Abstract
Numerous examples exist of how disrupting the actions of physiological regulators of blood cell development yields hematologic malignancies. The master regulator of hematopoietic stem/progenitor cells GATA-2 was cloned almost 20 years ago, and elegant genetic analyses demonstrated its essential function to promote hematopoiesis. While certain GATA-2 target genes are implicated in leukemogenesis, only recently have definitive insights emerged linking GATA-2 to human hematologic pathophysiologies. These pathophysiologies include myelodysplastic syndrome, acute myeloid leukemia and an immunodeficiency syndrome with complex phenotypes including leukemia. As GATA-2 has a pivotal role in the etiology of human cancer, it is instructive to consider mechanisms underlying normal GATA factor function/regulation and how dissecting such mechanisms may reveal unique opportunities for thwarting GATA-2-dependent processes in a therapeutic context. This article highlights GATA factor mechanistic principles, with a heavy emphasis on GATA-1 and GATA-2 functions in the hematopoietic system, and new links between GATA-2 dysregulation and human pathophysiologies.
INTRODUCTION
Drilling into mechanisms governing the control of hemoglobin synthesis led to the discovery in the 1980s of a new class of transcription factors containing a highly conserved Cys4 dual zinc finger DNA-binding module. These proteins were deemed GATA factors based on the nucleotide composition of their cognate DNA-binding motif (1). The discovery of GATA-1 was followed by the cloning of five additional mammalian GATA factors (GATA-2–6) (2–9). Historically, GATA-1, GATA-2 and GATA-3 are deemed the hematopoietic GATA factors (10), while GATA-4, GATA-5 and GATA-6 are termed the cardiac GATA factors (11,12). Extensive biological and genetic analyses have revealed exceptions to this generalization, including expression of the hematopoietic GATA factors in endothelium (9,13,14), breast and prostate (15,16) and neurons (17,18).
Loss-of-function analyses established the essential GATA-1 functions to promote erythrocyte, megakaryocyte, mast cell and eosinophil development (19–25) and GATA-3 functions to promote specific aspects of T-cell lymphopoiesis (26,27). GATA-2 is uniquely essential for the genesis and/or function of hematopoietic stem/progenitor cells (28–30). Gata2-null mouse embryos are severely anemic and die at approximately embryonic day (E) 10 (28). Despite the critical GATA-2 requirement for the formation of all lineages of blood cells, some primitive erythroblasts exist in Gata2-null embryos. The absence of these cells in Gata1−/––Gata2−/– compound mutants indicate that GATA-1 and GATA-2 can function redundantly in the genesis and/or survival of primitive erythroblasts (31). The GATA-2 requirement for the control of hematopoietic stem/progenitor cells is dose dependent, as Gata2+/– HSCs are functionally impaired, even though the mice are viable (30,32). GATA-2 overexpression in murine bone marrow is also inhibitory for hematopoiesis (33). While this result has potentially important pathophysiological implications, transcription factor overexpression studies are difficult to interpret, given ample opportunities for overexpressed proteins to aberrantly engage cellular regulatory factors. As GATA-2 is also expressed in endothelial cells, placenta, prostate, pituitary and select neurons, it will be instructive to compare GATA-2 mechanisms in hematopoietic versus non-hematopoietic systems, although this is currently a virgin territory. Clearly, many unanswered questions remain regarding cell type-specific GATA factor mechanisms and biological actions.
GATA FACTOR MECHANISMS: FUNDAMENTAL PRINCIPLES
The purification and cloning of GATA-1 (34,35) ushered in studies that elucidated mechanistic principles governing GATA factor function (36). The zinc finger residing closest to the carboxy-terminus (C-finger) mediates sequence-specific DNA binding to WGATAR motifs (37,38), while the zinc finger proximal to the amino-terminus (N-finger) mediates an important protein–protein interaction with the nine zinc finger-containing coregulator Friend of GATA-1 (39–42). The N-finger may also stabilize DNA binding in certain contexts (43). Additional interactions involving the zinc fingers have been documented (44,45), including binding to the myeloid transcription factor PU.1 (46), the erythroid transcription factor ELKF (47) and the mediator complex component Med1 (48). Much less is known about the structural basis and biological implications of these interactions. The broad GATA-1 N-terminus enhances endogenous target gene activation in a context-dependent manner (49). Missense mutations in the N-terminus trigger the usage of an alternative translational start site, yielding a mutant that is strongly associated with the development of transient myeloproliferative disease and acute megakaryoblastic leukemia (50–52).
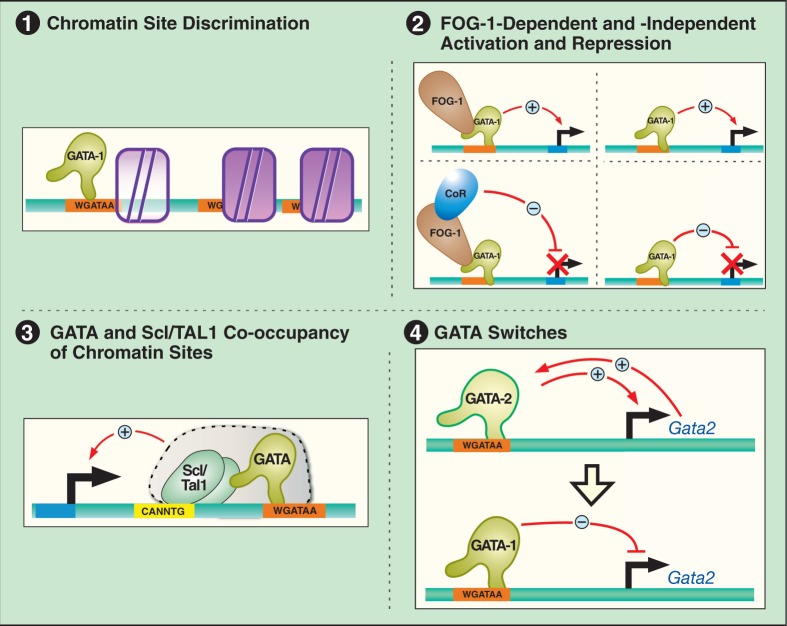
Despite approximately 7 million GATA motifs in the human genome, all capable of forming high-affinity complexes with GATA factors and naked DNA in vitro, GATA-1 and GATA-2 occupy only 0.1–1% of these motifs in erythroblasts, based on chromatin immunoprecipitation coupled with massively parallel sequencing and real-time PCR validation (14,53,54). While the molecular determinants for this exquisite discrimination are not fully understood (55), FOG-1 facilitates GATA-1 occupancy at a subset of chromatin sites (56,57). Genome-wide analysis of cis-elements residing at endogenous GATA-1 and GATA-2 occupancy sites led to refinement of the GATA consensus from WGATAR to WGATAA. However, the percent of total WGATAA motifs occupied remains very low. Beyond GATA motif sequence composition, the most rudimentary determinant of chromatin occupancy, diagnostic patterns of histone posttranslational modifications demarcate occupied versus unoccupied sites, both containing conserved GATA motifs (53,58–60) (Figure 1, Principle 1). In principle, the unique epigenetic signature of occupied sites may represent primed chromatin structures recognized by GATA-1 as a pivotal determinant of site selection. Alternatively, the signature may arise as a consequence of GATA-1 chromatin occupancy, followed by recruitment of GATA-1 coregulators that modify chromatin surrounding the occupancy site.
Figure 1.
GATA factor mechanistic principles. The models depict mechanistic principles derived from studies of GATA-1 and GATA-2. While the fundamental nature of these principles is likely to be shared by other GATA factors, additional GATA factor-specific mechanistic permutations are expected. Principle 1: GATA factors occupy a very small percent of the WGATAA motifs in a genome (<1%), suggesting that crucial mechanisms exist that control the discrimination among these highly abundant motifs. However, such mechanisms are not firmly established. The model depicts the occlusion of select GATA motifs, thus creating an obligate requirement for chromatin remodeling/modification reactions to increase access of the WGATAA residues required for GATA factor binding and/or to selectively occlude the vast majority of sites. At certain sites, FOG-1 (56,57) and GATA-1 acetylation (95) enhance chromatin access. Presumably, a host of regulatory factors mediate the essential process of establishing/maintaining accessible and occluded sites. Principle 2: GATA factors activate and repress target genes via multiple mechanisms, including with or without FOG-1 (36). Presumably, this mechanistic diversity reflects the specific chromatin architecture at a genetic locus, the subnuclear environment in which the locus resides and the regulatory mileau characteristic of the specific environment. Principle 3: GATA-1 and GATA-2 commonly co-localize with Scl/TAL1, another master regulator of hematopoiesis (96), at chromatin sites. The model illustrates GATA factor and Scl/TAL1 occupancy of a composite element consisting of an E-box and a WGATAA motif, which was originally described by Wadman et al. (76). Similar to the description above, only a very small percentage of composite elements are occupied by GATA factors in cells (53,58). As co-localization does not require the E-box (72), there is much to be learned about the biochemical nature of the GATA factor and Scl/TAL1 interaction. However, the co-localization measured by ChIP often correlates with transcriptional activity (54,58,72). Principle 4: GATA switches are defined as a molecular transition in which one GATA factor replaces another from a chromatin site, which is often associated with an altered transcriptional output. The GATA switch depicted reflects that occurring at the Gata2 locus during erythropoiesis, in which GATA-1 displaces GATA-2 from chromatin, which rapidly instigates repression (87). Context-dependent GATA switches may either activate or repress transcription and, in certain cases, may sustain the original transcriptional output (36).
