it has been generally accepted and propagated as a universal mitochondrial mechanism (13,17,20,36,74–76).
In the most parsimonious scenario, the supposed IF1-replacing insertion in vertebrate IF2 occurred secondarily to the loss of IF1, and therefore may be the result of ongoing optimization of the system, subsequent to IF1 loss. This and other independent insertions in this region of mIF2 may have arisen simply because there is no disadvantage in adding sequence in that region, filling the free space in the A site on the ribosome which would otherwise be occupied by IF1 (77,78). The insertion appears to have a positive effect on IF1-less translation, increasing IF2 affinity to the ribosome (35) possibly via its biased amino acid composition. The insertion may secondarily serve as an additional anchoring point for mIF2, which could be the reason why in the E. coli system, bovine mtIF2 rescues the IF1-knock out strain (17).
The lack of IF1 raises the question of whether the mitochondrial translational system has a reduced requirement for classical IFs. Even in the absence of universal initiation factor mIF2, yeast still can translate a subset of mRNAs (16). We propose that this phenomenon may be the combined result of a reduction in the set of translated mRNAs, along with lineage-specific evolution of activators, specific for just one or two mRNAs. Such dedication of initiation-associated factors to specific mRNAs would not be feasible for a genome with a large protein-coding component. When the activators Aep3p and Rsm28p were found to affect mIF2 binding and dependence on initiator tRNA formylation for initiation (21,23), this raised the question of whether these proteins could be IF1 and/or IF3 functional analogues. We analysed the distribution of Aep3p, Rsm28p and other activator proteins and found that these particular means of regulating of initiation appear to be peculiar to distinct lineages. While it is likely that other mRNA-specific activators are also found in other organisms, the S. cerevisiae activators we searched for are fungi-specific, and mostly limited to Saccharomycetales.
Identification and verification of the missing Saccharomycetale mIF3 (Aim23p)
Through PSI-Blast, we have identified Aim23p as the previously undiscovered S. cerevisiase mtIF3 orthologue. An earlier attempt to find the missing mIF3 gene identified Ygl159wp and not Aim23p as a possible homologue (19). Ygl159wp does not posses a mitochondrial transit peptide and has not been identified in mitochondrial proteomes. We find no sequence homology between Ygl159wp and mIF3, and searching Ygl159wp against the Pfam domain database suggests it, in fact, belongs to the Ornithine cyclodeaminase family, unrelated to IF3 (33).
While no specific function has previously been assigned to Aim23p, it has been identified in the S. cerevisiae mitochondrial proteome, and deletion of Aim23p in high-throughput screening analyses results in abnormal mitochondrial genome maintenance and the absence of respiratory growth (37–39). Through in vivo complementation analyses, we show that S. pombe mIF3 can compensate for a deletion of Aim23p in S. cerevisiae (Figure 5), confirming that Aim23p is the functional, as well as evolutionary orthologue of mIF3. S. cerevisiae is a promising model system for the study of human mitochondrial disorders, as has recently been exemplified in a screen for compounds able to suppress the respiratory growth disorder caused by a defect in an ATP synthase assembly protein (79). A polymorphism in human mIF3 that results in reduced levels of the protein is associated with onset of Parkinson’s disease (80–82). With the identification of Aim23p as yeast mIF3, a similar screen could be carried out to find compounds that suppress the phenotypic effect of defects in Aim23p. However, while S. cerevisiae is a very promising model system, it is important to keep in mind the significant differences between the mammalian and yeast mitochondrial systems, such as the contribution of translational activators in yeast that are absent in animal lineages.
