Abstract
Small RNAs (sRNAs) are a class of short (20–25 nt) non-coding RNAs that play important regulatory roles in gene expression. An essential first step in understanding their function is to confidently identify sRNA targets. In plants, several classes of sRNAs such as microRNAs (miRNAs) and trans-acting small interfering RNAs have been shown to bind with near-perfect complementarity to their messenger RNA (mRNA) targets, generally leading to cleavage of the mRNA. Recently, a high-throughput technique known as Parallel Analysis of RNA Ends (PARE) has made it possible to sequence mRNA cleavage products on a large-scale. Computational methods now exist to use these data to find targets of conserved and newly identified miRNAs. Due to speed limitations such methods rely on the user knowing which sRNA sequences are likely to target a transcript. By limiting the search to a tiny subset of sRNAs it is likely that many other sRNA/mRNA interactions will be missed. Here, we describe a new software tool called PAREsnip that allows users to search for potential targets of all sRNAs obtained from high-throughput sequencing experiments. By searching for targets of a complete ‘sRNAome’ we can facilitate large-scale identification of sRNA targets, allowing us to discover regulatory interaction networks.
INTRODUCTION
RNA silencing is a phenomenon that was independently discovered in animals and plants in the early 1990s. The core RNA silencing machinery is now known to be highly conserved between eukaryotic kingdoms, and the common feature of all RNA silencing pathways is the production of non-coding small RNAs (sRNAs), mostly in the size range of 20–25 nt. These sRNAs are excised from longer, double-stranded or hairpin precursors by RNaseIII-type enzymes called Dicers (1). One strand of the initial sRNA duplex is recruited into a member of the Argonaute protein family, which can be part of a larger complex known as the RNA Induced Silencing Complex (RISC). The sRNA component confers sequence specificity to RISC by establishing Watson–Crick base pairs with potential target RNA or DNA molecules. Having bound its target, the effector complex can silence it at the transcriptional or translational level by employing one of the following mechanisms: (i) cleavage and degradation, (ii) translational repression, (iii) DNA methylation and heterochromatin formation (2). This highly versatile machinery plays important roles in gene regulation, defence against pathogens and genome maintenance (3,4). In plants, sRNA-mediated post-transcriptional gene regulation generally leads to messenger RNA (mRNA) cleavage and degradation due to a high degree of sequence complementarity between the sRNA and its mRNA target (5). This cleavage is highly specific and the mRNA is sliced between positions 10 and 11 of the bound sRNA (6). Computational prediction methods such as (ref. 7–9) have been successfully employed to find sRNA targets in plants but tend to suffer from a high number of false positive predictions (10) and therefore usually require further experimental validation.
Next generation sequencing has become a de-facto standard for the analysis of sRNA samples in recent years (11–13). Typically, a single experiment will produce millions of sRNA reads capturing a snapshot of the expression profile of the ‘sRNAome’ in a single sample (14,15). Recent technological advances have enabled researchers to conduct high-throughput target identification experiments in plants by using an approach called ‘Parallel Analysis of RNA Ends’ (PARE). This approach sequences the 5′-ends of uncapped mRNAs including all transcripts targeted by sRNAs and subjected to endonucleolytic cleavage, i.e. it captures a snapshot of the ‘degradome’ of an organism (16–18). The sRNA and degradome data can be used to identify interactions between sRNAs and their target mRNAs. Degraded mRNA fragments provide support for the interaction between sRNAs and their complementary mRNA targets that lead to cleavage and degradation of the mRNA (16).
Computational tools to analyse such data are both scarce and limited in functionality. CleaveLand (19) was the first tool developed specifically to analyse degradome data, and it has been successfully used to identify micro RNA (miRNA) targets in a variety of organisms (18,20–22). Due to the algorithms implemented in CleaveLand and the size of sRNA and degradome data sets (typically millions of sequences) it is impractical to analyse all possible sRNA/degradome interactions using this software in a reasonable timescale without a large degree of parallelization across multiple machines. As a consequence the tool is generally used to find cleaved targets of a small number of sRNAs, such as known or candidate miRNAs. This means that users typically have to ignore the vast majority of sRNA reads in such analyses and have to assume some prior knowledge of which sRNAs are likely to have targets. As a result many legitimate sRNA-mediated mRNA cleavages could potentially be missed. While this is acceptable for users interested in looking for targets of known miRNAs, it greatly restricts the possibility to get a sense of all of the sRNA regulatory interactions leading to mRNA cleavage. In addition, CleaveLand is a command-line-based application that can only be used in a Linux/UNIX environment. This excludes a large number of potential users who do not have access to, or expertise in, such environments.
To the best of our knowledge, only two other methods have been developed for identifying sRNA/target interactions evidenced through the degradome in addition to CleaveLand; SoMART (23) and SeqTar (24). SoMART is a collection of web server tools for processing sRNAs. To process degradome data, the user first needs to predict sRNAs that could potentially target a user-supplied transcript with the Slicer detector tool. The dRNA mapper tool can then be used to align degradome sequences to the transcript sequence. The user then has to manually compare the output from Slicer detector and dRNA mapper to identify cleaved targets. To automatically process more than one transcript the user would therefore have to develop additional methods and post-processing software. In addition, the SoMART website is restricted to a prescribed list of sRNA and degradome databases. SeqTar attempts to broaden the alignment rules used in CleaveLand between sRNAs and their potential targets so as to identify miRNA targets. As with CleaveLand, SeqTar suffers from the fact that its underlying algorithms make it impractical to analyse all possible sRNA/degradome interactions in a reasonable timescale without a large degree of parallelization across multiple machines. Moreover, SeqTar is not available in a publicly downloadable package, which greatly reduces its potential user base.