GATA-1 chromatin occupancy leads to either activation or repression of target genes, both of which can be mediated by FOG-1 (36) (Figure 1, Principle 2). One mode of FOG-1 function involves interaction of its N-terminus with the NuRD chromatin remodeling complex (61–63), which can mediate both repression and activation. GATA-1 utilizes FOG-1 to induce higher order chromatin loops, based on chromosome conformation capture (3C) data (64–66). In principle, such loops can mediate activation or repression, dependent upon the physical relationship between the loop and functional features of a gene and the precise nature of the structure formed. GATA-1 also recruits the chromatin remodeler BRG1 to chromatin, which can mediate higher order looping (67–69).
Additional GATA-1 mechanisms exist, including FOG-1-independent activation and repression (41,70,71), although these mechanisms remain poorly understood. GATA-1 commonly co-localizes on chromatin with the stem cell leukemia/T-cell acute lymphocytic leukemia-1 (Scl/TAL1) protein (58,59,72), and the co-localization commonly correlates with transcriptional activity (54,58,72) (Figure 1, Principle 3). Scl/TAL1 is a master regulator of hematopoiesis that binds E-boxes and non-DNA-binding components including LMO2, LDB1, ETO2, and single-stranded DNA-binding proteins (73–79). In the context of naked DNA, optimal composite elements that support complex formation contain an E-box, a downstream GATA motif, and an 8-bp spacer (76). The 8-bp spacing is crucial for GATA-2-dependent enhancer activity in a transient transfection assay using cells expressing endogenous GATA-2 (58). However, GATA-1 and Scl/TAL1 also co-localize at certain chromatin sites lacking composite elements (59,72). Notably, the additional protein constituents of the complex modulate its transcriptional regulatory activity in a context-dependent manner (80) and are linked to the development and/or progression of human hematologic malignancies (81–84). Sophisticated ChIP-seq analyses in the HPC-7 multipotent hematopoietic cell line demonstrated that additional components co-localized with GATA-2 and Scl/TAL1 (85). This analysis revealed 1015 regions of 200 bp or less in which Scl/TAL1, LYL1, LMO2, GATA-2, ERG, FLI-1 and RUNX1 occupancy was detected. As each of these factors is likely to engage additional important partners, considerably more work is required to understand the structure/function of these higher order chromatin complexes containing multiple master regulators of hematopoiesis.
GATA factor interplay appears to be a common mechanism for controlling developmental processes (36,86). During the development of erythrocytes, GATA-1 displaces GATA-2 from chromatin sites at target genes, and this GATA switch (defined as an exchange of different GATA factors at a chromatin site) is tightly coupled to an altered transcriptional output (53,87–90) (Figure 1, Principle 4). GATA switches were first described at the Gata2 locus, at which GATA-1 binding instigates repression, thus explaining the differential GATA-1 and GATA-2 expression pattern during erythropoiesis (36,87). GATA-1 utilizes FOG-1 to displace GATA-2 from chromatin (56). The capacity of FOG-1 to bind the NuRD complex is required for the GATA switch, as GATA switches were impaired in a knock-in mouse strain expressing FOG-1 defective in NuRD complex binding (62). Ectopic FOG-1 expression in mast cell progenitors induces a GATA switch in which GATA-1 replaces GATA-2 from the –2.8 kb GATA switch site of the Gata2 locus, which was linked to Gata2 repression and generation of erythroid, megakaryocytic and granulocytic progeny (97). During the differentiation of trophoblast giant cells, GATA-2 displaces GATA-3 at Gata2, which is associated with transcriptional activation (91). Though GATA switches have not been studied in many systems, it is attractive to propose that they represent common devices to change transcriptional activity in diverse biological contents.
Two aspects of the GATA switch paradigm merit careful consideration. First, erythroid GATA switches inform us that different GATA factors can exert qualitatively distinct functions through an identical chromatin site; one GATA factor mediates target gene activation, while the other confers repression or vice versa. Thus, while different GATA factors share certain biochemical attributes, including their highly conserved zinc finger module (92), intrinsic differences underlie the qualitatively distinct activities. A notable difference is the relative high and low stabilities of GATA-1 and GATA-2, respectively (93,94). As proteasome inhibition stabilizes GATA-2 and blocks GATA switches, the low stability appears to be an important determinant of GATA switches (93). Another important implication of the GATA switch paradigm is that GATA switches and the requisite factors/signals that control the switches represent a novel tool to control developmental processes. Since certain non-hematopoietic cell types can express multiple GATA factors, it would not be surprising if the erythroid GATA switch mechanism were applicable to non-hematopoietic contexts. Despite major progress in elucidating GATA factor mechanistic principles, many questions remain unanswered regarding how cellular signaling pathways dynamically control GATA factor activities and GATA factor-dependent biological processes.
In summary, GATA factor mechanistic principles (Figure 1) include: (1) GATA factors target a small subset of chromatin sites containing a cis-element with the consensus sequence WGATAA; (2) GATA-1 activates or represses target genes in a FOG-1-dependent or -independent manner; (3) GATA-1 and GATA-2 commonly co-occupy chromatin sites with Scl/TAL1, and members of the Scl/TAL1 complex promote or suppress GATA factor-regulated transcription in a context-dependent manner; and (4) GATA switches can involve qualitatively distinct activities of different GATA factors through an identical chromatin site.
REGULATING GATA FACTORS POSTTRANSLATIONALLY
While multiple posttranslational modifications are implicated in regulating GATA factor function, progress on defining the respective mechanisms does not seem to be commensurate with the level of activity in the field. Common themes have not emerged regarding how posttranscriptional mechanisms regulate different GATA factors. Furthermore, the precise impact of most posttranslational modifications on GATA factor activities, including chromatin occupancy, coregulator recruitment, GATA switches and higher order chromatin transitions at endogenous loci is unknown.
GATA-1 harbors seven serines that can be phosphorylated in cultured cells (98). Six of these serines (S26, S49, S72, S142, S178 and S187) reside in the N-terminal region, while another (S310) is near the C-finger. S72, S142 and S310 are conserved among multiple species. Whereas six serines in the N-terminal region are constitutively phosphorylated, S310 phosphorylation is elevated upon dimethyl sulfoxide (DMSO)-induced differentiation of mouse erythroleukemia (MEL) cells (98). Substitution of all seven serines with alanines does not affect GATA-1 binding to naked DNA or transactivation activity in a non-erythroid cell transient transfection assay (98). S310 resides in the region implicated in DNA bending, based on GATA-1 C-finger peptide binding to DNA (38), but S310 mutations do not affect DNA bending (98). Though mutation of S310 blocks fetal liver erythroid progenitor cell maturation (99), mice bearing alanine substitutions at S72, S142 and S310 exhibit a normal phenotype, save moderately decreased erythroid burst-forming unit (BFU-E) and erythroid colony-forming unit (CFU-E) in bone marrow (100). Phosphorylation of these residues is therefore either not essential for murine erythropoiesis or undefined mechanisms compensate for loss of phosphorylation sites in vivo. Treatment of K562 cells with hemin, sodium butyrate (NaB) or N-acetylcysteine increases GATA-1 phosphorylation and enhances DNA binding in vitro, but the phosphorylated residues mediating this effect are unknown (101). Mitogen-activated protein kinase (MAPK)-mediated phosphorylation of S26 in interleukin 3 (IL-3)-dependent Ba/F3 hematopoietic cells increases expression of E4bp4 and Bcl-XL survival genes in a transient transfection assay (102). Erythropoietin induces S310 phosphorylation via phosphatidylinositol 3-kinase (PI3K)/Akt (103), and this enhances expression of TIMP-1, which encodes tissue-inhibitor of metalloproteinase-1 (103). Since multiple kinases phosphorylate GATA-1, and GATA-1 phosphorylation is regulated in distinct contexts, it is attractive to consider how extracellular stimuli, such as hematopoietic cytokines, instigate cellular signaling mechanisms that orchestrate GATA factor function in physiological and pathophysiological states. However, the triple phosphorylation site knockin mouse described above did not reveal compelling insights in this regard.
