Abstract
In the past decades, RNA molecules have emerged as important players in numerous cellular processes. To understand these processes at the molecular and atomic level, large amounts of homogeneous RNA are required for structural, biochemical and pharmacological investigations. Such RNAs are generally obtained from laborious and costly in vitro transcriptions or chemical synthesis. In 2007, a recombinant RNA technology has been described for the constitutive production of large amounts of recombinant RNA in Escherichia coli using a tRNA-scaffold approach. We demonstrate a general applicable extension to the described approach by introducing the following improvements: (i) enhanced transcription of large recombinant RNAs by T7 RNA polymerase (high transcription rates, versatile), (ii) efficient and facile excision of the RNA of interest from the tRNA-scaffold by dual cis-acting hammerhead ribozyme mediated cleavage and (iii) rapid purification of the RNA of interest employing anion-exchange chromatography or affinity chromatography followed by denaturing polyacrylamide gel electrophoresis. These improvements in the existing method pave the tRNA-scaffold approach further such that any (non-)structured product RNA of a defined length can cost-efficiently be obtained in (multi-)milligram quantities without in vitro enzymatic manipulations.
INTRODUCTION
Systematic studies of the structure and function of RNA molecules, ligand screening in pharmaceutical drug development and several other biophysical techniques usually require milligram quantities of homogeneous and pure RNA. Such studies applied to proteins greatly benefited from the development of various bacterial and eukaryotic recombinant expression systems. In contrast to proteins, such recombinant systems for RNA are not readily available, and RNA has therefore mostly been produced by in vitro transcription from DNA templates (1) and via chemical synthesis (2). Both production methods are costly, laborious and have their drawbacks with respect to sequence requirements, variations in yield, non-templated nucleotide additions and/or the maximum length of the oligonucleotide. Recombinant RNA synthesis in bacteria could be a suitable alternative to overcome these problems; however, the production of homogeneous RNA in vivo has been impeded by host nuclease degradation and the inability to purify the RNA of interest from the cellular RNAs. Until recently, the only recombinant RNAs successfully produced were 5S ribosomal RNA (5S rRNA) (3), transfer RNAs (tRNAs) (4–7) and (derivatives of) natural RNAs that occur as stable molecules (8–10). This has been possible because these RNAs are recognized by cellular machinery, and the primary transcript is precisely processed, post-transcriptionally modified and prevented from 3′-polyadenylation that triggers RNA degradation (11–13). The ability of these RNAs to escape nuclease degradation has been adapted by Ponchon and Dardel to produce recombinant RNA by substituting the anticodon stem-loop of a tRNA for a structured RNA molecule, a strategy which they called the tRNA-scaffold approach (14–16). The tRNA-scaffold in the chimera possesses the same properties as a regular tRNA and is being recognized and processed by the cellular machinery in a similar fashion. The tRNA-chimers are protected from breakdown by host nucleases and accumulate to copious amounts inside the bacterial cells. Although this approach yields large amounts of pure and homogeneous recombinant RNA, the RNA fragment of interest is covalently attached to the tRNA-scaffold. To release this RNA, Ponchon and Dardel employed in vitro RNase H cleavage directed by two DNA oligonucleotides complementary to the invariable part of the tRNA-scaffold. The released RNA of interest was purified from the DNA oligonucleotides and the remainder of the tRNA-scaffold by anion-exchange chromatography under denaturing conditions. More recently, a strategy similar to the tRNA-scaffold approach was demonstrated by Liu et al. (17) who inserted a 71-nt product RNA in place of helix III-loop C segment of the 5S ribosomal RNA of Vibrio proteoliticus and produced this recombinant RNA constitutively in Escherichia coli. In this approach, excision of the product RNA from the 5S rRNA scaffold is mediated via biotinylated DNAzyme cleavage.
In this contribution, a general applicable improvement of the existing tRNA-scaffold approach is described that allows for the production of large amounts of homogeneous product RNAs that are released from the protective tRNA-scaffold without the requirement of any in vitro enzymatic manipulation or input of (modified) DNA oligonucleotides. To accomplish this, the recombinant RNA molecule consists of the human tRNAlys-scaffold with a Sephadex aptamer fused to the anticodon stem, similar as has been demonstrated by Ponchon and Dardel. This construct is used to insert various RNA cassettes on top of the Sephadex aptamer (Figure 1A and Supplementary Figure S1) each consisting of the RNA of interest (product RNA) flanked with cis-acting hammerhead (HH) ribozymes. The DNA sequence coding for the recombinant RNA molecule is inserted downstream the T7 promotor sequence in pET23b (Novagen), a vector commonly used for recombinant protein expression (18,19), allowing transcription of RNA in E. coli BL21 (DE3) to be initiated upon the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG). The recombinant RNA is purified from the host RNAs by anion-exchange chromatography and/or by affinity chromatography using Sephadex G-200. The product RNA excised from the scaffold after HH ribozyme cleavage is separated from the scaffold moiety using preparative denaturing polyacrylamide gel electrophoresis (PAGE). The general applicability of this method is demonstrated by the production of a variety of RNA molecules (displayed in Figure 1B). We also show that mutants of the product RNAs can rapidly be obtained by using site-directed mutagenesis (20) and fusion-PCR (21), demonstrated by the production of several mutants of the Duck Hepatitis B Virus epsilon encapsidation signal RNA (22) (displayed in Figure 1C). Finally, we show that stable isotope labeling for NMR structural analysis is easily and cost-efficiently achieved by the example of the production on 15N-substituted minimal medium of three different sequences of the Duck HBVε RNA sequence.
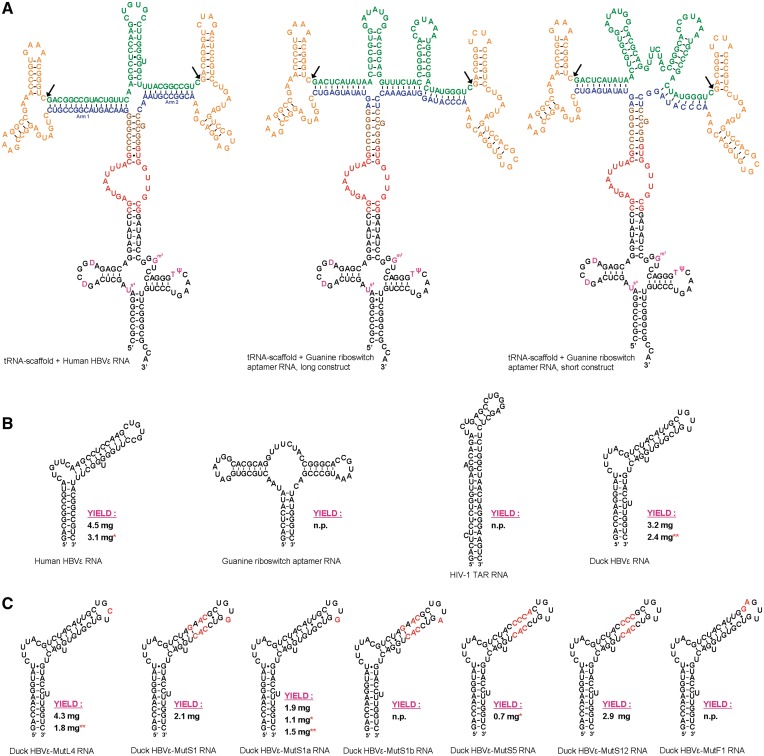
Figure 1.
Secondary structure representations of the recombinant (product) RNAs. (A) From left to right, the constructs for the Human HBVε RNA—the long construct for the B. subtilis guanine riboswitch aptamer RNA and the short construct for the B. subtilis guanine riboswitch aptamer RNA—are displayed. The tRNAlys-scaffold is shown in black (modified nucleotides in purple), the Sephadex aptamer in red, the HH ribozymes flanking the RNA of interest are in orange and the two variable arms base pairing with the ends of the RNA of interest in blue. The brown nucleotides are the restriction sites for subcloning. The bonds between the nucleotides that are cleaved by the HH ribozymes are indicated by arrows. The 5′-end and 3′-end of the tRNAlys-scaffold are depicted as matured termini. See Supplementary Figure S1 for representations of the additional produced full-recombinant RNAs. (B) Different types of recombinant product RNAs, from left to right: Human HBVε RNA (15,63–65), B. subtilis guanine riboswitch aptamer RNA (66), HIV-1 TAR (67) RNA and Duck HBVε RNA (22,35,68). (C) Produced mutants of the Duck HBVε RNA. The introduced mutations with respect to the Duck HBVε RNA are shown in red. The presented secondary structures of the mutants of Duck HBVε RNA are for visual clarification of the location of the introduced mutations and are, except for Duck HBVε-mutL4, not in accordance with the predicted secondary structures obtained with Mfold. The indicated yields are from 1 l cultures employing anion-exchange chromatography except for those indicated with a single red asterisk, which were purified with Sephadex G-200 affinity. Yields indicated with two red asterisks are obtained from 1 l of 15N-labeled minimal media, n.p., not produced on liter scale. See Supplementary Table S3 for the nucleotide sequence of all product RNAs.