Lineage-specific duplications of classical IFs
Other branches of the eukaryotic tree of life have experienced their own remodeling of the set of classical IFs. Our sequence searching has identified duplications of mIF2 in alveolates and kinetoplastida (mIF2-2; Figure 1) and mIF3 in kinetoplastida (Supplementary Table S2). It is not clear why these protists would require multiple mitochondrial initiation factors. However, their mitochondrial translational apparatuses have several distinctive features. Many alveolates and kinetoplastida translate as few as three proteins, have fragmented rRNA genes that are assembled post-transcriptionally, and import all or most tRNAs from the cytoplasm, possibly in their aminoacylated form in the case of apicomplexan alveolates (83). Kinetoplastida also have drastically reduced mitochondrial rRNAs (9S and 12S ribosomal RNAs in the case of Trypanosoma brucei) and have experienced a dramatic expansion in ribosomal protein content (84,85). Recent investigations of gene duplications of mitochondrial EF-Tu (86) and EF-G (32,87) have shed light on the subfunctionalization (i.e. functional divergence of two copies of a duplicated multifunctional protein) of these core components of translational machinery, and similar investigations of mIF2 and mIF3 would be very interesting. Indeed, both of these proteins are multifunctional; in addition to their roles in initiator tRNA recruitment to the ribosome, IF2 promotes subunit docking to the pre-initiation complex, while IF3 promotes premature subunit association in the recycling step of protein synthesis (3–11).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S3, Supplementary Figures S1–S3, Supplementary Text S1: SI Methods and Supplementary References [19,24–26,29,88–94].
FUNDING
The European Social Fund (MJD99, Mobilitas grant to G.C.A.); Estonian Science Foundation (ETF7616 and ETF9012 to V.H., ETF9020 to G.C.A.); the European Regional Development Fund through the Center of Excellence in Chemical Biology (code RLOTITIPP to V.H. and T.T.); Russian Foundation for Basic Research, Ministry of Education and Science of Russia, the President of Russian Federation Grant for Young Researchers and funds from International Associated Laboratory ‘RNA-mitocure’ (to P.K., A.K., V.L. and E.S.); SA Archimedes foundation, European Social Fund and DoRa program (to A.K. and S.T.); U.M.N.I.K. program (E.S. and A.K.); ANR (Agence Nationale de la Recherche to M.V.). Funding for open access charge: Estonian Science Foundation grant of G.C.A. and the Center of Excellence in Chemical Biology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Many thanks to Ivan Tarassov for providing yeast strain Y21294, Fedor Severin for the opportunity to use the micromanipulator for yeast tetrad dissection and Arvi Jõers and Antonio Caballero Reyes for advice in flow cytometry. V.H. and G.C.A. conceived of this study and GCA carried out sequence searching and phylogenetic analyses. G.C.A. and V.H. drafted the article with input from A.K. and T.T. T.T. helped draft the article and participated in the coordination of this study. A.S. performed in vitro 30S binding and 70S splitting experiments. A.K., P.K., V.L., M.Y.V., S.T. and E.S. performed in vivo complementation experiments. All authors read and approved the final article.
REFERENCES
- 1.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 2.Gray MW, Burger G, Lang BF. The origin and early evolution of mitochondria. Genome Biol. 2001;2:REVIEWS1018. doi: 10.1186/gb-2001-2-6-reviews1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall RA, Aitken CE, Puglisi JD. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol. Cell. 2009;35:37–47. doi: 10.1016/j.molcel.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milon P, Carotti M, Konevega AL, Wintermeyer W, Rodnina MV, Gualerzi CO. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep. 2010;11:312–316. doi: 10.1038/embor.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoun A, Pavlov M, Andersson K, Tenson T, Ehrenberg M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 2003;22:5593–5601. doi: 10.1093/emboj/cdg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol. Cell. 2006;23:183–193. doi: 10.1016/j.molcel.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 7.