IL-3 induces GATA-2 phosphorylation in hematopoietic progenitor cell lines, which is dependent upon MAPK. However, the phosphorylated residues were not described (104). In transiently transfected COS cells, GATA-2 phosphorylation does not affect reporter gene activity (104). Insulin treatment of HEK293 cells stimulates PI3-K/Akt signaling, which induces GATA-2 phosphorylation at serine 401 (105). Serine 401 phosphorylation was reported to impair nuclear translocation, based on overexpression of the mutant in HEK293 cells (105). In addition, naked DNA-binding studies suggested that serine 401 phosphorylation impairs GATA-2 DNA-binding activity (105). Additional work is required to discover the full ensemble of GATA-2 phosphorylation sites, relevant kinases and functional consequences of phosphorylation in distinct cell types in vivo.
Analogous to phosphorylation, posttranslational acetylation of the ε-amino group of lysine represents a common mode of controlling protein structure/function (106–109). Acetylation of histone and non-histone proteins (110) is mediated by a host of histone acetyltransferases (HATs) or histone deacetylases (HDACs). Through recruitment to chromatin via binding trans-acting factors (111), HATs acetylate the N-terminal flexible tails of core histones in nucleosomes at specific genetic loci. Molecular consequences of histone acetylation include neutralizing the lysine positive charge, which reduces the histone affinity for DNA and increases cis-element accessibility to their cognate binding protein. Histone acetylation can also increase chromatin accessibility by opposing higher order chromatin folding (112). Finally, acetyl-lysine binds a protein module termed a bromodomain (113), thus creating a platform for protein recognition (114,115).
GATA factors contain multiple acetylation sites located predominantly within their zinc finger regions. The Adenovirus E1A-binding region of the HATs CREB-binding protein (CBP) (116) and its paralog p300 (117) bind and acetylate the GATA-1 C-finger (118). Studies with the CBP/p300 inhibitor E1A provided evidence for an important role of CBP/p300 in erythroid maturation and gene regulation (118). Two lysine-rich motifs (amino acids 243–246 and 312–315) at the C-terminus of the GATA-1 zinc fingers are acetylated (119). GATA-1 acetylation facilitates transactivation in transient transfection assays (119) and promotes GATA-1 chromatin occupancy (95). Acetylated GATA-1 binds and recruits Bromodomain Protein 3 (BRD3) to chromatin (120). As a small molecule inhibitor that antagonizes this interaction reduces GATA-1 and BRD3 chromatin occupancy and decreases erythroid maturation of G1E-ER4 cells, it will be interesting to further explore the mechanistic and biological implications of this interaction. GATA-1 recruits CBP/p300 to chromatin sites, including the β-globin LCR and βmajor promoter, and presumably this underlies GATA-1-dependent induction of H3 and H4 acetylation at these sites (121–123).
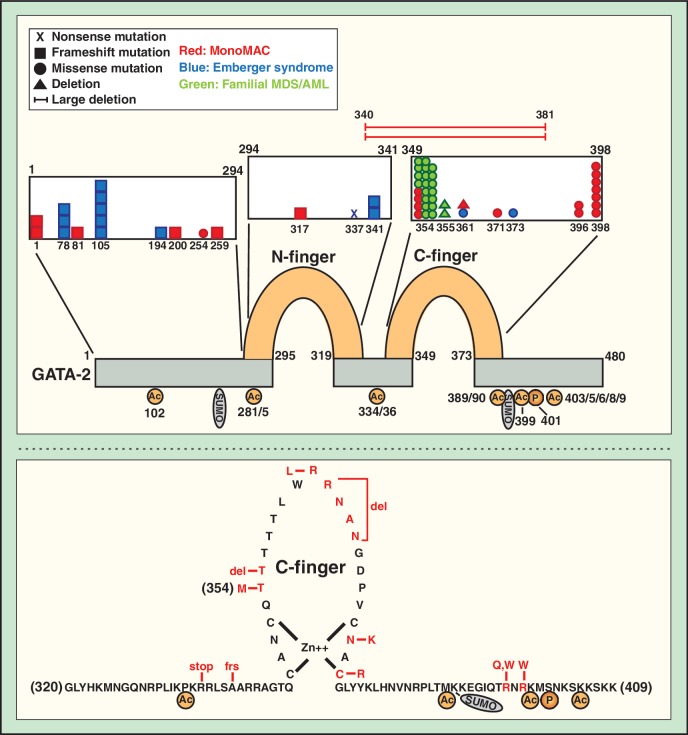
GATA-2 is acetylated at K102 within the N-terminal region and at multiple additional lysines within the zinc finger module including K281, 285, 334, 336, 389, 390, 399, 403, 405, 406, 408 and 409 (124) (Figure 2). p300-mediated acetylation of GATA-2 in hematopoietic cells enhances its DNA binding and transactivation activities in a transient transfection assay and inhibits GATA-2-mediated growth inhibition (124). A GATA-2 mutant lacking four lysine acetylation sites, C-terminal to the C-finger, was unable to rescue primitive erythropoiesis in GATA-2 morphant Xenopus tadpoles (125). In this system, Ca+2-calmodulin-dependent kinase-4 signaling inhibits GATA-2 acetylation and function (125). Thus, signal-dependent control of GATA-2 acetylation appears to represent an important mode of regulating GATA-2 activity. HDAC3 and HDAC5, but not HDAC1, bind GATA-2, suppressing GATA-2 transactivation activity in HEK293T cells (126).
Figure 2.
GATA-2 mutations in human hematologic disorders (143–146). Specific GATA-2 mutations in human patients are indicated, with details denoted in the legend. Each symbol represents a single patient with the particular mutation. The diagram of GATA-2 protein organization illustrates the N- and C-fingers, acetylation sites (124), the serine 401 phosphorylation site and two sumoylation consensus motifs. The diagram at the bottom illustrates the amino acid sequence composition of the C-finger and neighboring regions, with the positions of disease mutations highlighted. Stop, mutation that creates stop codon; frs, frameshift mutation; del, deletion.
Certain posttranslational modifications involve the conjugation of small proteins, including ubiquitin and related small ubiquitin-related modifier (SUMO) proteins, to recipient proteins. The four vertebrate SUMO proteins are ∼10 kDa and structurally resemble ubiquitin (127,128). While SUMO-2 and SUMO-3 share >90% sequence identity, SUMO-1 is only 50% identical to SUMO-2/3 (129). SUMO-4 has sequence similarity to SUMO-2, but endogenous SUMO-4 has not been detected (130). Sumoylation, which covalently links SUMO to a lysine within a target protein, is reversible and dynamically regulated (131). Sumoylation involves an enzymatic cascade, analogous to ubiquitination (132). The E1 activating enzyme Aos1-Uba2 forms a thioester bond with SUMO in an ATP-dependent reaction and subsequently transfers SUMO to the E2 conjugating enzyme Ubc9. An E3 ligase facilitates the transfer of SUMO to its substrate and an isopeptide bond is formed between the C-terminal glycine residue and the ε–amino group of a lysine residue of the acceptor protein. Conjugating enzymes and SUMO-specific proteases regulate the level of sumoylation. Six mammalian sentrin/SUMO-specific protease (SENP) homolog (SENP1–3, SENP5–7) have been identified (133). Whereas polyubiquitination triggers proteasome-mediated proteolysis, sumoylation commonly controls protein–protein interactions by regulating the activity, localization and stability of target proteins, masking an existing binding site, occluding a site for a distinct modification or providing an interface for interaction with proteins containing a SUMO-interacting/binding motif (SIM/SBM) (134).
