Abstract
The myotonic dystrophies (DM) are human diseases in which the accumulation of toxic RNA (CUG or CCUG) repeats in the cell causes sequestration of splicing factors, including MBNL1, leading to clinical symptoms such as muscle wasting and myotonia. We previously used Dynamic Combinatorial Chemistry to identify the first compounds known to inhibit (CUG)-MBNL1 binding in vitro. We now report transformation of those compounds into structures with activity in vivo. Introduction of a benzo[g]quinoline substructure previously unknown in the context of RNA recognition, as well as other modifications, provided several molecules with enhanced binding properties, including compounds with strong selectivity for CUG repeats over CAG repeats or CAG–CUG duplex RNA. Compounds readily penetrate cells, and improve luciferase activity in a mouse myoblast assay in which enzyme function is coupled to a release of nuclear CUG–RNA retention. Most importantly, two compounds are able to partially restore splicing in a mouse model of DM1.
INTRODUCTION
High affinity, sequence-selective recognition of RNA by synthetic molecules is increasingly recognized as a key strategic goal for the production of novel therapeutics and biochemical probes (1). The importance of this area is accentuated by the ever-increasing pace of discovery of new RNA sequences with biochemically important (and therefore potentially biomedically important) functions. Among the many recent advances in this area is the recognition that many non-coding RNA (ncRNA) elements present in the eukaryotic RNAome play a direct role in controlling cellular processes and disease (2). Human diseases believed to have an ncRNA origin include spinocerebellar ataxia, fragile X-syndrome, diabetes mellitus, myoclonus epilepsy and the myotonic dystrophies (DM) (3). However, to date only a relatively small number of compounds have been reported that bind specific RNA sequences and elicit a desired biological response. For these reasons, expanding the pool of sequence-selective RNA-targeted synthetic molecules presents a critically important but under-examined challenge in chemical biology.
The DM are central examples of a growing family of RNA-mediated diseases (3–5). Myotonic dystrophy type 1 (DM1) is the most common form of adult-onset muscular dystrophy, affecting ∼1 in 8000 people (6). An autosomal dominant inherited disease, DM1 results from a CTG repeat expansion (CTGexp) in the 3′-UTR of the DM protein kinase gene (DMPK) on chromosome 19q. The expanded CTG is transcribed into long CUGexp repeat mRNA. These RNA repeats sequester RNA-binding proteins such as the MBNL (muscleblind-like) family of splicing regulators, retaining them in the nucleus as foci. This in turn leads to misregulated alternative splicing, or spliceopathy. Myotonic dystrophy type 2 (DM2) is caused by an unstable expansion of a CCTG repeat in intron 1 of the cellular nucleic acid-binding protein gene (CNBP, also known as the zinc finger protein 9 gene or ZNF9) on chromosome 3q. Transcription produces toxic mRNA containing hundreds to thousands of CCUGexp. Like CUGexp, these are also sequestered into foci, and deplete MBNL1 protein from the affected cell (7,8). Currently, there is no pharmaceutical therapy for either DM1 or DM2. However, the molecular understanding of these diseases suggests that displacement of MBNL1 from its CUGexp or CCUGexp binding sites constitutes an attractive strategy for developing therapies targeting DM (8,9). Experiments in animal models support this hypothesis. For example, a morpholino antisense oligonucleotide (CAG-25) complementary to CUGexp RNA was able to hybridize with the CUGexp RNA in vitro, and displace MBNL1. Intramuscular injection of CAG-25 in a transgenic DM1 mouse model partially restored chloride channel-1 (Clcn-1) protein expression, and DM1 symptoms lessened (10). The high cost and challenging pharmacological properties of oligonucleotide-based drugs suggest, however, that alternative approaches to targeting CUGexp RNA are of value.
In the absence of well-defined rules guiding the design of sequence-selective RNA-binding molecules [such as those developed by Dervan et al. (11) for DNA recognition], the question arises as to how one successfully addresses the binding problem. Several years ago, a number of groups, including ours, developed the concept of Dynamic Combinatorial Chemistry (DCC) as a ‘rapid prototyping’ method for testing binding hypotheses and facilitating the identification of compounds with novel architectures and properties (12). Since that time, DCC has evolved in many directions, demonstrating its potential as a method for the identification of receptors for small-molecule analytes, catalysts, new materials, sensors and a broad range of compounds able to bind protein and nucleic acid targets. However, to our knowledge no DCC-derived hit compound has either directly or through analog production resulted in a structure with in vivo activity.
