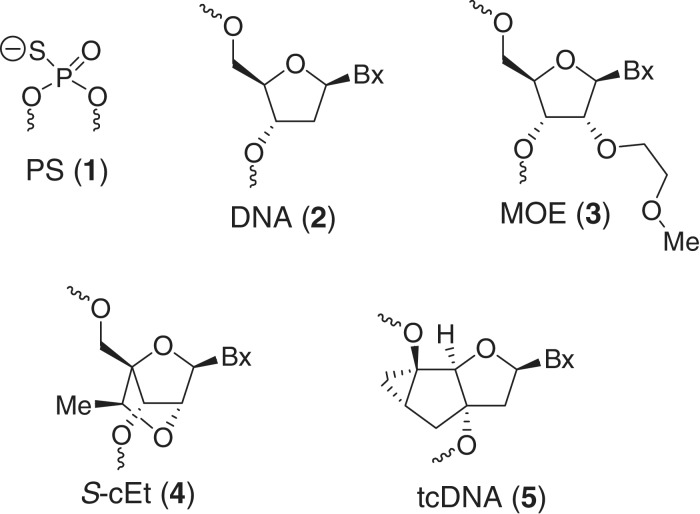
Abstract
We report the evaluation of 20-, 18-, 16- and 14-mer phosphorothioate (PS)-modified tricycloDNA (tcDNA) gapmer antisense oligonucleotides (ASOs) in Tm, cell culture and animal experiments and compare them to their gap-matched 20-mer 2′-O-methoxyethyl (MOE) and 14-mer 2′,4′-constrained ethyl (cEt) counterparts. The sequence-matched 20-mer tcDNA and MOE ASOs showed similar Tm and activity in cell culture under free-uptake and cationic lipid-mediated transfection conditions, while the 18-, 16- and 14-mer tcDNA ASOs were moderate to significantly less active. These observations were recapitulated in the animal experiments where the 20-mer tcDNA ASO formulated in saline showed excellent activity (ED50 3.9 mg/kg) for reducing SR-B1 mRNA in liver. The tcDNA 20-mer ASO also showed better activity than the MOE 20-mer in several extra-hepatic tissues such as kidney, heart, diaphragm, lung, fat, gastrocnemius and quadriceps. Interestingly, the 14-mer cEt ASO showed the best activity in the animal experiments despite significantly lower Tm and 5-fold reduced activity in cell culture relative to the 20-mer tcDNA and MOE-modified ASOs. Our experiments establish tcDNA as a useful modification for antisense therapeutics and highlight the role of chemical modifications in influencing ASO pharmacology and pharmacokinetic properties in animals.
INTRODUCTION
Over the recent years, chemically modified oligonucleotides have been extensively investigated as potential drugs (1). Currently, more than 30 candidates are in clinical trials targeting a large variety of diseases including cancer, metabolic diseases and genetic disorders. Mipomersen, a second generation antisense oligonucleotide that targets apolipoprotein B, has shown impressive reductions in serum apolipoprotein B, LDL-cholesterol and other atherogenic lipids in multiple phase III clinical trials (2). The general mode of action of antisense oligonucleotides is based on specific Watson–Crick recognition of target RNAs, which results in modulation of the function of the target RNA. In the original antisense approach, a single-stranded oligonucleotide targets a mRNA and suppresses its translation either via a steric block or by RNase H-mediated RNA degradation. Variations of steric block inhibitors include oligonucleotides that interfere with splicing of a pre-mRNA, leading to selective exon inclusion or exclusion, or alternative splicing. In addition, oligonucleotide-dependent degradation of target RNA can be induced by small double-stranded interfering RNAs (siRNAs) which guide an mRNA of interest into the RNA-induced silencing complex (RISC) where it is cleaved (3). Yet, another approach is based on targeting micro-RNAs (miRNAs) that are genetically encoded and function as natural regulators of translation, by single-stranded antisense oligonucleotides (4–6).
The specific chemical features of single-stranded antisense oligonucleotides fundamentally determine their biological function (7). The most important factors are affinity for target RNA, biostability against nuclease degradation and bioavailabilty in animals. First generation classical antisense oligodeoxyribonucleotide phosphorothioates (PS 1, Figure 1) elicit RNaseH activity and show increased biostability, however, their affinity to complementary RNA is moderate. On the other hand, high-affinity RNA binders such as 2′-modified RNA (8), 2′,4′-bridged nucleic acids (BNA, also known as locked nucleic acid or LNA) (9), hexitol nucleic acids (HNA) (10) or tricyclo-DNA (tcDNA) (11) do not elicit RNaseH activity when used as mixmers. To circumvent this drawback, oligonucleotide gapmers have been introduced that show a window of natural nucleotides in the center of the sequence flanked by chemically modified wings on either side (12). Such chimeric oligonucleotides combine the features of high-RNA affinity and biostability while maintaining the ability to degrade the targeted mRNA via an RNaseH mechanism.
Figure 1.
Structures of oligonucleotide modification evaluated in this study.
As part of a comprehensive program aimed at elucidating the structure–activity relationships (SAR) of chemically modified ASOs in animals, we have recently reported the evaluation of RNase H active gapmer ASOs containing 2′,4′-bridged nucleic acids and hexitol analogs (13–17). tcDNA represents another oligonucleotide scaffold that exhibits favorable physico-chemical properties, such as improved target RNA affinity and nuclease stability, for the antisense approach (18,19). Yet, gapmer ASOs containing PS-modified tcDNA have not been evaluated in cellular or in animal experiments. In this light, we present here a comprehensive analysis of the biophysical, pharmacological and tissue distribution properties of 14- to 20-mer phosphorothioate tcDNA gapmer ASOs and compare them to sequence matched MOE and 2′,4′-constrained ethyl BNA (cEt) control ASOs. We show that tcDNA-modified ASOs formulated in saline show potent and robust antisense effects in hepatic as well as extra-hepatic tissues without producing toxicity and highlight the role of chemical modifications in influencing ASO pharmacology and pharmacokinetic properties in animals.