GATA-1, GATA-2 and GATA-4 sumoylation have been described (135–137). Though most sumoylation substrates contain the consensus motif ΨKXE (138) (Ψ, large hydrophobic amino acid; X, any amino acid), some SUMO targets lack this consensus, and experimental analysis is required to determine whether a consensus is a bona fide sumoylation site in vivo. GATA-1 is sumoylated at K137, which is embedded in a sumoylation consensus, within the N-terminal region. The SUMO ligase PIASy can sumoylate K137 (136). Initial analyses using a transient transfection assay in non-erythroid cells and a Xenopus animal cap explant assay suggested that the K137R mutant and wild-type GATA-1 have similar activities (136). PIASy binds GATA-1 and was reported to repress GATA-1-mediated transactivation via a K137-independent mechanism in a transient transfection assay with overexpressed factors. In genetic complementation analysis in GATA-1-null erythroid precursor (G1E) cells expressing GATA-1 fused with an estrogen-receptor ligand-binding domain (ER-GATA-1) at near physiological levels, K137 sumoylation promotes GATA-1-mediated transcriptional regulation (both activation and repression) at a subset of endogenous GATA-1 target genes (139). SUMO-dependent genes are predominantly FOG-1-dependent targets. The GATA-1 V205G mutant, defective in FOG-1 binding, yields molecular phenotypes similar to the K137R mutant. Furthermore, SUMO-and FOG-1-dependent genes migrate away from the nuclear periphery upon GATA-1-induced erythroid maturation, while SUMO- and FOG-1-independent genes persist at the periphery (139). The use of tiled bacterial artificial chromosome probes revealed that sumoylation endows GATA-1 with the capacity to expel the β-globin locus from the nuclear periphery without inducing gross changes in the positioning of neighboring chromosomal regions (140). Given these mechanistic insights, it is of considerable interest to investigate how SUMO-specific proteases fit into the GATA factor regulatory circuitry. SENP1 knockout mice die from severe anemia between E13.5 and postnatal day 1 (141). SENP1 knockout mice exhibit hematopoietic defects in the fetal liver, which correlate with accumulation of sumoylated GATA-1, as well as hypoxia-inducible factor-1α. As SENP1 desumoylates a broad spectrum of substrates, the hematopoietic defects presumably reflect the aggregate actions of this broad activity, presumably including FOG-1, which is sumoylated in erythroid cells (142).
GATA-2 interacts with PIASy in transfected COS cells, which preferentially conjugates SUMO-2 to GATA-2 (135). In a transient transfection assay in endothelial cells, PIASy suppresses GATA-2 transcriptional activity at the endothelin-1 (ET-1) promoter. Whereas the repression requires the GATA-2-PIASy interaction, the PIASy RING-like domain with SUMO ligase activity is dispensable, indicating that PIASy regulates GATA factor activity independent of sumoylation. While GATA-2 contains two potential sumoylation sites (human amino acids 221–224 and 388–391) that conform to the consensus (Figure 2), the sumoylation site has not been described. Further analysis is required to elucidate the function of GATA-2 sumoylation at endogenous loci. GATA-4 is sumoylated at K366 in the C-terminal region (137). Based on the initial evidence for functional significance of at least certain GATA-1 and GATA-2 posttranslational modifications, it is attractive to propose that signal-dependent targeting of GATA factors represents a canonical mode of regulating hematopoiesis. By contrast to well-established cytoplasmic to nuclear signaling paradigms, many questions remain unanswered regarding the nature of the signaling pathways that target GATA factors, the precise molecular consequences of the posttranslational modifications and how dysregulated signaling, often a hallmark of hematologic malignancies, influences GATA factor activity.
HUMAN PATHOPHYSIOLOGIES CAUSED BY GATA-2 DYSREGULATION
Given the essential GATA-2 function to promote hematopoiesis, alterations in GATA-2 levels/activity would be expected to initiate and/or promote the development of hematologic malignancies. However, until recently, only circumstantial evidence implicated GATA-2 in human cancers. Four related human disease syndromes harboring germline mutations in GATA2 are associated with an increased incidence of myeloid neoplasia, either myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML). Three of the four syndromes—Monocytopenia/Mycobacterium avium complex (MonoMAC) (143,147), Dendritic cell, monocyte, B and Natural Killer Lymphoid deficiency (DCML deficiency) (144) and Emberger’s syndrome (145,148)—share the hallmark feature of immune dysfunction with an increased propensity to develop MDS or AML. The fourth, a familial MDS/AML, lacks the immune dysfunction and systemic symptomatology characteristic of the other cases (146). Though these entities appear to be rare, their study provides new insight into the role of GATA2 in immune function and in the regulation of growth, maturation and apoptosis in the myeloid compartment.
Sequencing of candidate gene exons in the genomic DNA of pedigrees of familial MDS/AML revealed a T354M GATA2 mutation in three pedigrees and a T355del in one pedigree (146). This analysis also revealed several pedigrees harboring mutations in RUNX1 or CEBPA, disease genes for familial AML (149). Molecular modeling suggested that T354 and T355 stabilize the GATA-2 C-finger (146). The T354M mutant exhibits lower affinity for DNA, reduced transactivation activity and is less effective in synergizing with PU.1 to activate the CSF1R (Fms) promoter in a transient transfection assay (146). When tested for the ability to block ATRA-induced maturation and apoptosis of HL-60 cells, the T355del mutant acted as a null, while the T354M mutant blocked ATRA effects, consistent with expectations for a leukemogenic oncogene. Based on gene expression profiling in HL-60 cells, the T354M and T355del mutants appeared to be null alleles (146). Two additional pedigrees harboring the T354M mutation were recently reported (150,151). Further mechanistic analysis is required to rigorously analyze the function of these disease mutants at endogenous loci and in diverse cellular contexts.
Holland’s group had previously described a syndrome of monocytopenia with susceptibility to opportunistic infections by the Mycobacterium avium complex, termed MonoMAC (143,147), which occurs in both sporadic and autosomal dominant familial form. The immune deficiencies were significant: patients suffered from disseminated cutaneous human papilloma virus infection, aspergillosis, histoplasmosis or cryptococcal meningitis. In addition, some suffered from pulmonary alveolar proteinosis, which is typically associated with macrophage dysfunction. Patients had markedly diminished circulating monocytes (10 cells per microliter, average), as well as B cells and natural killer (NK) cells; T cells were variable (147). Other causes typically associated with the particular spectrum of immune defect seen in these patients (e.g. HIV infection, IL12/IL23/IFNg or NF-κB dysfunction) were ruled out (147).
The frequency of myeloid neoplasia (MDS or AML) in the combined familial and sporadic cases of MonoMAC was 50%; these were associated with trisomy 8, monosomy 7 and dicentric chromosome 6 (147). Given the GATA2 mutations in familial MDS/AML (146), the authors investigated such mutations in the MonoMAC kindreds, 13 of the 16 they originally reported. This revealed frameshift mutations (G81fs, M1del290, D259fs and N317fs), a deletion spanning the N- and C-fingers (D340–381), a small deletion in the C-finger (Δ362–365) and missense mutations within the GATA-2 C-finger (T354M, N371K, R396W, R396Q, R398W) (143). In addition, one missense mutation occurred outside the finger region: P254L.
Bigley et al. (152) further characterized the immune deficiency in four patients with MonoMAC syndrome, referring to it as DCML deficiency. DCML is associated with the absence of granulocyte monocyte progenitors (GMPs) and common lymphoid progenitors (CLPs) (152). As expected, with long latency, DCML can lead to MDS (153,154). Exome sequencing revealed GATA2 mutations in these four patients: a frame shift after amino acid 200; a T354M missense mutation; a R398W missense mutation and a D340–381 deletion spanning the C-terminal end of the N-finger, extending into the C-finger (144).
Emberger’s syndrome is characterized by lymphedema with myelodysplasia progressing to AML, as well as immune dysfunction (widespread cutaneous warts and sensoneural deafness) (148). Whole-exome sequencing of three individuals with Emberger syndrome revealed GATA2 mutations, and further analysis of additional cases of Emberger syndrome (four more sporadic cases and additional individuals from the two affected kindreds) revealed additional GATA2 mutations (145). These mutations span from the N-terminus to the C-finger, and include five frame shift mutations, one nonsense mutation and two missense mutations within the C-finger (R361L and C373R) (145). Using a luciferase reporter bearing a GATA-2-responsive CD34 promoter construct transfected into HEK293 cells, they demonstrated that the R361L and C373R mutants have a decreased capacity to transactivate the reporter. However, the molecular basis for the defective activity was not established.
It is instructive to consider the mechanistic basis of the immune deficiency in MonoMAC/DCML deficiency and Emberger’s syndrome. It is unlikely that the myeloid neoplasia in these patients yields immune dysfunction, since the spectrum of opportunistic infections is distinct from that seen in the neutropenia of MDS, but is similar to that seen in IRF8 deficiency and IFNgamma/IL-12 deficiency. There are several clues that through its interaction with PU.1 (155), GATA-2 plays an important role in monocyte/macrophage/dendritic cell development. First, GATA-2 controls phagocytosis by pulmonary alveolar macrophages (156), and pulmonary alveolar proteinosis is a feature of the MonoMAC syndrome. Second, with haploinsufficient GATA-2 mice, there is loss of lymphoid and monocytic cells with retention of granulocytes (32). Furthermore, there are specific defects in the granulocyte/macrophage progenitor pool (157). GATA-2 interacts with and represses PU.1 (158), which induces c-Fms and Flt3 expression (159,160). The common developmental origin for macrophage/dendritic cells and lymphoid cells (161) may tie together the deficiencies in B and NK cells seen in MonoMAC patients. In addition to PU.1, GATA-2 can bind C/EBP-α (162), a key regulator of granulopoiesis. Whether GATA-2 disease mutations affect this interaction, thereby altering C/EBP-α function, has not been addressed.
