Abstract
X-ray crystallography provides excellent structural data on protein–DNA interfaces, but crystallographic complexes typically contain only small fragments of large DNA molecules. We present a new approach that can use longer DNA substrates and reveal new protein–DNA interactions even in extensively studied systems. Our approach combines rigid-body computational docking with hydrogen/deuterium exchange mass spectrometry (DXMS). DXMS identifies solvent-exposed protein surfaces; docking is used to create a 3-dimensional model of the protein–DNA interaction. We investigated the enzyme uracil-DNA glycosylase (UNG), which detects and cleaves uracil from DNA. UNG was incubated with a 30 bp DNA fragment containing a single uracil, giving the complex with the abasic DNA product. Compared with free UNG, the UNG–DNA complex showed increased solvent protection at the UNG active site and at two regions outside the active site: residues 210–220 and 251–264. Computational docking also identified these two DNA-binding surfaces, but neither shows DNA contact in UNG–DNA crystallographic structures. Our results can be explained by separation of the two DNA strands on one side of the active site. These non-sequence-specific DNA-binding surfaces may aid local uracil search, contribute to binding the abasic DNA product and help present the DNA product to APE-1, the next enzyme on the DNA-repair pathway.
INTRODUCTION
The determination of protein–DNA interactions can be instrumental for understanding function, for designing experiments to probe biological mechanisms and for developing new drugs. X-ray crystallography provides high-resolution structures of protein–DNA complexes, but these complexes are difficult to crystallize. When high-quality crystals of protein–DNA complexes are obtained, they typically contain only small DNA fragments due to constraints imposed by crystal packing (1). Therefore, there is a need for new methods that examine the complete protein–DNA interaction. Here, we use an innovative experimental/theoretical approach that combines rigid-body macromolecular docking with hydrogen/deuterium exchange mass spectrometry (DXMS). Computational docking guides the design of the DXMS experiment, which identifies exposed protein surfaces in a solution environment. Docking is then used to interpret the experimentally determined DNA footprint, producing a 3-dimensional model of the protein–DNA interaction.
Despite significant progress in applying macromolecular docking methods to protein–protein complexes (2–4), the prediction of protein–DNA interactions remains a largely unaddressed challenge (5). So far, there have been only a few applications of macromolecular docking methods to the prediction of protein–DNA complexes (6–14). We developed the global, systematic search program DOT (15,16), in which interaction energies are calculated as the sum of electrostatic and van der Waals components. Tests on transcription factor proteins demonstrated that rigid-body docking with DOT successfully identified dsDNA-binding sites and the orientation of the DNA at those sites (9). Furthermore, the ensembles of favorable DNA placements indicated the degree of bending of bound DNA over the protein surface.
DXMS has proved a powerful method for studying protein interactions. In DXMS, hydrogen/deuterium exchange is followed by protein proteolysis and characterization of the resulting peptides by mass spectrometry, revealing the degree of solvent exposure for backbone amide hydrogen atoms throughout the protein chain. By examining the change in solvent exposure between the unbound and DNA-bound protein, DXMS has the potential to reveal the DNA footprint on the protein surface (17,18).
We applied our combined computational docking/DXMS approach to the essential DNA-repair enzyme uracil-DNA glycosylase (UNG), which cleaves uracil from ssDNA and dsDNA by hydrolysis of the N-glycosylic bond between uracil and the deoxyribose. Extensive studies of UNG (19,20) include examination of its sequence specificity (21,22), its interactions with undamaged DNA (23–25) and with other proteins involved in DNA binding and repair (26–28), and its search mechanism (20,29,30). Crystallographic structures of the catalytic domain of human UNG bound to a 10 base pair abasic DNA product (26,31) and to dsDNA analogs (24,32) show that uracil and its associated sugar are rotated ≈180° out of the base stack. The UNG active site consists of a deep uracil-binding pocket with an overlying groove that binds one DNA strand (see Supplementary Figure S1A). The Leu 272 side chain inserts through the DNA minor groove to replace the flipped-out uracil nucleotide. Mutagenesis studies have identified active-site residues involved in catalysis and specificity of the uracil-binding pocket (31,33,34) and support the proposed role of residue Leu 272 (31).
Our goals were to evaluate the ability of our combined approach to identify the known UNG active site and to probe the full extent of the UNG–DNA contact surface. The solution DXMS studies allowed use of a 30 bp DNA fragment, considerably longer than the 10 bp DNA used in the X-ray crystallographic studies. The exhaustive search performed by the computational docking algorithm explored the entire surface of UNG. Together, these two techniques provide strong evidence that the DNA-binding surface on UNG extends considerably beyond the immediate active site.
MATERIALS AND METHODS
Protein and DNA preparation for DXMS
The full catalytic domain of human UNG (21) was expressed and purified. In this UNG construct, the 85 N-terminal residues were replaced by a 22 amino acid His tag (MGSSHHHHHHSSGLVPRGSHMG). The final UNG stock solution had a protein concentration of 9.9 mg/ml (0.36 mM) in a buffer of 10 mM Tris, 10mM NaCl, 1mM DTT, pH 7.5. Oligonucleotides were obtained from IDT (Integrated DNA Technologies). One strand contained deoxy-U: 5′-ctgtuatcttgatcgatcgatcgatcgatc-3′. The other strand was biotinylated at the 5′-end: 5′-biotin-gatcgatcgatcgatcgatcaagatgacag-3′. The two oligos were annealed to give 30 bp dsDNA with a U:G mismatch pair. The DNA stock solution had a DNA concentration of 56 mg/ml (2.97 mM) in 10 mM NaCl, 10 mM Tris, pH 7.5.
Characterization of the UNG–DNA complex
We verified that a 1:1 complex of UNG with product DNA was formed under the conditions and protein and DNA concentrations used in the DXMS experiments. Activity assays were performed with [3H]dUMP-containing DNA substrate in the presence or absence of the same amount of non-labeled DNA used in the DXMS experiments. The reactions (21 μl) contained 1.8 nmol UNG in 3.4 mM Tris, pH 7.4, 10 mM NaCl, 0.28 mM DTT and 1.8 μM [3H]dUMP-containing calf thymus DNA (sp. act. 0.5 mCi/mmol). Prior to addition of UNG, 56 μg of non-labeled calf thymus DNA was added to half of the reaction mixtures. After incubation for 90 min at room temperature, the amount of uracil released was measured as described (35). Notably, all DNA uracil was released from the substrate both in the presence and absence of high concentrations of non-labeled DNA (data not shown), demonstrating that UNG was active under the assay conditions.
The solution state of the UNG–DNA complex was assessed using multiangle light scattering (MALS) mass measurements (Supplementary Methods). UNG–DNA complexes were examined at 1:1 and 1:1.6 stoichiometric ratios. UNG at a concentration of 5 mg/ml (in 10 mM NaCl, 10 mM Tris, pH 7.5) was pre-incubated with either 1 or 1.6 equivalents of the 30 bp dsDNA at room temperature for 90 min. Notably, the two stoichiometric ratios gave similar results: a single elution peak with light scattering masses of 35.39 (±3%) and 38.97 (±4%) kDa for the 1:1 and 1:1.6 ratios, respectively. The DNA has a mass of 18.8 kDa, consistent with a 1:1 complex of UNG and DNA (36–38) at both a 1:1 ratio and the 1:1.6 ratio used in the DXMS experiments.