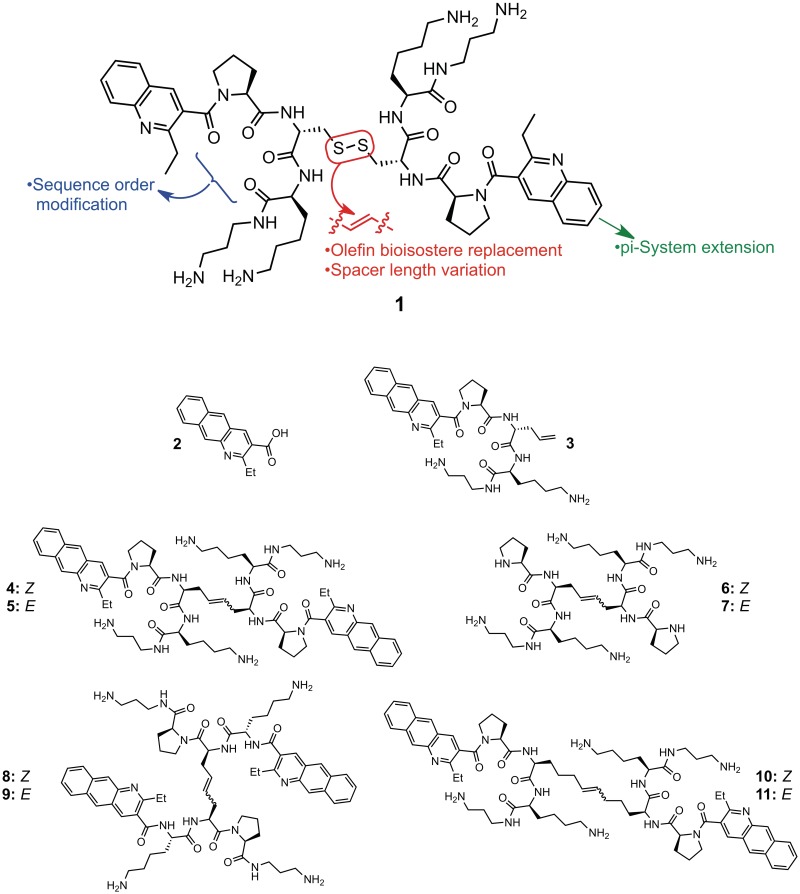
In 2008, we reported the first non-nucleic acid-based compounds (1 is a representative structure) capable of binding CUGexp RNA and competitively inhibiting CUGexp-MBNL1 binding in vitro (13). This work relied on a resin-bound form of DCC, termed RBDCC, that we developed to facilitate the identification of sequence-selective DNA (14) and RNA (13) binding compounds. Several groups have subsequently demonstrated elegant and structurally varied approaches to binding CUGexp and CCUGexp RNA (8,15–19). This recent upsurge of interest highlights the fact that DM1 and DM2 RNAs are important therapeutic targets, as well as valuable model systems for testing hypotheses regarding the factors influencing selectivity and affinity in RNA recognition. Despite these advances, demonstration of the restoration of MBNL1 activity in vivo by cell-permeable, highly selective CUGexp RNA binders remains an important goal.
RBDCC hit compound 1 (Figure 1) and related molecules identified in our initial work provided a useful demonstration of feasibility, and set the stage for building toward a compound that would be suitable for further evaluation in the biological context. To accomplish that goal, we anticipated that replacing the disulfide bridge with an olefin bioisostere would not have a dramatic impact on affinity, based on results from parallel efforts in our lab targeting an RNA sequence involved in regulating −1 ribosomal frameshifting in HIV (20). Since disulfides are easily reduced in the cytoplasm, replacing the disulfide with an olefin or alkane would facilitate cellular studies. Second, molecules containing hydrocarbon bridges of varied length would allow us to examine the effect of linker length and configuration on binding ability and selectivity. Third, we wished to explicitly examine the importance of the amino acid sequence order. Finally, as quinolines are known intercalators, at least in the DNA-binding context (21), we hypothesized that increasing the pi surface area of this group would enhance affinity. In this regard, we were surprised to discover that despite the vast amount of research conducted into the nucleic acid recognition properties and biological activity of acridine derivatives, including the use of several acridines in humans as antimicrobials (22) and chemotherapeutic agents (23), we are only aware of one mention of the closely related benzo[g]quinoline heterocycle (i.e. 2, Figure 1) in the nucleic acid recognition literature (24). Thus, synthesizing and testing derivatives incorporating this moiety would constitute the first examination of this heterocycle in the RNA binding context.