Regarding mechanisms underlying the pathogenic activities of mutant GATA2 alleles, do the proteins function as null alleles? Dickinson et al. posit that the frameshift and 42 amino acid deletions act as null alleles, though N-terminal frameshifts in GATA1 (associated with AML) can be translated through initiation at a downstream codon, resulting in the production of an N-terminally truncated mutant with reduced activity (50); whether this happens with GATA-2 frameshift mutations is not known. Strikingly, nearly all the point mutations identified in these four papers localize to the GATA2 C-finger. Initial mechanistic analyses (146) suggested that the T354M and T355del alleles exhibit certain functional differences. While we propose that these mutations lead to haploinsufficiency, considerably more work is required to evaluate functional consequences of the mutations.
Are there links between the immune deficiency and MDS/AML? It was suggested that the increased incidence of MDS in patients with GATA-2 mutation-induced MonoMAC syndrome may result from defective regulation of HSC proliferation: in the face of systemic immune dysfunction and recurrent infection, the stem cell compartment is stimulated to proliferate, and since committed progenitors are also dysfunctional due to GATA-2 haploinsufficiency, this leads to chronic stress on the HSC compartment, HSC exhaustion and MDS (163). GATA-2 haploinsufficient mice have a decreased stem cell pool, with a higher percentage of quiescent LSK cells and increased apoptosis (32). The immune deficiency and MDS/AML may not be linked, since GATA-2 mutations alone, independent of immune dysfunction, may lead to MDS/AML (146). The presence of chromosomal aberrations (trisomy 8 and 21, monosomy 7 and dicentric chromosome 6 (147) suggest that additional events are needed for malignancy to develop and/or GATA-2 mutations increase the likelihood of these mutagenic events occurring. These points may explain the long latency for the development of this set of syndromes and the low penetrance of MDS/AML development. Alternatively, the long latency and variable penetrance may be due to the time required for subtle deregulation of hematopoiesis to segue to a full-blown malignancy.
GATA-2 expression can be upregulated in AML, and this portends a poor prognosis (164–167). By contrast, GATA-2 can be downregulated in murine retroviral transplant models of AML, and forced GATA-2 expression did not sustain leukemic cell growth (168). GATA2 has been identified as a common site of proviral insertional activation in AMLs occurring in retrovirally infected NUP98-HOXD13 transgenic mice (169). Zhang et al. (170) identified a gain-of-function GATA-2 mutation occurring in the accelerated phase (AP) and blast crisis (BC) phase of CML (170). They analyzed 85 cases of CML in either accelerated phase or blast crisis for genetic alterations in any of 13 candidate genes of known importance to hematopoiesis or tumor progression. Eleven of these had mutations in Runx1 (AML1), while eight had a L359V mutation. Another case had a six amino acid internal deletion (D341–346). Both these reside in the C-finger. Kaplan–Meier-type analysis of patients with the L359V mutation showed a statistically significant shortening of time to disease progression. Though more work needs to be done to establish the mechanism, Zhang et al. provide initial evidence that the L359V mutant acts as a dominant-active allele: it appears to bind naked DNA more tightly; to bind to PU.1 more tightly, and to more strongly inhibit PU.1-mediated transactivation on target promoters; it had a modest inhibitory effect on vitamin D3 and all-trans retinoic acid differentiation of HL-60 cells and caused BCR-ABL-transduced primary hematopoietic cells to take on a more monocytic phenotype (170). Analysis of 688 DNA samples from patients with non-CML-AP/BC hematologic malignancies failed to identify sequence alterations in GATA2, emphasizing that the L359V mutation is specific to AP/BC phase of CML (171).
CONCLUDING REMARKS
Despite the emergence of compelling mechanistic principles, knowledge of how cellular signaling pathways regulate GATA factor activity remains primitive. Taken together with the many unanswered questions regarding how GATA-2 coding region mutations dysregulate GATA-2 function, major efforts are required to construct a lucid picture of how physiological GATA-2 activity suppresses the development of MDS and leukemia. Elucidating the cell type-specific regulatory circuitry in which GATA-2 is embedded, and careful scrutiny of the dynamics of this circuitry, will reveal key regulatory components/nodes that offer opportunities for innovative and efficacious targeted interventions. Given the rapidity of progress vis-à-vis linking GATA-2 dysregulation to hematologic pathophysiologies, and the ever-increasing ease in conducting whole-genome analyses, further rigorous explorations into this rich pipeline will almost certainly yield pivotal insights into the molecular basis of the human diseases discussed and more.
FUNDING
National Institutes of Health (NIH) (DK68034, DK50107 to E.H.B., CA120313 to A.S.P.). H.-Y.L. was the recipient of an American Heart Association Predoctoral Fellowship. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Evans T, Reitman M, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc. Natl Acad. Sci. USA. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto M, Ko LJ, Leonard MW, Beug H, Orkin SH, Engel JD. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 1990;4:1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- 3.Ho IC, Vorhees P, Marin N, Oakley BK, Tsai SF, Orkin SH, Leiden JM. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. 1991;10:1187–1192. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorfman DM, Wilson DB, Bruns GA, Orkin SH. Human transcription factor GATA-2. Evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J. Biol. Chem. 1992;267:1279–1285. [PubMed] [Google Scholar]
- 5.Joulin V, Bories D, Eleouet JF, Labastie MC, Chretien S, Mattei MG, Romeo PH. A T-cell specific TCR delta DNA-binding protein is a member of the human GATA family. EMBO J. 1991;10:1809–1816. doi: 10.1002/j.1460-2075.1991.tb07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zon LI, Mather C, Burgess S, Bolce ME, Harland RM, Orkin SH. Expression of GATA-binding proteins during embryonic development in Xenopus laevis. Proc. Natl Acad. Sci. USA. 1991;88:10642–10646. doi: 10.1073/pnas.88.23.10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol. Cell. Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-6: a zinc finger transcription factor that is expressed in multple cell lineages derived from lateral mesoderm. Dev. Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 9.Lee ME, Temizer DH, Clifford JA, Quertermous T. Cloning of the GATA binding protein that regulates endothelin-1 gene expression in endothelial cells. J. Biol. Chem. 1991;266:16188–16192. [PubMed] [Google Scholar]
- 10.Orkin SH. Transcription factors and hematopoietic development. J. Biol. Chem. 1995;270:4955–4958. doi: 10.1074/jbc.270.10.4955. [DOI] [PubMed] [Google Scholar]
- 11.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 12.Charron F, Nemer M. GATA transcription factors and cardiac development. Semin. Cell. Dev. Biol. 1999;10:85–91. doi: 10.1006/scdb.1998.0281. [DOI] [PubMed] [Google Scholar]
- 13.Song H, Suehiro J, Kanki Y, Kawai Y, Inoue K, Daida H, Yano K, Ohhashi T, Oettgen P, Aird WC, et al. Critical role for GATA3 is mediating Tie2 expression and function in large vessel endothelial cells. J. Biol. Chem. 2009;284:29109–29124. doi: 10.1074/jbc.M109.041145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linnemann AK, O'Geen H, Keles S, Farnham PJ, Bresnick EH. Genetic framework for GATA factor function in vascular biology. Proc. Natl Acad. Sci. USA. 2011;108:13641–13646. doi: 10.1073/pnas.1108440108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Li W, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependewnt prostate cancer growth. Mol. Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craven SE, Lim KC, Ye W, Engel JD, de Sauvage F, Rosenthal A. Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development. 2004;131:1165–1173. doi: 10.1242/dev.01024. [DOI] [PubMed] [Google Scholar]
- 18.Nardelli J, Thiesson D, Fujiwara Y, Tsai F-Y, Orkin SH. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev. Biol. 1999;210:305–321. doi: 10.1006/dbio.1999.9278. [DOI] [PubMed] [Google Scholar]
- 19.Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D'Agati V, Orkin SH, Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 20.Simon MC, Pevny L, Wiles MV, Keller G, Costantini F, Orkin SH. Rescue of erythroid development in gene targeted GATA-1- mouse embryonic stem cells. Nat. Genet. 1992;1:92–98. doi: 10.1038/ng0592-92. [DOI] [PubMed] [Google Scholar]
