Abstract
The structure-specific endonuclease activity of the human XPF–ERCC1 complex is essential for a number of DNA processing mechanisms that help to maintain genomic integrity. XPF–ERCC1 cleaves DNA structures such as stem–loops, bubbles or flaps in one strand of a duplex where there is at least one downstream single strand. Here, we define the minimal substrate requirements for cleavage of stem–loop substrates allowing us to develop a real-time fluorescence-based assay to measure endonuclease activity. Using this assay, we show that changes in the sequence of the duplex upstream of the incision site results in up to 100-fold variation in cleavage rate of a stem-loop substrate by XPF-ERCC1. XPF–ERCC1 has a preference for cleaving the phosphodiester bond positioned on the 3′-side of a T or a U, which is flanked by an upstream T or U suggesting that a T/U pocket may exist within the catalytic domain. In addition to an endonuclease domain and tandem helix–hairpin–helix domains, XPF has a divergent and inactive DEAH helicase-like domain (HLD). We show that deletion of HLD eliminates endonuclease activity and demonstrate that purified recombinant XPF–HLD shows a preference for binding stem–loop structures over single strand or duplex alone, suggesting a role for the HLD in initial structure recognition. Together our data describe features of XPF–ERCC1 and an accepted model substrate that are important for recognition and efficient incision activity.
INTRODUCTION
The human XPF–ERCC1 complex is a metal-dependent structure-specific endonuclease which is required for a number of pathways essential for genome maintenance (1). It has roles in several DNA repair processes including nucleotide excision repair (NER), interstrand crosslink (ICL) repair, double-strand break (DSB) repair and recombinational repair (2–5). Mutations in XPF (xeroderma pigmentosum complementation group F) in humans gives rise to a defect in the 5′-incision step of NER of bulky adducts in one strand of DNA and results in the UV sensitive cancer-prone syndrome xeroderma pigmentosum. A more severe manifestation of an XPF defect led to progeroid syndrome, a premature ageing disease with a similar phenotype to both XPF and ERCC1 knockout mice (6–8). ERCC1 (excision repair cross complementation group 1) was first identified as a human gene that restored the DNA repair capability to a UV-sensitive Chinese hamster ovary cell line (9). One case of a defect in ERCC1 in humans has been identified to date which gave rise to congenital cerebro-oculo-facio-skeletal syndrome (COFS) (10), a condition usually associated with a defect in repair of transcribing genes [for review see (11)].
The XPF and ERCC1 polypeptides both belong to the highly conserved Mus81–XPF structural family of proteins (12–15) possessing a common core containing a nuclease or nuclease-like domain and tandem C-terminal helix–hairpin–helix (HhH)2 domains (Figure 1A) which bind DNA (16–19). Human XPF also has a highly divergent helicase-like domain (HLD) which has apparently lost residues required for ATP hydrolysis (Figure 1A) (20) and may be involved in protein–protein interactions or DNA binding. XPF binds to SLX4, a component of the SLX4–SLX1 endonuclease complex, which recruits multiple structure-specific endonucleases to recombination and repair processes in the cell (21–24) but the interacting region on XPF has not been identified to date. Core catalytic and (HhH)2 domains of archaeal XPFs homodimerize and bind DNA asymmetrically such that only one catalytic site can be active at a time (16,17). Eukaryotic XPFs have diverged from this arrangement and form obligate heterodimers with structurally related, catalytically inactive partners via their C-termini (13). The catalytic motif resides in the XPF protomer of human XPF–ERCC1 (25) and ERCC1 instead binds XPA for recruitment to the NER process (26,27) (Figure 1A). In humans, members of the XPF family also include Mus81–Eme1/Eme2 and FancM/FAAP24, although the latter has lost essential residues for nuclease activity but has retained an active N-terminal translocase domain (28).
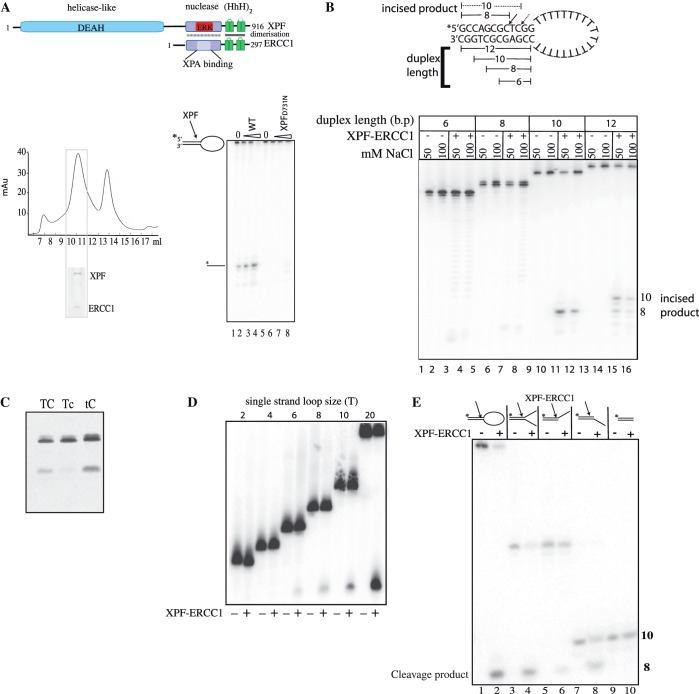
Figure 1.
Minimum substrate requirements for incision activity. (A) Top: domain organization of the human XPF–ERCC1 complex; bottom left panel: gel-filtration profile of XPF–ERCC1 expressed in E. coli after purification on nickel-agarose, and heparin–Sepharose. XPF–ERCC1 eluted where indicated. Fractions 7–8 is the void and the peak at 11 ml corresponded to a MW of ∼250 kDa, and 13 ml to 35 kDa. Inset shows 12% SDS–PAGE gel of fraction 12 stained with Coomassie blue. XPF migrates at 115 kDa and ERCC1 at 36 kDa. Bottom right panel: sequencing gel showing cleavage of 200 fmol stem–loop substrate by wild-type XPF–ERCC1 and XPF-D731N–ERCC1 mutant. Lanes 1 and 5 contained protein buffer from each preparation of protein, lanes 2–4 contained 12, 24 and 48 nM XPF–ERCC1, respectively. Lanes 6–8 contained 12, 24 and 48 nM XPF-D731N–ERCC1. (B) Sequencing gel showing cleavage of stem–loops with either 6 8, 10 or 12 bp duplex stems. Wells contained protein buffer (−) or 42.6 nM XPF–ERCC1 (+) in buffer containing either 50 or 100 mM NaCl as indicated. Cleavage of the 12-bp duplex yields products of 10 and 8 bp, and the 10-bp duplex is cleaved at one major site yielding an 8-bp product. Dual uncleaved bands in lanes 1–8 are full-length (FL) and FL-1 nucleotide oligonucleotides produced during synthesis of the shorter 6 and 8 bp substrates. (C) Separation of products of cleavage reaction on a 10% denaturing polyacrylamide gel. Stem–loops (100 nM) labelled 5′ with Cy-5 with a phosphorothioate bond replacing the phosphodiester bond 3′ to the base represented in lower case in the sequences 5′-CAGCGCTCGG-3′ (TC), 5′-CAGCGCTcGG-3′ (Tc), and 5′-CAGCGCtCGG-3′(tC) in the top strand of the duplex were incubated with 42.6 nM XPF-ERCC1 for 1 hour. (D) Ten percent sequencing gel showing incision reactions containing stem–loops with single-strand loops of increasing T loop size as indicated. An amount of 400 fmol labelled stem–loop substrates were incubated with buffer only (−) or 42.6 nM XPF–ERCC1 (+). (E) Sequencing gel showing incisions made by XPF–ERCC1 on stem–loop, splayed arm, 3′-overhangs, 5′-overhangs and duplex. The sequence of the duplex portion of the structure is 5′-CAGCGCTCGG-3′/5′CCGAGCGCTG-3′ and single-strand regions are 20T for stem–loop and 10T for splayed arms/overhangs. The top strand of each structure was labelled 5′ as indicated and oligonucleotides were annealed and purified at 15°C as described in experimental procedures. The cleavage product is the same eight-base oligo for each structure as indicated in the figure.