MATERIALS AND METHODS
Oligonucleotide synthesis and purification
The syntheses of the tc-DNA gapmer oligonucleotides were performed on a 10.0 μmol scale on a Gene Assembler Plus DNA synthesizer (Pharmacia/GE healthcare) using standard phosphoramidite chemistry. Tc-nucleoside-modified long chain alkylamino CPG (Link Technologies) was used as solid support. Tc-phosphoramidites [0.15 M in CH3CN for T, C and G and dichloroethane (DCE) for A] were coupled with 5-(ethylthio)-1H-tetrazole (ETT, 0.25 M in CH3CN) as activator with a coupling time of 12 min. Sulfurization was carried out using phenylacetyl disulfide (PADS, 0.2 M solution in dry pyridine/CH3CN 1:1, v/v) for 3.5 min. After synthesis was completed, the support-bound oligonucleotides were treated with a solution of Et3N/CH3CN (1:1, v/v) for 2 h and then deprotected and detached from solid support with 33% aqueous NH3 for 16 h at 55°C. The crude material was purified by ion-exchange HPLC (Source 30Q, GE Healthcare) with linear gradients (0–50%) of buffer B (0.1 M NH4OAc, H2O:CH3CN 7:3, pH 8.0, 1.5 M NaBr) in buffer A (0.1 M NH4OAc, H2O:CH3CN 7:3 pH 8.0) as eluent. Oligonucleotides were desalted over HiPrep™ 26/10 columns (GE Healthcare) and lyophilized. Sequences and analytical data are summarized in Supplementary Table S1. ASOs A5 and A6 were synthesized according to procedures described previously (16,20).
Cells and reagents
MHT cells (21) were cultured in DMEM supplemented with 10% fetal calf serum, streptomycin (0.1 μg/ml) and penicillin (100 U/ml). ASO transfection was performed using Opti-MEM containing 5 µg/ml Lipofectamine 2000 at the indicated amount of ASO for 4 h at 37°C, as described previously (22,23).
Taqman RT–PCR
Total mRNA was isolated using a QIAGEN RNAeasy kit (QIAGEN, Valencia, CA, USA). Reduction of target mRNA expression was determined by real time RT–PCR (1) using StepOne RT–PCR machines (Applied Biosystems). The sequences used in the RT–PCR reaction are 5′-TGACAACGACACCGTGTCCT-3′ for the forward primer, 5′-ATGCGACTTGTCAGGCTGG-3′ for the reverse primer and 5′-CGTGGAGAACCGCAGCCTCCATT-3′ for the probe. The expression data were normalized to ribogreen (Invitrogen). Data are mean values ± standard deviations of three replicates. IC50 values were calculated using GraphPad Prism 4 software.
Protocols for animal experiments
The Institutional Animal Care and Use committee (IACUC) approved all procedures. Male Balb/c mice were housed 4/cage on a 12:12-h light/dark cycle. For the first study, tcDNA, MOE, and cEt ASO solutions were prepared in PBS and injected subcutaneously (s.c) twice a week at a concentration of 25, 5, 1 or 0.5 mg/kg for 3 weeks. For the follow-up study, mice were injected s.c at 25 mg/kg twice a week for 3 weeks. Mice were sacrificed 48 h after the last dose. Blood samples were collected by cardiac puncture and plasma chemistries (alanine amino transferase, aspartate amino transferase, bloood urea nitrogen, total bilirubin, cholesterol and triglycerides) values were measured on the Olympus AU400 Clinical Analyzer (Beckman Coulter, CA, USA).
RNA analysis for animal experiments
Liver, kidney, diaphragm, lung, white adipose, heart, quadriceps and gastrocnemius muscle were dissected, weighed and immediately homogenized in 2 ml of PureLink RNA lysis buffer (Life Technologies, CA, USA). Total RNA was isolated using the Purlink RNA Mini Kit (Life Technologies). Reduction of target mRNA expression was determined by real time RT–PCR using 7700 RT–PCR sequence detector (Applied Biosystems). Data are mean values ± standard deviations of three replicates. The sequences used in the RT-PCR reaction are 5′-TGACAACGACACCGTGTCCT-3′ for the forward primer, 5′-ATGCGACTTGTCAGGCTGG-3′ for the reverse primer and 5′-CGTGGAGAACCGCAGCCTCCATT-3′ for the probe. RNA transcripts were normalized to total RNA levels using RiboGreen, RNA Quantitation Reagent (Molecular Probes). RiboGreen is an ultrasensitive flourescent nucleic acid stain which when bound to RNA has a maximum excitation/emission at ∼500 nm/525 nm. ED50 values were calculated using GraphPad Prism 4 software.