Establishing optimal proteolysis conditions for DXMS
We first determined the concentrations of the denaturant guanidine hydrochloride that gave overlapping peptides spanning the full UNG sequence. Peptides should be long enough to be uniquely identified but short enough to localize changes in solvent protection. Samples of the UNG stock solution (5 μl, 1.8 nmol) were diluted with 15 μl of 1.7 mM Tris (pH 7.1), 10 mM NaCl, and then mixed with 30 μl of quench solution [0.08 M, 0.8 M, 1.6 M, 3.2 M or 6.4 M guanidine hydrochloride in 0.8% (v/v) formic acid, 16.6% (v/v) glycerol] on ice. The UNG samples were then subjected to proteolysis, and the resulting peptides were separated and analyzed by mass spectrometry (39) (details in Supplementary Methods). The resulting fragmentation maps revealed that both 0.8 M and 3.2 M guanidine hydrochloride (0.5 M and 2.0 M final concentration) gave the best results.
DXMS
After establishing good fragmentation maps, UNG and the UNG–DNA complex were subjected to hydrogen/deuterium exchange experiments followed by mass spectrometry analysis to determine the degree of deuteration of UNG backbone amide hydrogen atoms. To prepare samples of the UNG–DNA complex, 5 μl of the UNG stock solution (1.8 nmol) was incubated with 1.0 μl (3.0 nmol) of the dsDNA stock solution (ratio of 1:1.6 UNG:DNA) at room temperature for 90 min then cooled to 0° C. Deuterium oxide (D2O) buffer at 0° C [15 μl, 1.7 mM Tris, 10 mM NaCl, pD (read) 7.1] was added to 5 μl samples (1.8 nmol) of UNG and to the prepared UNG–DNA samples. Samples were incubated for 10, 30, 100, 300, 1000, 3000 and 10 000 s at 0° C and for 1000, 3000, 10 000 and 30 000 s at room temperature. Hydrogen exchange rates are about 10 times faster at room temperature (40), so these experiments are equivalent to 10 000, 30 000, 100 000 and 300 000 s at 0° C. The data shown in Figure 2 and Supplementary Tables S1–S3 are given in terms of the deuteration times at 0° C. Samples were quenched with 30 μl of 0.5 M (final concentration) guanidine hydrochloride solution for UNG or with 30 μl of 2.0 M (final concentration) guanidine hydrochloride solution for the UNG–DNA complex, and then proteolyzed and analyzed by mass spectrometry (Supplementary Methods). Non-deuterated and fully deuterated samples were analyzed for comparison. UNG (5 μl) was mixed with 15 μl of 1.7 mM Tris (pH 7.1), 10 mM NaCl on ice (non-deuterated sample) or with 15 μl 0.5% formic acid in D2O overnight at room temperature (fully deuterated sample). To check for consistency and correct peptide identification, we examined all overlapping peptides within each data set. Each peptide typically was present in multiple charge states, with each identified and analyzed independently, providing a further check of consistency.
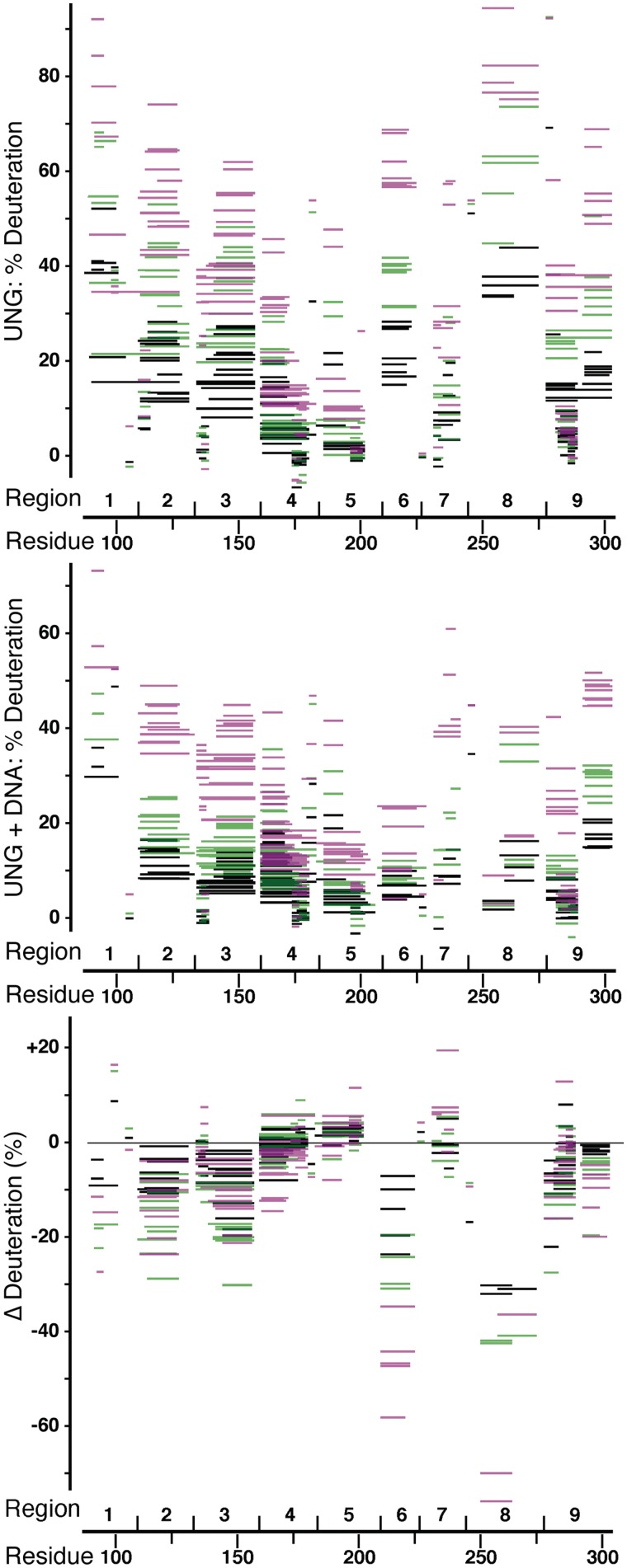
Figure 2.
Percent deuterium incorporation for peptides after 30 (black), 300 (green) or 10000 s (magenta) are shown for UNG (top) and the UNG–DNA complex (middle). The change in deuteration (bottom) is shown for peptides common to both data sets, where a negative percentage indicates less deuteration in the UNG–DNA complex. Regions are defined as in Supplementary Figure S3 and Supplementary Tables S1–S3.
Docking procedure and analysis
Coordinates of unbound UNG (PDB code 1AKZ, resolution 1.57 Å) (33) and of UNG bound to a 10 bp DNA fragment base (1SSP, resolution 1.9 Å) (26) were obtained from the Protein Data Bank (41). The human UNG construct in the crystallographic structures contains the full catalytic domain, with the N-terminal tail (84 residues) replaced by Met-Glu-Phe. In 1SSP, the DNA strand bound in the active site has the sequence 5′-ctgtuatctt-3′, but the uracil base is cleaved. The complementary strand has A opposite the U and an additional 5′ overhang of a single adenine. A linear 11 bp B-DNA model was built with the Nucleic Acid Builder (NAB) program (42) (see Supplementary Methods) with the sequences 5′-ctgtuatcttt-3′ and 5′-aaagatgacag-3′, creating a U:G mismatch pair. Minimization with AMBER 8 using the generalized Born model (43,44) gave a wobble geometry with two hydrogen bonds for the U:G pair.
Docking calculations were performed with the program DOT (15,16), which is part of the DOT2 Suite distributed by the Computational Center for Macromolecular Structure at the San Diego Supercomputer Center (URL: http://www.sdsc.edu/CCMS). The DNA molecule, represented by its atomic positions with partial atomic charges, was systematically moved within the shape and electrostatic potentials calculated for the stationary UNG molecule. Potentials were calculated using utilities in the DOT2 Suite (Supplementary Methods), including use of the program REDUCE (45) to add hydrogen atoms, determine His side chain protonation states, and correct the geometry of Asn, Gln and His side chains; the program MSMS (46) to calculate molecular surfaces that encompass the volumes defining the UNG shape potential; the AMBER library of heavy atoms with added polar hydrogens (47) to assign partial atomic charges; and the program UHBD (48) to calculate the electrostatic potential of UNG by finite difference methods to solve the linearized Poisson–Boltzmann equation. Poisson–Boltzmann methods take into account the effects of dielectric, solvation and ionic strength on the electrostatic potential. The continuous electrostatic potential was then modified (9,16) to be compatible with the discontinuous shape potential.