Here, we describe a new, user-friendly, cross-platform degradome analysis tool, PAREsnip, which enables flexible and comprehensive high-throughput target analysis, allowing users to identify genome-wide networks of sRNA/target interactions resulting in transcript cleavage. As well as being able to analyse data sets like CleaveLand PAREsnip is also able to process entire sRNAome and transcriptome data sets in a short timeframe on a typical desktop computer.
MATERIALS AND METHODS
Input
For a specific organism the inputs for PAREsnip are:
the mRNA data set (transcriptome),
the transcript degradation fragments obtained from a PARE experiment (degradome),
the sRNA data set (sRNAome) and
the genome sequence.
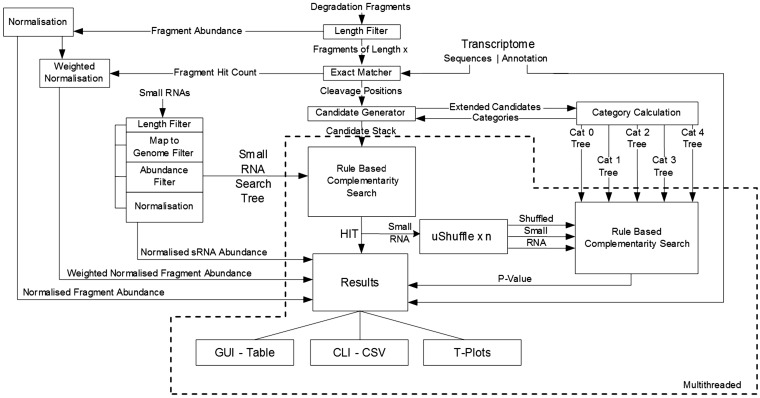
The first three inputs are required but the genome is optional. When included, the genome is used during the data-filtering process described later. All of the inputs must be in FASTA format and must only contain the characters ‘A’, ‘C’, ‘G’, ‘T’ and ‘U’. Sequences containing unknown characters and ambiguity codes are discarded as they cannot be accurately aligned later. FASTQ to FASTA and adaptor removal tools are provided within the UEA sRNA Workbench (7). An overview of the steps involved in processing the input data is shown in Figure 1.
Figure 1.
Schematic of PAREsnip. Boxes represent functions and solid arrowed lines represent data flow. The functions and dataflow operating concurrently using multithreading are enclosed with a dotted line.
Data filtering
Several user-configurable filters based on: sequence length, sequence abundance and sequence complexity may be applied to the sRNAome. If a sequence has an exact full-length match to known tRNA or rRNA, it will be omitted. T/rRNA sequences are obtained from Rfam (25) and EMBL/Genbank (26) sequence databases. If a genome is provided, sRNA sequences are mapped to it using PatMaN (27). Any sequences without a match to the genome are removed from further analysis, as they are likely to be either sequencing errors or sample contamination.
Signals of cleavage
Degradome fragments are exactly matched to the transcriptome and 5′-end alignment positions are recorded. The degradome fragment abundance at any given position could represent an sRNA cleavage event at that position (16,18). Potential cleavage sites on a single transcript can be categorized according to degradome read abundance. Higher abundance reads are more likely to be the result of endonucleolytic cleavage as opposed to random degradation products, which are more likely to accumulate at a lower background level. PAREsnip uses the 5-category system defined in CleaveLand (version 2) (19), which are:
Category 0 is defined as a signal having greater than one raw read at the position. The abundance at that position is equal to the maximum on the transcript, and there is only one maximum.
Category 1 is the same as Category 0 in all aspects except that more than one maximum is found on the transcript. This implies that there are two or more signals on the transcript with the same strength (abundance).
Category 2 is defined as a signal having greater than one raw read at the position. The abundance at that position is less than the maximum, but greater than the median abundance for that transcript.
Category 3 is defined as a signal having greater than one raw read at the position and the abundance at that position is less than or equal to the median value for that transcript.
Category 4 is defined as only one raw read at the position.
The categorization of the signal strength is based on either the raw abundance or weighted abundance of degradation fragments; the latter is the default PAREsnip setting. Weighted abundance is calculated by dividing the abundance of a degradome fragment (tag) by the number of positions across all transcripts to which the tag has aligned. The strongest signals, described as Categories 0, 1 and 2, convey the strongest empirical evidence for true cleavage products (18). The weaker Categories 3 and 4 signals could be difficult to distinguish from background noise and random degradation. It is therefore possible for the user to exclude any of the five categories before commencing an analysis in PAREsnip.