MATERIALS AND METHODS
Vector construction
Prior to vector construction, the folds of the full-recombinant RNAs were predicted using the program Mfold (23). The desired folds of the RNAs (Figure 1A and Supplementary Figure S1) were obtained by varying the length of the arms and the nucleotide composition in the four-way junction region (Figure 1A and Supplementary Figure S1, blue residues) until the desired structures were predicted to be more stable than the predicted alternative structures. The DNA sequence coding for the tRNAlys-scaffold with sephadex aptamer containing the Human HBVε RNA flanked with cis-acting HH ribozymes (Figure 1A) was purchased from Baseclear (Leiden, The Netherlands). This DNA template was cloned into the XbaI + XhoI restriction sites of vector pET23b (Novagen) (Figure 2) thereby retaining the purine-rich DNA sequence between the T7 RNA polymerase promoter sequence and the XbaI site to ensure efficient initiation of RNA transcription. This vector was subsequently used as a shuttle vector for subcloning various cassettes coding for HH ribozyme flanked product RNAs, listed in the upper section of Supplementary Table S3 (see Supplementary Figure S1 for all secondary structure representation of the full-recombinant RNAs). The cloning sites were SmaI (5′) and SacII (3′), located just above the Sephadex aptamer (Figure 1A, brown residues). This specific combination of restriction enzymes ensured that the stem above the Sephadex aptamer in the RNA transcript formed correctly and that the insert RNAs would not interfere with the fold of the protective tRNAlys-scaffold.
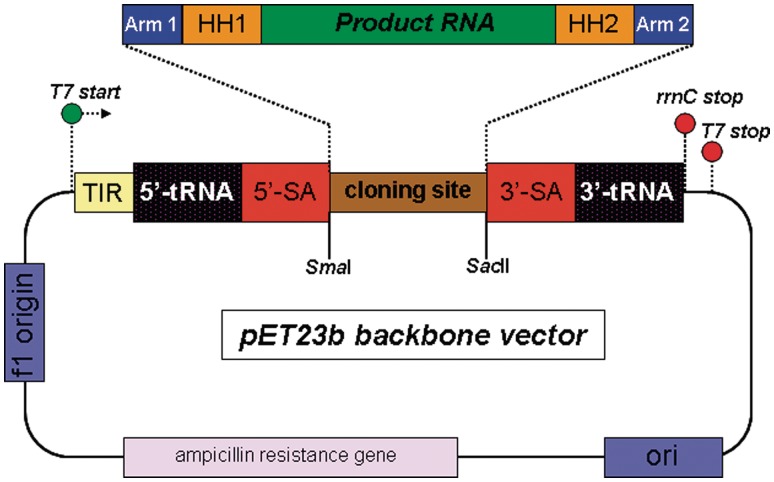
Figure 2.
Schematic of the used vector for recombinant RNA synthesis. The cloning site for the insertion of the various DNA sequences is flanked with a SmaI and a SacII site for directed cloning. The color coding of the blocks correspond with the sequence colors in the full-recombinant RNA transcript (Figure 1A). The 5′-transcription initiation region (5′-TIR) directly after the T7 promotor sequence ensures efficient transcription initiation by T7 RNA polymerase. Transcription is terminated by the presence of the rrnC terminator sequence at the 3′-end of the coding DNA sequence.
Site-directed mutations were introduced into the pET23b-vector containing the DNA sequence for the Duck HBVε RNA (or a mutant hereof) to obtain the desired mutant RNAs of the Duck HBVε RNA (displayed in Figure 1C) as described (20,21) (See Supplementary Tables S1 and S2 for used oligonucleotides and Supplementary Figure S2 for a more detailed description). The DNA sequence of each construct was verified (Baseclear) prior to recombinant RNA productions.
RNA production
Freshly transformed E. coli BL21 (DE3) cells or from glycerol stock were plated on LB-agar containing 50 μg/ml ampicillin and grown overnight at 37°C. For checking the level of recombinant RNA production, a single colony was picked and cultivated in 5 ml of TB medium or LB medium (24) containing 50 μg/ml ampicillin (TB-amp50/LB-amp50) at 37°C, 250 rpm. At OD600nm ∼0.6–0.8, IPTG was added to a concentration of 1 mM, and cultivation was continued for 3 h. Every hour post-induction, the cells from 1 ml of culture were pelleted and resuspended in 100 μl of 10 mM Tris-HCl (pH 7.4) + 10 mM magnesium acetate (buffer L). The suspension was extracted with 100 μl of phenol saturated with buffer L, and the aqueous phase was ethanol precipitated by adding 0.1 volume of 3 M sodium acetate (pH 5.2) and 3 volumes of ethanol. The pellet was dissolved in 20 μl of Milli-Q water and mixed with 20 μl of 50 mM EDTA in formamide containing 0.02% bromophenol blue and 0.02% xylene cyanol (2× LB). The RNA was analyzed by electrophoresis on an analytical 8% polyacrylamide gel in 1× TBE containing 8 M urea. For large-scale cultures of unlabeled RNA, 30 ml of overnight preculture was added to 1 l of TB-amp50 or LB-amp50 in a baffled 2 l Erlenmeyer. 15N-labeled RNA was produced by inoculation of 1 l minimal medium [125 mM potassium phosphate buffer (pH 7.2), 35 mM Na2SO4, 27.5 mM 15NH4Cl, 15 mM d-(+)-glucose, 1 mM MgSO4 and 50 μg/ml ampicillin] with 100 ml of overnight preculture grown on the same minimal medium. Cells were cultivated and induced as described for the 5 ml cultures.
RNA purification using anion-exchange chromatography
Cells from 1 l of culture were pelleted and resuspended in 40 ml of buffer L. The RNA was extracted from the cells by the addition of 20 ml of phenol saturated in buffer L and gentle agitation for 30 min at room temperature. The suspension was centrifuged at 7500 g in a JA-14 rotor (Beckman) at 4°C for 10 min; the aqueous phase was pipetted off, and the organic layer was re-extracted with another 20 ml of buffer L. The aqueous phases were pooled and dialyzed overnight at 4°C against 2.5 l of 20 mM Tris-HCl (pH 8.1) + 0.25 mM EDTA in a 6–8 kDa MWCO dialysis tubing (Spectra), refreshing the dialysis buffer once. The dialyzed RNA was diluted 4-fold with 20 mM Tris-HCl (pH 8.1), and MgCl2 was added to 10 mM. This mixture was incubated at 37°C for 1 h to cleave the residual intact HH ribozymes. Any formed precipitate was removed by centrifugation at 20 000 g in a JA-14 rotor at 4°C for 30 min, and the RNA was loaded with 5 ml/min onto a ResourceQ 6 ml column (GE Healthcare) equilibrated in 25 mM Tris-HCl (pH 8.1) + 1 mM EDTA (buffer C) connected to an ÅKTA basic (GE Healthcare). Bound RNA was eluted by applying a segmented linear gradient (Supplementary Table S4) to 1.5 M NaCl in buffer C. Eluate that showed UV absorbance was collected in fractions of 5 ml. Twenty microliters of each fraction was mixed with 20 μl 2× LB and checked on 8% PAGE. The fractions containing recombinant RNA were pooled, concentrated by ethanol precipitation and purified as described under preparative denaturing PAGE.
RNA purification using Sephadex G-200 affinity chromatography
Cells from 1 l of culture were pelleted and resuspended in 50 ml of binding buffer [buffer SB; 50 mM potassium phosphate buffer (pH 7.5) + 10 mM MgCl2 + 100 mM NaCl] containing 200 μg/ml lysozyme. The suspension was sonicated four times during 30 s with intervals of 2 min on ice. The cellular debris was removed by centrifugation at 30 000 g in a JA-20 rotor (Beckman) for 30 min at 4°C. One hundred milliliters of Sephadex G-200 swollen in buffer SB was added to the supernatant, and the RNA was allowed to bind at 4°C for 2 h under gentle agitation. The slurry was poured into a glass column (ø2.5 cm) and allowed to settle. The flow-through was collected, and the column was washed with portions of 15 ml buffer SB until the RNA concentration of the wash dropped to ∼100 ng/μl, which was determined with UV absorbance using a Nanodrop 1000 spectrophotometer (Isogen). The bound RNA was eluted with buffer SB containing 4 M urea and collected in fractions of 10 ml. Recombinant RNA containing fractions were pooled and dialyzed against 2.5 l of 20 mM Tris-HCl (pH 8.1) + 0.25 mM EDTA in a 6–8 kD MWCO dialysis tubing (Spectra) at 4°C, refreshing the dialysis buffer once. MgCl2 was added to the dialyzed RNA to 10 mM, and the residual intact HH ribozymes were cleaved by incubation at 37°C for 1 h. The RNA was concentrated by ethanol precipitation and purified using denaturing PAGE as described below.