Docking calculations (Supplementary Methods) used a cubic grid 128 Å on a side with 1 Å grid spacing (about 2.1 million points). The DNA was centered at each grid point in 54 000 distinct orientations, giving ≈108 billion placements of the DNA about UNG. The 2000 placements with the most favorable interaction energies, calculated as the sum of electrostatics and van der Waals intermolecular energy terms, were kept. These energies were mapped to the grid point at which the DNA was centered, allowing the distribution of the placements over the UNG to be visualized. The 30 top-ranked placements using coordinates from the UNG–DNA crystallographic complex were analyzed by calculating the rmsd between the docked DNA and the crystallographic DNA. Calculation of rmsd is a poor method for clustering B-DNA placements (6,9), because lack of sequence recognition results in shifts along the DNA axis by one or more base pairs within the same cluster. Instead, the 30 top-ranked placements and the distribution of the 2000 top-ranked placements over the UNG surface were analyzed with computer graphics (Figure 1C and Supplementary Figure S2A).
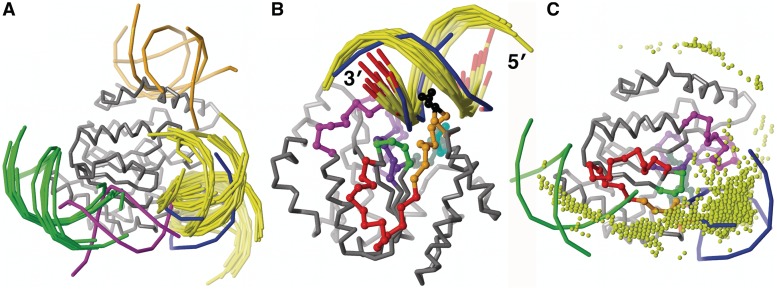
Figure 1.
B-DNA docked to the DNA-bound structure of UNG (gray Cα backbone). (A) The 30 top-ranked B-DNA placements compared with bound DNA from 1SSP (blue phosphate backbone): 21 (yellow) at the active site; 6 (green) tightly clustered at a secondary site; 1 (magenta) between the active site and the secondary site; and 2 (orange) near the UNG N-terminus. (B) The larger active-site cluster (14 placements) replicates the UNG–DNA active-site contacts found in the 1SSP complex, including insertion of Leu 272 (black) into the DNA minor groove. These dockings also show direct contact of the complementary strand with residues 210–220 (magenta). In all, the active-site strand has the same 5′ to 3′ direction as the crystallographic DNA, as indicated by red coloring of the 3′-ends. The UNG backbone is colored by the DXMS results (see Figure 3). (C) The 2000 top-ranked B-DNA placements, represented by their geometric centers (spheres), are concentrated over the active site (indicated by the crystallographic DNA, blue, right), at the secondary site (indicated by docked B-DNA, green, left), and between the two sites.
RESULTS
We applied computational docking and DXMS to the interaction of the catalytic domain of human UNG with DNA. Using the macromolecular docking program DOT (15,16), the DNA was systematically translated and rotated around UNG, resulting in ≈108 billion placements of the DNA, which were ranked by the sum of electrostatic and van der Waals energies. The electrostatic energy term is an essential component of the ranking function for highly polar interactions. For protein–DNA complexes, DOT provides a good approximation of the electrostatic energy calculated for the full complex by Poisson–Boltzmann methods (9).
Three computational dockings were done. First, UNG and DNA coordinates from the crystal complex (PDB code 1SSP) (26) were docked to test the docking parameters and energy evaluation. Second, the DNA-bound UNG coordinates and a linear B-form DNA model (B-DNA) were docked to evaluate the fit of B-DNA to the optimized active site. Third, B-DNA and the unbound human UNG structure (PDB code 1AKZ) (33) were docked to evaluate the ability to identify critical features of the biological interaction in the absence of the known structure of the complex.
We performed two DXMS experiments: UNG alone and UNG bound to a 30 bp dsDNA fragment that contained a U:G base pair. Before the hydrogen/deuterium exchange experiment, the DNA and UNG were preincubated, resulting in the formation of the complex of UNG with product DNA from which the uracil base had been cleaved.
UNG–DNA: docking coordinates from the crystal complex
Docking the abasic DNA product to the DNA-bound UNG coordinates reproduced the crystallographic UNG–DNA complex 1SSP. The 1SSP UNG construct (26) contains the full catalytic domain, but lacks 84 N-terminal residues of full-length UNG. In the 10 bp 1SSP DNA, the uracil has been cleaved from the strand bound in the UNG active site and there is a 1-nt overhang at the 5′-end of the complementary strand. Twenty-seven of the 30 top-ranked DNA placements docked close to the crystallographic position (rmsd <5Å; Table 1) and show an excellent fit to the 4 nt in the UNG active-site groove (Supplementary Figure S1A). As in 1SSP, neither the DNA major groove, which lies over residues 210–220 on the 3′ side of the active site (direction based on the active-site strand), nor the minor groove on the opposite (5′) side of the active site directly contacted UNG. Two of the 30 top-ranked DNA placements (Table 1 and Supplementary Figure S1B) bound the complementary DNA strand in the active-site groove, demonstrating that a DNA strand with geometry close to B-DNA and with stacked bases can fit into the UNG active site. These two placements showed potential new contacts with UNG residues 210–220 on the 3′ side of the active site. The final placement in the top 30 overlapped the 3′ side of the crystallographic DNA and extended over the surface created by residues 248–268 (Supplementary Figure S1B). Thus, this rigid-body docking unambiguously identified the crystallographic complex as the dominant cluster, but also suggested additional UNG–DNA contacts not present in 1SSP.
Table 1.
Distribution of top 30 DNA placements from computational docking over the UNG surface
| Molecules | Number | UNG region | Cluster description; new contactsa |
|---|---|---|---|
| UNG (bound, 1SSP) + | 27 | Active site | Rmsdb <5 Å with 1SSP DNA; contacts as in 1SSP complex |
| DNA (bound, 1SSP) | 2 | Active site | Complementary strand in active site; residues 210, 212–215, 218, 220 |
| 1 | Between sitesc | Partial overlap with active site; residues 248, 250–251, 253, 265–266 | |
| UNG (bound, 1SSP) + | 14 | Active site | Good alignment with 1SSP DNA; residues 210, 212–216, 218–220 |
| B-DNA model | 7 | Active site | Rotated ≈35° relative to 1SSP DNA toward secondary site; residues 248, 251 |
| 6 | Secondary site | Tight cluster; residues 258–265, 286, 293, 296 | |
| 1 | Between sites | Contacts active-site residues 247 and 276; residues 248, 251, 258–259, 262–265 | |
| 2 | UNG N-terminus | Variable orientation | |
| UNG (unbound, 1AKZ) + | 1 | Active site | Correct 5′ to 3′ direction for strand bound in active-site |
| B-DNA model | 22 | Active site | Reversed 5′ to 3′ direction for strand in active site |
| 5 | Secondary site | Tight cluster; residues 258–260, 286, 293, 296 | |
| 2 | Between sites | Variable orientation |
aUNG residues within 4.5 Å of docked DNA that are not listed in Parikh et al. (1998) (26) for the structure of the UNG–DNA complex (1SSP).
bThe rmsd values were calculated between the heavy (non-hydrogen) atoms of the docked DNA placements and the crystallographic position of the DNA, given a fixed position for UNG.
cDNA placements lying between the active site and the predicted secondary site, with partial overlap of one or both sites.