- 21.Weiss MJ, Orkin SH. Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc. Natl Acad. Sci. USA. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl Acad. Sci. USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in mekagaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Migliaccio AR, Rana RA, Sanchez M, Lorenzini R, Centurione L, Bianchi L, Vannucchi AM, Migliaccio G, Orkin SH. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. J. Exp. Med. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J. Exp. Med. 2002;195:1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 27.Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 28.Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 29.Tsai F-Y, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- 30.Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J. Exp. Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujiwara Y, Chang AN, Williams AM, Orkin SH. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103:583–585. doi: 10.1182/blood-2003-08-2870. [DOI] [PubMed] [Google Scholar]
- 32.Rodrigues NP, Janzen V, Forkert R, Dombkowski DM, Boyd AS, Orkin SH, Enver T, Vyas P, Scadden DT. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem cell homeostasis. Blood. 2005;106:477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- 33.Persons DA, Allay JA, Allay ER, Ashmun RA, Orlic D, Jane SM, Cunningham JM, Nienhuis AW. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood. 1999;93:488–499. [PubMed] [Google Scholar]
- 34.Evans T, Felsenfeld G. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- 35.Tsai SF, Martin DI, Zon LI, D'Andrea AD, Wong GG, Orkin SH. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 36.Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J. Biol. Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin DI, Orkin SH. Transcriptional activation and DNA binding by the erythroid factor GF- 1/NF-E1/Eryf 1. Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 38.Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- 39.Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 40.Tsang AP, Fujiwara Y, Hom DB, Orkin SH. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of altered specificity mutants to probe a specific protein–protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- 42.Fox AH, Liew C, Holmes M, Kowalski K, MacKay JP, Crossley M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 1999;18:2812–2822. doi: 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trainor CD, Omichinski JG, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high- affinity interaction. Mol. Cell. Biol. 1996;16:2238–2247. doi: 10.1128/mcb.16.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S-I, Bresnick EH. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene. 2007;26:6777–6794. doi: 10.1038/sj.onc.1210761. [DOI] [PubMed] [Google Scholar]
- 45.Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 2007;32:63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 46.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol. Cell. Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stumpf M, Waskow C, Krotschel M, van Essen D, Rodriguez P, Zhang X, Guyot B, Roeder RG, Borggrefe T. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc. Natl Acad. Sci. USA. 2006;103:18504–18509. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson KD, Kim S-I, Bresnick EH. Differential sensitivities of transcription factor target genes underlie cell type-specific gene expression patterns. Proc. Natl Acad. Sci. USA. 2006;103:15939–15944. doi: 10.1073/pnas.0604041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, LeBeau MM, Crispino JD. Acquired mutations in GATA-1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 51.Mundschau G, Gurbaxani S, Gamis AS, Greene ME, Arceci RJ, Crispino JD. Mutagenesis of GATA1 is an initiating event in Down syndrome leukemogenesis. Blood. 2003;101:4298–4300. doi: 10.1182/blood-2002-12-3904. [DOI] [PubMed] [Google Scholar]
- 52.Crispino JD. GATA-1 in normal and malignant hematopoiesis. Semin. Cell Dev. Biol. 2005;16:137–147. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann A, Kang Y-A, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bresnick EH, Johnson KD, Kim S-I, Im H. Establishment and regulation of chromatin domains: mechanistic insights from studies of hemoglobin synthesis. Prog. Nucl. Acids Res. Mol. Biol. 2006;81:435–471. doi: 10.1016/S0079-6603(06)81011-1. [DOI] [PubMed] [Google Scholar]
- 56.Pal S, Cantor AB, Johnson KD, Moran T, Boyer ME, Orkin SH, Bresnick EH. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc. Natl Acad. Sci. USA. 2004;101:980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letting DL, Chen YY, Rakowski C, Reedy S, Blobel GA. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc. Natl Acad. Sci. USA. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wozniak RJ, Keles S, Lugus JJ, Young K, Boyer ME, Tran TT, Choi K, Bresnick EH. Molecular hallmarks of endogenous chromatin complexes containing master regulators of hematopoiesis. Mol. Cell. Biol. 2008;28:6681–6694. doi: 10.1128/MCB.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen K-B, Zhang Y, Drautz D, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu W, Cheng Y, Keller CA, Ernst J, Kumar SA, Mishra T, Morrissey C, Dorman CM, Chen KB, Drautz D, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011;21:1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miccio A, Wang Y, Gregory GD, Wang H, Yu X, Choi JK, Shelat S, Tong W, Poncz M, Blobel GA. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 2010;29:442–456. doi: 10.1038/emboj.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao Z, Huang Z, Olivey HE, Gurbuxani S, Crispino JD, Svensson EC. FOG-1-mediated recruitment of NuRD is required for cell lineage re-inforcement during hematopoiesis. EMBO J. 2010;29:457–468. doi: 10.1038/emboj.2009.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24:67–78. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 65.Kim S-I, Bultman SJ, Jing H, Blobel GA, Bresnick EB. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol. Cell. Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol. Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S-I, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc. Natl Acad. Sci. USA. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu G, Schnoes DE, Cui K, Ybarra R, Northrup D, Tang Q, Gattinoni L, Restifo NP, Huang S, Zhao K. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 2011;21:1650–1658. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim SI, Bresnick EH, Bultman SJ. BRG1 directly regulates nucleosome structure and chromatin looping at the alpha globin locus to activate transcription. Nucleic Acids Res. 2009;37:6019–6027. doi: 10.1093/nar/gkp677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson KD, Boyer ME, Kim S-I, Kang SY, Wickrema A, Cantor AB, Bresnick EH. Friend of GATA-1-independent transcriptional repression: a novel mode of GATA-1 function. Blood. 2007;109:5230–5233. doi: 10.1182/blood-2007-02-072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pal S, Nemeth MJ, Bodine D, Miller JL, Svaren J, Thein SL, Lowry PJ, Bresnick EH. Neurokinin-B transcription in erythroid cells: direct activation by the hematopoietic transcription factor GATA-1. J Biol Chem. 2004;279:31348–31356. doi: 10.1074/jbc.M403475200. [DOI] [PubMed] [Google Scholar]
- 72.Tripic T, Deng W, Cheng Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated protein distinguish active from repressive GATA transcription factor complexes. Blood. 2008;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mikkola HK, Klintman J, Yang H, Hock H, Schlaeger TM, Fujiwara Y, Orkin SH. Hematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukemia SCL/tal-1 gene. Nature. 2003;421:547–551. doi: 10.1038/nature01345. [DOI] [PubMed] [Google Scholar]
- 74.Ema M, Faloon P, Zhang WJ, Hirashima M, Reid T, Stanford WL, Orkin S, Choi K, Rossant J. Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 2003;17:380–393. doi: 10.1101/gad.1049803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 76.Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu Z, Meng X, Cai Y, Liang H, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 2007;21:942–955. doi: 10.1101/gad.1528507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goardon N, Lambert JA, Rodriguez P, Nissaire P, Herblot S, Thibault P, Dumenil D, Strouboulis J, Romeo PH, Hoang T. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Begley CG, Aplan PD, Denning SM, Haynes BF, Waldmann TA, Kirsch IR. The gene SCL is expressed during early hematopoiesis and encodes a differentiation-related DNA-binding motif. Proc. Natl Acad. Sci. USA. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fujiwara T, Lee HY, Kumar S, Bresnick EH. Building multifunctionality in a complex containing master regulators of hematopoiesis. Proc. Natl Acad. Sci. USA. 2010;107:20429–20434. doi: 10.1073/pnas.1007804107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rabbitts TH, Axelson H, Forster A, Grutz G, Lavenir I, Larson R, Osada H, Valge-Archer V, Wadman I, Warren A. Chromosomal translocations and leukaemia: a role for LMO2 in T cell acute leukaemia, in transcription and in erythropoiesis. Leukemia. 1997;11(Suppl. 3):271–272. [PubMed] [Google Scholar]