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 2006;25:2539–2550. doi: 10.1038/sj.emboj.7601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grigoriadou C, Marzi S, Kirillov S, Gualerzi CO, Cooperman BS. A quantitative kinetic scheme for 70 S translation initiation complex formation. J. Mol. Biol. 2007;373:562–572. doi: 10.1016/j.jmb.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigoriadou C, Marzi S, Pan D, Gualerzi CO, Cooperman BS. The translational fidelity function of IF3 during transition from the 30 S initiation complex to the 70 S initiation complex. J. Mol. Biol. 2007;373:551–561. doi: 10.1016/j.jmb.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peske F, Rodnina M, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Zavialov AV, Hauryliuk VV, Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Pavlov MY, Antoun A, Lovmar M, Ehrenberg M. Complementary roles of initiation factor 1 and ribosome recycling factor in 70S ribosome splitting. EMBO J. 2008;27:1706–1717. doi: 10.1038/emboj.2008.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Martinez X, Funes S, Camacho-Villasana Y, Marjavaara S, Tavares-Carreon F, Shingu-Vazquez M. Protein synthesis and assembly in mitochondrial disorders. Curr. Top. Med. Chem. 2008;8:1335–1350. doi: 10.2174/156802608786141124. [DOI] [PubMed] [Google Scholar]
- 14.Towpik J. Regulation of mitochondrial translation in yeast. Cell Mol. Biol. Lett. 2005;10:571–594. [PubMed] [Google Scholar]
- 15.Naithani S, Saracco SA, Butler CA, Fox TD. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:324–333. doi: 10.1091/mbc.E02-08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tibbetts AS, Oesterlin L, Chan SY, Kramer G, Hardesty B, Appling DR. Mammalian mitochondrial initiation factor 2 supports yeast mitochondrial translation without formylated initiator tRNA. J. Biol. Chem. 2003;278:31774–31780. doi: 10.1074/jbc.M304962200. [DOI] [PubMed] [Google Scholar]
- 17.Gaur R, Grasso D, Datta PP, Krishna PD, Das G, Spencer A, Agrawal RK, Spremulli L, Varshney U. A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol. Cell. 2008;29:180–190. doi: 10.1016/j.molcel.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yassin AS, Haque ME, Datta PP, Elmore K, Banavali NK, Spremulli LL, Agrawal RK. Insertion domain within mammalian mitochondrial translation initiation factor 2 serves the role of eubacterial initiation factor 1. Proc. Natl Acad. Sci. USA. 2011;108:3918–3923. doi: 10.1073/pnas.1017425108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koc E, Spremulli L. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J. Biol. Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- 20.Christian BE, Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim. Biophys. Acta. 2011 doi: 10.1016/j.bbagrm.2011.11.009. Dec 7 (doi:10.1016/j.bbagrm.2011.11.009; epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C, Tibbetts AS, Kramer G, Appling DR. Yeast AEP3p is an accessory factor in initiation of mitochondrial translation. J. Biol. Chem. 2009;284:34116–34125. doi: 10.1074/jbc.M109.055350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams EH, Bsat N, Bonnefoy N, Butler CA, Fox TD. Alteration of a novel dispensable mitochondrial ribosomal small-subunit protein, Rsm28p, allows translation of defective COX2 mRNAs. Eukaryot. Cell. 2005;4:337–345. doi: 10.1128/EC.4.2.337-345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams EH, Butler CA, Bonnefoy N, Fox TD. Translation initiation in Saccharomyces cerevisiae mitochondria: functional interactions among mitochondrial ribosomal protein Rsm28p, initiation factor 2, methionyl-tRNA-formyltransferase and novel protein Rmd9p. Genetics. 2007;175:1117–1126. doi: 10.1534/genetics.106.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 26.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 27.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 28.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 29.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 30.Zuegge J, Ralph S, Schmuker M, McFadden GI, Schneider G. Deciphering apicoplast targeting signals—feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene. 2001;280:19–26. doi: 10.1016/s0378-1119(01)00776-4. [DOI] [PubMed] [Google Scholar]