Preparative denaturing PAGE and NMR spectroscopy
Ethanol precipitated RNA was dissolved with Milli-Q water to 2.5 ml; the RNA concentration was determined by UV spectroscopy and mixed with 2.5 ml of 2× LB. The sample was boiled at 95°C for 10 min and purified over preparative denaturing 8% polyacrylamide gel (19:1) in 1× TBE containing 8 M urea and 5 mM EDTA. Per preparative gel (435 × 380 × 3 mm) a maximum of 20 mg of column purified RNA was loaded and electrophorized for 16 h (overnight) at 300 V. The band of the product RNA was excised from the gel under UV shadowing on a TLC Silica gel 60 F254 plate (Merck) using a 254-nm wavelength UV lamp. The RNA was subsequently electro eluted from the gel bands at 180 V using an Elutrap device (Schleicher & Schuell) and subsequently ethanol precipitated. The RNAs were further purified by extensive dialysis in a centricon YM-10 device (Amicon) as described earlier (25) and lyophilized. For NMR structural analysis, unlabeled and 15N-labeled Duck HBVε RNA, Duck HBVε-mutS1a RNA and Duck HBVε-mutL4 RNA were reconstituted in 300 μl of NMR buffer [10 mM sodium phosphate (pH 6.7), 10 μM EDTA and 7% D2O] to obtain RNA concentrations in the range of 0.3–0.6 mM. The samples were heated at 95°C for 2 min, snap cooled on ice-water and transferred to a Shigemi NMR tube. NMR data were acquired at 800 MHz 1H frequency at 15°C on a Varian Unity Inova spectrometer equipped with a cryo-cooled probe. The 1H, 15N-HMQC-spectra were recorded with 4096 (t2) × 200 (t1) points for a spectral width of 20 kHz (1H) and 1.5 kHz (15N) [see e.g. (26,27)]. A 800 MHz (1H, 1H) NOESY spectrum of the unlabeled Duck HBVε-mutS1a RNA was recorded with 512 (t1) × 4096 (t2) points for a spectral width of 17 kHz (t1) by 20 kHz (t2) and a mixing time of 300 ms [see e.g. (26,27)]. Spectra were processed with NMRpipe (28), and resonances were assigned using the program SPARKY (T.D. Goddard and D.G. Kneller, University of San Francisco).
RESULTS
Principle of the method
Our approach for the synthesis of recombinant RNA is based on the tRNA-scaffold strategy that was first described by Ponchon and Dardel (14–16). Different from their method, our RNA is not constitutively produced in the E. coli cells, but transcription is initiated during growth at an OD600nm of 0.6–0.8 by the addition of IPTG, comparable with modern recombinant protein synthesis techniques. Furthermore, the termini of the product RNA are flanked with cis-acting hammerhead (HH) ribozymes (Figure 1A), leading to an efficient excision of the product RNA from the scaffold after the addition of MgCl2 to the recombinant RNA and a 1-h incubation at 37°C. This facile ribozyme-mediated cleavage circumvents the requirement of enzymatic in vitro RNase H treatment to release the product RNA from the tRNA-scaffold. After ribozyme cleavage, the global structure of the full RNA is retained through ∼10–15 base pairing interactions of the 5′-region and 3′-region of the product RNA with the arms of the scaffold. This allows for the separation of the large recombinant RNA from the smaller tRNAs, 5S rRNA and small RNA fragments by anion-exchange purification or alternatively, since the recombinant RNA molecules are equipped with a Sephadex aptamer (29,30), by affinity chromatography using Sephadex G-200. Finally, the product RNA is separated from the tRNA-scaffold on preparative denaturing PAGE. The difference in sequence length between the product RNA and the two scaffold moieties allows for efficient and clean excision of the product RNA band from the gel. The product RNA is further purified employing electro-elution and extensive ultra-filtration dialysis. Supplementary Table S4 gives an overview of the expected timing for each of the employed purification steps and the expected total production time of an NMR sample of RNA.
The feasibility of the method is demonstrated by the production of several RNA molecules (Figure 1 and Supplementary Figure S1), and NMR structural analysis is demonstrated on unlabeled and 15N-labeled Duck HBVε RNA and Duck HBVε-mutS1a RNA.
Construct design
The various RNA cassettes that are placed on top of the Sephadex aptamer are designed in such a way that they do not disturb the predicted secondary structure of the protective tRNA-scaffold (Figure 1A and Supplementary Figure S1). The SmaI (5′) and SacII (3′) restriction sites located in the stem above the Sephadex aptamer are successfully used for cloning the insert cassettes in the correct orientation and retaining the desired base pairings in this stem for stability (Figure 1, brown residues). Each RNA cassette consists of three basic elements, namely (i) the product RNA (green) flanked with (ii) cis-acting hammerhead ribozymes (orange) and (iii) two arms (blue) that base pair with the 10–15 terminal nucleotides of the product RNA facilitating efficient ribozyme cleavage and downstream purification. The desired fold of the recombinant RNA molecules is complex, and therefore representations of the predicted secondary structure were generated with in silico folding simulations using the Mfold web server (23). To obtain the desired fold predicted more stable than alternate structures, the length of the arms and the nucleotide composition of the junction regions (only in the blue residues) were altered.
RNA production
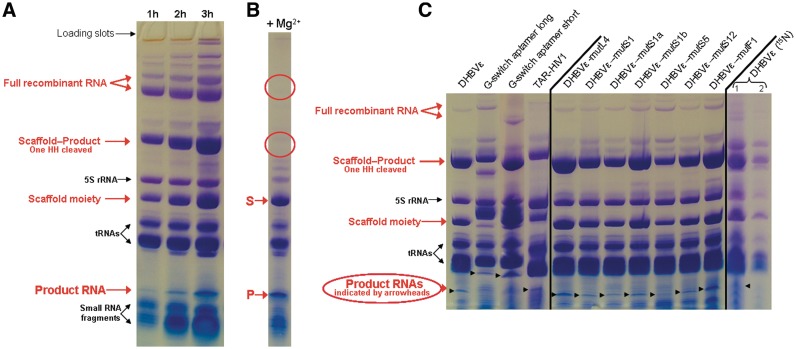
All unlabeled recombinant RNA transcriptions have been carried out in either TB medium or LB medium. The production yields differed not significantly on the type of employed growth medium. Prior to large-scale cultures, 5 ml cultures were grown to check RNA transcription levels. During 3 h post-induction, 1 ml samples were taken from the culture every hour and the RNA content was checked on 8% denaturing PAGE. Figure 3A shows an example of a time-course experiment for the production of the Human HBVε RNA (HHBVε). It is observed that recombinant RNA (indicated with red arrows) stably accumulates after induction. Generally, the plateau of RNA accumulation was reached 3 h post-induction. Incubations extended up to 6 h showed no difference on the amount of produced RNA, but overnight incubation resulted in degradation of the majority of the recombinant RNA (results not shown). The band of the full (i.e. without HH ribozymes cleaved) recombinant RNA is indicated with two arrows, of which the upper band contains the vector borne 5′ transcription initiation region (5′-TIR) RNA still attached to the tRNA-scaffold. The presence of this incompletely processed scaffold moiety depends on the produced RNA and is not always observed. The band pattern of the recombinant RNA shows that the HH ribozymes are already cleaved to some extent inside the bacterial cells during the time of production. The bands containing the majority of recombinant RNA are accredited to ‘Scaffold + Product − 1 HH cleaved off’ and to ‘Scaffold moiety’ (it is here not clear which HH has been cleaved and which side of the scaffold moiety is seen). The band indicated with ‘product RNA’ (i.e. the released RNA after dual HH cleavage) is located below the tRNAs. After dialysis of the aqueous phase, the residual intact HH ribozymes were readily cleaved after 4-fold dilution of the RNA in fresh dialysis buffer supplemented with 10 mM MgCl2 and 1-h incubation at 37°C (Figure 3B, HHBVε-RNA). The HH ribozymes cleaved to completion as can be observed from the disappearance of the bands accredited to ‘Full RNA’ and ‘Scaffold + Product-one HH cleaved’ from Figure 3a (red circled) and only the bands of the scaffold RNA (S) and product RNA (P) present as recombinant RNA.