Data structures
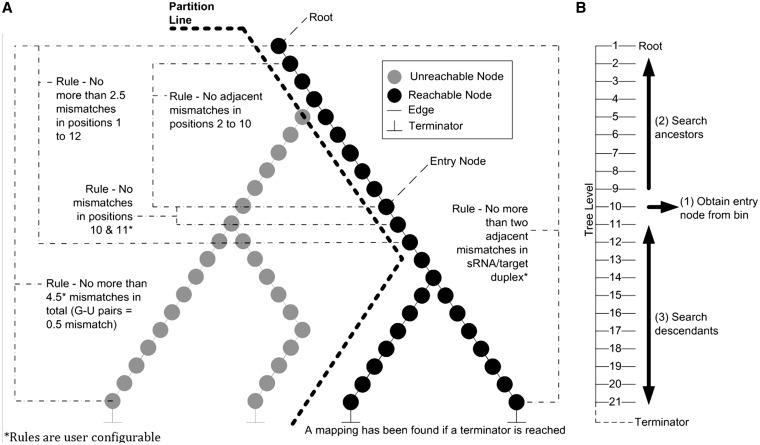
Small RNA sequences are encoded into unique paths within a trie (28), which is an m-way search tree data structure. Since RNA and DNA sequences are described by the symbols (‘A’,‘C’,‘G’,‘T’ or ‘A’,‘C’,‘G’,‘U’) we use a 4-way tree (Figure 2A). Edges represent nucleotide bases and nodes offer path choice through the tree. Many short sequences share a similar nucleotide composition. By encoding all sequences into a 4-way tree, those that share a similar composition will lie on the same path until the similarity ends and new branches are created. A terminator node marks the end of a path and therefore an sRNA sequence encoded within the tree. This structure allows us to remove sequence and subsequence redundancy, therefore reducing our search space and memory footprint. Also, the number of nucleotide/edge comparisons required when attempting to search for a sequence within the tree is reduced.
Figure 2.
(A) Applying the binding rules to the partitioned 4-way tree. Small RNAs are encoded into a 4-way tree. The tree is partitioned based on the nucleotides at positions 10 and 11 in the pattern sequence to be searched for. As the tree is searched, sRNA/target binding rules are applied. (B) Searching the partitioned 4-way tree. To search for a pattern within the tree we start at level 10 denoted as (1), which corresponds to the 10th nucleotide in a small RNA (counted from the 5′ end). The tree is followed towards the root performing Watson and Crick base pairing denoted as (2). At each traversal, the binding rules are checked. If the root is reached successfully the algorithm jumps back to (1) and begins a pre-order walk down the tree, denoted as (3). While walking down the tree, if the rules are broken, then the traversals of that branch stop. If a terminator node is reached, then a successful alignment has been made and an sRNA/target interaction discovered.
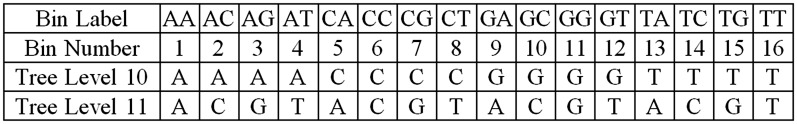
Once the sRNAs are encoded in the tree, target searches can be performed. The starting node for each search is the 10th node because we know that position 10 of the sRNA/target duplex must be complementary in order to cleave a target (6,29). Therefore pairs of nodes at levels 10 and 11 within the 4-way tree are collected and placed into labelled bins (Figure 3) according to the pair’s nucleotide composition. There are a total of 16 bins that correspond to the 16 possible dinucleotide combinations. Searches for sRNAs that could cause cleavage at a given degradome peak position are initiated by identifying the bin corresponding to nucleotides 10 and 11 of the candidate sequence. The tree is then traversed from nucleotide 10 towards the root. We place a restriction that once a walk up the tree from an entry point has occurred, the parent node of the entry point obtained from the bin may never be visited again during the current search and only descendent nodes of the entry point may be traversed. This restriction ensures that unnecessary nucleotide comparisons are not computed. We partition the tree by hiding all paths that have starting nodes in any of the other 15 labelled bins.
Figure 3.
Organization of partitioned 4-way tree entry points. Nodes at levels 10 and 11 within a 4-way tree data structure are collected and placed into labelled bins. There are a total of 16 bins as there are a total of 16 possible dinucleotide combinations. The label for each bin is the nucleotide at level 10 followed by the nucleotide at level 11. The bins hold entry points into the tree data structure. Entry nodes within a bin are used to partition the 4-way tree.
The organization of the data in this way lends itself to the fast mapping of sequences in an all-against-all search because only a small fraction of the millions of sequences obtained from a high-throughput sequencing experiment, that are encoded into the 4-way tree, have the potential to be aligned with the candidate pattern. This is possible as we know that the 10th and 11th nucleotides of the sRNA, which sit at levels 10 and 11 in the tree, must match the 10th and 11th nucleotide of the search pattern exactly (29). This contributes to the computational speed of PAREsnip.
Search algorithm
The core of PAREsnip’s operation is what we call the Rule-Based Complementarity Search algorithm. It is a method of traversing the partitioned 4-way tree, searching for sRNA sequences that could potentially cleave a transcript accounting for the degradome peak at a given position. The method is designed to make as few nucleotide comparisons as possible and will disregard the large sections of the 4-way tree that will never produce a valid alignment, based on a set of previously described targeting rules (29,30). The rules used by the search algorithm are user configurable and the default settings are:
No more than 4.5 mismatches between sRNA and target (G-U bases count as 0.5 mismatches).
No more than two adjacent mismatches in the sRNA/target duplex.
No adjacent mismatches in positions 2–12 of the sRNA/target duplex (5′ end of the sRNA).
No mismatches in positions 10–11 of sRNA/target duplex.
No more than 2.5 mismatches in positions 1–12 of the sRNA/target duplex (5′ of sRNA).