Docking B-DNA to the DNA-bound UNG structure
We next investigated if the pre-formed DNA-binding site on UNG could accommodate B-DNA. Our 11 bp B-DNA model matched the sequence of the crystallographic DNA, except that the U:A base pair was replaced with a U:G mismatch, the best substrate for UNG. Since UNG interrogates extrahelical bases (23,24), we did not expect specific recognition of U within our B-DNA model. Therefore, we defined clusters based on shared alignment of the DNA axis and phosphate groups, allowing shifts along the DNA axis by one or more base pairs.
Twenty-one of the 30 most favorable B-DNA placements bound in the active-site groove (Figure 1A) with the correct 5′ to 3′ direction of the active-site strand and Leu 272 inserted into the DNA minor groove. Two distinct active-site clusters were formed (Table 1). In the larger cluster (14 structures), one DNA strand fully occupied the active-site groove (Figure 1B). The complementary strand contacted residues 210–220 on the 3′ side of the UNG active site (Table 1). In the smaller cluster (seven structures), one DNA strand only partially filled the active-site groove and showed new contacts with the Tyr 248 and Lys 251 side chains.
Unexpectedly, a secondary DNA-binding site distant from the UNG active site was found (green, Figure 1A, Table 1). The docked cluster (six structures) showed tight alignment of the phosphate backbones and the DNA axes, with a maximum spread of 9°, which includes translation over the curved UNG surface. Potential DNA-contacting residues include the positively charged side chains of Lys 259, 286, 293 and 296 and Arg 258 and 260. Residues 258–260 are closest to the active site, about 30 Å from the uracil-binding pocket. The smaller active-site cluster is pointed towards this secondary DNA-binding site. One B-DNA placement (magenta, Figure 1A) partially overlapped this active-site cluster and the secondary-site cluster, suggesting a continuous DNA-binding surface from the active site to the secondary DNA-binding site.
Two structures (orange, Figure 1A) lay near the truncated N-terminus of the UNG catalytic domain. These would clash with the N-terminal region of full-length UNG. In 1SSP, bound DNA contacts the truncated N-terminus of a UNG molecule in a neighboring asymmetric unit. Thus, computational docking indicated a non-physiological UNG-DNA crystallographic interaction.
The distribution of the 2000 most favorable B-DNA placements (Figure 1C) supported a DNA-binding surface that extends from the active site to the predicted secondary DNA-binding site. The preponderance docked at the active site, at the well-defined secondary binding site, or between the two sites.
Docking B-DNA models to the free UNG structure
Finally, we applied DOT to the typical situation where only the isolated structure of the protein is known. The most favorable 30 and 2000 B-DNA placements showed the same distribution, with the majority docked at the active site and others at the secondary DNA-binding site or between the two sites (Table 1 and Supplementary Figure S2A). The majority at the active site showed a reversed 5′ to 3′ direction for the active-site DNA strand, positioning the DNA major groove, rather than the minor groove, over Leu 272 (Supplementary Figure S2B). Relaxing the shape fit, which can be useful for unbound protein–protein dockings (16), did not improve results (Supplementary Methods), as we also found when docking DNA to transcription factors (9).
DXMS of free and DNA-bound UNG
UNG alone and the pre-formed UNG–DNA complex were examined by DXMS. DXMS experiments were performed at low ionic strength (10 mM NaCl), where recombinant UNG shows high activity (21), good substrate binding (20,21), a highly efficient search mechanism (29,30,49), and efficient uracil excision (29). All of these functions become less efficient as the ionic strength is raised (29), with no detectable activity at 200 mM NaCl (21). The low ionic strength should also shift the equilibrium of the UNG–DNA product interaction to the bound state, enhancing our ability to detect changes in solvent protection.
For the UNG–DNA complex, we designed a 30 bp dsDNA with the potential to reach from the active site to the secondary DNA-binding site predicted by computational docking. One end of the DNA matched the sequence of the 10 bp fragment in 1SSP, except that the U:A pair was replaced by a U:G pair. Twenty base pairs were added to the 3′-end of the U-containing strand. To form the UNG–DNA complex, a 1:1.6 ratio of UNG and the designed U-containing DNA were preincubated for 90 min. After this time, uracil cleavage was complete, as shown by activity assays, and a 1:1 complex was formed, as determined by MALS (‘Materials and Methods’ section).
In the DXMS studies, UNG alone and the pre-formed UNG–DNA complex were exposed to deuterium for 10 time points ranging from 10 to 300000 s. The solutions were then quenched and subjected to protease digestion. The resulting peptides were separated by HPLC and analyzed by electrospray mass spectrometry. For UNG alone, 128 overlapping peptides were identified. For the UNG–DNA complex, 138 overlapping peptides were identified. In both cases, peptides spanned the entire UNG sequence. The two data sets shared 108 peptides, allowing direct comparisons of the change in deuteration. The percentage of deuteration for each peptide was determined by comparison with undeuterated and fully deuterated UNG samples that were subjected to the same quench and proteolysis conditions (‘Materials and Methods’ section).
The first two backbone nitrogen atoms of each peptide exchange rapidly under the experimental conditions following the deuteration step (40), so neither contributes to the deuteration count. For example, the mass envelope corresponding to peptide 140–157 provides deuteration information only for residues 142–157. We use the residue range for which there is deuteration information in all Figures and Tables, for example residues 142–157, rather than the full peptide.
Peptides with 4–25 amides common to UNG (Supplementary Table S1) and the UNG–DNA complex (Supplementary Table S2) were analyzed to obtain the change in deuteration (Supplementary Table S3). In these Tables and Figure 2, which shows the 30, 300 and 10000 s deuteration times, peptides are assigned to nine distinct regions following the fragmentation pattern. UNG showed no continguous unstructured regions, which would be fully deuterated within 10 s (50). In free UNG, most peptides showed <40% deuteration at the 30 s deuteration time (Figure 2, top), with deuteration gradually increasing at longer times. At 10 000 s, regions 4 and 5 showed the least deuteration (10–50%) and regions 1 and 8 showed the most deuteration (75–100%).
Examination of non-overlapping peptides at all deuteration times (Supplementary Figure S3) revealed three regions with significant decreases in deuteration: residues 142–157 (region 3), 210–220 (region 6) and 245–274 (region 8). Together with active-site residues 160–170 (region 4), which showed a subtle change in deuteration, these regions include all of the DNA- and uracil-contacting UNG residues in the UNG–DNA crystallographic structures (26), except for residues 275 and 276. In addition, residues 111–131 (region 2) and 277–290 (region 9) showed small decreases in deuteration upon DNA binding.
Active site residues 142–157 (region 3) and 160–170 (region 4)
Residues 142–158 play a central role in binding the uracil ring and the catalytic water molecule (26). Residues 142–157 showed a decrease of 1–3 deuterons in the presence of DNA (Figure 3A). Overlapping residues 142–158 and 145–157 in region 3 showed consistent decreases (Supplementary Table S3).
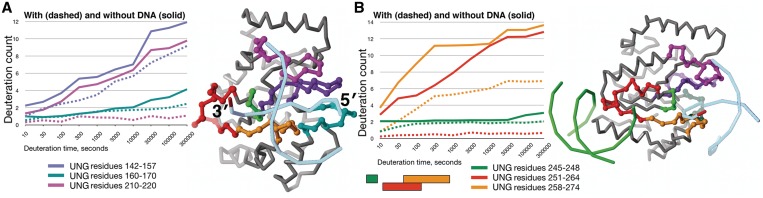
Figure 3.