Figure 3.
Denaturing 8% polyacrylamide gels of several recombinant RNA transcriptions in E. coli BL21 (DE3) using the employed tRNA-scaffold. (A) Recombinant Human HBVε RNA production (Figure 1A). RNA content of cells from 1 ml of culture at time = 1, 2 and 3 h post-induction is visualized. The produced recombinant RNA is indicated with red arrows, and cellular RNAs and small RNA fragments are indicated with black arrows. (B) Result of the HH ribozyme cleavage of the dialyzed extract of the recombinant RNA containing the Human HBVε RNA. The scaffold (S) and the released product (P) are indicated by the red arrows. The red circles indicate the locations of the RNA bands that have disappeared after HH ribozyme cleavage. (C) Various transcribed RNA sequences at 3 h post-induction (RNA content from 1 ml of culture), employing the described tRNA-scaffold (see Figure 1A and Supplementary Figure S1 for a representation of the fold of the different constructs). The lanes indicated DHBVε (15N) 1 and 2 show the RNA contents of the first extract and the re-extract of 1 ml of cells from the 15N-labeled minimal media culture for production of Duck HBVε RNA, respectively. Bands are visualized with Stains-All (Acros organics, Geel, Belgium).
For all designed RNA constructs and/or mutants, similar band patterns were obtained (Figure 3C). As can be observed from Figure 3C, the production efficiencies are different for each construct. For instance, the constructs for Duck HBVε-mutL4 RNA and Duck HBVε-mutF1 RNA produce better than the long construct for the guanine riboswitch aptamer (GRA) and the construct for the HIV-1 TAR RNA. We generally obtained yields in the range of 10–30 mg of unlabeled full-recombinant RNA per liter (corresponding to ∼2–4.5 mg of purified product RNA). The yields of full-recombinant RNA and purified product RNA obtained from minimal media cultures were somewhat lower, ∼7.5−15 and ∼1.5–2.5 mg/l, respectively.
RNA purification
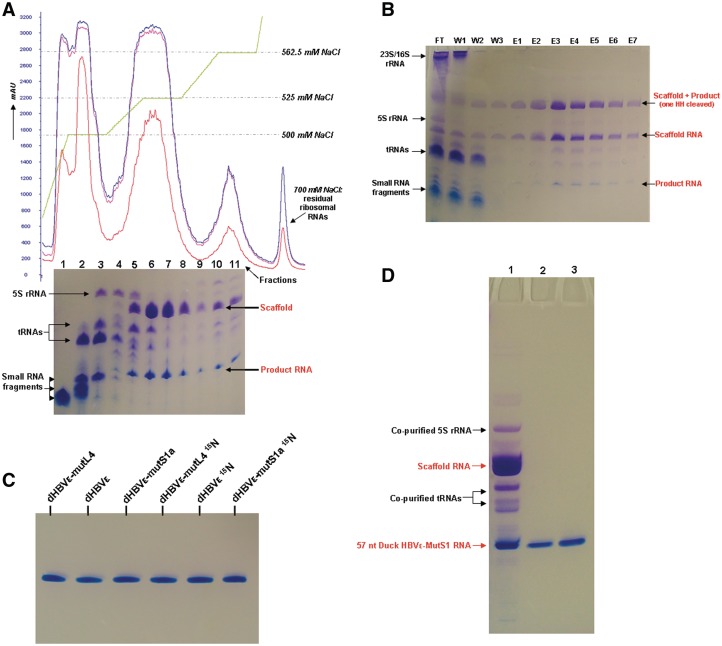
After extraction of the bacterial cells using phenol saturated with 10 mM Tris-HCl (pH 7.5) containing 10 mM magnesium acetate, the majority of the large 23S/16S ribosomal RNAs, proteins and the chromosomal DNA precipitate or are located in the organic phase, whereas the recombinant RNA (and also tRNAs, 5S rRNA and small RNAs) are located in the aqueous phase. The implementation of a re-extraction step of the organic phase is to ensure that the organic phase is completely cleared from recombinant RNA that might be trapped in aggregates or in the residual aqueous layer from the first extraction that could not be harvested without phenol. The effect of re-extraction is shown in Figure 3C under DHBVε RNA 15N-labeled, Lane 2. Additional re-extraction steps yielded no substantial amounts of RNA. Omitting magnesium acetate from buffer L results in co-extraction of the majority of the 23S/16S ribosomal RNAs. After HH ribozyme cleavage, the recombinant RNA still resembles the full-recombinant RNA molecule through base pairing interactions in the arms of the construct (Supplementary Figure S3) and is separated from the small RNA fragments, tRNAs and 5S rRNA, by anion-exchange chromatography on a ResourceQ 6 ml column (Figure 4A, chromatogram). The small RNAs, tRNAs and 5S rRNA, eluted from the column at ∼500 mM NaCl, whereas the recombinant RNA eluted from 525 to 562.5 mM NaCl.
Figure 4.
Purification of recombinant (product) RNAs. (A) FPLC elution profile of the anion-exchange purification of phenol extracted and HH ribozyme cleaved RNA, here shown for unlabeled Duck HBVε-mutL4 RNA. UV absorbance is monitored at 280 nm (red), 260 nm (blue) and 254 nm (purple); the gradient profile is shown in green. Small RNA fragments, tRNAs and 5S rRNA, elute at ∼500 mM NaCl from the column (Fractions 1–4). The large recombinant RNA construct elutes from 525–562.5 mM NaCl (Fractions 5–11). The RNA content of 20 μl of each of the collected fractions is checked on 8% denaturing PAGE (lane numbers correspond with the collected fractions). (B) Course of the Sephadex G-200 purification of unlabeled Duck HBVε-mutS1a RNA visualized on 8% denaturing PAGE. Twenty microliters of each fraction is loaded onto the gel. FT is the column flowthrough during settlement of the Sephadex beads. W1, W2 and W3 are the consecutive washing steps with buffer SB to wash out unbound RNAs. E1–E7 are the elution fractions with buffer SB containing 4 M urea. (C) Final purity of the produced NMR samples of the Duck HBVε RNA, Duck HBVε-mutL4 RNA and Duck HBV-mutS1a RNA, unlabeled and 15N-labeled. Bands are visualized with stains-all. (D) Result of the anion-exchange column purification and denaturing PAGE purification. Lane 1 contains 30 μg of recombinant Duck HBVε-mutS1 RNA after anion-exchange column purification. Lanes 2 and 3 contain 2 and 4 μg, respectively, of purified end product Duck HBVε-mutS1 RNA.
Affinity purification of the recombinant RNA with Sephadex G-200 has been employed alternatively to the phenol-based purification. To this extent, the bacterial cells were disrupted by sonication in a phosphate binding buffer (buffer SB) containing lysozyme. The cleared cell lysate was mixed with Sephadex G-200 beads equilibrated in buffer SB, binding the majority of the recombinant RNA as can be observed from Figure 4B, Lane FT (flow-through). Cellular components as tRNAs, rRNA, proteins, etc., are washed out with buffer SB (Figure 4B, Lanes W1–W3), and the bound RNA is then eluted with buffer SB containing 4 M urea (Figure 4B, Lanes E1–E7). As can be observed from the Lanes FT and W1–W3, some leakage of the bound recombinant RNA from the Sephadex beads occurred during flowthrough and column washing.
Although small amounts of tRNA and 5S rRNA co-eluted in the early fractions with the recombinant RNA during both anion-exchange purification of the recombinant RNA and, albeit to a lesser extent, the elution of the recombinant RNA from the Sephadex G-200 beads (Figure 4A, gel Lanes 5 and 6 correspond to Fractions 5 and 6), it was not problematic to distinguish between the product RNA and the small amount of contaminating RNAs during excision of the product RNA from preparative denaturing PAGE, because highly pure product RNA was obtained in the end (Figure 4C). Lane 1 of Figure 4D shows a sample of the pooled fractions after anion-exchange purification of the recombinant RNA. As can be observed, the majority of the degradation fragments, tRNAs and 5S rRNA, are removed. Lanes 2 and 3 contain 2 and 4 μg, respectively, of the denaturing PAGE-purified RNA, showing the absence of admixtures in the isolate.