The algorithm requires a candidate pattern on which to execute its rules. The pattern is the reverse complement of the first 11-nt downstream and up to 15-nt upstream from the position of a categorized degradome cleavage signal on the transcript. The algorithm looks at the two nucleotides either side of the cleavage position in the pattern and identifies the appropriate bin (Figure 3). The algorithm retrieves a starting node from the bin and traverses a single path up the tree to the root (Figure 2B). As it does so, it makes a nucleotide comparison between the pattern and the edge in the path and tests the rule set (Figure 2A). If at any point one of the rules is broken, the search is aborted, the starting node discarded and the next starting node is obtained from the bin. If, on the other hand, the algorithm successfully reaches the root of the tree without breaking any of the rules, then it returns to the entry point and begins a pre-order walk through the tree. A history of alignment records is kept while the tree is traversed. Each record is composed of nucleotide matches, mismatches and single gaps along with a running alignment score. A mismatch contributes 1.0 to the score, unless it is a G-U (wobble) pair in which case it contributes 0.5 to the score. A gap in the alignment contributes a value of 1.0 to the score. If a terminator node is found, then the algorithm must have reached it without breaking the rules in one or more of the alignment records kept in its history. In this case the algorithm examines its history of alignment records and selects the alignment with the lowest score and places it onto a communal stack of identified valid alignments. If at any point a rule is broken during a traversal and there is no valid alignment in its maintained history, the algorithm no longer continues down its current path. When there are no more paths to traverse, the algorithm looks in the bin and if there are any remaining starting nodes, it will obtain the next starting node from the bin and repeat the procedure until the bin is empty. The stack of valid alignments represents possible sRNA/target interactions. Each interaction within the stack is passed on to the system to calculate the P-value before being reported to the user.
Calculating P-values
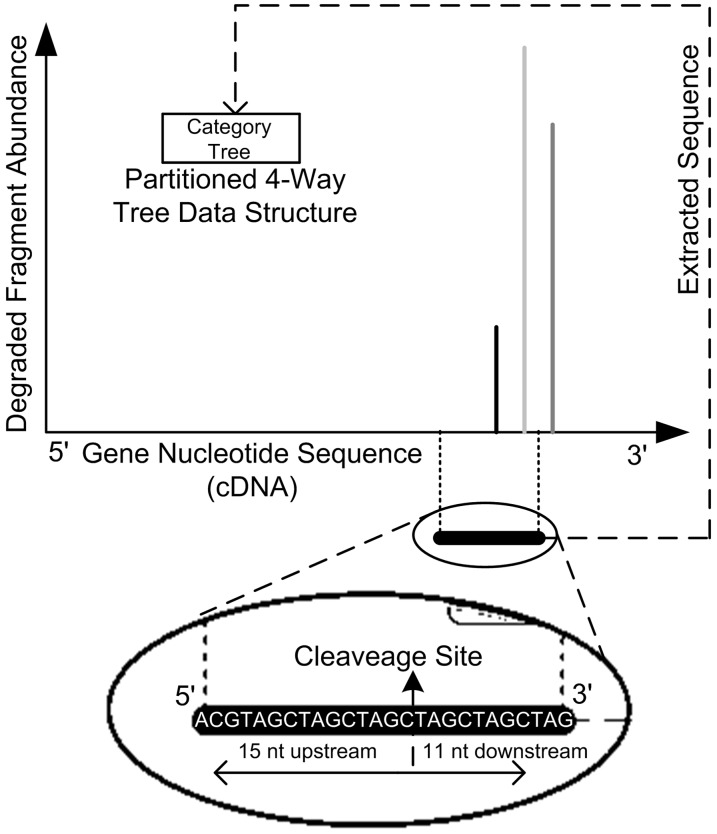
For each sRNA/target duplex reported by PAREsnip, a P-value is calculated. The P-value gives us a score that indicates how likely the reported duplex occurred by chance. The P-value calculation methods are based on those published in CleaveLand (version 2.0) (19) but use our Rule-Based Complementarity Search algorithm and partitioned 4-way trees during the calculation. For every position, on every mRNA containing a cleavage signal a 26-nt sequence representing the sRNA-binding site is extracted and placed into one of five possible category trees (Figure 4). The category trees are the same in structure and function to the partitioned 4-way tree used to encode sRNAs, but instead contain sections of mRNAs where cleavage has occurred.
Figure 4.
Data structure created from degradome fragments mapped to transcripts. Bars represent 5′ ends of degradome fragments aligned to a transcript. Degradome signals are characterized by category. A sub-sequence of 26 nt is extracted from the transcript based on the cleavage site. The sub-sequence is encoded into a partitioned 4-way tree according to the assigned category.
The sRNA for each sRNA/target alignment on the stack of valid alignments is randomly shuffled and mapped to all target sites encoded into a 4-way tree (Figure 2A). The chosen 4-way tree corresponds to the category given to the output sRNA/target record. The random shuffles of the sRNA preserve dinucleotide frequency and are generated by the third-party Java programme uShuffle (31). The user may define the number of shuffles to be used (the default is 100) and the resulting P-value is the number of times the randomly shuffled sRNA aligns to a target site encoded within the category tree. The P-value is provided as a decimal. For example, if 100 shuffles were used and 5 of those aligned to a target site of the same category, then the resulting P-value would be 0.05. An alignment below the user-specified P-value cut-off is accepted as valid and output to file or to the user interface.