Figure 1.
Hit compound 1 identified via RBDCC and molecules (2–11) synthesized in this work.
MATERIALS AND METHODS
Benzo[g]quinoline 2 was synthesized by condensation of methyl acetoacetate and 3-nitro-2-naphthaldehyde (25), using a one-pot procedure originally developed by us in the context of quinoline synthesis (26). Compounds 3–9 were synthesized on solid phase by analogy to methods previously reported by our group (20). For compounds 10 and 11, l-pentenyl glycine was synthesized via asymmetric alkylation of pseudoephedrine glycinamide (27). Complete synthesis procedures and compound characterization are provided in Supplementary Data.
Surface plasmon resonance (SPR) analysis
All SPR experiments were conducted using a Biacore-X instrument (Biacore, Inc.). Both flow cells (FC1 and FC2) of a research grade carboxymethyl dextran coated sensor chip (CM5, GE Healthcare) were functionalized with streptavidin following activation by EDC/NHS. Unreacted NHS-ester was deactivated with ethanolamine. Next, a known amount (response unit, RU, between 200–1000) of 5′-biotin labeled RNA (IDT Inc.) in running buffer was captured in FC2; FC1 was blocked with biotin and used as a reference. The level of RNA immobilized was limited to reduce mass transfer effects on the association phase. Binding analyses were carried out by flowing various concentrations of compound in HBS-N buffer (0.01 M HEPES, 0.15 M NaCl, pH = 7.4) at 30 or 60 µl/min over the immobilized RNA and recording the reference-subtracted sensorgrams. For kinetic experiments, dissociation was monitored for at least 200 s after which the surface was regenerated, where necessary, with a pulse of either 0.5 or 1 M NaCl, followed by buffer wash to reestablish baseline. Each compound concentration was injected twice to verify consistency. Blank subtraction was performed by subtracting the result obtained from the injection of buffer alone. Equilibrium binding constants (KD) and kinetic rate constants (kon, koff) were obtained by globally fitting sensorgrams to the 1:1 Langmuir equation using Biaevaluation software (Biacore, Inc.). Use of the 1:1 Langmuir equation assumes that each binding site on the RNA is identical; this is of course an oversimplification. Reported apparent KD values are an average of two separate SPR experiments on different sensor chips, with different surface densities of immobilized RNA to verify that measured values were not impacted by surface RNA density. Dissociative half-lives t1/2 were calculated from the dissociation rate constants kd (t1/2 = ln2/kd). Dissociative rates were calculated using a single-exponential decay. This methodology has been proven robust even for complexes with slower off rates than those observed in this study, and in cases where the SPR signal does not return to baseline within the measurement time frame (28). In part, this is because competing fast processes manifest as changes to early time points in the SPR dissociation curve. Stoichiometry n was assessed by repeating each SPR experiment with increasingly higher ligand concentration to saturation (RUmax), followed by Scatchard analysis (r/Cfree versus r) of the steady-state RUs. All error measurements were calculated as described in the literature (29,30) (Supplementary Methods, and Supplementary Tables S1–S44).
Fluorescence titrations
RNA (10 µM stock) in HBS-N buffer was titrated into a solution of 1 µM compound in HBS-N. After each addition of RNA, 10 min were allowed for equilibration before recording the change in fluorescence at 468.5 nm. Fluorescence units (FU) were then corrected for dilution and the FU after each addition was subtracted from the FU at zero RNA concentration to give ΔFU. Following saturation, ΔFU was plotted against RNA concentration using Origin 7 (OriginLab, Inc.), and fitted to a 1:1 Langmuir binding model to obtain apparent dissociation constants (KD). The reported KD are a composite fit from at least two repetitions.