- 31.Bender A, van Dooren GG, Ralph SA, McFadden GI, Schneider G. Properties and prediction of mitochondrial transit peptides from Plasmodium falciparum. Mol. Biochem. Parasitol. 2003;132:59–66. doi: 10.1016/j.molbiopara.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Atkinson GC, Baldauf SL. Evolution of elongation factor G and the origins of mitochondrial and chloroplast forms. Mol. Biol. Evol. 2011;28:1281–1292. doi: 10.1093/molbev/msq316. [DOI] [PubMed] [Google Scholar]
- 33.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma J, Farwell MA, Burkhart WA, Spremulli LL. Cloning and sequence analysis of the cDNA for bovine mitochondrial translational initiation factor 2. Biochim. Biophys. Acta. 1995;1261:321–324. doi: 10.1016/0167-4781(95)00041-e. [DOI] [PubMed] [Google Scholar]
- 35.Spremulli LL, Coursey A, Navratil T, Hunter SE. Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 2004;77:211–261. doi: 10.1016/S0079-6603(04)77006-3. [DOI] [PubMed] [Google Scholar]
- 36.Spencer AC, Spremulli LL. The interaction of mitochondrial translational initiation factor 2 with the small ribosomal subunit. Biochim. Biophys. Acta. 2005;1750:69–81. doi: 10.1016/j.bbapap.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Hess DC, Myers CL, Huttenhower C, Hibbs MA, Hayes AP, Paw J, Clore JJ, Mendoza RM, Luis BS, Nislow C, et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 39.Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 40.Lin K, Kuang Y, Joseph JS, Kolatkar PR. Conserved codon composition of ribosomal protein coding genes in Escherichia coli, Mycobacterium tuberculosis and Saccharomyces cerevisiae: lessons from supervised machine learning in functional genomics. Nucleic Acids Res. 2002;30:2599–2607. doi: 10.1093/nar/30.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biou V, Shu F, Ramakrishnan V. X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix. EMBO J. 1995;14:4056–4064. doi: 10.1002/j.1460-2075.1995.tb00077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sette M, Spurio R, van Tilborg P, Gualerzi CO, Boelens R. Identification of the ribosome binding sites of translation initiation factor IF3 by multidimensional heteronuclear NMR spectroscopy. RNA. 1999;5:82–92. doi: 10.1017/s1355838299981487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 45.Haque ME, Grasso D, Spremulli LL. The interaction of mammalian mitochondrial translational initiation factor 3 with ribosomes: evolution of terminal extensions in IF3mt. Nucleic Acids Res. 2008;36:589–597. doi: 10.1093/nar/gkm1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christian BE, Spremulli LL. Evidence for an active role of IF3mt in the initiation of translation in mammalian mitochondria. Biochemistry. 2009;48:3269–3278. doi: 10.1021/bi8023493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Bellis D, Liveris D, Goss D, Ringquist S, Schwartz I. Structure-function analysis of Escherichia coli translation initiation factor IF3: tyrosine 107 and lysine 110 are required for ribosome binding. Biochemistry. 1992;31:11984–11990. doi: 10.1021/bi00163a005. [DOI] [PubMed] [Google Scholar]
- 48.Sacerdot C, de Cock E, Engst K, Graffe M, Dardel F, Springer M. Mutations that alter initiation codon discrimination by Escherichia coli initiation factor IF3. J. Mol. Biol. 1999;288:803–810. doi: 10.1006/jmbi.1999.2737. [DOI] [PubMed] [Google Scholar]
- 49.Petrelli D, Garofalo C, Lammi M, Spurio R, Pon CL, Gualerzi CO, La Teana A. Mapping the active sites of bacterial translation initiation factor IF3. J. Mol. Biol. 2003;331:541–556. doi: 10.1016/s0022-2836(03)00731-9. [DOI] [PubMed] [Google Scholar]
- 50.O'Connor M, Gregory ST, Rajbhandary UL, Dahlberg AE. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA. 2001;7:969–978. doi: 10.1017/s1355838201010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maar D, Liveris D, Sussman JK, Ringquist S, Moll I, Heredia N, Kil A, Blasi U, Schwartz I, Simons RW. A single mutation in the IF3 N-terminal domain perturbs the fidelity of translation initiation at three levels. J. Mol. Biol. 2008;383:937–944. doi: 10.1016/j.jmb.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Payne MJ, Finnegan PM, Smooker PM, Lukins HB. Characterization of a second nuclear gene, AEP1, required for expression of the mitochondrial OLI1 gene in Saccharomyces cerevisiae. Curr. Genet. 1993;24:126–135. doi: 10.1007/BF00324676. [DOI] [PubMed] [Google Scholar]