- 82.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 83.Chyla BJ, Moreno-Miralles I, Steapleton MA, Thompson MA, Bhaskara S, Engel M, Hiebert SW. Deletion of Mtg16, a target of t(16:21), alters hematopoietic progenitor cell proliferation and lineage allocation. Mol. Cell. Biol. 2008;28:6234–6247. doi: 10.1128/MCB.00404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mellentin JD, Murre C, Donlon TA, McCaw PS, Smith SD, Carroll AJ, McDonald ME, Baltimore D, Cleary ML. The gene for enhancer binding proteins E12/E47 lies at the t(1;19) breakpoint in acute leukemias. Science. 1989;246:379–382. doi: 10.1126/science.2799390. [DOI] [PubMed] [Google Scholar]
- 85.Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem-progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 86.Bresnick EH, Martowicz ML, Pal S, Johnson KD. Developmental control via GATA factor interplay at chromatin domains. J. Cell. Physiol. 2005;205:1–9. doi: 10.1002/jcp.20393. [DOI] [PubMed] [Google Scholar]
- 87.Grass JA, Boyer ME, Paul S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl Acad. Sci. USA. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martowicz ML, Grass JA, Boyer ME, Guend H, Bresnick EH. Dynamic GATA factor interplay at a multi-component regulatory region of the GATA-2 locus. J. Biol. Chem. 2005;280:1724–1732. doi: 10.1074/jbc.M406038200. [DOI] [PubMed] [Google Scholar]
- 89.Grass JA, Jing H, Kim S-I, Martowicz ML, Pal S, Blobel GA, Bresnick EH. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol. Cell. Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lugus JJ, Chung YS, Mills JC, Kim SI, Grass JA, Kyba M, Doherty JM, Bresnick EH, Choi K. GATA2 functions at multiple steps in hemangioblast development and differentiation. Development. 2007;134:393–405. doi: 10.1242/dev.02731. [DOI] [PubMed] [Google Scholar]
- 91.Ray S, Dutta D, Rumi MA, Kent LN, Soares MJ, Paul S. Context-dependent function of regulatory elements and a switch in chromatin occupancy between GATA3 and GATA2 regulate Gata2 transcription during trophoblast differentiation. J. Biol. Chem. 2009;284:4978–4988. doi: 10.1074/jbc.M807329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liew CK, Simpson RJY, Kwan AHY, Crofts LA, Loughlin FE, Matthews JM, Crossley M, MacKay JP. Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction. Proc. Natl Acad. Sci. USA. 2005;102:583–588. doi: 10.1073/pnas.0407511102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lurie LJ, Boyer ME, Grass JA, Bresnick EH. Differential GATA factor stabilities: implications for chromatin occupancy by structurally similar transcription factors. Biochem. 2008;47:859–869. doi: 10.1021/bi701692p. [DOI] [PubMed] [Google Scholar]
- 94.Minegishi N, Suzuki H, Kawatani Y, Shimizu R, Yamamoto M. Rapid turnover of GATA-2 via ubiquitin-proteasome protein degradation pathway. Genes Cells. 2005;10:693–704. doi: 10.1111/j.1365-2443.2005.00864.x. [DOI] [PubMed] [Google Scholar]
- 95.Lamonica JM, Vakoc CR, Blobel GA. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 2006;108:3736–3738. doi: 10.1182/blood-2006-07-032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 97.Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, Akashi K, Orkin SH. Antagonism of F0G-1 and GATA factors in fate choice for the most cell lineage. J. Exp. Med. 2008;205:611–624. doi: 10.1084/jem.20070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crossley M, Orkin SH. Phosphorylation of the erythroid transcription factor GATA-1. J. Biol. Chem. 1994;269:16589–16596. [PubMed] [Google Scholar]
- 99.Zhao W, Kitidis C, Fleming MD, Lodish HF, Ghaffari S. Erythropoietin stimulates phosphorylation and activation of GATA-1 via the PI3-kinase/AKT signaling pathway. Blood. 2006;107:907–915. doi: 10.1182/blood-2005-06-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rooke HM, Orkin SH. Phosphorylation of Gata1 at serine residues 72, 142, and 310 is not essential for hematopoiesis in vivo. Blood. 2006;107:3527–3530. doi: 10.1182/blood-2005-10-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Partington GA, Patient RK. Phosphorylation of GATA-1 increases its DNA-binding affinity and is correlated with induction of human K562 erythroleukaemia cells. Nucleic Acids Res. 1999;27:1168–1175. doi: 10.1093/nar/27.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu YL, Chiang YJ, Chen YC, Papetti M, Juo CG, Skoultchi AI, Yen JJ. MAPK-mediated phosphorylation of GATA-1 promotes Bcl-XL expression and cell survival. J. Biol. Chem. 2005;280:29533–29542. doi: 10.1074/jbc.M506514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kadri Z, Maouche-Chretien L, Rooke HM, Orkin SH, Romeo PH, Mayeux P, Leboulch P, Chretien S. Phosphatidylinositol 3-kinase/Akt induced by erythropoietin renders the erythroid differentiation factor GATA-1 competent for TIMP-1 gene transactivation. Mol. Cell. Biol. 2005;25:7412–7422. doi: 10.1128/MCB.25.17.7412-7422.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Towatari M, May GE, Marais R, Perkins GR, Marshall CJ, Cowley S, Enver T. Regulation of GATA-2 phosphorylation by mitogen-activated protein kinase and interleukin-3. J. Biol. Chem. 1995;270:4101–4107. doi: 10.1074/jbc.270.8.4101. [DOI] [PubMed] [Google Scholar]
- 105.Menghini R, Marchetti V, Cardellini M, Hribal ML, Mauriello A, Lauro D, Sbraccia P, Lauro R, Federici M. Phosphorylation of GATA2 by Akt increases adipose tissue differentiation and reduces adipose tissue-related inflammation: a novel pathway linking obesity to atheroschlerosis. Circulation. 2005;111:1946–1953. doi: 10.1161/01.CIR.0000161814.02942.B2. [DOI] [PubMed] [Google Scholar]
- 106.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 108.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 109.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell. Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 110.Allfrey V, Faulkner RM, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 112.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 114.Marmorstein R. Protein modules that manipulate histone tails for chromatin regulation. Nat. Rev. Mol. Cell. Biol. 2001;2:422–432. doi: 10.1038/35073047. [DOI] [PubMed] [Google Scholar]
- 115.Fischle W, Wang Y, Allis CD. Histone and chromatin crosstalk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 116.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 117.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 118.Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl Acad. Sci. USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hung HL, Lau J, Kim AY, Weiss MJ, Blobel GA. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, Cheng Y, Billin AN, Hardison RC, Mackay JP, et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc. Natl Acad. Sci. USA. 2011;108:E159–E168. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kiekhaefer CM, Grass JA, Johnson KD, Boyer ME, Bresnick EH. Hematopoietic activators establish an overlapping pattern of histone acetylation and methylation within a tissue-specific chromatin domain. Proc. Natl Acad. Sci. USA. 2002;99:14309–14314. doi: 10.1073/pnas.212389499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol. Cell. Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Im H, Grass JA, Johnson KD, Kim S-I, Boyer ME, Imbalzano AN, Bieker JJ, Bresnick EH. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl Acad. Sci. USA. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hayakawa F, Towatari M, Ozawa Y, Tomita A, Privalsky ML, Saito H. Functional regulation of GATA-2 by acetylation. J. Leukoc. Biol. 2004;75:529–540. doi: 10.1189/jlb.0603389. [DOI] [PubMed] [Google Scholar]
- 125.Dalgin G, Goldman DC, Donley N, Ahmed R, Eide CA, Christian JL. GATA-2 functions downstream of BMPs and CaM KIV in ectodermal cells during primitive hematopoiesis. Dev. Biol. 2007;310:454–469. doi: 10.1016/j.ydbio.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ozawa Y, Towatari M, Tsuzuki S, Hayakawa F, Maeda T, Miyata Y, Tanimoto M, Saito H. Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood. 2001;98:2116–2123. doi: 10.1182/blood.v98.7.2116. [DOI] [PubMed] [Google Scholar]
- 127.Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 128.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 130.Guo D, Li M, Zhang Y, Yang P, Eckenrode S, Hopkins D, Zheng W, Purohit S, Podolsky RH, Muir A, et al. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 2004;36:837–841. doi: 10.1038/ng1391. [DOI] [PubMed] [Google Scholar]
- 131.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell. Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 132.Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr. Opin. Struct. Biol. 2007;17:726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yeh ET. SUMOylation and De-SUMOylation: wrestling with life's processes. J. Biol. Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kerscher O. SUMO junction – what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chun TH, Itoh H, Subramanian L, Ingiguez-Lluhi JA, Nakao K. Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASgamma. Circ. Res. 2003;92:1201–1208. doi: 10.1161/01.RES.0000076893.70898.36. [DOI] [PubMed] [Google Scholar]