Cell permeation
C2C12 mouse myoblasts grown to 80% confluence were exposed to compounds for 12 h in a 96-well tissue culture plate. After removal of the culture media [DMEM containing 10% FBS, 1% penicillin–streptomycin (GIBCO)], the cells were washed twice with PBS to remove excess compounds. Cells were then imaged while in buffer under a fluorescence microscope (Olympus IX70) in the 96-well plate using a 460-nm emission filter.
Cell toxicity
C2C12 mouse myoblasts were plated in a 96-well tissue culture plate in DMEM (10% fetal bovine serum, 1% penicillin–streptomycin) and allowed to grow to ∼80% confluence at 37°C under CO2. Varying compound concentrations (up to 1 mM) were incubated with cells (48 h, 37°C). Media was then removed, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) in media was added to each well and incubated at 37°C for 4 h. After removal of the MTT media, isopropanol (100 µl) was added, and absorbance was measured at 600 nm on a Modulus microplate reader (Turner Biosystems).
Activity in transformed mouse myoblasts
C2C12 mouse myoblasts were engineered to stably express a firefly luciferase transcript with or without ∼800 uninterrupted CUG repeats in an hDMPK 3′-UTR (clones C5-14 and C1-S, respectively). Cells were grown in 100 µl growth media in 96-well plates to ∼10% confluency. Compound was added at various concentrations in triplicate to both C1-S and C5-14 cultures, and incubated for 3 days. Culture media was replaced with fresh media containing 1% WST-1 reagent (Roche). After ∼15–20 min of incubation, the WST-1 containing media was transferred to a clear 96-well plate, and the absorbances at 450 and 690 nm measured with a PerkinElmer EnVision Plate Reader. Cells were gently rinsed (1× PBS) before incubation at −20°C in 100 µl 1× Passive Lysis Buffer (Promega) for 10 min. Immediately before luminescence detection, 20 µl of each lysate was mixed with 50 µl of Luciferase Assay Reagent (Promega) in a fresh opaque-white 96-well plate. Luminescence values were normalized for well-to-well variations in viable cell numbers by dividing by the corresponding (A450–A690 nm) values of the WST-1 containing media.
Mouse studies
Mouse handling and experimental procedures were conducted in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care. HSALR transgenic mice in line 20 b expressing human skeletal actin RNA with 220 CUG repeats in the 3′-UTR were previously described (while initially incorporating 250 CUG repeats, these have subsequently shortened) (31). Initial experiments were conducted with age- and gender-matched HSALR mice (12–16 weeks old), while follow-up experiments on compound 11 used age-matched mice, with four males and one female in each group. In each case, experimental animals were injected intraperitoneally with 40 mg/kg of compounds or saline alone once per day for 5 days. Mice were sacrificed 1 day after the last injection, and vastus muscle was obtained for splicing analysis. RNA extraction, cDNA preparation and RT-PCR were performed as described previously (31,32). Each RT-PCR reaction was performed in triplicate. The PCR products were separated on agarose gels, and scanned with a laser fluorimager (Typhoon, GE Healthcare). Quantitative analysis of amplified products was performed by ImageQuant software (Molecular Dynamics). Differences between two groups were evaluated by Student's t-test.