NMR spectroscopy experiments
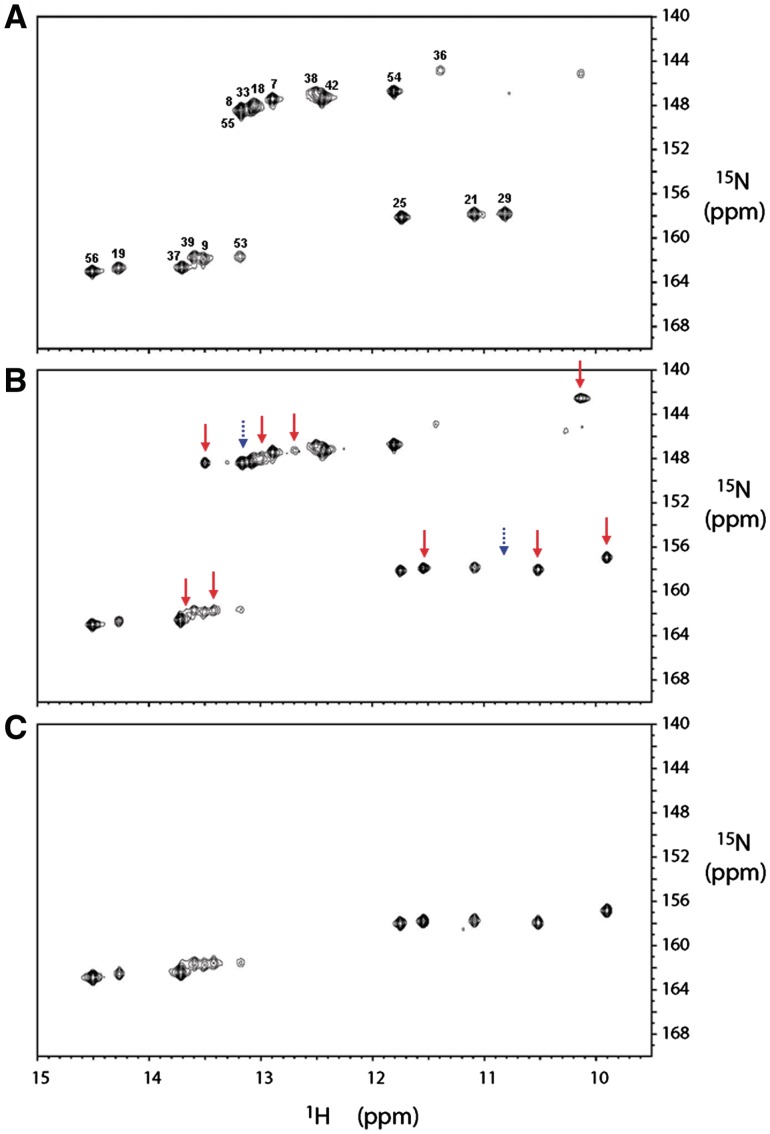
Figures 5A–C show the (1H-imino, 15N) HMQC spectra of the 15N-labeled Duck HBVε RNA, the 15N-labeled Duck HBVε-mutS1a RNA and the 13C/15N-uridine labeled Duck HBVε-mutS1a RNA, respectively. The HMQC spectrum of the recombinant uniformly 15N-labeled Duck HBVε RNA is confirmed by comparison with spectra obtained from in vitro produced Duck HBVε RNA and its fragments (31–35). Resonances assignments are based on an imino proton sequential walk in the (1H,1H) NOESY spectrum combined with the (1H-imino, 15N) HMQC spectrum and confirmed by comparison with the earlier assignments obtained for Duck HBVε RNA and fragments thereof (31–35). The (1H-imino, 15N) HMQC spectrum of uniformly 15N-labeled Duck HBVε-mutS1a RNA (Figure 5B) shows, despite only a single mutation in the apical loop (U32 to G32), a significant number of new resonances as well as resonances that have disappeared in comparison with the Duck HBVε RNA spectrum (Figure 5A). This indicates that the structure of Duck HBVε RNA has changed as a result of the introduced point mutation in the apical loop (U32 to G32). In Figure 5C, which shows the (1H-imino, 15N) HMQC spectrum of 13C/15N-uridine labeled Duck HBVε-mutS1a RNA, the G-imino resonances have indeed disappeared compared to Figure 5B. The additional resonances in the spectral region between 10 and 12 ppm indicate that additional G:U base pairs are formed as well as U:U base pairs. The presence of an alternative structure in Duck HBVε-mutS1a RNA is substantiated by the Mfold prediction of a shift in base pairings in the primer binding loop and in the apical loop region (Supplementary Figure S4a). The ΔG of this structure is predicted to be 2.2 kcal/mol lower than the structure predicted for the Duck HBVε RNA. UV melting studies carried out on Duck HBVε-mutS1a RNA, Duck HBVε RNA and Duck HBVε-mutL4 RNA (Supplementary Figure S4b), resulted in melting temperatures of 70, 68 and 68°C, respectively, confirming the predicted thermodynamic higher stability of Duck HBVε-mutS1a RNA. Analysis of the folded state of these Duck HBVε RNAs assessed by native polyacrylamide gel electrophoresis (Supplementary Figure S4c) revealed that the Duck HBVε-mutS1a RNA migrated slightly slower compared to the Duck HBVε RNA and the Duck HBVε-mutL4 RNA suggesting a more stretched out structure for Duck HBVε-mutS1a RNA. At present, the NMR and related biophysical data are further analyzed to fully establish the nature of the alternate structure.
Figure 5.
(A) The (1H-imino, 15N) HMQC spectrum of the 15N-labeled recombinant Duck HBVε RNA. The sequence specific assignment for the imino protons was achieved from 2D (1H,1H) NOESY and (1H-imino,15N) HMQC experiments as described earlier (see the text). The imino resonances are labeled with their corresponding uridine residues (resonances between 156 and 164 ppm of the 15N-axis) and guanosine residues (resonances between 142 and 150 ppm of the 15N-axis) when assigned. One resonance in the spectral region of imino protons of unpaired or mismatched guanosines remains unassigned. (B) The (1H-imino,15N) HMQC spectrum of the 15N-labeled recombinant Duck HBVε-mutS1a RNA. New appeared imino proton resonances (9) compared with spectrum (A) are indicated with red arrows whereas the positions of disappeared imino resonances (2) are indicated with dashed blue arrows. (C) The (1H-imino,15N) HMQC spectrum of the selective 13C/15N-uridine labeled in vitro produced Duck HBVε-mutS1a RNA. The observed imino resonances of the uridine residues of in vitro produced Duck HBVε-mutS1a RNA correspond with those observed in the recombinant Duck HBVε-mutS1a RNA.
DISCUSSION
We have shown that several structurally interesting product RNAs are efficiently produced in E. coli BL21 (DE3) using the proposed recombinant RNA production strategy. These product RNAs are fast and efficiently released from the protective scaffold by the action of the cis-acting hammerhead ribozymes flanking the product’s termini. The inexhaustible source of the recombinant RNA and the employment of common chromatographic purification methods provide a cheap and efficient way of producing multi-milligram quantities of pure recombinant product RNAs. The RNAs can be used for experiments varying from biochemical assays to structural analyses and drug screenings. We have shown that multi-milligram amounts of (stable isotope-labeled) RNA sufficient for NMR structural analyses can be obtained from single liter bacterial cultures. These recombinant RNAs have all been obtained within the time frame of one work week (Supplementary Table S5). Since it is possible to run productions in parallel, several samples can be produced simultaneously. Our proposed recombinant RNA in vivo production system is considerably less time consuming than enzymatic in vitro transcriptions because the latter requires additional handlings such as laborious and costly preparations of linearized plasmid DNA for in vitro transcriptions and optimization reactions. In addition, enzymatic in vitro transcriptions are more costly, because expensive (stable isotope) labeled rNTPs and large amounts of highly pure T7 RNA polymerase are needed. Finally, the straightforward bacterial replication adds another advantage to the in vivo system. Further, chemical synthesis of (stable isotope labeled) large RNAs (>50 nt) in high amounts is to date not achievable. In comparison with other in vivo methods, namely of that Ponchon et al. (14–16) and Liu et al. (17), our system does not require stoichiometric amounts of DNA splints to release the product RNA [RNase H (14–16) or DNAzyme (17)]. This makes our method somewhat less time consuming. In addition, no optimization of cleavage reactions is needed and/or recovery of DNA splints. For instance, the RNase H likely requires tests in advance to preparative cleavages [e.g. because unspecific cleavage may occur (36)]. In contrast, ribozymes, as we use, are highly specific and the cleavage initiator Mg2+ is cheap and can directly be added to the dialyzed RNA without any prior testing or heat annealing. It should further be mentioned that in our approach no protein (e.g. RNAse H) has to be purified. Finally, the RNAase H and DNA oligos required fit less optimally with scaling up of the RNA production. Initially, we attempted to produce the full-recombinant RNA construct containing the Human HBVε RNA (Figure 1A) sequence under the control of the E. coli lpp promoter (14–16). For this construct, the yield of accumulated recombinant RNA was very low (results not shown) which was not compatible with our aim to obtain sufficient amounts of RNA for NMR experiments. We therefore decided to bring the recombinant RNA production under the control of T7 RNA polymerase, which is known to display high transcription rates (18,19,37). After IPTG induction in E. coli BL21 (DE3), efficient and high accumulation of the recombinant Human HBVε RNA construct was observed, and we therefore continued to use this system to obtain the other presented product RNAs. It should be noted that the use of T7 RNA polymerase-driven expression for recombinant RNA synthesis in vivo has been previously extensively demonstrated by several research groups for the purpose of a variety functional studies [see e.g. (8,38,39)], e.g. for the production of large quantities of aptamer RNA for screening of aptamer libraries in vivo (8).