Output
PAREsnip displays results in a tabular format where each row in the table shows an sRNA/target interaction. The columns show alignment category, P-value, binding score and abundance information along with a visual sequence alignment of sRNA and target mRNA. Statistics relating to the input data set are provided such as sequence count and sequence length distribution. When the tool is operated in GUI mode, a results table is displayed and updated as interactions are found. Columns and rows may be sorted and re-arranged and the data in the table may be saved as comma separated value (csv) format. If the user operates the tool from the command line, the table is saved straight to disk in csv format, which can be imported directly into most spreadsheet and statistical packages. PAREsnip lets the user generate and investigate publication quality t-plots through the UEA sRNA Workbench tool called VisSR (7).
Availability
PAREsnip is a multi-platform, multi-threaded (Figure 1) application written in Java and is released as part of the UEA sRNA Workbench (7) (http://srna-workbench.cmp.uea.ac.uk). It may be run from the command line or a graphical user interface (GUI).
RESULTS
Benchmarking
To measure the runtime performance of PAREsnip we simulated 10 sRNA data sets of increasing size. The sRNAs were generated by extracting 19–24 nt sequences centred on cleavage positions within the Arabidopsis thaliana transcriptome (TAIR 10 representative gene model) (32). Transcripts, cleavage positions and sRNA sequence lengths were selected at random. The performance of PAREsnip was measured by using the simulated sRNAs with the A. thaliana transcriptome and the publicly available PARE degradome library GSM278370 A. thaliana Col-0 wild-type seedlings (18,33). We observe a linear time operation with a peak memory requirement of 5.5 gigabytes.
We also benchmarked the performance of CleaveLand (version 2) and compared the runtime with that of PAREsnip (Table 1). We found that PAREsnip significantly outperformed CleaveLand for the considered data sets. Note that, even though there is a version 3 of CleaveLand, we compared PAREsnip with version 2 since the target prediction step of version 3 only receives a single sRNA sequence for analysis, and therefore cannot be practically used on larger numbers of sRNAs without developing additional software. Even so, to get a rough idea of the performance of CleaveLand (version 3), we obtained an average runtime of 87 s per sRNA sequence for 10 simulated sRNAs, which is roughly 3 times faster than version 2, but still significantly slower than PAREsnip.
Table 1.
Run time for PAREsnip and CleaveLanda
| Number of sRNAs | CleaveLand Timing | PAREsnip Timing |
|---|---|---|
| 10 | 46 min 6s | 29s |
| 25 | 1 h 55 min 25s | 30s |
| 50 | 3 h 51 min 35s | 31s |
| 1000 | – | 2 min 3s |
| 10 000 | – | 10 min 14s |
| 20 000 | – | 19 min 11s |
| 40 000 | – | 39 min 8s |
| 60 000 | – | 53 min 9s |
| 80 000 | – | 73 min 24s |
| 100 000 | – | 87 min 16s |
aThe number of sRNAs processed. The amount of time taken to process the sRNAs in hours (h), minutes (min) and seconds (s). A desktop PC was used with the following specification: Intel i7 960 (3.20 GHz) CPU with 24 Gb of RAM using Windows 7 (64 bit) native and Linux (Ubuntu) virtualized. Transcripts used were A. thaliana (TAIR 10 representative gene model) consisting of 33 602 sequences. The degradome library used was GSM278370 consisting of 5 639 743 degradation tags.
Comparison with CleaveLand
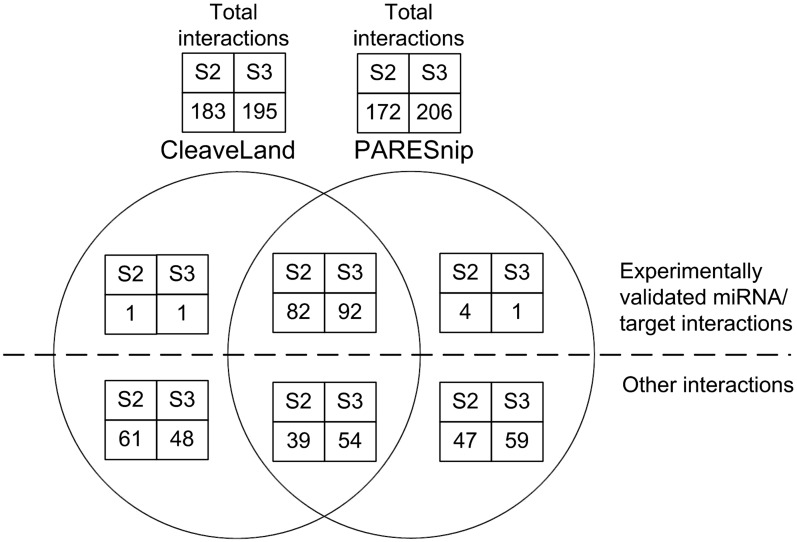
As CleaveLand is currently the only publicly available tool for degradome analysis, we compared all miRNA targets reported by CleaveLand (version 2) (19) with those reported by PAREsnip using two data sets. We obtained all known mature A. thaliana miRNAs from miRBase (release 17) (34) and analysed them using both tools, seeking targets within the transcriptome (A. thaliana representative gene model TAIR release 10) (32) using two publicly available degradome libraries: GSM278335 and GSM278370 A. thaliana Col-0 wild-type inflorescence tissue taken from Gene Expression Omnibus (18,33). A collection of previously validated miRNA targets obtained from the literature (16,35–38) and the MPSS database (39) (Supplementary Table S1) were used to identify previously validated miRNA targets reported by both tools.