Distribution of UNG regions showing significant solvent protection in the presence of DNA. (A) Peptides on the active-site face of UNG. Residues 142–157 (purple), 160–170 (blue green) and 210–220 (magenta) are highlighted on the UNG Cα backbone (right) and correspond to the deuteration profiles (left). Residues 210–220 show the greatest change in solvent protection, but have no contact with the bound DNA product (light blue phosphate backbone) in the 1SSP crystallographic structure. (B) Peptides that span the region between the active site and the secondary DNA-binding site predicted by computational docking. Residues 245–248 (green) and residues 251–264 (red) are highlighted on the UNG structure (right) and correspond to deuteration profiles. Residues 265–274 (orange) are shown on the UNG structure (left), but the deuteration profile is for residues 258–274, which partially overlap residues 251–264 (red). Residues 251–264 show the greatest change in solvent protection and make up part of the predicted secondary DNA-binding site (indicated by the docked DNA, green phosphate backbone), but have no DNA contacts in 1SSP.
Residues 160–170 showed a subtle difference in deuteration in the free and DNA-bound UNG states that appeared with deuteration times longer than 10000 s (Figure 3A). With five Pro residues within this segment, at most six amides can exchange. At long deuteration times, free UNG picked up two additional deuterons, while DNA-bound UNG had little change. This pattern was consistent with the deuteration profiles of residues 160–171, 161–170 and 160–174 in region 4. In bound UNG, a hydrogen bond is formed between the Ser 169 amide and a DNA phosphate group (26). In free UNG (33), the Ser 169 amide forms a hydrogen bond with a water molecule that fits into a pocket on the UNG surface and therefore may exchange slowly with bulk solvent. A plausible explanation of the deuteration profile for residues 160–170 is that two amides among residues 160–162 and 164, which are away from the active site, exchange with solvent similarly in free and DNA-bound UNG, whereas slowly exchanging residues 169 and 170 exchange even more slowly in DNA-bound UNG.
Strong protection of residues 210–220 (region 6) and 251–264 (region 8) in the presence of DNA
The striking decrease in deuteration in the presence of DNA found for residues 210–220 (Figure 3A) was also seen for residues 210–222 and 210–224 (Figure 2, Supplementary Table S3). This strong protection is inconsistent with the UNG–DNA crystallographic structures, in which the DNA major groove lies over residues 213–219, but the molecular surfaces are separated by at least 7 Å as a result of packing interactions in the crystal (see ‘Discussion’ section). Residues 114–126 (region 2), of which residues 116–119 contact residues 210–220, were responsible for the decrease of 1–2 deuterons seen for residues 111–131 in the presence of DNA.
Analysis of overlapping peptides within residues 245–274 revealed that the region farthest from the active site, residues 251–264, was primarily responsible for the observed decrease in deuteration upon DNA binding (Figure 3B). As observed for residues 210–220, residues 251–264 showed a striking decrease in deuteration in the presence of DNA, with one deuteron incorporated. Residues 251–264 include the solvent-exposed loop 258–262 (sequence Arg-Lys-Arg-His-His), which forms part of the secondary DNA-binding site predicted by computational docking. The decrease of 1–2 deuterons in the presence of DNA for residues 277–290 can be explained by the decrease seen for residues 277–280. These residues contact the β-strand formed by residues 262–267 and immediately follow residues Tyr 275 and Arg 276, which contact the DNA minor groove on one side of the active site.
Despite the DNA contacts with UNG seen in 1SSP, the beginning and ending segments of residues 245–274 showed smaller changes in their deuteration profiles in the presence of DNA. Residues 258–274 had a decrease of 6–7 deuterons at deuteration times of 300 s or more, but due to overlap with residues 251–264, at most three amides within 265–274 could be protected in the presence of DNA. This increased solvent protection is likely due to DNA contacts with loop 268–274 (5 amides, 2 Pro), including insertion of Leu 272 into the DNA base stack and hydrogen bonding of the backbone amide of residue 268 with a DNA phosphate group (26).
Residues 245–248 showed a decrease in deuteration at short and long deuteration times (Figure 3B). In bound UNG (26), the amide of residue 247 forms a hydrogen bond with a bound DNA phosphate oxygen atom. Residues 245 and 246 are the most buried of the four residues, with both amides hydrogen-bonded to other residues, while residues 247 and 248 are on the UNG surface. A plausible explanation for the change in deuteration profile is that partial burial of amide 247 by DNA slows the exchange of both amide 247 and the two buried amides.
DISCUSSION
The combination of hydrogen/deuterium exchange data and computational modeling has proven useful for constructing models of amyloid peptide oligomerization (51–55) and the assembly of pilin proteins into bacterial filaments (56). Here, we have demonstrated that the combination of computational docking and DXMS is a powerful tool for revealing the DNA footprint on a protein. Our initial goal was to test the ability of this approach to distinguish the UNG active site. Our results went far beyond this: we found a significantly larger DNA-binding surface on UNG than seen in crystallographic structures. Computational docking guided our choice of a 30 bp dsDNA fragment for our DXMS study, considerably longer than the 10 bp DNA used in crystallographic studies. DXMS supported the computational docking results, showing interactions of the abasic DNA product with two distinct regions on the 3′ side of the UNG active site.
Identification of the UNG active site
Computational docking of the unbound structure of UNG and a B-DNA model found the largest concentration of favorable-energy DNA placements at the UNG active site, identifying the active-site groove and loop residues 268–276 as important DNA-contacting regions. Essential Leu 272 stands out as a surprisingly hydrophobic side chain with direct contact to DNA (Supplementary Figure S2B). Computational docking was less successful in identifying the DNA surface that contacts the UNG active site. Reversal of the active-site strand packed the DNA major groove, rather than the minor groove, against the Leu 272 side chain. Strand direction reversal was also seen in DNA dockings to the transcription factor FadR (9), which, like UNG, causes a widening of the minor groove of the bound DNA. Therefore, the switching of minor and major groove surfaces needs to be considered when interpreting the docking of DNA models to unbound protein structures.
DXMS supported the computational docking but, by itself, did not definitively identify the active site. Peptides away from the immediate active site showed more pronounced solvent protection in the presence of DNA than peptides forming the immediate active site. This is a consequence of the fragmentation pattern. Active-site peptides included unprotected segments outside the active site, but two peptides outside the active site created continuous surfaces that were almost completely protected from solvent in the presence of DNA. Computational rigid-body docking provided essential structural interpretation of the DXMS data by unambiguously identifying the active site.
Two DNA-binding regions on the 3′ side of the UNG active site
Computational docking and the dramatic change in solvent protection found by DXMS support two distinct DNA-contact surfaces, created by residues 210–220 and 251–264, on the 3′ side of the UNG active site. Although DNA binding can indirectly increase solvent protection of amide protons by formation of large assemblies (39), conformational change (50), or stabilization of unstructured regions (57,58), direct DNA contact is the most plausible mechanism for UNG. UNG is a single-domain protein that forms a 1:1 complex with the 30 bp DNA product. Large conformational changes in UNG are unlikely, given the strong conservation of the UNG backbone found in crystallographic structures of human (26,31,33,59), Escherichia coli (60) and Herpes simplex virus UNG (61), alone or in complex with DNA or the inhibitor protein UGI. Active-site residues of UNG show induced dynamics upon binding to undamaged DNA (25) that may reflect the clamping movement that occurs upon binding both non-target (24) and target DNA (26). In this movement, the two lobes on either side of the active site, which include residues 210–220 and 251–264, close down to narrow the active-site groove by about 2 Å. These concerted movements do not alter residue interactions, secondary structure, or hydrogen-bonding patterns within each lobe. DXMS (Figure 2) demonstrates that the catalytic domain of UNG contains no unstructured regions, ruling out an unstructured to structured transition. In free UNG, the only solvent-exposed peptides that show strong solvent protection encompass well-defined helices (105–108, 173–177, 225–235, 281–290). All other strongly protected regions are buried and include β-strands (136–139, 197–203). The structures of residues 210–220 and 251–264 contain a variety of amide proton environments (Supplementary Table S4), including amide protons with no hydrogen bonds. These irregular, solvent-exposed structures are unlikely to have the capability to form hydrogen bonds sufficiently strong to cause the protection seen at long deuteration times.