RESULTS
Oligonucleotide design considerations
The prototypical design of a second generation antisense oligonucleotide employs a 8–14 base PS-modified deoxynucleotide ‘gap’ flanked on either end with 2–5 MOE nucleotides (MOE gapmer) (24–26). Typically, each MOE nucleotide confers + 1–2°C/mod. towards the overall duplex thermal stability depending on the position and sequence context of the incorporation. The PS backbone promotes binding to plasma proteins thereby reducing renal excretion of the oligonucleotide (27). This allows the ASO to distribute to peripheral tissues such that one observes reproducible and robust antisense effects in animal models especially when targeting genes expressed in the liver. More recently, we and others have shown that replacing MOE with high-affinity BNA nucleotides in the ‘wings’ of second generation ASOs allows for the use of oligonucleotides as short as 12- to 14-mer in length (13,28). In these designs, the BNA nucleotide contributes +3–4°C/mod. towards duplex thermal stability. However, the reduced PS content of the shorter ASOs designs results in reduced exposure to peripheral tissues. Despite this, somewhat counterintuitively, these ASOs exhibit improved activity in animal experiments although antisense effects towards gene targets expressed in tissues other than liver have not been well-characterized to date. In comparison to BNA, tcDNA exhibits intermediate improvement (ΔTm + 2–4°C/mod.) in duplex thermal stability as measured by incorporation in the interior of a phosphodiester deoxyribo-oligonucleotide (29). However, RNase H-active gapmer ASOs are typically uniformly PS modified and position the modified nucleotides in the wings where the effects of tcDNA on duplex thermal stability have not been well-characterized. Given this background, it was difficult to ascertain a priori what ASO length and tcDNA content would be optimal. As a result, we prepared the gap-matched 20-, 18-, 16- and 14-mer tcDNA (ASOs A1–A4, Table 1) versions of a previously identified potent 5-10-5 MOE gapmer (A5) targeting scavenger receptor B1 (SR-B1), a ubiquitously expressed gene whose physiological role is related to cholesterol uptake into tissues (30). The SR-B1 receptor has also been implicated as an entry point for viruses such as HCV (31) and other pathogens (32) and its down-regulation could provide a therapeutic benefit by preventing entry of infectious pathogens into host cells. In addition to the 5-10-5 MOE control, we also evaluated a gap-matched 2-10-2 14-mer cEt ASO A6 as an additional control for the comparative study.
Table 1.
Sequence, design features, Tm, activity for reducing SR-B1 mRNA in MHT cells and in mouse liver for tcDNA, MOE and cEt-modified PS gapmer ASOs
| ASO | Sequence (5′to 3′)a | Mod. | Length | Design | Tm (°C)b | IC50 (nM)c | IC50 (nM)d | ED50 (mg/kg)e |
|---|---|---|---|---|---|---|---|---|
| A1 | p-GCTTCAGTCATGACTTCCTT | tcDNA | 20-mer | 5-10-5 | 71.2 | 0.57 | 54.5 | 3.9 |
| A2 | p-CTTCAGTCATGACTTCCT | tcDNA | 18-mer | 4-10-4 | 63.9 | 0.73 | 238.0 | 4.5 |
| A3 | p-TTCAGTCATGACTTCC | tcDNA | 16-mer | 3-10-3 | 58.2 | 5.34 | 407.7 | 7.1 |
| A4 | p-TCAGTCATGACTTC | tcDNA | 14-mer | 2-10-2 | 49.7 | 11.58 | >1000 | 16.4 |
| A5 | GmCTTmCAGTmCATGAmCTTmCmCTT | MOE | 20-mer | 5-10-5 | 69.7 | 0.63 | 45.0 | 3.5f |
| A6 | TmCAGTmCATGAmCTTmC | S-cEt | 14-mer | 2-10-2 | 59.0 | 2.48 | 138.5 | <1.0g |
aBold and underlined alphabet indicates modified nucleotides.
bTm values were measured in 10 mM sodium phosphate buffer (pH 7.2) containing 100 mM NaCl and 0.1 mM EDTA, Sequence of RNA complement 5′-r(UUGAAAGGAAGTCATGACTGAAGC)-3′; all internucleosidic linkages in ASOs A1–A6 are phosphorothioate except the 5′-terminal phosphate in ASOs A1–A4.
cIC50 values for reducing SR-B1 mRNA in MHT cells after transfection of ASO with lipofectamine.
dIC50 values for reducing SR-B1 mRNA in MHT cells under free-uptake conditions.
eED50 values for reducing SR-B1 mRNA in mouse liver.
fED50 values obtained from a different experiment.
gEstimated ED50 based on reduction of SR-B1 mRNA (54 ± 6.6% at 1 mg/kg dose) observed in this study. See Supplementary Figures S1, S2, S4 and S5 for dose–response curves.
Duplex thermal stability measurements of modified ASOs with RNA
We first evaluated all the ASOs in thermal stability experiments using a 24-mer RNA complement (Table 1). We chose the longer complement since the biological target for these ASOs, i.e. the mRNA, is not length matched to the ASOs (33). As would be expected, increasing the length and tcDNA content in the modified ASOs led to increases in duplex thermal stability. However, somewhat surprisingly, the increase in affinity was highly dependent on the context of the incorporation. For example, the 14-mer 2-10-2 tcDNA ASO A4 showed a Tm of 49.7°C, but the Tm of the 16-mer 3-10-3 ASO A3 was 58.2°C, corresponding to a ΔTm of +4°C for each of the added nucleotide. For the 18-mer 4-10-4 (Tm 63.9°C) and the 20-mer 5-10-5 (Tm 71.2°C) tcDNA ASOs A2 and A1, respectively, each tcDNA nucleotide contributed roughly +2.5°C and +3.5°C, respectively, towards the increase in overall duplex thermal stability. In comparison, the Tm of the 5-10-5 MOE and the 2-10-2 cEt ASOs A5 and A6 were 69.7 and 59.0°C, respectively. It should be noted that ASOs A5 and A6 were synthesized using 5-Me groups (ΔTm + 0.5°C/mod.) (34) on the cytosine nucleobases in the gap and in the wings (6 for MOE ASO A5 and 4 for cEt ASO A6) while the tcDNA ASOs were not. Thus, it is anticipated that, if needed, the RNA affinity of the tcDNA ASOs can be further increased by introducing 5-Me groups on the cytosine nucleobases.