XPF–ERCC1 cleaves one strand of DNA a few bases into a duplex upstream from a junction, containing at least one downstream single strand. These structures arise in the cell during DNA processing and repair in the form of bubbles, stem–loops, splayed arms, 3′-overhangs, 5′-overhangs and 3′-flaps. In contrast, Mus81–Eme1/Eme2 shares the substrate specificity displayed by the archaeal homodimeric XPF homologues and cleaves junctions with a downstream duplex including nicked Holliday junctions, forks, 3′-flaps and D loops (29).
In order to understand more about the interaction of the full-length recombinant human XPF–ERCC1 complex with its substrate we have established the minimum DNA junction requirements for incision on a generally accepted stem–loop model substrate (2,19,25). We have used this information to develop a fluorescence-based assay to measure the kinetic constants of the incision reaction at a single incision site. Using this assay, we show that the duplex sequence at the incision site influences the incision rate and XPF–ERCC1 prefers to cleave the phosphodiester bond on the 3′-side of a T which is flanked on the 5′-side by a T or an A. Substitution of these bases with deoxy-uracil paired with A increased the rate of incision by ≤5-fold, suggesting that a pocket exists in the nuclease domain which accommodates pyrimidines, and the exocyclic groups present on C and T inhibit entry into the pocket. We then demonstrate that the HLD of XPF is necessary for activity, in contrast to a previous study which used a different truncation mutant (19). We show that recombinant purified XPF–HLD preferentially binds the stem–loop structure implicating a role for the XPF–HLD in structure recognition and stabilization of the core domains at the junction.
MATERIALS AND METHODS
DNA substrates
PAGE-purified DNA oligonucleotides were obtained from Sigma with the following sequences:
Stem12–loop20: 5′-GCCAGCGCTCGG(20T)CCGAGCGCTGGC-3′;
Stem10–loop 20:5′-CAGCGCTCGG(20T)CCGAGCGCTG-3′;
Stem8–loop20:5′-GCGCTCGG(20T)CCGAGCGC-3′
Stem6–loop 20: 5′-GCTCGG(20T)CCGAGC-3′;
Control stem10–loop 2:5′-CAGCGCTCGGTTCCGAGCGCTG-3′;
F1: 5′-CAGCGCTCGG-3′; F2: 5′-CCGAGCGCTG-3′; F3: 5′-CAGCGCTCGG(11T)-3′;F4: 5′-(11T)CCGAGCGCTG-3′.
For fluorescence polarization:
15-mer single strand: 6-FAM-5′-GCGCTCGGCGTTTTT-3′; 2T stem–loop: 6-FAM 5′-CAGCGC
TCGG(2T)CCGAGCGCTG-3′
Other sequences were used as described in the results.
Proteins
Human XPF–ERCC1 wild-type and mutant proteins were produced in Escherichia coli strain FB810 BL21 (DE3) using a dicistronic expression plasmid derived from pET30b. ERCC1 (residues 1–297) was expressed first with a polyhistidine tag (His6) at the C-terminus. XPF (residues 12–916) had a His-6 at the N-terminus. Recombinant protein was extracted from harvested E. coli cells in 20 mM HEPES/KOH pH 7.4 containing 500 mM NaCl, 2 mM β-mercaptoethanol, 2 mM MgCl2, 10% glycerol, 0.01% CHAPS, 5 mM imidazole and protease inhibitors. The resulting extract was incubated with Ni–NTA resin for 30 min, poured into a disposable column and washed with extract buffer containing 40 mM imidazole. XPF–ERCC1 was eluted in buffer containing 500 mM imidazole. Fractions from the Ni–NTA column were dialysed into heparin buffer; 20 mM HEPES/KOH pH 7.4 containing 50 mM NaCl, 2 mM β-mercaptoethanol, 10% glycerol, 0.01% CHAPS and 0.5 mM EDTA before concentrating and loading onto a Hi-trap heparin column and eluted in heparin buffer containing 500 mM NaCl. Fractions containing XPF–ERCC1 were concentrated to 0.5 ml and loaded onto a Superose 12 column (GE Healthcare) previously equilibrated in 20 mM HEPES pH 7.4 containing 50 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 2 mM MgCl2 10% glycerol and 0.01% CHAPS. Fractions from the Superose 12 column were stored at −80°C in 10 mM HEPES pH 7.4, 2.5 mM β-mercaptoethanol, 50% glycerol, 0.25 mM EDTA and 25 mM NaCl.
The Δ640XPF–ERCC1 truncation mutant was purified by the same method as wild-type XPF–ERCC1, except that the pooled fractions from the Ni–NTA column were diluted 1:20 in heparin buffer before loading onto the Hi-trap heparin column. The gel filtration was carried out using a 16/60 prep grade Superose 12 column. The XPFD731N–ERCC1 mutations were made using Quikchange site-directed mutagenesis kit (Stratagene) and the above WT construct as template, expressed and purified using exactly the same procedure as wild-type. XPF–HLD was produced in E. coli strain FB810 using vector derived from pET41b (Novagen) containing an N-terminal GST tag and a 3C protease cleavage site. A stop codon was introduced after A672 in XPF. Cells were induced with IPTG at a concentration of 50 µM for 24 h at 18°C. Protein was extracted from harvested E. coli cells in 50 mM Tris pH8 containing 500 mM NaCl, 1 mM DTT, 10% glycerol, 0.1% TX-100, and the protease inhibitors AEBSF, benzamidine and Complete inhibitor cocktail (Roche). Glutathione beads (Pierce) were added and incubated for 1 h at 4°C. Protein was cleaved with 3C protease and loaded onto a Superose 12 column previously equilibrated in elution buffer. Fractions were dialysed into elution buffer containing 50 mM NaCl and stored at −80°C. Protein measurements were carried out using the Bio Rad protein assay kit (Bio-Rad).
Gel-based nuclease assays
Stem–loops and oligonucleotides F1 and F3 (sequences described above and used to make the structures in Figure 3E as described below) were labelled on the 5′-terminus with [γ32P]ATP using T4 polynucleotide kinase (NEB) for 30 min at 37°C, and the reaction stopped by heating to 95°C for 3 min. F1 was mixed with 2 M excess of F2 for double-strand (duplex), and F4 for 5′-overhang. F3 was mixed with F2 for 3′-overhang, and F4 for splayed arm.