The production of the 13 RNAs was very efficient for all of them (Figure 3, Supplementany Table S3), although for some the production efficiencies were somewhat lower, i.e. for HIV-1 TAR RNA and Duck HBVε-mutS5 RNA as well as for the long construct containing the GRA RNA. Optimization of the production may however be of interest and can be achieved by considering a number of factors.
(i) The lower production yield of the Duck HBVε-mutS5 RNA is unexpected since the construct differs only in a predicted structural silent point mutation from the construct for Duck HBVε-mutS12 RNA (see Supplementary Figure S1, structures J + K) which produces much better (for unknown reason, the Sephadex aptamer is predicted to fold differently in Duck HBV-mutS12). Such differences in production efficiency are also observed between the construct for Duck HBVε RNA and its predicted structural silent point mutants Duck HBVε-mutS1a RNA and Duck HBVε-mutL4 RNA (Supplementary Figure S1, structures E, F and H). The effect of such small variations in nucleotide composition on the production efficiency of the recombinant RNA may be exploited to increase the efficiency of constructs that produce moderately.
(ii) The second aspect that appears to affect production is conformational strain at the junction of the insert (Figure 1A). The positive effect of a looser fold in this region is substantiated by the higher production efficiency of the short construct containing the GRA RNA, which lacks seven base pairing interactions between the scaffold and the insert compared with the long construct of the GRA RNA (Figure 1A and 3C).
(iii) Improvements in the yield can potentially also be achieved by shortening the full-recombinant RNA construct. For the production of recombinant Duck HBVε RNA and Human HBVε RNA covalently attached to the tRNAlys-Sephadex aptamer scaffold (15) (i.e. constructs without flanking HH ribozymes), we observed that these RNAs accumulated to significantly higher amounts (Supplementary Figure S5) compared to the much larger constructs containing HH ribozymes. This has also been observed by Ponchon and Dardel, and they suggested that smaller constructs accumulate more efficiently (i.e. to higher number of copies) or are less susceptible to nuclease degradation. The Sephadex aptamer might be removed to possibly improve production; purification using anion-exchange chromatography is anyhow faster and yields more product RNA than purification using Sephadex affinity. The 5′-TIR could also be removed as long as two or three guanosine residues remain to facilitate effective initiation of transcription by T7 RNA polymerase.
We have tried to further increase the transcription yield of the recombinant RNAs (without modifying the sequences of the full-recombinant RNA constructs) by transcribing them from the high copy number vector pUC18 (whereas the pET23b vector is a low copy number). In our hands, the opposite effect was achieved and only negligible amounts of recombinant RNA were produced (results not shown). In addition, lowering the growth temperature to 30°C or 25°C, which is common practice in recombinant protein expression, had no positive effect on the produced amounts of these and other recombinant RNAs tested using either pET23b or pUC18 as the transcription vector.
(iv) We have briefly examined the effect of DNA templated differences in length of the 5′- and 3′-termini of the full-recombinant RNA on the observed band pattern and yield of a recombinant RNA construct lacking the Sephadex aptamer and containing a HH ribozyme flanked Duck HBVε RNA (Supplementary Figure S6). We observe that the overall band pattern of the recombinant RNA is identical for all three constructs, indicating precise recognition and processing of the tRNA-scaffold moiety by host mechanisms. However, the yield was significantly lower for the construct that lacks the 5′-TIR and contains a long 3′-tail of 136 nt compared to the yield for the construct that lacks both 5′-TIR and 3′-tail and the construct that contains the 5′-TIR and lacks a 3′-tail; the latter constructs were in turn comparable in yield.
In all cases for which we demonstrated the fast RNA production using the HH-tRNA scaffold, the product RNAs terminate in a closing stem. The sequences at the termini of these stems are 5′-GAC∼∼ and ∼∼GUC-3′ and are designed to be compatible with the UX (X ≠ G) sequence requirements of the HH ribozyme preceding 5′ to the cleavage site at the 3′-terminus of the product RNA (40–42). If desired, the sequence restrictions at the 3′-terminus can be overcome by placing the hepatitis δ virus ribozyme or the hairpin ribozyme (43,44) at this position instead. The 5′-terminus of the product RNA is not impeded by sequence restrictions of the HH ribozyme and can be tailored to ones wishes.
During the time of recombinant RNA production in E. coli BL21 (DE3), the HH ribozymes are already cleaved to some extent because of the presence of Mg2+ (and/or other divalent metal ions catalyzing cleavage, such as Mn2+ and Ca2+) in the growth medium. However, we can conclude from native gel experiments (Supplementary Figure S3) that the ‘product RNA’ does not (fully or partially) dissociate from the scaffold at room temperature after HH-ribozyme cleavage and thus also not during the in-cell HH-ribozyme cleavage. DINAmelt (23) simulations confirm that the integrity of the recombinant RNA is maintained and results from the highly complementary sequence of the product RNA and scaffold arms. Furthermore, note that short RNA degradation fragments are observed during all recombinant RNA productions. These short RNA degradation fragments are also seen in productions of recombinant RNAs without flanking hammerhead ribozymes (Supplementary Figure S5) as well as in productions with an inactive ribozyme, as employed in the construct for Human HBVε RNA (results not shown). These short RNA degradation fragments probably originate from the processed recombinant RNA and are efficiently removed by both proposed purification methods and do not contaminate the end-product RNA. Thus, it appears that although cleavage takes place, the recombinant RNA (scaffold and product RNA) stays intact in the cell. This also implies that the Mg2+-activated HH-ribozyme cleavage does not hamper up-scaling of the RNA production. On the other hand, if for some reason in-cell cleavage is undesired, e.g. that apart from the product RNA also the full-recombinant RNA is required, the constructs can potentially be equipped with allosterically regulated ribozymes (45). This allows cleavage to be initiated only upon the addition of the ligand that induces the required conformational change to activate the ribozyme, provided that this ligand is not naturally occurring in E. coli. These ligands are often readily available small molecules, so that scaling up of the production appears straightforward. The proposed method is demonstrated here on product RNAs in the range of 55–70 nt in length. Production and purification of product RNAs in the range of 70–200 nt is possible with the same methodology. Note, however, that care should be taken in the design of the full-recombinant RNA molecule that the sizes of product RNA and scaffold moieties are such that their bands do not overlap on the denaturing gel during separation (e.g. tailor the length of the scaffold arms).
After ribozyme cleavage, the product RNA contains a 5′-hydroxyl and a 2′3 ′-cyclic phosphodiester at its termini. These groups are protected against ligation catalyzed by nucleic acid ligases. This opens the route for cost-efficient segmental stable isotope labeling by cross ligation of RNA segments obtained by site-specific RNase H digestion (36,46). The segments can either be purified using preparative denaturing PAGE or using denaturing anion-exchange HPLC/FPLC (14–16,36). Ligation can be performed via DNA splinted ligation using T4 DNA ligase (25,36,47) or via hybridization of the individual RNA segments and T4 RNA ligase type 1 or 2 (25,36,48–50).