Docking B-DNA to the DNA-bound UNG structure demonstrated how DNA can extend from the active site over residues 210–220 and 251–264. In the larger active-site cluster, DNA bound in the UNG active site and contacted residues 210–220 with just a small change in orientation relative to the crystallographic DNA. Potential DNA contacts include main-chain atoms, the side chains of residues Gln 213, Asn 215, Lys 218 and Glu 219, and the Arg 210 and Arg 220 guanidinium groups, which are spaced to interact with two adjacent phosphate groups (Supplementary Figure S4A). The smaller active-site cluster contacted residues 248 and 251 in an orientation appropriate for extending over residues 251–264, with additional DNA placements supporting a continuous DNA-binding region that extends to the predicted secondary DNA-binding site (Supplementary Figure S4B). Potential DNA contacts include main-chain atoms of residues 251–264, and side chains of Lys 251, Ser 254, residues 258–260 (sequence Arg-Lys-Arg, part of the secondary DNA-binding site), His 262 and Leu 264, which together form a surface groove between the active and secondary sites. DNA contacts at the secondary site include the positively charged side chains of residues 258–260 and Lys 286, 293 and 296 (Supplementary Figure S4C). The one studied mutation within residues 251–264, replacement of His 261 by Leu, shows a 71% drop in activity (33), which is significant, given that His 261 is about 30 Å from the active site. The His 261 side chain extends into the UNG interior (Supplementary Figure S4B), so the mutation acts indirectly, perhaps through changes in adjacent His 262 or solvent-exposed loop 258–260.
In contrast with the docking and DXMS results, three human UNG–DNA structures [1SSP (26), 4SKN (31) and 2OYT (24)] show no DNA contact with residues 210–220. A fourth structure, 2OXM (24), has a single contact of the His 212 side chain with an extrahelical thymine. However, all four structures suggest the potential for DNA interaction with residues 210–220. The DNA major groove lies over the surface created by these residues, but a 6–10 Å layer of water molecules separates the protein and DNA surfaces. This water layer is connected to bulk solvent, so from the crystallographic structures, we would predict that surface residues 210–220 would show significant deuteration in the presence of the bound DNA product.
Our examination of the full crystal environment resolved the apparent inconsistency between our DXMS results on residues 210–220 and the four human UNG–DNA crystallographic complexes, all of which have a single UNG–DNA pair in the asymmetric unit. In all cases a neighboring UNG molecule creates a wedge between UNG and its bound DNA (Supplementary Figure S5A). This neighboring UNG molecule also forms a pocket near its N-terminus that completely envelopes the exposed surface of the 5′ overhanging adenine of the complementary DNA strand, putting strong constraints on the DNA geometry. The result is virtually identical DNA structures on the 3′ side of the active site (Supplementary Figure S5B). These interactions may be required for successful crystallization of the UNG–DNA complex, but they are clearly not physiological, since they involve an end of the DNA fragment and the truncated N-terminus of UNG, both artifacts. Crystal packing may also influence the DNA geometry on the 5′ side of the active site, where both the DNA geometry and type of crystal contacts vary among the four structures (Supplementary Figure S5B). The strong influence of crystal packing raises doubts that the DNA geometry away from the immediate active site reflects the biological interaction, particularly given the DXMS results for residues 210–220.
The four UNG–DNA crystallographic structures show no evidence for interaction of the bound product DNA with residues 251–264. With the abasic site bound at the active site, the 10 bp crystallographic DNA fragment is not long enough to reach this region.
An expanded picture of DNA binding
With little change in UNG structure, it is difficult to envision how the bound dsDNA product can maintain Watson–Crick base pairing and simultaneously contact the two surfaces created by residues 210–220 and 251–264. The simplest explanation for strong protection of both surfaces is separation of the two DNA strands on the 3′ side of the UNG active site.
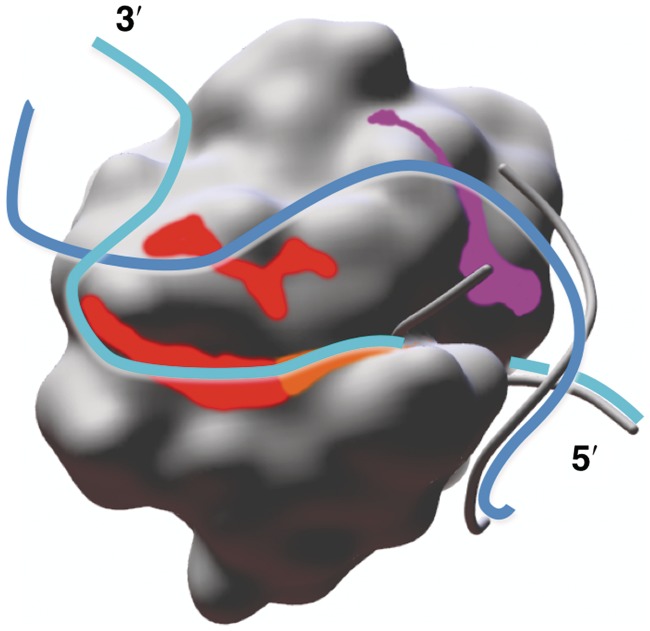
In our proposed model (Figure 4), the DNA is bound with the abasic site in the UNG active site, matching the position of the crystallographic 10 bp DNA on the 5′ side of the active site and in the active site. On the 3′ side, the active-site strand (light blue) extends from the active site toward the predicted secondary site, contacting the shallow surface groove created by residues 251–274. The complementary strand (blue), with a small shift from its crystallographic position, contacts the shallow groove created by residues 210–220 and then extends over residues 251–258 to meet the active-site strand. Interestingly, the surfaces at the bottom of both shallow grooves are created by main-chain atoms: the exposed edge strand (residues 262–266) of the central β-sheet (Figure 4, red and orange) and the interlocking β-turn structure of residues 210–220 (Figure 4, magenta). Main-chain atoms frequently form hydrogen bonds with DNA phosphate groups (62) and here may provide an organized hydrogen-bonding platform for non-sequence-specific DNA interactions.
Figure 4.
Model of the 30-bp product dsDNA bound to UNG. Both DNA strands in the model align with the crystallographic DNA (gray phosphate backbone) on the 5′ side of the active site. On the 3′ side of the active site, the active-site strand (light blue) contacts the groove created by residues 251–274, including the continuous surface formed by main-chain atoms of residues 251–264 (red) and 265–274 (orange). The complementary strand (blue) contacts the groove created by residues 210–220, including the surface created by the main-chain atoms (magenta), and may also contact the surface created by main-chain atoms of residues 251–258 (red).
The extensive DNA-binding surfaces on the 3′ side of the active site may have important functional roles. Extensive weak interactions with product DNA could contribute incrementally to the strong affinity of the product [KD = 6 nM (26)], and compensate for the energy needed for UNG-induced strand separation. In 1SSP, the distortions of the active-site strand around the abasic site appear hidden by the overlying complementary strand (Supplementary Figure S5A). Strand separation of the dsDNA product on the 3′ side of the active site could help expose the conformational changes of the processed active-site strand, providing a mechanism for recognition of the UNG-bound dsDNA product by APE-1, the next enzyme in the base-excision repair pathway. These DNA-binding surfaces may also assist the local search for uracil. NMR studies find that UNG has a passive role in dsDNA base pair opening, but substantially increases the lifetime of an extrahelical base (23,63). Our predicted DNA-binding surfaces provide a large region for trapping a spontaneously opened base during loose association of UNG and dsDNA, initiating the local search for uracil. With this mechanism, UNG takes advantage of the faster spontaneous opening rates of A:T, A:U and G:U base pairs relative to G:C, focusing the local search on dsDNA regions most likely to contain U.