Figure 3.
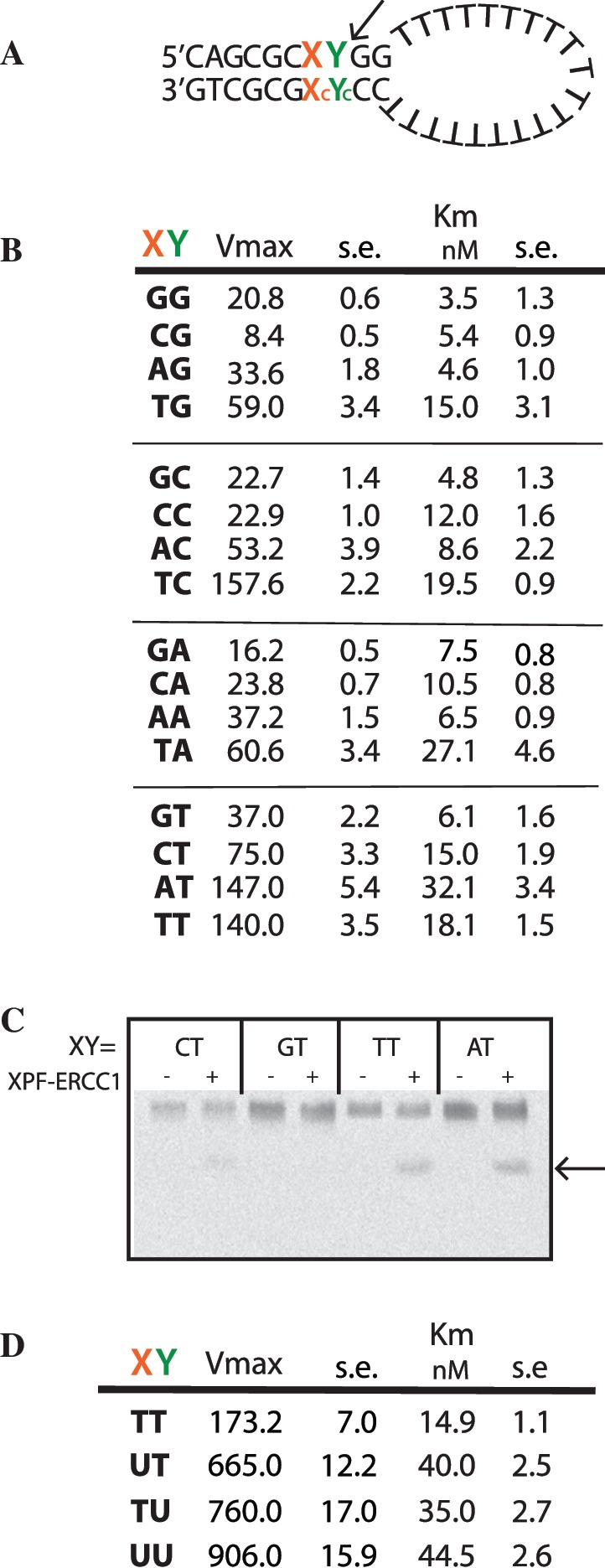
Local sequence preferences at the XPF–ERCC1 incision site on the substrate affects the rate of cleavage. (A) Stem–loop structure showing the positions of substituted bases X and Y in the duplex. Xc and Yc are the complementary bases to X and Y, A is used complementary to U. (B) Kinetic data where X and Y were substituted with each of the four bases. Data are mean of triplicate samples, ±1 standard error. (C) Incisions produced by 4.27 nM XPF–ERCC1 on 5′ 6-FAM-labelled stem–loop substrates with bases at X and Y as indicated. Arrow shows cleavage product. Reactions were incubated for 15 min at 25°C and therefore did not run to completion. (D) Kinetic data where X and Y were substituted with the indicated bases. Data are mean of triplicate samples +/− standard error.
The DNA was annealed by heating to 95°C for 15 min, followed by 60°C, 37°C, and 25°C for 10 min each, and kept on ice before purifying on a 10% non-denaturing polyacrylamide gel in TBE (Tris, borate, EDTA electrophoresis buffer) and diffusion overnight at 4°C into T.E. (10 mM Tris, 1 mM EDTA) containing 50 mM NaCl, and stored at −20°C. Reactions were carried out in a volume of 20 µl at 25°C for 90 min in reaction buffer containing 50 mM Tris pH 8, 20 mM NaCl, 0.5 mM DTT (dithiothreitol), 10% glycerol, 0.1 mg/ml bovine serum albumin (BSA) and 0.75 mM MnCl2 (unless indicated). Incision products were separated on a 12% sequencing gel (Sequagel-National Diagnostics) for 2 h. The gel was removed and dried and products visualized by auroradiography, or on a STORM phosphorimager (Molecular Dynamics).
Fluorescent gel-based nuclease assays contained 100 nM oligonucleotide (obtained from Sigma) labelled on the 5′-terminus with 6-FAM (an isomer of carboxyfluorescein) or Cy5 (fluorescent dye from the cyanine family). These were incubated for 1 h with 12 nM XPF–ERCC1 in reaction buffer. Products were separated on a 12% sequencing gel. One plate was removed from the gel and the fluorescence measured directly on a STORM phosphorimager (blue channel for 6-FAM and red channel for Cy5).
Microplate fluorescence incision assay
Polyacrylamide gel electrophoresis (PAGE)-purified oligonucleotides labelled 5′ with 6-FAM and 3′-with dabcyl ([4-((4-(dimethyllamino) phenyl)azo)benzoic acid) (pre-labelled molecular beacons from Sigma). These were diluted to 100 µM in T.E containing 50 mM NaCl and stored at −20°C. After thawing, substrates were annealed by heating to 95°C for 1 min, and cooling to room temperature. Reactions contained the appropriate concentration of substrate, 50 mM Tris pH 8, 20 mM NaCl, 0.5 mM DTT, 10% glycerol and 0.75 mM MnCl2 (unless indicated) in a total volume of 20 µl. XPF–ERCC1 was thawed immediately before use and diluted in protein storage buffer for assay. Reactions were carried out in 384-well black, flat-bottomed microtitre plates (Corning 3854) and readings were taken in a Tecan Safire II fluorimeter (Tecan group LTD) using Megellen software at an excitation and emission wavelengths of 483 and 525 nm, respectively. For kinetic runs, 30 readings were collected at 2-min intervals.
Kinetics
The rate of increase in fluorescence units produced by the enzyme per minute during the initial phase of the reaction (0–10 min) was measured in triplicate. The Vmax and Km values were calculated using Sigmaplot software (Systat Software Inc) and the Michaelis–Menten equation:
where x = substrate concentration (nM) and Y = reaction velocity (relative fluorescence units/min).