Cell culture evaluation of ASOs A1–A6
We next evaluated all the ASOs in MHT cells under free-uptake conditions and with cationic lipid transfection to deliver the oligonucleotides (21). Using cationic lipid transfection, all the tcDNA ASOs showed activity in cell culture consistent with their duplex thermostability measurements (Table 1 and Supplementary Figure S1). The 20-mer tcDNA ASO A1 had the highest Tm and showed the best activity in cell culture (IC50 = 0.57 nM), followed by the 18-mer ASO A2 (IC50 = 0.73 nM), 16-mer ASO A3 (IC50 = 5.34 nM) and the 14-mer ASO A4 (IC50 = 11.6 nM). Also consistent with the Tm data, the 5-10-5 MOE ASO A5 (IC50 = 0.63 nM) showed activity comparable to 20-mer tcDNA ASO A1. The 14-mer cEt ASO A6 showed ∼5-fold reduced activity (IC50 = 2.48 nM) relative to the 20-mer tcDNA or MOE ASO A1 and A5, respectively. However, the activity of ASO A6 was 2-fold better than that of the 16-mer tcDNA ASO A3 even though both the ASOs had almost identical Tm.
In the absence of the cationic lipid transfection, all the ASOs showed almost 100-fold reduction in activity but the overall potency trends were similar to those observed in the transfection assay (Table 1 and Supplementary Figure S2). In the tcDNA series, the 20-mer ASO A1 showed the best activity (IC50 = 55 nM) followed by A2 (IC50 = 238 nM) and A3 (IC50 = 408 nM), while A4 showed very poor activity (IC50 > 10 000 nM). As before, the MOE 20-mer ASO A5 (IC50 = 45 nM) showed activity comparable to ASO A1 while the 14-mer cEt ASO A6 (IC50 = 139 nM) was ∼3-fold less active. Once again, ASO A6 showed 3-fold better activity as compared to the 16-mer ASO A3 despite similar overall Tm.
We also measured oligonucleotide copy numbers in each cell to ascertain if ASOs with different lengths and chemical modifications in the flanks were taken up differentially into MHT cells under free-uptake conditions (Supplementary Figure S3). Somewhat surprisingly, we found that all the tcDNA ASOs showed very similar uptake into MHT cells irrespective of oligonucleotide length. Thus, it appears that introducing more than four tcDNA monomers into an ASO does not increase cellular uptake or accumulation, but the increased tcDNA content is required to boost RNA affinity to produce an antisense effect. In contrast to the tcDNA ASOs, the MOE and cEt ASOs A5 and A6, respectively, showed almost 2-fold lower ASO copy numbers/cell. Thus, the improved activity of the cEt ASO A6 relative to the tcDNA 14-mer, 16-mer or the 18-mer ASOs cannot be explained by increased uptake of this ASO into MHT cells under free-uptake conditions.
Evaluation of ASOs A1–A6 in animal experiments
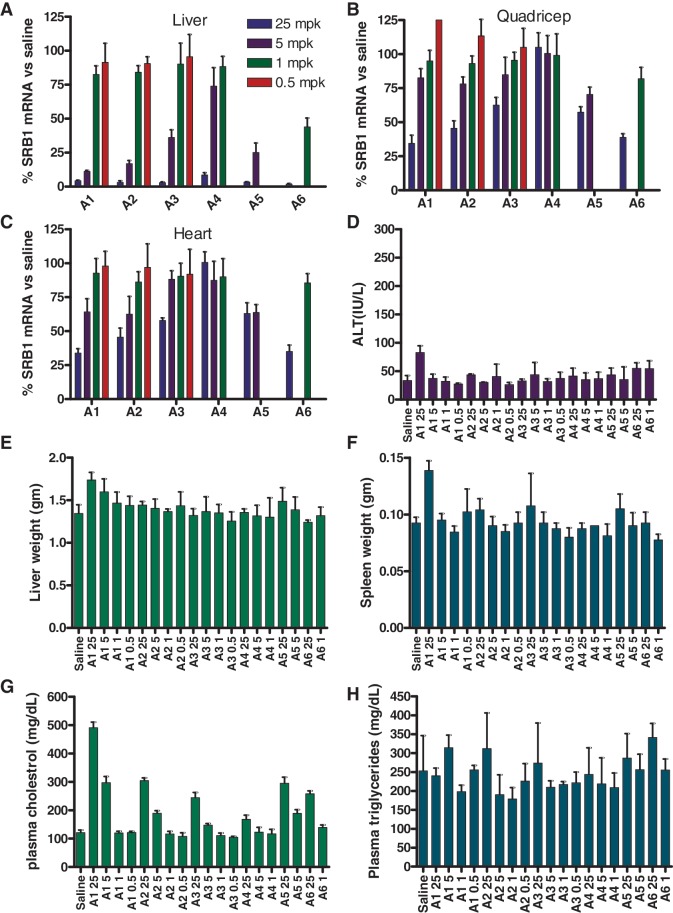
We next evaluated the ASOs A1–A6 in animal experiments. Mice (n = 4/group) were injected sub-cutaneously (s.c.) with 0.5, 1.0, 5.0 and 25.0 mg/kg of tcDNA ASOs, 5 and 25 mg/kg of MOE ASO A5 and 1 and 25 mg/kg of cEt ASO A6 formulated in saline twice a week for 3 weeks (Figure 2). While the MOE and cEt ASOs A5 and A6 were only evaluated at two doses in this study, ASO A5 was previously evaluated in a separate dose-reponse experiment where it showed good potency (ED50 = 3.5 mg/kg; Supplementary Figure S4). While a complete dose–response for the cEt ASO A6 is not available, the ED50 can be estimated (ED50 < 1 mg/kg) based on the reduction of SR-B1 mRNA (54 ± 6.6% at the 1 mg/kg dose) observed in this study (Table 1). The animals were sacrificed 48 h after the last ASO dose and the SRB1 mRNA in liver was measured by quantitative RT–PCR and normalized to the saline treated group (Figure 2A and Supplementary Figure S5). In addition, we also recorded other parameters, such as plasma transaminases and organ weights, as gross indicators of ASO tolerability. As seen in the cell culture experiments, the tcDNA ASOs showed dose-dependent reductions of the SR-B1 mRNA in liver with the 20-mer A1 exhibiting the best potency (ED50 = 3.9 mg/kg) followed by the 18-mer A2 (ED50 = 4.5 mg/kg), 16-mer A3 (ED50 = 7.1 mg/kg) and 14-mer A4 (ED50 = 16.4 mg/kg). Thus, in the tcDNA length series, the animal experiments recapitulated the potency trend observed in the free-uptake cell culture study even though the magnitude of the potency difference was not similar.