RESULTS
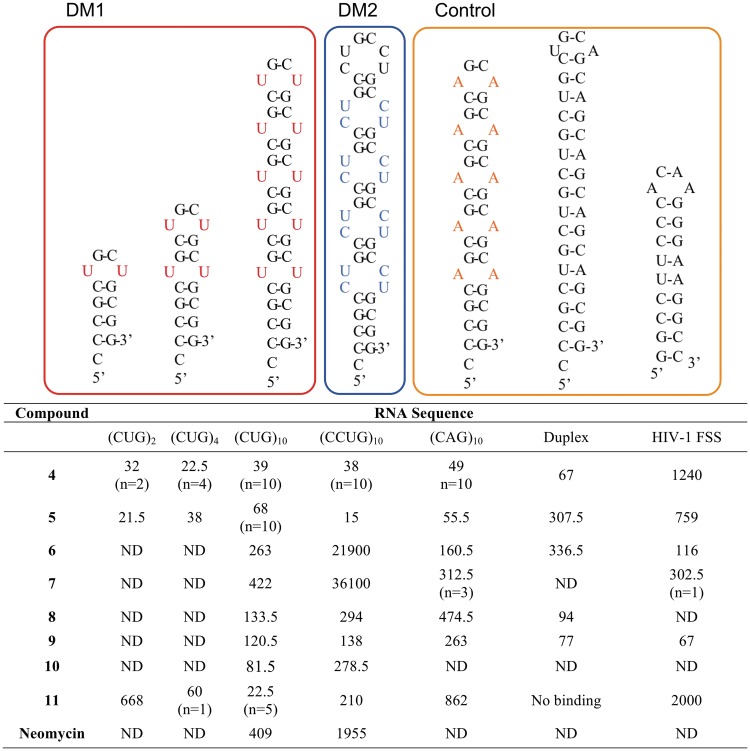
The binding characteristics of 3–11 were evaluated against pathogenic CUGexp RNA and other RNA sequences by surface plasmon resonance (SPR). This technique allows the association and dissociation rates (ka and kd), equilibrium binding constants (KD) and the binding stoichiometry (n) to be measured in a label-free format (33). RNA sequences, shown in Figure 2 along with their respective abbreviated names, were designed in order to test the effect of increasing numbers of CUG repeats [‘(CUG)2’, ‘(CUG)4’ and ‘(CUG)10’], differences between recognition of DM1 and DM2 RNA [‘(CCUG)10’], U–U versus A–A mismatches [‘(CAG)10’] and mismatched RNA versus a CUG–CAG duplex (‘Duplex’). An RNA sequence derived from the gag/pol frameshift-stimulating region of HIV-1 (‘HIV-1 FSS’) (34) was employed as an off-target control.
Figure 2.
Thermodynamic affinities (apparent KD, nM) and selected stoichiometries (n) as measured by SPR. Binding constants are reported as an average of two complete titrations determined at differing surface RNA density; measured KD values for each run and associated errors are provided in Supplementary Data.
Equilibrium affinities
Compound 3 showed no measurable affinity for (CUG)10. In contrast, 4 and 5 bound (CUG)10 with apparent KD values of 39 and 68 nM, respectively, in both cases representing a roughly 50-fold improvement in affinity relative to 1 (6.7 µM, as measured by filter binding) (14). Scatchard analysis of the steady-state SPR response units (RU) for the binding between 4 or 5 and (CUG)10 yielded a binding stoichiometry of 10:1 for each isomer. This implies that one molecule of compound binds one (CUG) trinucleotide in the (CUG)10 RNA hairpin stem and loop. The Scatchard plot for these data displays a strong convex curvature (Supplementary Data), and Hill coefficients of 1.9 (compound 4) or 1.5 (compound 5) were calculated, all consistent with positively cooperative binding (35,36). Both the apparent affinity and binding stoichiometry (the latter as a function of number of CUG repeats) remained consistent for 4 binding to (CUG)4 and (CUG)2, confirming that one molecule of 4 binds to each CUG. In contrast, the affinity of 5 actually appeared to strengthen slightly for (CUG)4 and (CUG)2. Reducing the number of CUG repeats in the target RNA yielded linear Scatchard plots for 4 (Supplementary Tables S3 and S4). This confirms an expected loss of cooperativity as the number of CUG repeats decreases. Likewise, calculated Hill coefficients were consistent with non-cooperative binding to short repeats. One possible explanation for this is that the longer (CUG)10 repeat RNA has greater flexibility, and ligand binding induces a conformational change which then affects the mode of binding of subsequent ligand molecules. While 4 and 5 are strongly selective for binding CUG repeat RNAs relative to unrelated hairpins such as the HIV-1 FSS (apparent KD 1240 and 759 nM, respectively), both (CCUG)10 and (CAG)10 were bound with similar affinities to the CUG repeat sequences. It is not surprising that differentiation among these sequences is particularly challenging. MBNL1 itself has been reported to bind CUG and CCUG repeats with only a 2-fold difference in affinity as measured by gel-shift assay (37). Likewise, MBNL1 was found to bind (CUG) and (CAG) repeats with similar affinity in a filter binding assay (38). The X-ray crystal structures of a (CUG)6 repeat duplex and a (CAG) repeat containing RNA duplex showed that both adopt A-form RNA conformation with alternating stripes of positive and negative potential due to the G–C pairs (39–41). It has been predicted that the (CCUG) repeat RNA may also adopt a similar conformation (38). Binding of 4 to duplex CUG–CAG appears to be ∼4.6-fold stronger than the E isomer, 5.