Femtomol (fmol) product release was quantified by plotting the relative fluorescence units (RFU) produced by known amounts of the oligonucleotides 6-FAM-5′-CAGCGCTC and GG(20T)CCGAGCGCTG-3′-dabcyl corresponding to the cleavage products. A standard curve and predicted fmol cleavage product released was calculated from this.
Fluorescence polarization
Assays were carried out and equilibrium dissociation constants (KD) calculated using PRISM (Graphpad Software Ltd) as described in (28). Reactions contained 5 µM DNA substrate and 0.005% Tween-20. The oligonucleotides used were: Stem10–loop 20 (above with sequence substitutions as described in the text), or 15T single stranded oligonucleotide labelled 5′ with 6-FAM, or indicated in the text. The binding reactions were incubated for 30 min at 25°C and read in a Tecan Saffire 2 fluorimeter, using Megellen software (Tecan group Ltd).
Biacore surface plasmon resonance
Experiments were carried out on a T200 Biacore analyser (GE Healthcare). PAGE-purified biotinylated oligonucleotides were obtained from Sigma, diluted to 10 nM in T.E containing 50 mM NaCl and bound to a Series S Sensor SA sensor chip (GE Healthcare) according to manufacturers’ instructions at a flow rate of 10 µl/min to give a final theoretical Rmax of 100 after analyte binding. XPF–ERCC1 superose 12 fractions were pooled and dialysed overnight into 10 mM HEPES, 50 mM NaCl, 0.5 mM DTT and 0.05% P20 (GE Healthcare) surfactant. Single cycle kinetic analysis was carried out according to the Biacore software using 2-fold dilutions of XPF–ERCC1 in the above buffer. The data from a reference cell without DNA was subtracted and the apparent association (Ka) and dissociation (Kd) rate constants were measured using the equation for 1:1 Langmuir binding (BIA Evaluation 4.1 software). The apparent equilibrium dissociation constants (KD) were calculated from these values.
RESULTS
Minimum substrate requirements for incision activity
Previous attempts to purify full-length recombinant human XPF–ERCC1 from E. coli resulted in aggregated protein consisting mainly of ERCC1, and only a small amount of active heterodimer (25). We found that the addition of 0.01% CHAPS to the buffers considerably increased the yield of soluble, heterodimeric XPF–ERCC1 (Figure 1A). After purification on Ni–agarose, heparin sepharose and Superose 12-gel filtration the yield was 100–150 µg of pure ERCC1–XPF per 2 l of culture (Figure 1A). This protocol separated high and low molecular weight bacteria-derived nucleases from the XPF–ERCC1 heterodimer (Supplementary Figure S2), (Figure 1A, bottom left). Pooled fractions 10–12 incised a stem–loop substrate in 7.5 mM MnCl2 (Figure 1A, bottom right and Supplementary Figure S2, lanes 6 and 8). The catalytically impaired mutant XPF-D731N–ERCC1 (25,30,31) was prepared using exactly the same protocol to control for the presence of bacterial nuclease contamination and showed very little specific cleavage (Figure 1A) (Supplementary Figures S1A and S2: lanes 18 and 20). This is an important control as other nucleases can mimic structure specific activity (Supplementary Figure S2, lanes 25–30).
We used gel-filtered recombinant XPF–ERCC1 to define the minimal substrate requirements for making incisions at a single defined position in the duplex using a gel-based assay. The generally accepted model XPF–ERCC1 substrate used in previous studies is a stem–loop substrate consisting of a 12-bp stem and a 22-base single-strand loop (2,19,25). We reduced the duplex length from 12, to 10, 8 and 6 bp. All of these substrates were gel purified by non-denaturing PAGE before use, which ensured that they formed the stem–loop structure at 50 mM NaCl. Two major incisions were made within the 12-bp duplex (Figure 1B, lanes 15 and 16) which have been previously mapped to the phosphodiester bond 3′ to the third and fifth bases upstream from the duplex single-strand junction (2,32,33). We found that reduction of the duplex to 10 bp by removing the two pairs at the free end of the duplex resulted in only one incision and this yielded a product of eight bases (Figure 1B, lanes 11 and 12). The incision position was verified by replacing the cleaved phosphodiester bond with a non-cleavable phosphorothioate link in which a non-bridging oxygen (PO) is replaced by sulphur (PS), and this prevented incisions only when it was placed 3′ to the third base upstream from the duplex single-strand junction but not at the fourth (Figure 1C). No cleavage occurred on the substrates containing 6- or 8-bp stems (Figure 1B, lanes 4, 5, 7 and 8). We therefore had produced conditions for a single incision within the stem–loop substrate. Catalytic activity was reduced at 100 mM NaCl indicating that DNA binding by the enzyme may be reduced at higher salt, or the increase in duplex stability inhibits the enzyme.
The minimal single-strand length was examined using the 10-bp duplex stem–loop substrate. The loop consists solely of Ts to avoid internal base pairing which may occur if mixed sequences are used, or tracts of Gs and Cs, which may base pair with the GC-rich duplex. The loop was reduced to 20, 10, 8, 6, 4 and 2 Ts (Figure 1D). The minimum single-strand loop requirement was between 4 and 6 thymidines and this confirms a previous finding where the single-strand loop was reduced on substrates with concomitantly increasing duplex regions (32). Incision activity increased up to a loop of 20 Ts and consequently the 10 duplex–20 loop structure incised at a single site by XPF–ERCC1 was used for all subsequent activity assays.
Oligonucleotide structures were made with the identical 10-bp duplex and either 5′ or 3′ 10-T single-strand overhangs, or a splayed arm containing both. XPF–ERCC1 cleaved all the structures at exactly the same position in the duplex, yeilding an 8-base labelled incision product, (Figure 1E, lanes 1–4) irrespective of the position of the single-strand region. A 3′-single-strand arm on the cleaved strand is not required for cleavage when a 5′-single-strand arm is present on the uncleaved strand and vice versa. We found that duplex DNA is not cleaved at all under these conditions (Figure 1E, lanes 9 and 10).
Kinetics of XPF–ERCC1 incision activity
We then developed a real-time fluorescence-based microtitre plate assay using the 10-duplex 20-T single-strand stem–loop model substrate (Figure 2A, left). This was labelled on the 5′-termini with 6-FAM and 3′ with the quencher, dabcyl. In the annealed state, the dabcyl quenches the fluorescent signal. The 10-bp duplex region of this stem–loop has a melting temperature of 29°C at 20 mM NaCl. Cleavage in the 10-bp duplex region by XPF–ERCC1 generates an eight-base oligonucleotide, which has a melting temperature of ∼23°C at 20 mM NaCl that diffuses into solution at the incubation temperature of 25°C, resulting in an increase in fluorescent signal with time (Figure 2A, right). An amount of 1.5 nM (30 fmol) of XPF–ERCC1 gave reproducible results with a low signal/noise ratio and the incision rate (relative fluorescence units (RFU)/min) increased with increasing XPF–ERCC1 concentration. The XPFD731A–ERCC1 point mutation has been shown to abolish incision activity whilst retaining DNA binding (25) although some residual NER restoration activity has been noted in one study (30). The catalytically impaired XPFD731N–ERCC1 showed considerably less incision activity than the wild-type endonuclease (Supplementary Figure S1A) although this was not completely abrogated at very high concentrations.