Our results bring up the question of whether strand separation might apply to dsDNA substrate as well as product. Strand separation or partial melting of dsDNA substrate induced by UNG has been suggested as a binding mechanism (21,64) based on the sequence-dependent rate of uracil removal in dsDNA and the lack of sequence dependence in ssDNA (21,22). Strand separation allows UNG to use the same mechanism for local uracil search in both dsDNA and ssDNA substrates (64), explaining their similar rates of uracil cleavage [≈3-fold faster in ssDNA (21)]. On the other hand, NMR studies find no evidence for strand separation for an undamaged 10-bp DNA fragment (63). Furthermore, a high local GC content generally causes a slower rate of uracil cleavage (21), so dsDNA flexibility may be a major source of the sequence-dependent effect (65). However, some substrates with a low GC content show reduced uracil cleavage rates (21), so DNA flexibility alone does not completely explain the observed sequence specificity.
The combination of DXMS and computational docking has revealed new aspects of the well-characterized interaction of UNG and DNA. Computational docking provided the initial evidence for a more extensive DNA-binding surface than seen in crystallographic structures. DXMS, as the experimental technique to test this hypothesis, has two key advantages over X-ray crystallography: it uses a solution environment and puts no constraints on the length of the DNA. The results found by DXMS and computational docking for active-site peptides are consistent with the crystallographic structures of UNG and other base-repair enzymes (66,67), but extend this information in important ways. The two DNA-binding surfaces adjacent to the active site found by DXMS cannot be identified in the UNG–DNA crystallographic structures because of the influence of crystal packing and the short, 10 bp DNA. The overall UNG mechanism requires capabilities beyond the catalytic reaction—the impressive detection of uracil amid vast numbers of undamaged bases and the delivery of the toxic abasic product to the apurinic/apyrimidinic endonuclease, APE-1. Although our methods are lower resolution than the crystallographic structures, they have defined new DNA-binding regions at the UNG residue level that may be essential for these critical functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S4, Supplementary Figures S1–S5, Supplementary Materials and Methods, Supplementary References [9,15,16,33,36–39,42,44–48].
FUNDING
The National Science Foundation (DBI 99-04559 to V.A.R. and L.F.T); the California HIV/AIDS Research Program (187637 to V.A.R.); the National Institutes of Health (GM070996 to V.A.R. and L.F.T, AI076961, AI081982, AI2008031, AI072106, AI068730, GM037684, GM020501, GM066170 to V.L.W., GM46312 to J.A.T); the Innovative Technologies for the Molecular Analysis of Cancer (IMAT) program (CA099835, CA118595 to V.L.W.); and the University of California IUCRP program, BiogenIDEC corporate sponsor (UC10591 to V.L.W.). Funding for open access charge: National Institutes of Health (GM46312 to J.A.T.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ottar Sundheim of the Norwegian University of Science and Technology for the UNG protein and Louis Noodleman (TSRI) for helpful discussions. Arthur J. Olson (TSRI) provided physical molecular models produced on a 3-D printer that were invaluable for studying UNG crystal packing.
REFERENCES
- 1.Tan S, Hunziker Y, Pellegrini L, Richmond TJ. Crystallization of the yeast MATα2/MCM1/DNA ternary complex: general methods and principles for protein/DNA cocrystallization. J. Mol. Biol. 2000;297:947–959. doi: 10.1006/jmbi.2000.3606. [DOI] [PubMed] [Google Scholar]
- 2.Janin J, Henrick K, Moult J, Ten Eyck L, Sternberg MJE, Vajda S, Vakser I, Wodak SJ. CAPRI: A Critical Assessment of PRedicted Interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 3.Méndez R, Leplae R, De Maria L, Wodak SJ. Assessment of blind predictions of protein-protein interactions: Current status of docking methods. Proteins. 2003;52:51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- 4.Méndez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3-5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 5.Giudice E, Lavery R. Simulations of nucleic acids and their complexes. Acc. Chem. Res. 2002;35:350–357. doi: 10.1021/ar010023y. [DOI] [PubMed] [Google Scholar]
- 6.Aloy P, Moont G, Gabb HA, Querol E, Aviles FX, Sternberg MJE. Modelling repressor proteins docking to DNA. Proteins. 1998;33:535–549. [PubMed] [Google Scholar]
- 7.De Luca L, Pedretti A, Vistoli G, Barreca ML, Villa L, Monforte P, Chimirri A. Analysis of the full-length integrase-DNA complex by a modified approach for DNA docking. Biochem. Biophys. Res. Commun. 2003;310:1083–1088. doi: 10.1016/j.bbrc.2003.09.120. [DOI] [PubMed] [Google Scholar]
- 8.Adesokan AA, Roberts VA, Lee KW, Lins RD, Briggs JM. Prediction of HIV-1 integrase/viral DNA interactions in the catalytic domain by fast molecular docking. J. Med. Chem. 2004;47:821–828. doi: 10.1021/jm0301890. [DOI] [PubMed] [Google Scholar]
- 9.Roberts VA, Case DA, Tsui V. Predicting interactions of winged-helix transcription factors with DNA. Proteins. 2004;57:172–187. doi: 10.1002/prot.20193. [DOI] [PubMed] [Google Scholar]
- 10.Zhu HM, Chen WZ, Wang CX. Docking dinucleotides to HIV-1 integrase carboxyl-terminal domain to find possible DNA binding sites. Bioorg. Med. Chem. Lett. 2005;15:475–477. doi: 10.1016/j.bmcl.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Fan L, Roberts VA. Complex of linker histone H5 with the nucleosome and its implications for chromatin packing. Proc. Natl Acad. Sci. USA. 2006;103:8384–8389. doi: 10.1073/pnas.0508951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fanelli F, Ferrari S. Prediction of MEF2A-DNA interface by rigid body docking: a tool for fast estimation of protein mutational effects on DNA binding. J. Struct. Biol. 2006;153:278–283. doi: 10.1016/j.jsb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 15.Ten Eyck LF, Mandell JG, Roberts VA, Pique ME. Surveying molecular interactions with DOT. In: Hayes A, Simmons M, editors. Proceedings of the 1995 ACM/IEEE Supercomputing Conference, San Diego. Los Alamitos, CA: IEEE Computer Society Press; 1995. p. 22. www.sdsc.edu/CCMS/Papers/DOT_sc95.html. [Google Scholar]
- 16.Mandell JG, Roberts VA, Pique ME, Kotlovyi V, Mitchell JC, Nelson E, Tsilgeny I, Ten Eyck LF. Protein docking using continuum electrostatics and geometric fit. Prot. Eng. 2001;14:105–113. doi: 10.1093/protein/14.2.105. [DOI] [PubMed] [Google Scholar]
- 17.Sperry JB, Shi X, Rempel DL, Nishimura Y, Akashi S, Gross ML. A mass spectrometric approach to the study of DNA-binding proteins: interaction of human TRF2 with telomeric DNA. Biochemistry. 2008;47:1797–1807. doi: 10.1021/bi702037p. [DOI] [PubMed] [Google Scholar]
- 18.Black BE, Brock MA, Bedard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl Acad. Sci. USA. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 22.Nilsen H, Yazdankhah SP, Eftedal I, Krokan HE. Sequence specificity for removal of uracil from U·A pairs and U·G mismatches by uracil-DNA glycosylase from Escherichia coli and correlation with mutational hotspots. FEBS Lett. 1995;362:205–209. doi: 10.1016/0014-5793(95)00244-4. [DOI] [PubMed] [Google Scholar]