In the protocol by Ponchon and Dardel for recombinant RNA production, freshly transformed bacterial cells and a strict growth protocol are required to avoid possible formation of mutations in the RNA (14,15). By using our method, the cells can be stored in glycerol at −80°C and cultivation can be initiated by inoculation from this stock or, alternatively, from plate/small cultures. The recombinant RNA does not have to accumulate during overnight growth, but the plateau is generally reached 3 h post-induction, which makes the production less time consuming. It should be mentioned that the employed T7 system is prone to basal transcription during bacterial growth in the absence of the inducer (IPTG). If the transcribed recombinant RNA is detrimental to the host, the gene coding for the recombinant RNA might be lost or inactivated. In the case of problems (e.g. mutations or gene loss) with recombinant RNA productions, E. coli Bl21 (DE3) strains expressing T7 lysozyme (e.g. from the plasmids pLysS or pLysE) (51) or attenuation of basal T7 RNA polymerase expression (52) might be used to restrict basal transcription and the possible acquisition of mutations. Thus far, we have not encountered such production failures with the employed system, but this might occur in productions for other RNA constructs in which case the above suggested work around may form a viable option.
Furthermore, with the presented RNA production system, perdeuterated recombinant RNA can potentially be produced by exchanging the growth medium with D2O (53–55) and/or site-specific labeling patterns can be introduced in the nucleotides (55–57). Additionally, the scaffold RNA, 5S rRNA and tRNAs (and also the 23S/16S rRNA where Sephadex G-200 is used), can be co-purified during purification of the product RNA. These RNAs contain the same labeling pattern as the product RNA and can be enzymatically digested into rNMPs. Phosphorylation of the rNMPs into their corresponding rNTPs (58) makes them useful for the production of labeled RNA transcribed in vitro (1) or, after conversion into their corresponding dNTPs, for the production of labeled DNA (59–62).
Scalability of RNA production and purification
The described protocol for laboratory scale purification of the full-recombinant RNA and also of the product RNAs from the scaffold RNA involves production of milligram quantities (corresponding to 1 l cultures). We routinely purify (stable isotope labeled) nucleic acids for NMR structural analysis via this protocol, which employs large-scale (preparative) PAGE purification. We find that it is reliable, relatively fast and reproducible. In addition, it has high resolution (1 nt) and leads to highly pure RNAs. Scaling up of the production up to 10 l appears doable with this PAGE purification. Alternatively, for the separation of the product RNA from the scaffold RNA moieties, more specialized equipment can be used such as preparative denaturing PAGE cells (Bio-Rad).
Beyond the 10 l scale, other purification methods should be considered. Potentially, alternate methods such as e.g. denaturing reversed phase HPLC or FPLC or denaturing anion-exchange HPLC or FPLC may then be employed, although they may also be applicable at the laboratory scale. However, these column methods may suffer from lower resolution. It depends on the application whether the final-product RNA needs to be highly pure. For recombinant RNA productions and purifications beyond laboratory scale, more development is clearly required.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–6.
FUNDING
The 6th framework program of the EU; 6th framework STREP project FSG-V-RNA. Funding for open access charge: Department of Biophysical Chemistry of the Radboud University Nijmegen, The Netherlands.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA-polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall WS, Kaiser RJ. Recent advances in the high-speed solid phase synthesis of RNA. Curr. Opin. Chem. Biol. 2004;8:222–229. doi: 10.1016/j.cbpa.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Moore PB, Abo S, Freeborn B, Gewirth DT, Leontis NB, Sun G. Preparation of 5S RNA-related materials for nuclear magnetic resonance and crystallography studies. Methods Enzymol. 1988;164:158–174. doi: 10.1016/s0076-6879(88)64041-9. [DOI] [PubMed] [Google Scholar]
- 4.Gaudin C, Nonin-Lecomte S, Tisné C, Corvaisier S, Bordeau V, Dardel F, Felden B. The tRNA-like domains of E. coli and A. aeolicus transfer-messenger RNA: structural and functional studies. J. Mol. Biol. 2003;331:457–471. doi: 10.1016/s0022-2836(03)00760-5. [DOI] [PubMed] [Google Scholar]
- 5.Tisné C, Rigourd M, Marquet R, Ehresmann C, Dardel F. NMR and biochemical characterization of recombinant human tRNA (Lys) (3) expressed in Escherichia coli: identification of posttranscriptional nucleotide modifications required for efficient initiation of HIV-1 reverse transcription. RNA. 2000;6:1403–1412. doi: 10.1017/s1355838200000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallis NG, Dardel F, Blanquet S. Heteronuclear NMR studies of the interactions of 15N-labeled methionine specific transfer RNAs with methionyl-tRNA transformylase. Biochemistry. 1995;34:7668–7677. doi: 10.1021/bi00023a013. [DOI] [PubMed] [Google Scholar]
- 7.Xue H, Shen W, Wong JTF. Purification of hyperexpressed Bacillus subtilis tRNA (Trp) cloned in Escherichia coli. J. Chromatogr. B Biomed. Appl. 1993;613:247–255. doi: 10.1016/0378-4347(93)80139-u. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Potty AS, Jackson GW, Stepanov V, Tang A, Liu Y, Kourentzi K, Strych U, Fox GE, Willson RC. Engineered 5S ribosomal RNAs displaying aptamers recognizing vascular endothelial growth factor and malachite green. J. Mol. Recognit. 2009;22:154–161. doi: 10.1002/jmr.917. [DOI] [PubMed] [Google Scholar]
- 9.Pitulle C, Hedenstierna KO, Fox GE. A novel approach for monitoring genetically engineered microorganisms by using artificial, stable RNAs. Appl. Environ. Microbiol. 1995;61:3661–3666. doi: 10.1128/aem.61.10.3661-3666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitulle C, Dsouza L, Fox GE. A low molecular weight artificial RNA of unique size with multiple probe target regions. Syst. Appl. Microbiol. 1997;20:133–136. doi: 10.1016/S0723-2020(97)80057-4. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Deutscher MP. Analyzing the decay of stable RNAs in E. Coli. Methods Enzymol. 2008;447:31–45. doi: 10.1016/S0076-6879(08)02202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li ZW, Reimers S, Pandit S, Deutscher MP. RNA quality control: degradation of defective transfer RNA. EMBO J. 2002;21:1132–1138. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F. A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat. Protoc. 2009;4:947–959. doi: 10.1038/nprot.2009.67. [DOI] [PubMed] [Google Scholar]
- 15.Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat. Methods. 2007;4:571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 16.Ponchon L, Dardel F. Large scale expression and purification of recombinant RNA in Escherichia coli. Methods. 2011;54:267–273. doi: 10.1016/j.ymeth.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Stepanov VG, Strych U, Willson RC, Jackson GW, Fox GE. DNAzyme-mediated recovery of small recombinant RNAs from a 5S rRNA-derived chimera expressed in Escherichia coli. BMC Biotechnol. 2010;10:1–9. doi: 10.1186/1472-6750-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 19.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 20.Fisher CL, Pei GK. Modification of a PCR-based site-directed mutagenesis method. Biotechniques. 1997;23:570–571, 574. doi: 10.2144/97234bm01. [DOI] [PubMed] [Google Scholar]
- 21.Yon J, Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu KH, Beck J, Nassal M. SELEX-derived aptamers of the duck hepatitis B virus RNA encapsidation signal distinguish critical and non-critical residues for productive initiation of reverse transcription. Nucleic Acids Res. 2004;32:4377–4389. doi: 10.1093/nar/gkh772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 25.Nelissen FH, van Gammeren AJ, Tessari M, Girard FC, Heus HA, Wijmenga SS. Multiple segmental and selective isotope labeling of large RNA for NMR structural studies. Nucleic Acids Res. 2008;36:e89. doi: 10.1093/nar/gkn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wijmenga SS, van Buuren BNM. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Mag. Res. Sp. 1998;32:287–387. [Google Scholar]
- 27.Cromsigt J, van Buuren B, Schleucher J, Wijmenga S. Resonance assignment and structure determination for RNA. Methods Enzymol. 2001;338:371–399. doi: 10.1016/s0076-6879(02)38229-6. [DOI] [PubMed] [Google Scholar]