Figure 2.
Reduction of SR-B1 mRNA and tolerability profile after treatment with ASOs A1–A6. Mice (BalbC, n = 4/group) were dosed s.c. with 0.5, 1.0, 5.0 and 25.0 mg/kg of ASOs A1–A4, 5.0 and 25.0 mg/kg of ASO A5 and 1.0 and 25.0 mg/kg of ASO A6, twice a week for 3 weeks and animals were sacrificed 48 h after last dose. Organs were harvested and SR-B1 mRNA was quantified by qRT–PCR and normalized to the saline-treated group. The 0.5 mg/kg dose group for ASO A4 was not counted in the final analysis due to a dosing error. SR-B1 mRNA reduction in (A) liver (B) quadriceps and (C) heart. Plasma levels post sacrifice for (D) ALT. Organ weights for (E) liver (F) spleen. Plasma levels post-sacrifice for (G) cholesterol and (H) triglycerides. Error bars indicate ±standard deviation.
To see if the activity seen in liver could be extended to other tissues, we also measured the down-regulation of SR-B1 mRNA in quadriceps and heart muscle (Figure 2B and C). Once again, we observed dose-dependent reductions in SR-B1 mRNA in both heart and quadriceps although the potency relative to liver was reduced for all the ASOs tested. The down-regulation of SR-B1 gene expression results in an increase in the plasma cholesterol levels of the treated mice. The high-dose ASO-treated group showed between 3- and 5-fold increases in plasma cholesterol levels (Figure 2G) relative to saline which were consistent with the expected phamacological response for reducing SR-B1 gene expression. In contrast, there were no noticiable effects on the levels of plasma triglycerides (Figure 2H) indicating that the increase in plasma cholesterol levels were indeed related to SR-B1 down-regulation. In general, all the ASOs were well-tolerated with no elevations in transaminase levels (ALT) and liver weights (Figure 2D and E). Slight elevations in spleen weights were observed for the high-dose group ASO A1-treated mice although these could be, in part, related to the pharmacological effects of the ASO or to the lack of 5-Me groups on the cytosine nucleobases in the tcDNA-modified ASOs (Figure 2F). 5-Me groups on the deoxycytidine nucleobase are known to mitigate the immune stimulation properties of CpG oligonucleotides (35).
To ascertain if tcDNA 14-mers were generally less active as compared to 14-mer ASOs with other higher affinity modifications in the wings, we evaluated a 14-mer tcDNA ASO A7 targeting mouse phosphatase and tensin homologue (PTEN) and compared it to a 14-mer LNA benchmark ASO A8 (Supplementary Figure S6). This sequence has been used extensively by us to profile the antisense properties of ASOs modified with various BNA and HNA analogs (13–17). We found that the tc-DNA 14-mer showed lower Tm and reduced activity in cell culture and in the animal experiment as compared to the LNA benchmark ASO. However, unlike the LNA ASO A8, the tcDNA ASO did not show elevations in ALT levels for the high-dose group-treated animals.
We also quantified the level of ASOs A1–A6 in liver tissue (Supplementary Figure S7). Consistent with previous studies of PS-modified gapmer ASOs, drug accumulation in liver was dependent on dose and the overall PS content of the ASO. Thus, both the 20-mer ASOs A1 and A5 showed very similar levels of accumulation at the 25 and 5 mg/kg dose. However, the effect of wing chemistry on tissue accumulation was more evident for the shorter tcDNA ASOs which showed higher drug levels in liver tissue. For example, the16-mer and 18-mer tcDNA ASOs A2 and A3 showed similar liver accumulation as the 20-mer ASOs A1 and A6 but the 14-mer tcDNA ASO A4 showed almost 3-fold higher accumulation in liver tissue as compared to the 14-mer cEt ASO A6. It is possible that the more hydrophobic nature of the tcDNA monomers enhances binding to plasma proteins but this effect is most prominent for shorter ASOs as compared to the longer designs where increased PS content dominates ASO distribution.
To further understand ASO distribution between different cell types in the liver and kidney, we stained tissue sections from mice treated with the 20-mer tcDNA and MOE ASOs A1 and A5, respectively, and 14-mer cEt ASO A6 using a rabbit polyclonal antibody raised against the MOE ASO A5 (Supplementary Figure S8). We found no qualitative changes in ASO distribution between the different cell types in the liver but darker staining for oligonucleotide was observed in the non-parenchymal cells of the liver for all the ASOs evaluated. However, the differential recognition of the antibody for ASOs A1, A5 and A6 does not allow for any quantitative estimation of changes in sub-organ distribution to be made at this point.