- 53.Finnegan PM, Ellis TP, Nagley P, Lukins HB. The mature AEP2 gene product of Saccharomyces cerevisiae, required for the expression of subunit 9 of ATP synthase, is a 58 kDa mitochondrial protein. FEBS Lett. 1995;368:505–508. doi: 10.1016/0014-5793(95)00727-q. [DOI] [PubMed] [Google Scholar]
- 54.Helfenbein KG, Ellis TP, Dieckmann CL, Tzagoloff A. ATP22, a nuclear gene required for expression of the F0 sector of mitochondrial ATPase in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:19751–19756. doi: 10.1074/jbc.M301679200. [DOI] [PubMed] [Google Scholar]
- 55.Zeng X, Hourset A, Tzagoloff A. The Saccharomyces cerevisiae ATP22 gene codes for the mitochondrial ATPase subunit 6-specific translation factor. Genetics. 2007;175:55–63. doi: 10.1534/genetics.106.065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krause K, Lopes de Souza R, Roberts DG, Dieckmann CL. The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol. Biol. Cell. 2004;15:2674–2683. doi: 10.1091/mbc.E04-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rödel G. Translational activator proteins required for cytochrome b synthesis in Saccharomyces cerevisiae. Curr. Genet. 1997;31:375–379. doi: 10.1007/s002940050219. [DOI] [PubMed] [Google Scholar]
- 58.Ellis TP, Schonauer MS, Dieckmann CL. CBT1 interacts genetically with CBP1 and the mitochondrially encoded cytochrome b gene and is required to stabilize the mature cytochrome b mRNA of Saccharomyces cerevisiae. Genetics. 2005;171:949–957. doi: 10.1534/genetics.104.036467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rieger KJ, Aljinovic G, Lazowska J, Pohl TM, Slonimski PP. A novel nuclear gene, CBT1, essential for mitochondrial cytochrome b formation: terminal processing of mRNA and intron dependence. Curr. Genet. 1997;32:163–174. doi: 10.1007/s002940050262. [DOI] [PubMed] [Google Scholar]
- 60.Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wallis MG, Groudinsky O, Slonimski PP, Dujardin G. The NAM1 protein (NAM1p), which is selectively required for cox1, cytb and atp6 transcript processing/stabilisation, is located in the yeast mitochondrial matrix. Eur. J. Biochem. 1994;222:27–32. doi: 10.1111/j.1432-1033.1994.tb18837.x. [DOI] [PubMed] [Google Scholar]
- 62.Camougrand N, Pelissier P, Velours G, Guerin M. NCA2, a second nuclear gene required for the control of mitochondrial synthesis of subunits 6 and 8 of ATP synthase in Saccharomyces cerevisiae. J. Mol. Biol. 1995;247:588–596. doi: 10.1006/jmbi.1995.0165. [DOI] [PubMed] [Google Scholar]
- 63.Pelissier P, Camougrand N, Velours G, Guerin M. NCA3, a nuclear gene involved in the mitochondrial expression of subunits 6 and 8 of the Fo-F1 ATP synthase of S. cerevisiae. Curr. Genet. 1995;27:409–416. doi: 10.1007/BF00311209. [DOI] [PubMed] [Google Scholar]
- 64.Dunstan HM, Green-Willms NS, Fox TD. In vivo analysis of Saccharomyces cerevisiae COX2 mRNA 5′-untranslated leader functions in mitochondrial translation initiation and translational activation. Genetics. 1997;147:87–100. doi: 10.1093/genetics/147.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Costanzo MC, Fox TD. Specific translational activation by nuclear gene products occurs in the 5′ untranslated leader of a yeast mitochondrial mRNA. Proc. Natl Acad. Sci. USA. 1988;85:2677–2681. doi: 10.1073/pnas.85.8.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohmen JD, Kloeckener-Gruissem B, McEwen JE. Molecular cloning and nucleotide sequence of the nuclear PET122 gene required for expression of the mitochondrial COX3 gene in S. cerevisiae. Nucleic Acids Res. 1988;16:10783–10802. doi: 10.1093/nar/16.22.10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manthey GM, McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 69.Shoubridge EA, Sasarman F. In: Translational Control in Biology and Medicine. Mathews BM, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 775–801. [Google Scholar]
- 70.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl Acad. Sci. USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iraqui I, Vissers S, Cartiaux M, Urrestarazu A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 1998;257:238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- 72.Sorensen HP, Hedegaard J, Sperling-Petersen HU, Mortensen KK. Remarkable conservation of translation initiation factors: IF1/eIF1A and IF2/eIF5B are universally distributed phylogenetic markers. IUBMB Life. 2001;51:321–327. doi: 10.1080/152165401317190842. [DOI] [PubMed] [Google Scholar]