The results are summarized in Figure 5 (see full results in Supplementary Tables S2 and S3). As can be seen, PAREsnip reports either the same number or slightly more previously validated targets than CleaveLand. The interactions reported by PAREsnip and not by CleaveLand or vice versa are due to the random factor within the P-value systems used by both tools. For example, in contrast to CleaveLand, PAREsnip uses dinucleotide random shuffles when calculating a P-value through the use of uShuffle (31). Furthermore, differences between the interactions predicted by the two tools are probably also due to the reporting of hits that contain a mismatch at position 10 (from 5′ of sRNA), multiple gaps within a duplex and more than 2.5 mismatches or adjacent mismatches within the seed region (positions 1–12 5′ of sRNA) of the duplex. Again, in contrast to CleaveLand, these features within a duplex are not permitted by the Rule-Based Complementarity Search algorithm used by PAREsnip.
Figure 5.
Venn diagram showing the comparison of results produced by CleaveLand and PAREsnip. The Venn diagram shows the intersection of predictions made by PAREsnip and CleaveLand and is a summary of the results within Supplementary Tables S2 and S3.
Filtering by P-value
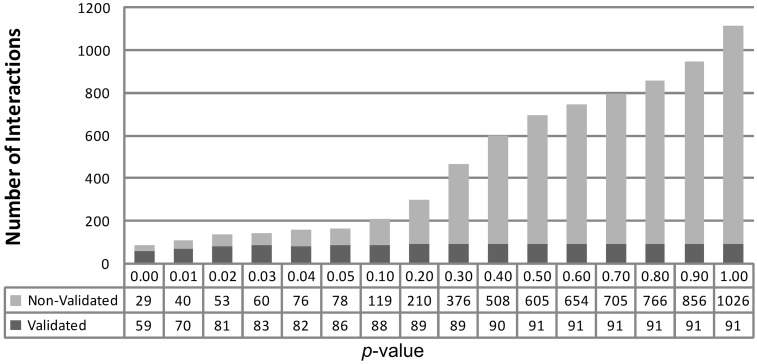
To examine the usefulness of the P-value computed by PAREsnip as a confidence score upon which predicted interactions can be excluded, we ran it on all known mature A. thaliana miRNAs, GSM278370 (18,33) degradome and the A. thaliana transcriptome (representative gene model, TAIR release 10) (32) with increasing P-value thresholds. The predictions were compared with previously validated interactions (Supplementary Table S1) to provide an insight into the number of validated interactions retained along with the number of other interactions reported in relation to the increasing threshold (Figure 6). Note that a P-value cut-off of 1 captures all possible predictions. PAREsnip reported a total of 91 validated and 1026 non-validated interactions using a P-value cut-off of 1. We find that a threshold of 0.05 captures 94.5% of possible validated interactions (a loss of 5.5% validated interactions) while capturing 7.6% of the total non-validated interactions. In light of this and other similar experiments we have chosen a default P-value setting for PAREsnip of 0.05.
Figure 6.
Interactions reported by PAREsnip with P-value increases. Starting from the smallest P-value of 0.00, we see a progressive increase in the number of small RNA/mRNA interactions reported. The P-value cut-off of 0.05 captures 94.5% of total validated interactions reported by PAREsnip and is the default setting.
Genome-wide discovery of sRNA/target interactions
Small RNA sample libraries obtained from a high-throughput sequencing experiment typically contain millions of sequences. To look for interactions on a genome-wide scale, including all sRNAs obtained from a high-throughput sequencing experiment, we used PAREsnip to analyse the following data sets: sRNAome GSM342999 A. thaliana Col-0 biological replicate 1 inflorescence tissue (33,40); degradome GSM278335; transcripts: A. thaliana (representative gene model TAIR release 10) (32). For this and every subsequent analysis the following settings were used: a maximum of 4.0 mismatches, 100 dinucleotide shuffles and a P-value threshold of 0.05. Within these data, PAREsnip reported 36 351 interactions. Despite the support found for these interactions, in particular the degradation signal, observed sRNA, sequence specificity within each duplex and low P-value, it is difficult to believe that so many interactions are genuine. Therefore the combined restrictions of mismatch positions, the number of permitted mismatches and P-value filter, on their own, do not appear to be sufficient measures to extract valid interactions above the noise when performing an analysis on such a large scale. It is likely that many degradome signals are not the product of sRNA-induced cleavage but are instead random degradation fragments that happen to also be complementary to one or more of the millions of sRNA inputs. To address this problem we employed cross-sample conservation with the aim of reducing the number of reported targets. The rationale behind this approach is that both degradome fragments and sRNA sequences that are products of random degradation are unlikely to be conserved between biological replicates whereas bona fide cleavage signals and functional sRNAs are likely to be present across samples.