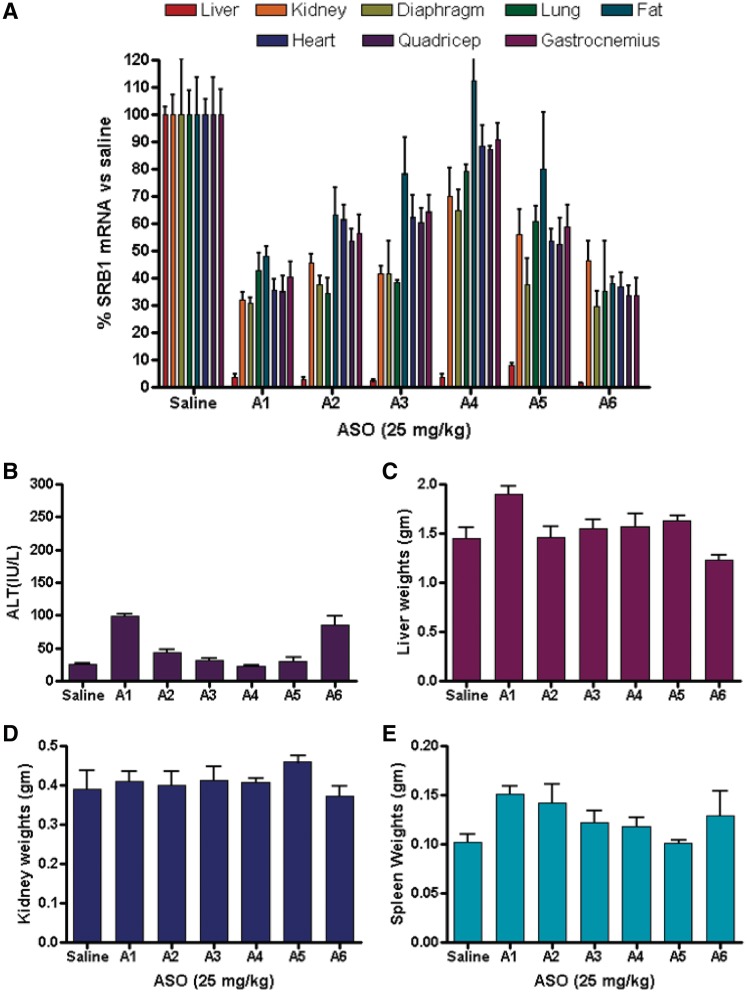
To confirm the observations from the first study and to examine the effect of the ASO treatment on SR-B1 mRNA down-regulation in tissues other than liver, heart and quadriceps, we carried out a follow-up study (Figure 3). Mice (n = 4/group) were injected s.c. with ASOs A1–A6 formulated in saline at 25 mg/kg twice a week for 3 weeks. Animals were sacrified 48 h after the last ASO dose and down-regulation of SR-B1 mRNA in liver, kidney, diaphragm, lung, fat, heart, quadriceps and gastrocnemius was measured by quatitative RT-PCR and the results were normalized to saline-treated animals (Figure 3A). As seen in the previous study, we observed >95% down-regulation of the SR-B1 mRNA for all the ASO-treated groups in liver tissue. In kidney, ASOs A1–A3 and A6 showed similar activity for reducing SR-B1 mRNA while A4 and A5 were less active. A similar trend was observed in lung tissue while in the diaphragm all but ASO A4 showed similar activity. In fat, heart, quadriceps and gastrocnemius, the tcDNA ASOs A1–A4 showed activity directly propotional to ASO length. The 14-mer cEt ASO A6 showed activity comparable to the 20-mer tcDNA A1 in all the tissues evaluated. Once again, all the ASOs were well-tolerated with no elevations in plasma ALT (Figure 3B) or organ weights (Figure 3C–E).
Figure 3.
Reduction of SR-B1 mRNA and tolerability profile after treatment with ASOs A1–A6. Mice (BalbC, n = 4/group) were dosed s.c. with 25.0 mg/kg of ASOs A1–A6, twice a week for 3 weeks and animals were sacrificed 48 h after last dose. Organs were harvested and SR-B1 mRNA in was quantified by qRT–PCR and normalized to the saline-treated group. SR-B1 mRNA reduction in (A) liver, kidney, diaphragm, lung, fat, heart, quadriceps and gastrocnemius. Plasma levels post-sacrifice for (B) ALT. Organ weights for (C) liver, (D) kidney and (E) spleen. Error bars indicate ± standard deviation.