- 73.Allen GS, Frank J. Structural insights on the translation initiation complex: ghosts of a universal initiation complex. Mol. Microbiol. 2007;63:941–950. doi: 10.1111/j.1365-2958.2006.05574.x. [DOI] [PubMed] [Google Scholar]
- 74.Smits P, Smeitink J, van den Heuvel L. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J. Biomed. Biotechnol. 2010;2010:737385. doi: 10.1155/2010/737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Myasnikov AG, Simonetti A, Marzi S, Klaholz BP. Structure-function insights into prokaryotic and eukaryotic translation initiation. Curr. Opin. Struct. Biol. 2009;19:300–309. doi: 10.1016/j.sbi.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 76.Benelli D, Londei P. Begin at the beginning: evolution of translational initiation. Res. Microbiol. 2009;160:493–501. doi: 10.1016/j.resmic.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 77.Carter AP, Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 78.Julian P, Milon P, Agirrezabala X, Lasso G, Gil D, Rodnina MV, Valle M. The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol. 2011;9:e1001095. doi: 10.1371/journal.pbio.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Couplan E, Aiyar RS, Kucharczyk R, Kabala A, Ezkurdia N, Gagneur J, St Onge RP, Salin B, Soubigou F, Le Cann M, et al. A yeast-based assay identifies drugs active against human mitochondrial disorders. Proc. Natl Acad. Sci. USA. 2011;108:11989–11994. doi: 10.1073/pnas.1101478108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abahuni N, Gispert S, Bauer P, Riess O, Kruger R, Becker T, Auburger G. Mitochondrial translation initiation factor 3 gene polymorphism associated with Parkinson's disease. Neurosci Lett. 2007;414:126–129. doi: 10.1016/j.neulet.2006.12.053. [DOI] [PubMed] [Google Scholar]
- 81.Anvret A, Ran C, Westerlund M, Thelander AC, Sydow O, Lind C, Hakansson A, Nissbrandt H, Galter D, Belin AC. Possible involvement of a mitochondrial translation initiation factor 3 variant causing decreased mRNA levels in Parkinson's disease. Parkinsons Dis. 2010;2010:491751. doi: 10.4061/2010/491751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Behrouz B, Vilarino-Guell C, Heckman MG, Soto-Ortolaza AI, Aasly JO, Sando S, Lynch T, Craig D, Uitti RJ, Wszolek ZK, et al. Mitochondrial translation initiation factor 3 polymorphism and Parkinson's disease. Neurosci Lett. 2010;486:228–230. doi: 10.1016/j.neulet.2010.09.059. [DOI] [PubMed] [Google Scholar]
- 83.Pino P, Aeby E, Foth BJ, Sheiner L, Soldati T, Schneider A, Soldati-Favre D. Mitochondrial translation in absence of local tRNA aminoacylation and methionyl tRNA Met formylation in Apicomplexa. Mol. Microbiol. 2010;76:706–718. doi: 10.1111/j.1365-2958.2010.07128.x. [DOI] [PubMed] [Google Scholar]
- 84.Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35:4686–4703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zikova A, Panigrahi AK, Dalley RA, Acestor N, Anupama A, Ogata Y, Myler PJ, Stuart K. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol. Cell. Proteomics. 2008;7:1286–1296. doi: 10.1074/mcp.M700490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ohtsuki T, Watanabe Y. T-armless tRNAs and elongated elongation factor Tu. IUBMB Life. 2007;59:68–75. doi: 10.1080/15216540701218722. [DOI] [PubMed] [Google Scholar]
- 87.Tsuboi M, Morita H, Nozaki Y, Akama K, Ueda T, Ito K, Nierhaus KH, Takeuchi N. EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis. Mol. Cell. 2009;35:502–510. doi: 10.1016/j.molcel.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 88.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 91.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. New York: CSHL Press; 1994. [Google Scholar]
- 92.Ocampo A, Zambrano A, Barrientos A. Suppression of polyglutamine-induced cytotoxicity in Saccharomyces cerevisiae by enhancement of mitochondrial biogenesis. FASEB J. 2010;24:1431–1441. doi: 10.1096/fj.09-148601. [DOI] [PubMed] [Google Scholar]
- 93.Ludovico P, Sansonetty F, Corte-Real M. Assessment of mitochondrial membrane potential in yeast cell populations by flow cytometry. Microbiology. 2001;147:3335–3343. doi: 10.1099/00221287-147-12-3335. [DOI] [PubMed] [Google Scholar]
- 94.Shapiro HM. Membrane potential estimation by flow cytometry. Methods. 2000;21:271–279. doi: 10.1006/meth.2000.1007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.