- 28.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe: a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 29.Srisawat C, Engelke DR. RNA affinity tags for purification of RNAs and ribonucleoprotein complexes. Methods. 2002;26:156–161. doi: 10.1016/S1046-2023(02)00018-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srisawat C, Goldstein IJ, Engelke DR. Sephadex-binding RNA ligands: rapid affinity purification of RNA from complex RNA mixtures. Nucleic Acids Res. 2001;29:e4. doi: 10.1093/nar/29.2.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ampt KA, van der Werf RM, Nelissen FH, Tessari M, Wijmenga SS. The unstable part of the apical stem of duck hepatitis B virus epsilon shows enhanced base pair opening but not pico- to nanosecond dynamics and is essential for reverse transcriptase binding. Biochemistry. 2009;48:10499–10508. doi: 10.1021/bi9011385. [DOI] [PubMed] [Google Scholar]
- 32.van der Werf RM, Girard FC, Nelissen F, Tessari M, Wijmenga SS. H-1, C-13 and N-15 NMR assignments of Duck HBV primer loop of the encapsidation signal epsilon. Biomol. NMR Assign. 2008;2:143–145. doi: 10.1007/s12104-008-9106-8. [DOI] [PubMed] [Google Scholar]
- 33.Ampt KAM, Ottink OM, Girard FC, Nelissen F, Tessari M, Wijmenga SS. H-1, C-13 and N-15 NMR assignments of Duck HBV apical stem loop of the epsilon encapsidation signal. Biomol. NMR Assign. 2008;2:159–162. doi: 10.1007/s12104-008-9110-z. [DOI] [PubMed] [Google Scholar]
- 34.Van der Werf RM. Ph.D. Thesis. Nijmegen: Radboud University; 2011. [Google Scholar]
- 35.Girard FC, Ottink OM, Ampt KA, Tessari M, Wijmenga SS. Thermodynamics and NMR studies on duck, heron and human HBV encapsidation signals. Nucleic Acids Res. 2007;35:2800–2811. doi: 10.1093/nar/gkm131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duss O, Maris C, von Schroetter C, Allain FHT. A fast, efficient and sequence-independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucleic Acids Res. 2010;38:e188. doi: 10.1093/nar/gkq756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golomb M, Chamberl M. Characterization of T7-specific ribonucleic-acid polymerase: IV. Resolution of major invitro transcripts by gel electrophoresis. J. Biol. Chem. 1974;249:2858–2863. [PubMed] [Google Scholar]
- 38.Seyhan AA, Amaral J, Burke JM. Intracellular RNA cleavage by the hairpin ribozyme. Nucleic Acids Res. 1998;26:3494–3504. doi: 10.1093/nar/26.15.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yadava RS, Kumar R, Yadava PK. Expression of lexA targeted ribozyme in Escherichia coli BL-21 (DE3) cells. Mol. Cell. Biochem. 2005;271:197–203. doi: 10.1007/s11010-005-6340-6. [DOI] [PubMed] [Google Scholar]
- 40.Perriman R, Delves A, Gerlach WL. Extended target-site specificity for a hammerhead ribozyme. Gene. 1992;113:157–163. doi: 10.1016/0378-1119(92)90391-2. [DOI] [PubMed] [Google Scholar]
- 41.Sheldon CC, Symons RH. Mutagenesis analysis of a self-cleaving RNA. Nucleic Acids Res. 1989;17:5679–5685. doi: 10.1093/nar/17.14.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price SR, Ito N, Oubridge C, Avis JM, Nagai K. Crystallization of RNA-protein complexes: I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol. 1995;249:398–408. doi: 10.1006/jmbi.1995.0305. [DOI] [PubMed] [Google Scholar]
- 43.Hampel A, Tritz R. RNA catalytic properties of the minimum (-)sTRSV sequence. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- 44.Pérez-Ruiz M, Barroso-DelJesus A, Berzal-Herranz A. Specificity of the hairpin ribozyme. Sequence requirements surrounding the cleavage site. J. Biol. Chem. 1999;274:29376–29380. doi: 10.1074/jbc.274.41.29376. [DOI] [PubMed] [Google Scholar]
- 45.Zivarts M, Liu Y, Breaker RR. Engineered allosteric ribozymes that respond to specific divalent metal ions. Nucleic Acids Res. 2005;33:622–631. doi: 10.1093/nar/gki182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, Lapham J, Crothers DM. Determining RNA solution structure by segmental isotopic labeling and NMR: Application to Caenorhabditis elegans spliced leader RNA 1. Proc. Natl Acad. Sci. USA. 1996;93:44–48. doi: 10.1073/pnas.93.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore MJ, Query CC. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- 48.Bullard DR, Bowater RP. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem. J. 2006;398:135–144. doi: 10.1042/BJ20060313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim I, Lukavsky PJ, Puglisi JD. NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J. Am. Chem. Soc. 2002;124:9338–9339. doi: 10.1021/ja026647w. [DOI] [PubMed] [Google Scholar]
- 50.Tzakos AG, Easton LE, Lukavsky PJ. Complementary segmental labeling of large RNAs: economic preparation and simplified NMR spectra for measurement of more RDCs. J. Am. Chem. Soc. 2006;128:13344–13345. doi: 10.1021/ja064807o. [DOI] [PubMed] [Google Scholar]
- 51.Studier FW. Use of bacteriophage-T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 52.Mertens N, Remaut E, Fiers W. Tight transcriptional control mechanism ensures stable high-level expression from T7 promoter-based expression plasmids. Bio-Technol. 1995;13:175–179. doi: 10.1038/nbt0295-175. [DOI] [PubMed] [Google Scholar]
- 53.Pochapsky SS, Pochapsky TC, Wei JW. A model for effector activity in a highly specific biological electron transfer complex: the cytochrome P450(cam)-putidaredoxin couple. Biochemistry. 2003;42:5649–5656. doi: 10.1021/bi034263s. [DOI] [PubMed] [Google Scholar]
- 54.Zwahlen C, Vincent SJF, Gardner KH, Kay LE. Significantly improved resolution for NOE correlations from valine and isoleucine (C-gamma 2) methyl groups in N-15,C-13- and N-15,C-13,H-2-labeled proteins. J. Am. Chem. Soc. 1998;120:4825–4831. [Google Scholar]
- 55.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 56.Dayie TK, Thakur CS. Site-specific labeling of nucleotides for making RNA for high resolution NMR studies using an E. coli strain disabled in the oxidative pentose phosphate pathway. J. Biomol. NMR. 2010;47:19–31. doi: 10.1007/s10858-010-9405-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee KM, Androphy EJ, Baleja JD. A novel method for selective isotope labeling of bacterially expressed proteins. J. Biomol. NMR. 1995;5:93–96. doi: 10.1007/BF00227474. [DOI] [PubMed] [Google Scholar]
- 58.Batey RT, Inada M, Kujawinski E, Puglisi JD, Williamson JR. Preparation of isotopically labeled ribonucleotides for multidimensional NMR-spectroscopy of RNA. Nucleic Acids Res. 1992;20:4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Louis JM, Martin RG, Gronenborn AM, Clore GM. Preparation of uniformly isotope-labeled DNA oligonucleotides for NMR spectroscopy. J. Biol. Chem. 1998;273:2374–2378. doi: 10.1074/jbc.273.4.2374. [DOI] [PubMed] [Google Scholar]
- 60.MacDonald D, Lu PZ. Determination of DNA structure in solution: enzymatic deuteration of the ribose 2 ' carbon. J. Am. Chem. Soc. 2002;124:9722–9723. doi: 10.1021/ja026678r. [DOI] [PubMed] [Google Scholar]
- 61.Nelissen FH, Girard FC, Tessari M, Heus HA, Wijmenga SS. Preparation of selective and segmentally labeled single-stranded DNA for NMR by self-primed PCR and asymmetrical endonuclease double digestion. Nucleic Acids Res. 2009;37:e114. doi: 10.1093/nar/gkp540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zimmer DP, Crothers DM. NMR of enzymatically synthesized uniformly 13C15N-labeled DNA oligonucleotides. Proc. Natl Acad. Sci. USA. 1995;92:3091–3095. doi: 10.1073/pnas.92.8.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flodell S, Petersen M, Girard F, Zdunek J, Kidd-Ljunggren K, Schleucher J, Wijmenga S. Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal. Nucleic Acids Res. 2006;34:4449–4457. doi: 10.1093/nar/gkl582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flodell S, Schleucher J, Cromsigt J, Ippel H, Kidd-Ljunggren K, Wijmenga S. The apical stem-loop of the hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidine bulges. Nucleic Acids Res. 2002;30:4803–4811. doi: 10.1093/nar/gkf603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petzold K, Duchardt E, Flodell S, Larsson G, Kidd-Ljunggren K, Wijmenga S, Schleucher J. Conserved nucleotides in an RNA essential for hepatitis B virus replication show distinct mobility patterns. Nucleic Acids Res. 2007;35:6854–6861. doi: 10.1093/nar/gkm774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ottink OM, Rampersad SM, Tessari M, Zaman GJ, Heus HA, Wijmenga SS. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined-induced fit mechanism. RNA. 2007;13:2202–2212. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–2365. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ampt KAM, van der Werf RM, Nelissen FHT, Tessari M, Wijmenga SS. The unstable part of the apical stem of Duck Hepatitis B virus epsilon shows enhanced base pair opening but not pico- to nanosecond dynamics and is essential for reverse transcriptase binding. Biochemistry. 2009;48:10499–10508. doi: 10.1021/bi9011385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.