DISCUSSION
Our evaluation of tcDNA gapmer ASOs presents several interesting observations. When combined with the phosphorothioate backbone modification, tcDNA gapmer ASOs show similar to slightly improved duplex thermal stability relative to length-matched MOE gapmer ASO but reduced thermostability relative to length-matched cEt or LNA gapmer ASOs. This observation is generally in line with the duplex stabilizing properties reported for these modifications in oligonucleotide sequences used for biophysical studies. In cell culture, tcDNA ASOs showed activity which was consistent with their duplex thermostability measurements under transfection and free-uptake conditions. For example, the 20-mer tcDNA and MOE ASOs A1 and A5, respectively, showed the highest Tm and best activity for reducing SR-B1 mRNA in cell culture followed by the 18-mer A2, 16-mer A3 and 14-mer A4 tcDNA ASOs. In general, the Tm and cell culture observations were recapitulated in the animal experiments where the 20-mer tcDNA and MOE ASOs A1 and A5, respectively, exhibited better activity relative to the 18-mer, 16-mer and 14-mer tcDNA ASOs A2, A3 and A4. However, somewhat surprisingly, the shorter 14-mer cEt ASO A6 showed the best overall activity in the animal experiments despite lower Tm and ∼5-fold reduced potency relative to the 20-mer tcDNA and MOE ASOs A1 and A5, respectively, in cell culture. The 14-mer cEt ASO A6 also showed ∼3-fold improved activity in cell culture as compared to 16-mer tcDNA ASO A3 even though both ASOs had identical RNA-binding affinity. Moreover, improved activity in animal experiments was observed despite reduced overall liver accumulation for the 14-mer cEt ASO A6. ASOs, unlike small molecules drugs, cannot distribute to all the different cellular compartments by passive diffusion. ASOs are known to enter cells via multiple pathways some of which are productive and lead to sequence specific gene down-regulation of the targeted mRNA in the nucleus (21,36). In contrast, non-productive pathways lead to ASO accumulation into endosomal and lysosomal compartment where they are most likely metabolized by nuclease-mediated digestion and excreted. It is also known that phosphorothioate-modified single-stranded ASOs interact with several proteins within the cell some of which might be responsible for differential trafficking of the ASO to different sub-cellular compartments leading to different pharmacological and toxicological properties (21,36). It is possible that ASO length and chemical modification can have a differential impact on these interactions resulting in the observed effects. Another interesting result from this study was the robust antisense activity seen in multiple tissues with several of the ASOs evaluated. Perhaps, more importantly, these effects were observed in the absence of elevated plasma transaminases indicating that it is possible to achieve robust antisense effects in tissues other than liver without producing hepatoxicity. Thus, our data show that phosphorothioate-modified gapmer ASOs can down-regulate gene expression in a therapeutically relevant manner in hepatic and extra-hepatic tissues and this can be further modulated by ASO length and wing chemistry.
CONCLUSIONS
In conclusion, we show for the first time that tcDNA phosphorothioate-modified gapmer ASOs show robust antisense effects for reducing gene expression in multiple tissues without producing hepatotoxicity. With the exception of the cEt ASO A6, all ASOs evaluated showed activity consistent with their Tm and cell culture profile. The origins of the improved potency despite lower Tm, cell culture activity and reduced tissue exposure observed with the cEt ASO A6 in liver are not properly understood as of yet. However, they are likely related to differential sub-cellular ASO distribution which is in turn influenced by ASO length and specific structural features of the modified nucleotides in the flanks of the ASO. Our data provides an impetus to further explore phosphorothioate-modified tcDNA and related modifications for therapeutic antisense applications using the RNase H and other antisense mechanisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–8.
FUNDING
Funding for open access charge: Isis Pharmaceuticals.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Hans Gaus and Dr Balkrishen Bhat for helpful discussions and Dr Gene Hung for the histology analysis.
REFERENCES
- 1.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 2.Raal FJ, Santos RD, Blom DJ, Marais AD, Charng MJ, Cromwell WC, Lachmann RH, Gaudet D, Tan JL, Chasan-Taber S, et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 3.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;43:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 5.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 6.Steffy K, Allerson C, Bhat B. Perspectives in MicroRNA therapeutics. Pharm. Technol. 2011;s18:s20–s22, s24. [Google Scholar]
- 7.Swayze EE, Bhat B. The Medicinal Chemistry of Oligonucleotides. In: Crooke ST, editor. Antisense Drug Technology: Principles, Strategies, and Applications. 2nd edn. Boca Raton: CRC Press; 2008. pp. 143–182. [Google Scholar]
- 8.Prakash TP, Bhat B. 2'-Modified oligonucleotides for antisense therapeutics. Curr. Top. Med. Chem. 2007;7:641–649. doi: 10.2174/156802607780487713. [DOI] [PubMed] [Google Scholar]
- 9.Veedu RN, Wengel J. Locked nucleic acids: promising nucleic acid analogs for therapeutic applications. Chem. Biodivers. 2010;7:536–542. doi: 10.1002/cbdv.200900343. [DOI] [PubMed] [Google Scholar]
- 10.Herdewijn P. Nucleic acids with a six-membered carbohydrate' mimic in the backbone. Chem. Biodiversity. 2010;7:1–59. doi: 10.1002/cbdv.200900185. [DOI] [PubMed] [Google Scholar]
- 11.Leumann CJ. DNA analogues: from supramolecular principles to biological properties. Bioorg. Med. Chem. 2002;10:841–854. doi: 10.1016/s0968-0896(01)00348-0. [DOI] [PubMed] [Google Scholar]