- 136.Collavin L, Gostissa M, Avolio F, Secco P, Ronchi A, Santoro C, Del Sal G. Modification of the erythroid transcription factor GATA-1 by SUMO-1. Proc. Natl Acad. Sci. USA. 2004;101:8870–8875. doi: 10.1073/pnas.0308605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang J, Feng XH, Schwartz RJ. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J. Biol. Chem. 2004;279:49091–49098. doi: 10.1074/jbc.M407494200. [DOI] [PubMed] [Google Scholar]
- 138.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 139.Lee H-Y, Johnson KD, Fujiwara T, Boyer ME, Kim S-I, Bresnick EH. Controlling hematopoiesis through sumoylation-dependent regulation of a GATA factor. Mol. Cell. 2009;36:984–995. doi: 10.1016/j.molcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee HY, Johnson KD, Boyer ME, Bresnick EH. Relocalizing genetic loci into specific subnuclear neighborhoods. J. Biol. Chem. 2011;286:18834–18844. doi: 10.1074/jbc.M111.221481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yu L, Ji W, Zhang H, Renda MJ, He Y, Lin S, Cheng EC, Chen H, Krause DS, Min W. SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis. J. Exp. Med. 2010;207:1183–1195. doi: 10.1084/jem.20092215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Snow JW, Kim J, Currie CR, Xu J, Orkin SH. Sumoylation regulates interaction of FOG1 with C-terminal-binding protein (CTBP) J. Biol. Chem. 2010;285:28064–28075. doi: 10.1074/jbc.M109.096909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hsu AP, Sampaio EP, Khan J, Calvo KR, Lemieux JE, Patel SY, Frucht DM, Vinh DC, Auth RD, Freeman AF, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118:2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dickinson RE, Griffin H, Bigley V, Reynard LN, Hussain R, Haniffa M, Lakey JH, Rahman T, Wang X-N, McGovern N, et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood. 2011;118:2656–2658. doi: 10.1182/blood-2011-06-360313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ostergaard P, Simpson MA, Connell FC, Steward CG, Brice G, Woollard WJ, Dafou D, Kilo T, Smithson S, Lunt P, et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome) Nat. Genet. 2011;43:929–931. doi: 10.1038/ng.923. [DOI] [PubMed] [Google Scholar]
- 146.Hahn CH, Chong C-E, Carmichael CL, Wilkins EJ, Brautigan PJ, Li X-C, Babic M, Lin M, Carmagnac A, Lee YK, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat. Genet. 2011;43:1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vinh DC, Patel SY, Uzel G, Anderson VL, Freeman AF, Olivier KN, Spalding C, Hughes S, Pittaluga S, Raffeld M, et al. Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood. 2010;115:1519–1529. doi: 10.1182/blood-2009-03-208629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mansour S, Connell F, Steward C, Ostergaard P, Brice G, Smithson S, Lunt P, Jeffery S, Dokal I, Vulliamy T, et al. Emberger syndrome—Primary lymphedema with myelodysplasia: Report of seven new cases. Am. J. Med. Gen. Part A. 2010;152A:2287–2296. doi: 10.1002/ajmg.a.33445. [DOI] [PubMed] [Google Scholar]
- 149.Owen C, Barnett M, Fitzgibbon J. Familial myelodysplasia and acute myeloid leukaemia - a review. Br. J. Haematol. 2008;140:123–132. doi: 10.1111/j.1365-2141.2007.06909.x. [DOI] [PubMed] [Google Scholar]
- 150.Kazenwadel J, Secker GA, Liu YJ, Rosenfeld JA, Wildin RS, Cuellar-Rodriguez J, Hsu AP, Dyack S, Fernandez CV, Chong CE, et al. Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood. 2012;119:1283–1291. doi: 10.1182/blood-2011-08-374363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bodor C, Renneville A, Smith M, Charazac A, Iqbal S, Etancelin P, Cavenagh J, Barnett MJ, Kramarzova K, Krishnan B, et al. Germ-line GATA2 p.THR354MET mutation in familial myelodysplastic syndrome with acquired monosomy 7 and ASXL1 mutation demonstrating rapid onset and poor survival. Hematologica. 2012;97:890–894. doi: 10.3324/haematol.2011.054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bigley V, Haniffa M, Doulatov S, Wang X-N, Dickinson R, McGovern N, Jardine L, Pagan S, Dimmick I, Chua I, et al. The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J. Exp. Med. 2011;208:227–234. doi: 10.1084/jem.20101459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Calvo KR, Vinh DC, Maric I, Wang W, Noel P, Stetler-Stevenson M, Arthur DC, Raffeld M, Dutra A, Pak E, et al. Myelodysplasia in autosomal dominant and sporadic monocytopenia immunodeficiency syndrome: diagnostic features and clinical implications. Haematologica. 2011;96:1221–1225. doi: 10.3324/haematol.2011.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bigley V, Collin M. Dendritic cell, monocyte, B and NK lymphoid deficiency defines the lost lineages of a new GATA-2 dependent myelodysplastic syndrome. Haematologica. 2011;96:1081–1083. doi: 10.3324/haematol.2011.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Walsh JC, DeKoter RP, Lee H-J, Smith ED, Lancki DW, Gurish MF, Friend DS, Stevens RL, Anastasi J, Singh H. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 156.Lasbury ME, Tang X, Durant PJ, Lee C-H. Effect of transcription factor GATA-2 on phagocytic activity of alveolar macrophages from Pneumocystis carinii-infected hosts. Infect. Immun. 2003;71:4943–4952. doi: 10.1128/IAI.71.9.4943-4952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rodrigues NP, Boyd AS, Fugazza C, May GE, Guo Y, Tipping AJ, Scadden DT, Vyas P, Enver T. GATA-2 regulates granulocyte-macrophage progenitor cell function. Blood. 2008;112:4862–4873. doi: 10.1182/blood-2008-01-136564. [DOI] [PubMed] [Google Scholar]
- 158.Kitajima K, Tanaka M, Zheng J, Yen H, Sato A, Sugiyama D, Umehara H, Sakai E, Nakano T. Redirecting differentiation of hematopoietic progenitors by a transcription factor, GATA-2. Blood. 2006;107:1857–1863. doi: 10.1182/blood-2005-06-2527. [DOI] [PubMed] [Google Scholar]
- 159.Carotta S, Dakic A, D'Amico A, Pang SHM, Greig KT, Nutt SL, Wu L. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 160.Krysinska H, Hoogenkang M, Ingram R, Wilson N, Tagoh H, Laslo P, Singh H, Bonifer C. A two-step, PU.1-dependent mechanism for developmentally regulated chromatin remodeling and transcription of the c-fms gene. Mol. Cell. Biol. 2006;27:878–887. doi: 10.1128/MCB.01915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 162.Tong Q, Tsai J, Tan G, Dalgin G, Hotamisligil GS. Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol. Cell. Biol. 2005;25:706–715. doi: 10.1128/MCB.25.2.706-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Migliaccio AR, Bieker JJ. GATA2 finds its macrophage niche. Blood. 2011;118:2647–2649. doi: 10.1182/blood-2011-06-362772. [DOI] [PubMed] [Google Scholar]
- 164.Vicente C, Vazquez I, Marcotegui N, Conchillo A, Carranza C, Rivell G, Bandres E, Cristobal I, Lahortiga I, Calasanz MJ, et al. JAK2-V617F activating mutation in acute myeloid leukemia: prognostic impact and association with other molecular markers. Leukemia. 2007;21:2386–2390. doi: 10.1038/sj.leu.2404812. [DOI] [PubMed] [Google Scholar]
- 165.Shimamoto T, Ohyashiki K, Ohyashiki JH, Kawakubo K, Fujimura T, Iwama H, Nakazawa S, Toyama K. The expression pattern of erythrocyte/megakaryocyte-related transcription factors GATA-1 and the stem cell leukemia gene correlates with hematopoietic differentiation and is associated with outcome of acute myeloid leukemia. Blood. 1995;86:3173–3180. [PubMed] [Google Scholar]
- 166.Ayala RM, Martínez-López J, Albízua E, Diez A, Gilsanz F. Clinical significance of Gata-1, Gata-2, EKLF, and c-MPL expression in acute myeloid leukemia. Am. J. Hematol. 2009;84:79–86. doi: 10.1002/ajh.21332. [DOI] [PubMed] [Google Scholar]
- 167.Vicente C, Vazquez I, Conchillo A, Garcia-Sanchez MA, Marcotegui N, Fuster O, Gonzalez M, Calasanz MJ, Lahortiga I, Odero MD. Overexpression of GATA2 predicts an adverse prognosis for patients with acute myeloid leukemia and it is associated with distinct molecular abnormalities. Leukemia. 2011;9:1–5. doi: 10.1038/leu.2011.235. [DOI] [PubMed] [Google Scholar]
- 168.Bonadies N, Foster SD, Chan W-I, Kvinlaug BT, Spensberger D, Dawson MA, Spooncer E, Whetton AD, Bannister AJ, Huntly BJ, et al. Genome-wide analysis of transcriptional reprogramming in mouse models of acute myeloid leukaemia. PLoS One. 2011;6:e16330. doi: 10.1371/journal.pone.0016330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Slape C, Hartung H, Lin Y-W, Bies J, Wolff L, Aplan PD. Retroviral insertional mutagenesis identifies genes that collaborate with NUP98-HOXD13 during leukemic transformation. Cancer Res. 2007;67:5148–5155. doi: 10.1158/0008-5472.CAN-07-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang SJ, Ma LY, Huang QH, Li G, Gu BW, Gao XD, Shi JY, Wang YY, Geo L, Cai X, et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc. Natl Acad. Sci. USA. 2008;105:2076–2081. doi: 10.1073/pnas.0711824105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang SJ, Shi JY, Li JY. GATA-2 L359V mutation is exclusively associated with CML progression but not other hematological malignancies and GATA-2 P250A is a novel single nucleotide polymorphism. Leukemia Res. 2009;33:1141–1143. doi: 10.1016/j.leukres.2009.02.025. [DOI] [PubMed] [Google Scholar]