To explore this approach we used PAREsnip to independently analyse two sRNA biological replicates GSM342999 (set B1) and GSM343000 A. thaliana Col-0 biological replicate 2 inflorescence tissue (set B2) (33,40) along with the degradome GSM278335. The results were compared and only the conserved interactions across the two samples were retained. For an interaction to be conserved the interaction must share the same target transcript, cleavage site and sRNA sequence. In set B1 36 351 interactions were identified (Supplementary Table S4a and b) and in set B2 26 098 interactions (Supplementary Table S5a–c). By comparing the interactions between the sets we found 7273 conserved interactions. To ascertain whether such a result could occur by chance, we carried out the same experiment again but using simulated sRNA sets containing randomly generated sequences. The simulated sets (set R1 and R2) maintained the same characteristics as the real sRNA libraries, including unique and redundant sequence count and sequence length distribution. The sequences themselves were randomly generated by sampling from the Arabidopsis genome sequence. Set R1 identified a total of 21 783 interactions and R2 identified 21 862 interactions. Comparing the interactions of R1 and R2 using the same conservation criteria we found that no interactions were conserved. This indicates that sRNAs being observed in multiple samples (biological replicates) could provide a method for extracting reliable hits above noise with some measure of confidence.
We extended the conservation method to include signals of degradation so that a reliable interaction should contain degradation products that are conserved across multiple degradome library samples as well as the sRNA being conserved across multiple sRNAomes. We analysed two data sets: Set D1 comprised sRNAome-GSM342999 and degradome-GSM280226 A. thaliana Col-0 inflorescence tissue (16,33) and set D2 comprised sRNAome-GSM343000 and degradome-GSM280227 A. thaliana xrn4 inflorescence tissue (16,33). Reference transcripts were the A. thaliana representative gene model (TAIR release 10) (32). Within sets D1 and D2 we found a total of 65 110 and 49 938 interactions, respectively. The 65 110 interactions are shown in Supplementary Table S6a–d, and the 49 938 interactions are shown in Supplementary Table S7a–c. Based on the previously validated interactions (Supplementary Table S1), 163 and 179 interactions within the total number of interactions found in sets D1 and D2, respectively, had been previously experimentally validated. When comparing the results of sets D1 and D2 we found a total of 4466 conserved interactions. Of the validated interactions, 149 were conserved giving an above 80% retention rate. The 4466 conserved interactions meet the binding rules criteria for mismatch positioning within the sRNA/mRNA duplex and have a mismatch score of 4 or less. They have a P-value of 0.05 or less and the sRNA and positional cleavage signal are conserved across multiple samples.
DISCUSSION
We have described a novel, freely-available application called PAREsnip, designed for the identification of cleaved targets from sRNA and degradome data sets generated using next-generation sequencing technologies. The tool can also be used on small-scale experiments. PAREsnip is a user-friendly GUI-based, cross-platform (Windows, Linux, MacOS) application that enables biologists to run the application and analyse their data without the need for dedicated bioinformatics support or specialized computer hardware. We have also made a command-line version of the tool available for users who wish to incorporate PAREsnip into computational pipelines.
We have shown that PAREsnip performs at least as well as current methods in detecting validated miRNA–mRNA interactions in published data sets and that it runs significantly faster than the competition on a standard desktop computer. The speed of PAREsnip opens up new avenues in the sRNA field as it enables users to look for targets of all sequenced sRNAs rather than a subset of sequences that they suspect might have a target (such as annotated miRNAs and trans-acting small interfering RNAs).
We have demonstrated that degradome and sRNA data are inherently noisy (probably due to background mRNA degradation) and that searching a random sRNA data set with the same properties as a real input data set against the degradome can lead to a comparable number of predicted target interactions. This makes it difficult to separate real targets from false positives when running on high-throughput data. However, by using biological replicates of sRNA and degradome data sets we appear to be able to remove spurious degradation products, as they are highly unlikely to be conserved between two or more samples. We show that by using this conservation method on a random sRNA set no targets are predicted (resulting in zero false positives), whereas when applying it to a real set we retrieve 4466 high-confidence interactions and recover ∼80% of the previously validated targets present.
PAREsnip is extensively user-configurable; this allows users to customize search parameters and binding rules in order to make searches more liberal or stringent. It was recently reported that several new miRNA targets were discovered and validated using more relaxed binding rules implemented in the SeqTar algorithm (24). By relaxing the stringency of the binding rules PAREsnip can also be used to search more deeply for individual miRNA targets. Conversely, tightening the rules will lead to a reduction in the number of candidates reported when run across entire sRNA sets. This flexibility also allows users to customize searches and could allow them to optimize parameters for searching degradome data sets such as those published by Bracken (41) and Karginov (42).
While the use of published binding rules and P-value filtering provides a strong set of predicted sRNA/target interactions it is difficult to estimate an accurate false positive rate. One of the reasons is that currently there is no experimental method to directly test sRNA/target interactions. The only method is the 5′RACE to map the non-capped 5′ end of individual mRNA fragments. However, this method is based on the same principle as the PARE/degradome library generation and so it is questionable whether it can be used to validate the high-throughput results. In fact, since 5′RACE experiments focus on a small region of an mRNA, it is more likely to yield an artefact than the unbiased PARE/degradome library approach.
CONCLUSION
PAREsnip can be used to search for genome-wide interactions between all sRNAs and transcripts as well as predicting targets of small groups of miRNAs. This high-throughput approach to degradome analysis opens a new avenue for researchers interested in identification of sRNA targets. Due to its speed and efficiency PAREsnip removes the need for users to know in advance which sequences are likely to have a target and instead allows users to generate complete networks of sRNA target interactions. By using replicates and applying a conservation rule we predict over 4000 putative sRNA/mRNA interactions in the Arabidopsis sets we analysed. This suggests that sRNA-mediated targeting and cleavage of transcripts may be even more widespread than previously anticipated and provides a useful new tool for experimentalists to study such interactions in more depth.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–7 and Supplementary References [16,34–39,43–46,12].