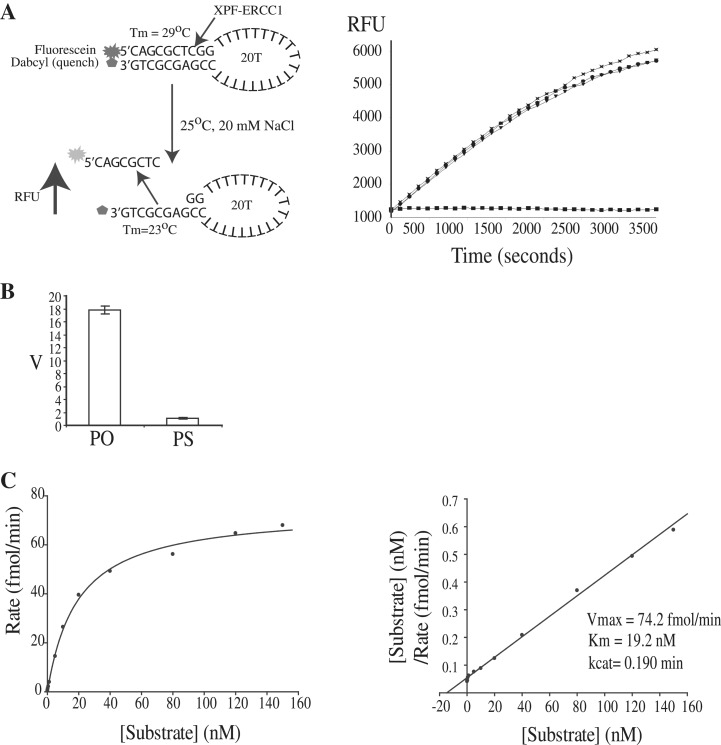
Figure 2.
A microplate fluorescence-based assay to measure XPF–ERCC1 incision activity. (A) Left panel: principle of the fluorescence-based assay. RFU is relative fluorescence units. Incubation of intact stem–loop with XPF–ERCC1 in 0.75 mM MnCl2 and 20 mM NaCl results in the cleaved oligonucleotide diffusing into solution at 25°C. Right panel: increase in fluorescence (RFU) with time. Reactions are shown in triplicate and contained: 3 nM XPF–ERCC1, 100 nM standard stem–loop substrate, 0.75 nM MnCl2, 0.5 mM DTT and 20 mM NaCl2 in 50 mM Tris buffer pH 8 at 25°C. Solid squares: buffer only (B) Graph showing rate of incisions (v) of 62.5 nM stem–loop by 3 nM XPF–ERCC1 where the scissile phosphate contains non-bridging oxygens (PO) or a non-bridging sulphur (PS). Average of three points ± SD. (C) Left panel: Michaelis–Menten plot showing rate versus substrate concentration for full-length XPF–ERCC1. Mean of three points. Right panel: Hanes–Woolf plot.
Replacement of a non-bridging oxygen on the scissile phosphate (PO) with a phosphorothioate (PS) (Figure 2B) eliminated nearly all of the endonuclease activity confirming that the increase in signal was due to incisions by the enzyme at this position, in agreement with the gel-based assay (Figure 1C). 0.75 mM MnCl2 was found to be optimal giving a 4-fold higher rate of incisions than the maximal MgCl2 concentration of 5 mM in accordance with previous work (2,25,32) (Supplementary Figure S1B). The incision reaction was sensitive to pH, which peaked at pH 8 with a sharp fall-off in activity at pH 7.5 and 8.5 (Supplementary Figure S1C). The kinetic constants for XPF–ERCC1 activity were then measured under steady-state conditions where substrate was in excess (Figure 2C). Representative Michaelis–Menten and Hanes–Woolf graphs are shown. The kcat and Km for human XPF–ERCC1 under these conditions was calculated to be 0.19/min ± 0.04 SE (standard error) and 19.2 ± 0.76 nM, respectively, for three independent experiments. This was consistent with a previous study where the kcat was found to be 0.17 ± 0.04 min−1 on the stem–loop substrate with a 12-bp duplex measured using a gel-based assay system (19).
Local incision site sequence preferences
Cleavage of synthetic substrates by XPF–ERCC1 has generally been regarded as sequence independent, although preference for a pyrimidine 5′ to the scissile phosphate at the incision site has been observed (32–36). In an effort to understand the basis for this apparent discrimination, we used the fluorescence assay to measure the kinetics of cleavage of stem–loops where the nucleotides at positions X and Y, immediately upstream of the single scissile phosphate (Figure 3A), were substituted with each of the four bases and their corresponding complementary bases in the opposite strand. The tandem G:C base pairs at the junction were kept constant in order to maintain its stability. The base substitutions resulted in differences in Vmax, which ranged 18.7-fold from 8.4 to 157 RFU/min (Figure 3B). Km values only varied 3-fold indicating similar binding levels to all sequence variants at X and Y. (Figure 3B). As a control, an abasic site at Y was not cleaved at all (data not shown).
Substitutions of the base at position X clearly have an impact on the incision rate (Figure 3B). There was a 2-fold preference for cleaving on the 3′-side of a pyrimidine at Y when X was a T. However, where X was A, C, or G there was a preference for a T at Y, but not for C. The presence of a G or C at position X resulted in ∼2-fold reduction in the Vmax for each substitution at Y (Figure 3B). One possible reason for this difference is that the cleaved oligonucleotide product containing G/C may have dissociated from the duplex at a lower rate in the assay. To test for this, the products of a reaction were denatured and separated on a denaturing polyacrylamide gel (Figure 3C). After 15-min incubation [during the linear part of the reaction (Figure 2A)], the substrates containing A or T at X produced more cleavage products than those with G or C at X demonstrating that this was not an artefact of the assay itself. Another possibility is that the enzyme may be separating the duplex to the base at X before cleavage and the presence of the more stable G/C pair at X may hinder this. XPF–ERCC1 cannot hydrolyse ATP so this would be a passive unwinding event, using the energy of binding of the enzyme to the substrate, or as a result of bending the substrate upon binding. To test this possibility, a mismatch was introduced opposite a T at X by substituting Xc with G. The duplex containing the mismatch annealed to form the stem–loop structure and migrated in the correct position in a non-denaturing gel (data not shown). The T/G mismatch reduced the Vmax to 75 RFU/min without a significant change in Km (16.2 nM). This suggests that either an intact duplex is required at this position for efficient substrate recognition or cleavage, or the presence of a G at Xc is inhibitory, possibly because the 2-amino group of G projects into the minor groove of the duplex and may hinder access of an amino acid side chain in the catalytic site.
Surprisingly, substitution of deoxyuridine at Y resulted in a 4.4-fold increase in Vmax over thymine, and at both X and Y resulted in a 5.2 increase in incision rate (Figure 3D) suggesting that exocyclic groups on C and T may be inhibiting entrance of these bases into a binding pocket. A 4-fold increase in Km also occurred indicating that efficient incision promoted release of the complex from its substrate. There was no difference in the incision rate when the C at position Y (and X = T) was replaced by 5-methyl C, which disrupts base stacking. Overall our data show that there was a 100-fold sequence-dependent variation in Vmax from 8.4 for CG to 906 RFU/min for UU.