- 23.Cao C, Jiang YL, Krosky DJ, Stivers JT. The catalytic power of uracil DNA glycosylase in the opening of thymine base pairs. J. Am. Chem. Soc. 2006;128:13034–13035. doi: 10.1021/ja062978n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman JI, Majumdar A, Stivers JT. Nontarget DNA binding shapes the dynamic landscape for enzymatic recognition of DNA damage. Nucleic Acids Res. 2009;37:3493–3500. doi: 10.1093/nar/gkp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagelhus TA, Haug T, Singh KK, Keshav KF, Skorpen F, Otterlei M, Bharati S, Lindmo T, Benichou S, Benarous R, et al. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34 kDa subunit of replication protein A. J. Biol. Chem. 1997;272:6561–6566. doi: 10.1074/jbc.272.10.6561. [DOI] [PubMed] [Google Scholar]
- 28.Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–458. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 29.Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc. Natl Acad. Sci. USA. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sidorenko VS, Mechetin GV, Nevinsky GA, Zharkov DO. Correlated cleavage of single- and double-stranded substrates by uracil-DNA glycosylase. FEBS Lett. 2008;582:410–414. doi: 10.1016/j.febslet.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 32.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Uracil-DNA glycosylase–DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl Acad. Sci. USA. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA. Crystal-structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 34.Kavli B, Slupphaug G, Mol CD, Arvai AS, Petersen SB, Tainer JA, Krokan HE. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 35.Krokan HE, Wittwer CU. Uracil DNA-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res. 1981;9:2599–2613. doi: 10.1093/nar/9.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers JC, Moore SA, Shamoo Y. Structure-based incorporation of 6-methyl-8-(2-deoxy-beta-ribofuranosyl)isoxanthopteridine into the human telomeric repeat DNA as a probe for UP1 binding and destabilization of G-tetrad structures. J. Biol. Chem. 2003;278:42300–42306. doi: 10.1074/jbc.M306147200. [DOI] [PubMed] [Google Scholar]
- 37.Sun S, Shamoo Y. Biochemical characterization of interactions between DNA polymerase and single-stranded DNA-binding protein in bacteriophage RB69. J. Biol. Chem. 2003;278:3876–3881. doi: 10.1074/jbc.M210497200. [DOI] [PubMed] [Google Scholar]
- 38.Godderz LJ, Peak MM, Rodgers KK. Analysis of biological macromolecular assemblies using static light scattering methods. Curr. Org. Chem. 2005;9:899–908. [Google Scholar]
- 39.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 40.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 42.Macke T, Case DA. Modeling unusual nucleic acid structures. In: Leontes NB, Santa Lucia J Jr, editors. Molecular Modeling of Nucleic Acids. Vol. 682. Washington, DC: American Chemical Society; 1998. pp. 379–393. [Google Scholar]
- 43.Onufriev A, Bashford D, Case DA. Modification of the generalized Born model suitable for macromolecules. J. Phys. Chem. B. 2000;104:3712–3720. [Google Scholar]
- 44.Tsui V, Case DA. Molecular dynamics simulations of nucleic acids with a generalized Born solvation model. J. Am. Chem. Soc. 2000;122:2489–2498. [Google Scholar]
- 45.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 46.Sanner MF, Olson AJ, Spehner J-C. Reduced surface: An efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 47.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Jr, Weiner P. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984;106:765–784. [Google Scholar]
- 48.Gilson MK, Davis ME, Luty BA, McCammon JA. Computation of electrostatic forces on solvated molecules using the Poisson-Boltzmann equation. J. Phys. Chem. 1993;97:3591–3600. doi: 10.1063/1.1924448. [DOI] [PubMed] [Google Scholar]
- 49.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem. Soc. Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 50.Mendillo ML, Putnam CD, Mo AO, Jamison JW, Li S, Woods VL, Jr, Kolodner RD. Probing DNA- and ATP-mediated conformational changes in the MutS family of mispair recognition protein using deuterium exchange mass spectrometry. J. Biol. Chem. 2010;285:13170–13182. doi: 10.1074/jbc.M110.108894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olofsson A, Sauer-Eriksson AE, Öhman A. The solvent protection of Alzheimer amyloid-β(1-42) fibrils as determined by solution NMR spectroscopy. J. Biol. Chem. 2006;281:477–483. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- 52.Olofsson A, Lindhagen-Persson M, Sauer-Eriksson AE, Öhman A. Amide solvent protection analysis demonstrates that amyloid-β(1-40) and amyloid-β(1-42) form different fibrillar structures under identical conditions. Biochem. J. 2007;404:63–70. doi: 10.1042/BJ20061561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olofsson A, Sauer-Eriksson AE, Öhman A. Amyloid fibril dynamics revealed by combined hydrogen/deuterium exchange and nuclear magnetic resonance. Anal. Biochem. 2009;385:374–376. doi: 10.1016/j.ab.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 54.Ma B, Nussinov R. Polymorphic C-terminal β-sheet interactions determine the formation of fibril or amyloid β-derived diffusible ligand-like globulomer for the Alzheimer Aβ42 dodecamer. J. Biol. Chem. 2010;285:37102–37110. doi: 10.1074/jbc.M110.133488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma B, Nussinov R. Polymorphic triple β-sheet structures contribute to amide hydrogen/deuterium (H/D) exchange protection in the Alzheimer amyloid β42 peptide. J. Biol. Chem. 2011;286:34244–34253. doi: 10.1074/jbc.M111.241141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Lim MS, Li S, Brock M, Pique ME, Woods VL, Jr, Craig L. Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure. 2008;16:137–148. doi: 10.1016/j.str.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen JC, Wexler BB, Rogers DJ, Hite KC, Panchenko T, Ajith S, Black BE. DNA binding restricts the intrinsic conformational flexibility of methyl CpG binding protein 2 (MeCP2) J. Biol. Chem. 2011;286:18938–18948. doi: 10.1074/jbc.M111.234609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalodimos CG, Biris N, Bonvin AMJJ, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 59.Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 60.Putnam CD, Shroyer MJN, Ludquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. Protein mimicry of DNA from crystal structures of the uracil glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 61.Savva R, Pearl LH. Nucleotide mimicry in the crystal structure of the uracil-DNA glycosylase-uracil glycosylase inhibitor protein complex. Nat. Struct. Biol. 1995;2:752–757. doi: 10.1038/nsb0995-752. [DOI] [PubMed] [Google Scholar]
- 62.Hoffman MM, Khrapov MA, Cox JC, Yao J, Tong L, Ellington AD. AANT: the amino acid-nucleotide interaction database. Nucleic Acids Res. 2004;32:D174–D181. doi: 10.1093/nar/gkh128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao C, Jiang YL, Stivers JT, Song F. Dynamic opening of DNA during the enzymatic search for a damaged base. Nat. Struct. Mol. Biol. 2004;11:1230–1236. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 64.Vassylyev DG, Morikawa K. Precluding uracil from DNA. Structure. 1996;4:1381–1385. doi: 10.1016/s0969-2126(96)00145-1. [DOI] [PubMed] [Google Scholar]
- 65.Seibert E, Ross JBA, Osman R. Role of DNA flexibility in sequence-dependent activity of uracil DNA glycosylase. Biochemistry. 2002;41:10976–10984. doi: 10.1021/bi026121o. [DOI] [PubMed] [Google Scholar]
- 66.Hitomi K, Shigenori I, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair. 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 67.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.