- 12.Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, Kawasaki AM, Cook PD, Freier SM. Evaluation of 2'-modified oligonucleotides containing 2'-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 13.Seth PP, Siwkowski A, Allerson CR, Vasquez G, Lee S, Prakash TP, Wancewicz EV, Witchell D, Swayze EE. Short antisense oligonucleotides with novel 2'-4' conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 2009;52:10–13. doi: 10.1021/jm801294h. [DOI] [PubMed] [Google Scholar]
- 14.Seth PP, Allerson CR, Berdeja A, Siwkowski A, Pallan PS, Gaus H, Prakash TP, Watt AT, Egli M, Swayze EE. An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA. J. Am. Chem. Soc. 2010;132:14942–14950. doi: 10.1021/ja105875e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seth PP, Allerson CR, Siwkowski A, Vasquez G, Berdeja A, Migawa MT, Gaus H, Prakash TP, Bhat B, Swayze EE. Configuration of the 5′-methyl group modulates the biophysical and biological properties of locked nucleic acid (LNA) oligonucleotides. J. Med. Chem. 2010;53:8309–8318. doi: 10.1021/jm101207e. [DOI] [PubMed] [Google Scholar]
- 16.Prakash TP, Siwkowski A, Allerson CR, Migawa MT, Lee S, Gaus HJ, Black C, Seth PP, Swayze EE, Bhat B. Antisense oligonucleotides containing conformationally constrained 2',4'-(N-methoxy)aminomethylene and 2',4'-aminooxymethylene and 2'-O,4'-C-aminomethylene bridged nucleoside analogues show improved potency in animal models. J. Med. Chem. 2010;53:1636–1650. doi: 10.1021/jm9013295. [DOI] [PubMed] [Google Scholar]
- 17.Egli M, Pallan PS, Allerson CR, Prakash TP, Berdeja A, Yu J, Lee S, Watt A, Gaus H, Bhat B, et al. Synthesis, improved antisense activity and structural rationale for the divergent RNA affinities of 3′-fluoro hexitol nucleic acid (FHNA and ara-FHNA) modified oligonucleotides. J. Am. Chem. Soc. 2011;133:16642–16649. doi: 10.1021/ja207086x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renneberg D, Leumann CJ. Watson-Crick base-pairing properties of tricyclo-DNA. J. Am. Chem. Soc. 2002;124:5993–6002. doi: 10.1021/ja025569+. [DOI] [PubMed] [Google Scholar]
- 19.Ittig D, Liu S, Renneberg D, Schumperli D, Leumann CJ. Nuclear antisense effects in cyclophilin A pre-mRNA splicing by oligonucleotides: a comparison of tricyclo-DNA with LNA. Nucleic Acids Res. 2004;32:346–353. doi: 10.1093/nar/gkh187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seth PP, Vasquez G, Allerson CA, Berdeja A, Gaus H, Kinberger GA, Prakash TP, Migawa MT, Bhat B, Swayze EE. Synthesis and biophysical evaluation of 2',4'-constrained 2'O-methoxyethyl and 2',4'-constrained 2'O-ethyl nucleic acid analogues. J. Org. Chem. 2010;75:1569–1581. doi: 10.1021/jo902560f. [DOI] [PubMed] [Google Scholar]
- 21.Koller E, Vincent TM, Chappell A, De S, Manoharan M, Bennett CF. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dean NM, McKay R, Condon TP, Bennett CF. Inhibition of protein kinase C-alpha expression in human A549 cells by antisense oligonucleotides inhibits induction of intercellular adhesion molecule 1 (ICAM-1) mRNA by phorbol esters. J. Biol. Chem. 1994;269:16416–16424. [PubMed] [Google Scholar]
- 23.Dean NM, Griffey RH. Identification and characterization of second-generation antisense oligonucleotides. Antisense Nucleic Acid Drug Dev. 1997;7:229–233. doi: 10.1089/oli.1.1997.7.229. [DOI] [PubMed] [Google Scholar]
- 24.Altmann KH, Martin P, Dean NM, Monia BP. Second generation antisense oligonucleotides - inhibition of pkc-alpha and c-raf kinase expression by chimeric oligonucleotides incorporating 6'-substituted carbocyclic nucleosides and 2'-O-ethylene glycol substituted ribonucleosides. Nucleosides Nucleotides Nucleic Acids. 1997;16:917–926. [Google Scholar]
- 25.Teplova M, Minasov G, Tereshko V, Inamati GB, Cook PD, Manoharan M, Egli M. Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA. Nat. Struct. Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 26.Martin P. Ein neuer Zugang zu 2'-O-(2-Methoxyethyl)ribonucleosiden ausgehend von D-Glucose Helv. Chim. Acta. 2003;86:204. [Google Scholar]
- 27.Geary RS, Yu RZ, Levin AA. Pharmacokinetics of phosphorothioate antisense oligodeoxynucleotides. Curr. Opin. Invest. Drugs. 2001;2:562–573. [PubMed] [Google Scholar]
- 28.Straarup EM, Fisker N, Hedtjarn M, Lindholm MW, Rosenbohm C, Aarup V, Hansen HF, Orum H, Hansen JB, Koch T. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38:7100–7111. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ittig D, Gerber A-B, Leumann CJ. Position-dependent effects on stability in tricyclo-DNA modified oligonucleotide duplexes. Nucleic Acids Res. 2010;39:373–380. doi: 10.1093/nar/gkq733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 31.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset F-L. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 2003;278:41624–41630. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 32.Rodrigues CD, Hannus M, Prudencio M, Martin C, Goncalves LA, Portugal S, Epiphanio S, Akinc A, Hadwiger P, Jahn-Hofmann K, et al. Host scavenger receptor SR-BI plays a dual role in the establishment of malaria parasite liver infection. Cell Host Microbe. 2008;4:271–282. doi: 10.1016/j.chom.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Freier SM, Alkema D, Sinclair A, Neilson T, Turner DH. Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry. 1985;24:4533–4539. doi: 10.1021/bi00338a008. [DOI] [PubMed] [Google Scholar]
- 34.Freier SM, Altmann KH. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krieg AM. Mechanisms and applications of immune stimulatory CpG oligodeoxynucleotides. Biochim. Biophys. Acta. 1999;1489:107–116. doi: 10.1016/s0167-4781(99)00147-5. [DOI] [PubMed] [Google Scholar]
- 36.Geary RS, Wancewicz E, Matson J, Pearce M, Siwkowski A, Swayze E, Bennett F. Effect of dose and plasma concentration on liver uptake and pharmacologic activity of a 2'-methoxyethyl modified chimeric antisense oligonucleotide targeting PTEN. Biochem. Pharmacol. 2009;78:284–291. doi: 10.1016/j.bcp.2009.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.