Full-length XPF–ERCC1 preferentially binds to the stem–loop structure
The kinetics of stem–loop binding for full-length XPF–ERCC1 was measured by Biacore surface plasmon resonance. Five 2-fold dilutions from 30 nM were applied over an SA sensor chip with 11 RU of DNA bound. The data was analysed using the single-cycle kinetics wizard (Table 1). The high chi-square parameter indicates that the global 1:1 binding fit was not optimal. XPF–ERCC1 did not bind to the duplex containing a 2T loop, which is consistent with the gel-based nuclease assay in Figure 1. The equilibrium dissociation constants in Table 1 show that binding to the stem–loop was 7-fold tighter than to the 20T single strand representing the loop (KD 2.19 × 10−9 and 1.37 × 10−8, respectively) which indicates that the single-strand binding is a dominant contributor to the substrate interaction of full-length XPF–ERCC1.
Table 1.
Binding constants for XPF–ERCC1 binding to stem–loop and single-strand DNA
| Ka(1/Ms) | Kd (1/s) | KD (M) | χ2 (RU2) | |
|---|---|---|---|---|
| Single strand (20T) | 3.51 × 105 | 4.80 × 10−3 | 1.37 × 10−8 | 2.12 |
| Stem–loop | 5.17 × 105 | 1.14 × 10−3 | 2.19 × 10−9 | 2.72 |
| Duplex (2T loop) | No binding |
The Biacore evaluation software generated Ka and Kd by fitting the data to a 1:1 Langmuir interaction model. The equilibrium dissociation constant KD is Kd/Ka.
The HLD of XPF is required for incision activity
In order to study the contribution made by the XPF–HLD towards incision activity, an amino-terminal truncation mutant complex Δ640XPF–ERCC1 was constructed which lacked the entire XPF–HLD. This heterodimer consists of the nuclease and (HhH)2 domains of XPF with a short portion of an amino-terminal linker (30.7 kDa), dimerized with the full-length ERCC1 polypeptide (32.6 kDa) (Figure 4A, top). The truncation produced a heterodimeric complex in much greater yields (∼1 mg/l of culture) than full-length XPF-ERCC1 and had relatively little aggregate (Figure 4A, middle). No structure-specific activity was seen with the Δ640XPF–ERCC1 mutant at either low or high enzyme concentrations (50 nM and 1.3 µM) (Figure 4A, bottom), even after 16 h (data not shown).
Figure 4.
XPF-HLD is required for activity and preferentially binds stem–loops. (A) Top; gel-filtration profile of Δ640XPF–ERCC1 after separation on Superose 12 16/60. Line indicates fractions pooled for analysis. Bottom; 12% SDS–PAGE gel of fractions from gel-filtration column stained with Coomassie blue. Lanes show fractions from a Superose 12 16/60 column. Graph shows incisions by wild-type, catalytically impaired and Δ640-XPF–ERCC1 complexes at 50 nM or 1.3 µM, as indicated. (B) Top: 12% SDS–PAGE gel of purified recombinant XPF–HLD stained with Coomassie blue. Top graph: binding of purified XPF–HLD to stem loops with TT and GG substitutions at X and Y (Figure 3). Bottom graph: binding of XPF–HLD to stem loop with CT substitutions at X and Y, a 15-mer single-strand oligonucleotide, and duplex corresponding to the duplex in the stem–loop with a 2T loop, as indicated.
The HLD of XPF preferentially binds DNA structures
Purified, recombinant XPF HLD (Figure 4B, top) bound stem–loop structures without regard to sequence at the incision site (Figure 4B, middle). At 50 mM NaCl, XPF–HLD bound stem loops with an apparent KD of 63 nM (Figure 4B, bottom) but much weaker binding to a 15-mer single-strand oligonucleotide and duplex DNA with a 2T loop was observed (KD 263 and 217 nM, respectively). We conclude that XPF–HLD is necessary for measurable XPF–ERCC1 activity and that this requirement is due to its ability to bind the single strand–duplex junction.
DISCUSSION
Here we report a detailed characterization of human XPF–ERCC1 endonuclease using a model stem-loop substrate that enabled the minimal substrate requirements to be defined for a single site incision and binding of the XPF–ERCC1 complex to a DNA junction structure. We used these data to develop the first real-time fluorescence-based assay for human XPF–ERCC1 activity, with the potential to facilitate a high-throughput screening for small molecule modulators of XPF–ERCC1. The utility of such chemical inhibitors of XPF–ERCC1 would be to enhance the efficacy of treatment regimes involving cisplatin-based drugs (37,38).
XPF–ERCC1 footprint on the duplex of the stem–loop structure
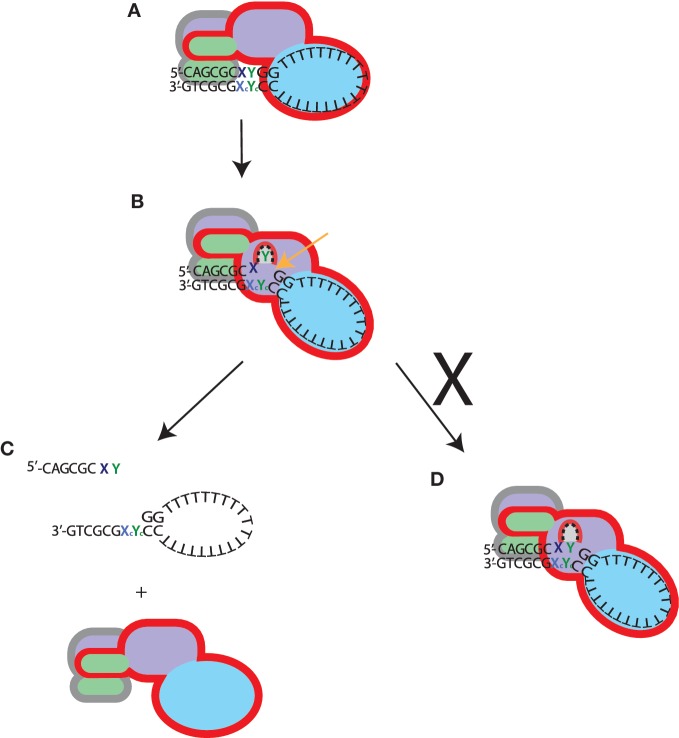
Much of our understanding of substrate interaction by XPFs comes from structural and biochemical studies of archaeal homodimers that recognize and cleave DNA structures with a downstream duplex such as 3′-flaps and nicked Holliday junctions. Models derived from these studies predict that the (HhH)2 domains of both XPF protomers bind ∼5 bp of duplex DNA upstream and downstream from the junction, with a further 5 bp required to extend into the catalytic cleft (16,17). This accounts for the observed requirement for a 10 base-pair duplex for one incision to be made. However, the second hairpin of the human XPF (HhH)2 domain is degenerate and no binding to DNA has been demonstrated. In contrast, the highly conserved ERCC1 (HhH)2 domain is capable of binding stem–loop DNA (18), and is likely to have retained the upstream duplex binding mode predicted by the A.pernix crystal structure (16). In addition, we have recently found a similar mode of double-strand DNA engagement for the homologous FancM–Faap24 complex (Coulthard et al., manuscript submitted). We anticipate that the 10-bp footprint on the duplex seen here would comprise the ERCC1 (HhH)2 domain binding the upstream 5 bp combined with sufficient DNA to enter the catalytic site of XPF (Figure 5) as described in Tripsianes et al. (26).