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) (V.M., H.W., and T.D., BB/H023895/1 and M.S., grant number BB/I00016X/1); and BBSRC studentship (to L.F.). Funding for open access charge: BBSRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank Frank Schwach for his contributions.
REFERENCES
- 1.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 2.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 3.Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 5.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 7.Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics. 2008;24:2252–2253. doi: 10.1093/bioinformatics/btn428. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnet E, He Y, Billiau K, Van de Peer Y. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. 2010;26:1566–1568. doi: 10.1093/bioinformatics/btq233. [DOI] [PubMed] [Google Scholar]
- 10.Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18:1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzker ML. Sequencing technologies – the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 12.Fahlgren N, Sullivan CM, Kasschau KD, Chapman EJ, Cumbie JS, Montgomery TA, Gilbert SD, Dasenko M, Backman TWH, Givan SA, et al. Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA. 2009;15:992–1002. doi: 10.1261/rna.1473809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horner DS, Pavesi G, Castrignanò T, De Meo PD, Liuni S, Sammeth M, Picardi E, Pesole G. Bioinformatics approaches for genomics and post genomics applications of next-generation sequencing. Brief. Bioinformatics. 2010;11:181–197. doi: 10.1093/bib/bbp046. [DOI] [PubMed] [Google Scholar]
- 14.Pais H, Moxon S, Dalmay T, Moulton V. Small RNA discovery and characterization in eukaryotes using high-throughput approaches. Adv. Exp. Med. Biol. 2011;722:239–254. doi: 10.1007/978-1-4614-0332-6_16. [DOI] [PubMed] [Google Scholar]
- 15.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 16.German MA, Pillay M, Jeong D-H, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 17.German MA, Luo S, Schroth G, Meyers BC, Green PJ. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 2009;4:356–362. doi: 10.1038/nprot.2009.8. [DOI] [PubMed] [Google Scholar]
- 18.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25:130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 2010;62:960–976. doi: 10.1111/j.0960-7412.2010.04208.x. [DOI] [PubMed] [Google Scholar]
- 21.Addo-Quaye C, Snyder JA, Park YB, Li Y-F, Sunkar R, Axtell MJ. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA. 2009;15:2112–2121. doi: 10.1261/rna.1774909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y-F, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010;62:742–759. doi: 10.1111/j.1365-313X.2010.04187.x. [DOI] [PubMed] [Google Scholar]
- 23.Li F, Orban R, Baker B. SoMART: a web server for plant miRNA, tasiRNA and target gene analysis. The Plant Journal. 2012;70:891–901. doi: 10.1111/j.1365-313X.2012.04922.x. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Li Y-F, Sunkar R, Zhang W. SeqTar: an effective method for identifying microRNA guided cleavage sites from degradome of polyadenylated transcripts in plants. Nucleic Acids Research. 2012;40:e28. doi: 10.1093/nar/gkr1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karsch-Mizrachi I, Nakamura Y, Cochrane G. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2012;40:D33–D37. doi: 10.1093/nar/gkr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prüfer K, Stenzel U, Dannemann M, Green RE, Lachmann M, Kelso J. PatMaN: rapid alignment of short sequences to large databases. Bioinformatics. 2008;24:1530–1531. doi: 10.1093/bioinformatics/btn223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodrich MT, Tamassia R. Data Structures and Algorithms in Java. 4th edn. USA: John Wiley and Sons; 2005. [Google Scholar]
- 29.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 30.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Jiang M, Anderson J, Gillespie J, Mayne M. uShuffle: a useful tool for shuffling biological sequences while preserving the k-let counts. BMC Bioinformatics. 2008;9:192. doi: 10.1186/1471-2105-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2010;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2010;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, Twell D, Dickinson H. MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics. 2009;10:643. doi: 10.1186/1471-2164-10-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsieh L-C, Lin S-I, Shih AC-C, Chen J-W, Lin W-Y, Tseng C-Y, Li W-H, Chiou T-J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moldovan D, Spriggs A, Yang J, Pogson BJ, Dennis ES, Wilson IW. Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J. Exp. Bot. 2010;61:165–177. doi: 10.1093/jxb/erp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM, Alexander A, Nguyen G, Allen E, Ahn JH, et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl Acad. Sci. USA. 2008;105:20055–20062. doi: 10.1073/pnas.0810241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bracken CP, Szubert JM, Mercer TR, Dinger ME, Thomson DW, Mattick JS, Michael MZ, Goodall GJ. Global analysis of the mammalian RNA degradome reveals widespread miRNA-dependent and miRNA-independent endonucleolytic cleavage. Nucleic Acids Res. 2011;39:5658–5668. doi: 10.1093/nar/gkr110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karginov FV, Cheloufi S, Chong MMW, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borges F, Pereira PA, Slotkin RK, Martienssen RA, Becker JD. MicroRNA activity in the Arabidopsis male germline. J. Exp. Bot. 2011;62:1611–1620. doi: 10.1093/jxb/erq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible W-R. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009;150:1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res. 2010;38:6883–6894. doi: 10.1093/nar/gkq590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Backman TWH, Sullivan CM, Cumbie JS, Miller ZA, Chapman EJ, Fahlgren N, Givan SA, Carrington JC, Kasschau KD. Update of ASRP: the Arabidopsis Small RNA Project database. Nucleic Acids Res. 2008;36:D982–D985. doi: 10.1093/nar/gkm997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.