Figure 5.
Proposed model for stem–loop substrate recognition by XPF–ERCC1. The XPF protomer has a red outline, and ERCC1, grey outline. The domain colouring is as Figure 1: (HhH)2 domains; green, nuclease or nuclease-like domains; mauve and HLD; blue. (A) The XPF–HLD binds to the junction containing single-strand stretches greater than six bases and the hairpins of ERCC1 occupy five bases of the duplex. (B) A domain movement occurs resulting in bending of the DNA, and the nuclease domain of XPF engages the duplex downstream from the ERCC1 (HhH)2 domain. A pocket in the catalytic centre of XPF becomes occupied by the base 5′ to the scissile phosphate, and cleavage occurs. (C) After incision the complex is released from the substrate. (D) Failure of cleavage due to lack of entry of an appropriate base into the binding pocket, results in the complex remaining bound and slow dissociation from the substrate.
Role for XPF–HLD in catalytic activity and substrate binding
In our study, highly pure preparations of recombinant Δ640 XPF–full lengthERCC1 complex lacking the HLD were found to be catalytically inactive. However, a previous study concluded that a shorter recombinant Δ666 XPF–Δ95ERCC1 complex (lacking the HLD and a connecting linker to the catalytic domain) exhibited some structure-specific nucleolytic activity in a gel-based assay, despite the absence of a control for non-specific nuclease activity (19). The discrepancy between our results and those of Tsodikov et al. (19) might be explained by the differences in the XPF truncation mutants used. One explanation could be that the acidic linker spanning residues (641–666) present in our Δ640 XPF–ERCC1 complex may inhibit activity by binding DNA or blocking the catalytic site. Alternatively, we cannot rule out the possibility that a contaminating bacterial nuclease in Δ666 XPF–Δ95ERCC1 complex preparations was responsible for the observed nucleolytic activity given the long incubation periods (30 min to 11 h).
Our findings suggest a model where the stem loop is bound and possibly distorted by the XPF–HLD into a more favourable conformation for engagement of the ERCC1 (HhH)2 and XPF nuclease domains with the upstream duplex and the junction, respectively (Figure 5). Although XPF–HLD binds both single-strand and duplex DNA weakly, a 4-fold increased binding was observed on the stem–loop substrate and consequently there may be two binding sites each accommodating duplex or single-strand DNA which co-operate to bind and possibly distort the DNA around the junction. The tighter binding displayed by the full-length complex (KD 2 nM) could be due to co-operativity between the XPF–HLD (KD 60–70 nM, Figure 5) and the C-terminal complex, which have previously been shown bind a stem–loop in an electromobility-shift assay with an apparent KD of 39 ± 13 µM (18).
There are no structures available for the nuclease domain of XPF bound to DNA and little is known about how the DNA is led into the active site. This may be because substrate binding within the catalytic centre requires remodelling of the substrate or stabilization of the core endonuclease domains on the DNA by the XPF–HLD. A remodelling role has been described for crenarchael PCNA which is required to interact with XPF for activity, and co-operates with XPF to bind and kink the DNA (39). The structure of the A. pernix XPF bound to duplex DNA suggests that the duplex downstream from a nick would need to bend by 90° in order to make contact with the HhH2 domains of both protomers (16). A similar mechanism of DNA discontinuity recognition was demonstrated in the structures of Fen1 and its homologue Exo1 bound to their substrates. These proteins recognized a distortion caused by a nick in duplex DNA and this enabled binding to both upstream and downstream regions via domains conserved in this superfamily, resulting in a bend angle of 100° (40,41). The ATPase activity of the N-terminal helicase domain of Hef is stimulated by fork DNA, and its presence increases the incision activity of the core nuclease and (HhH)2 domains (42). Since human XPF–HLD does not require ATP for incision activity, alterations to the DNA structure could result from DNA binding and subsequent protein domain movements. Stable binding and distortion of the DNA structure would allow limited movement of the core domains on the duplex so that the catalytic domain is positioned at an optimal sequence and would explain why multiple incisions are made in longer duplexes. Re-incision of the same substrate is unlikely to be responsible for incisions further upstream from the junction because the same pattern of cleavage is maintained on 3′-labelled stem–loops (2,32).
Substrate interactions at the XPF active site
It is not known whether the DNA enters the XPF catalytic site as a duplex or is separated into single strands. Single-strand DNA-binding regions have been identified in both the XPF nuclease and ERCC1 nuclease-like domains, which could interact with the nucleotides downstream from the scissile phosphate (26,27). A hydrophobic patch was described in the A. pernix model, which could accommodate single-strand DNA (16). These regions may stabilize an intermediate resulting from opening of the duplex from the junction so that the DNA enters the catalytic site as a single strand. The (HhH)2 domains of both protomers of the Hef homodimer were shown to mediate opening of the duplex in a potassium permanganate footprinting study containing tracts of TA base pairs at the junction of the structure (17). The structures of the Fen1 and Exo1 5′-flap nucleases with their substrates revealed that a terminal T in the incised strand of the 5′-flap is unpaired by superfamily conserved domains in order to engage the scissile phosphodiester bonds with the two metal ion catalytic site and seven highly conserved carboxylates (40,41). The Fen1 homologues cleave 1 nt into the duplex, in contrast to XPF, which incises the phosphodiester bond between the second and the third nucleotide suggesting XPF would need to separate the terminal 2 bp. The duplex terminus of the substrates used for the Fen1 structures contained a TA pair in contrast to the stem–loop exploited for XPF–ERCC1 which has a more stable GC pair. Opening of the duplex by the XPF–HLD might occur as a result of DNA bending, or by pushing a wedge or pin into the duplex terminus. However, we were unable to demonstrate opening at the junction by potassium permanganate footprinting or 2-aminopurine fluorescence studies (data not shown). Therefore, it cannot be ruled out that the duplex remains intact at the DNA junction and the scissile base is flipped out. In a previous study in which the duplex at the junction was crosslinked by a psoralen interstrand crosslink, incisions by XPF–ERCC1 showed that they occurred at the same position as uncrosslinked duplex supporting an intact duplex model (33). In this case, the duplex may enter the active site intact and become distorted in such a way as to flip a base into a binding pocket in the XPF nuclease domain. This would explain the apparent reliance of incision rate on the stability of the base pair at X. The bases at X/Xc may be separated by a component of the XPF catalytic domain entering the duplex to flip out the base. A candidate for this is the R689 of XPF, which is absolutely required for activity (25). The equivalent R26 in A. pernix was part of a functionally important region of the catalytic site and not directly involved in catalysis or protein folding (16). It was postulated to bind to assist in anchoring the backbone of the DNA at the junction but it may be important for intercalating into the duplex and distortion of the duplex or base flipping.
Implications for cellular XPF–ERCC1 substrates
It is unclear what role sequence preferences displayed here by XPF–ERCC1 might play in NER as filling in of the excised patch replaces lost bases, but it may be significant for other processes that XPF–ERCC1 is involved with in the cell. For instance, it may serve to regulate the activity of XPF–ERCC1 on GC rich regions of the genome, or have a role in directing incisions of the TTAGGG 3′-telomere overhangs. In a cellular context, structures that XPF–ERCC1 cleaves includes bubbles, flaps, splayed arm and overhangs and possibly more complex structures depending on the process. The local cellular conditions will affect the activity of XPF–ERCC1 in vivo and evidently it co-operates with many other protein partners in carrying out its endonucleolytic function in NER, ICL repair and other genome maintenance processes. The accepted model stem–loop substrate used in our present studies has enabled us to examine the conditions under which one incision was made and appears to be a good surrogate for the distorted physiological junction substrates presented to XPF–ERCC1 within a cell nucleus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary figures 1 and 2.
FUNDING
Funding for open access charge: Cancer Research UK core funding to the London Research Institute (to N.Q.M.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Rick Wood for the original pET 30b ERCC1-XPF dicistronic construct (33), Rachel Coulthard for help with fluorescence polarization experiments, Judith Murray-Rust for advice on design of the HLD-XPF construct, and Tamar Shafir and Catherine McDonnell at GE Healthcare for help with the collection and analysis of Biacore experiments. The authors also acknowledge access to the T200 Biacore facility at the Wolfson Institute, UCL.
REFERENCES
- 1.Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, Lindahl T, Lowndes N, Sarasin A, Wood RD. DNA repair: from molecular mechanism to human disease. DNA Repair. 2006;5:986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 3.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sargent RG, Meservy JL, Perkins BD, Kilburn AE, Intody Z, Adair GM, Nairn RS, Wilson JH. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 2000;28:3771–3778. doi: 10.1093/nar/28.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 7.Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol. Cell Biol. 2004;24:1200–1205. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McWhir J, Selfridge J, Harrison DJ, Squires S, Melton DW. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat.Genet. 1993;5:217–224. doi: 10.1038/ng1193-217. [DOI] [PubMed] [Google Scholar]
- 9.Westerveld A, Hoeijmakers JH, van Duin M, de Wit J, Odijk H, Pastink A, Wood RD, Bootsma D. Molecular cloning of a human DNA repair gene. Nature. 1984;310:425–429. doi: 10.1038/310425a0. [DOI] [PubMed] [Google Scholar]
- 10.Jaspers NG, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, Robinson AR, Giglia-Mari G, Hoogstraten D, Kleijer WJ, Hoeijmakers JH, et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am. J. Hum. Genet. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregg SQ, Robinson AR, Niedernhofer LJ. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair. 2011;10:781–791. doi: 10.1016/j.dnarep.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White MF. Archaeal DNA repair: paradigms and puzzles. Biochem. Soc. Trans. 2003;31:690–693. doi: 10.1042/bst0310690.. [DOI] [PubMed] [Google Scholar]
- 13.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 14.Macaisne N, Novatchkova M, Peirera L, Vezon D, Jolivet S, Froger N, Chelysheva L, Grelon M, Mercier R. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr. Biol. 2008;18:1432–1437. doi: 10.1016/j.cub.2008.08.041. [DOI] [PubMed] [Google Scholar]
- 15.Gaillard PH, Wood RD. Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res. 2001;29:872–879. doi: 10.1093/nar/29.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman M, Murray-Rust J, Lally J, Rudolf J, Fadden A, Knowles PP, White MF, McDonald NQ. Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. EMBO J. 2005;24:895–905. doi: 10.1038/sj.emboj.7600581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishino T, Komori K, Ishino Y, Morikawa K. Structural and functional analyses of an archaeal XPF/Rad1/Mus81 nuclease: asymmetric DNA binding and cleavage mechanisms. Structure. 2005;13:1183–1192. doi: 10.1016/j.str.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 18.Tripsianes K, Folkers G, Ab E, Das D, Odijk H, Jaspers NG, Hoeijmakers JH, Kaptein R, Boelens R. The structure of the human ERCC1/XPF interaction domains reveals a complementary role for the two proteins in nucleotide excision repair. Structure. 2005;13:1849–1858. doi: 10.1016/j.str.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Tsodikov OV, Enzlin JH, Scharer OD, Ellenberger T. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc. Natl Acad. Sci. USA. 2005;102:11236–11241. doi: 10.1073/pnas.0504341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sgouros J, Gaillard PH, Wood RD. A relationship betweena DNA-repair/recombination nuclease family and archaeal helicases. Trends Biochem. Sci. 1999;24:95–97. doi: 10.1016/s0968-0004(99)01355-9. [DOI] [PubMed] [Google Scholar]
- 21.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 23.Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol. Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enzlin JH, Scharer OD. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 2002;21:2045–2053. doi: 10.1093/emboj/21.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tripsianes K, Folkers GE, Zheng C, Das D, Grinstead JS, Kaptein R, Boelens R. Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization. Nucleic Acids Res. 2007;35:5789–5798. doi: 10.1093/nar/gkm503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsodikov OV, Ivanov D, Orelli B, Staresincic L, Shoshani I, Oberman R, Scharer OD, Wagner G, Ellenberger T. Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. EMBO J. 2007;26:4768–4776. doi: 10.1038/sj.emboj.7601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 30.Tapias A, Auriol J, Forget D, Enzlin JH, Scharer OD, Coin F, Coulombe B, Egly JM. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J. Biol. Chem. 2004;279:19074–19083. doi: 10.1074/jbc.M312611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Scharer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Laat WL, Appeldoorn E, Jaspers NG, Hoeijmakers JH. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- 33.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 34.Roberts JA, White MF. DNA end-directed and processive nuclease activities of the archaeal XPF enzyme. Nucleic Acids Res. 2005;33:6662–6670. doi: 10.1093/nar/gki974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts JA, White MF. An archaeal endonuclease displays key properties of both eukaryal XPF-ERCC1 and Mus81. J. Biol. Chem. 2005;280:5924–5928. doi: 10.1074/jbc.M412766200. [DOI] [PubMed] [Google Scholar]
- 36.Roberts JA, Bell SD, White MF. An archaeal XPF repair endonuclease dependent on a heterotrimeric PCNA. Mol. Microbiol. 2003;48:361–371. doi: 10.1046/j.1365-2958.2003.03444.x. [DOI] [PubMed] [Google Scholar]
- 37.Kirschner K, Melton DW. Multiple roles of the ERCC1-XPF endonuclease in DNA repair and resistance to anticancer drugs. Anticancer Res. 2010;30:3223–3232. [PubMed] [Google Scholar]
- 38.Wood RD. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environ. Mol. Mutagen. 2010;51:520–526. doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutton RD, Craggs TD, White MF, Penedo JC. PCNA and XPF cooperate to distort DNA substrates. Nucleic Acids Res. 2010;38:1664–1675. doi: 10.1093/nar/gkp1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orans J, McSweeney EA, Iyer RR, Hast MA, Hellinga HW, Modrich P, Beese LS. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.