To ascertain the importance of the benzo[g]quinoline moiety, we examined cis (6) and trans (7) peptides lacking the heterocycle. While both compounds bound (CUG)10 RNA with modest affinity, neither displayed selectivity for the CUG repeat. Likewise, benzo[g]quinoline-bearing compounds with a ‘scrambled’ peptide sequence (8 and 9) showed reduced affinity to (CUG)10, and no selectivity for CUG or CCUG repeats relative to duplex CUG–CAG or HIV-1 FSS RNAs. Both observations suggest that a substantial fraction of the functionality in 4 and 5 must be present in the correct relative spatial orientation in order for high affinity recognition to be achieved.
A key aspect of the original RBDCC library design was that the disulfide was incorporated only to reversibly link RNA-binding modules together, rather than participating in RNA binding itself. Such a reversible linkage is a critical feature of any dynamic combinatorial library, but depending on the exchange reaction used may require subsequent re-engineering of the compound in a form not subject to exchange under physiological conditions. In this case, replacement of the labile disulfide bridge with an olefin allowed us to not only improve the biostability of the compound, but also provided an opportunity to test the effect of varying the spacing between modules on binding. This was also motivated by the complete lack of binding by monomer 3 and benzo[g]quinoline 2, suggesting that a bidentate mode of binding stabilized by the two heterocyclic handles is required. For this hypothesis to hold, one would anticipate that changing the separation between the two halves of the ligand would alter selectivity and affinity. Thus, we synthesized ‘extended’ linker compounds 10 and 11. These compounds were found to bind (CUG)10 with an affinity similar to that displayed by 4 and 5; however, affinity for 11 decreased in tandem with decreasing hairpin length [from an apparent KD of 22.5 nM for (CUG)10 to 668 nM for (CUG)2]. Binding stoichiometry also changed, with 5:1 binding observed for (CUG)10 and 1:1 binding observed for (CUG)4. This more stringent structural requirement also manifested as a dramatic increase in sequence selectivity: 11 has a significant preference for (CUG)10 over (CCUG)10, and a 38-fold preference versus (CAG)10. No measurable binding was observed by SPR to the duplex CUG–CAG sequence, while binding to the HIV hairpin was reduced 89-fold. As 10 was isolated as only a very minor product of the metathesis reaction, only a limited number of experiments were possible with this compound. However, SPR suggests it is also somewhat selective for (CUG)10 over (CCUG)10. Likewise, 10 binds (CUG)10 with a 5:1 stoichiometry. Binding of 11 to (CUG)10 is cooperative (Hill coefficient of 1.9). The 2-fold difference in stoichiometry for ‘extended’ compounds 10 and 11 relative to 4 and 5 is readily observable in the SPR trace. For example, an injection of excess 10 produces roughly half the steady-state response of the injection of an equivalent concentration of 5 (Supplementary Figure S2). These data are consistent with the requirement of a more distributed binding site. Neomycin, a well-studied aminoglycoside antibiotic with relatively low sequence selectivity, has been reported to bind (CUG) repeats (9), and represents a useful positive control from another structural class. We found that neomycin binds (CUG)10 and (CCUG)10 with much weaker affinity (apparent KD of 409 and 1955 nM, respectively) than the best compounds described earlier.
Selected equilibrium affinities for (CUG)10 were confirmed by fluorescence titration. The benzo[g]quinoline moiety is fluorescent, with excitation and emission maxima at 362 and 439 nm, respectively, in methanol. The quantum yield of the ethylbenzo[g]quinoline carboxylic acid (2), relative to quinine sulfate, was determined to be 0.64 (Supplementary Figure S3). This unique feature allowed us to directly monitor RNA binding, and mammalian cell penetration and localization without the need for additional labeling. In all cases, compound binding resulted in saturable quenching of fluorescence (Supplementary Figures S4–S6). Apparent KD values measured in this manner are consistent with those obtained by SPR (Table 1); that they are not identical to SPR-measured values likely results from the differing formats of the two methods. As a control, yeast tRNA was titrated into a mixture of 4 and 5; no binding was observed.
Table 1.
Binding constants as measured by fluorescence titration
| Compound | Sequence | Apparent KD (nM) |
|---|---|---|
| 3 | (CUG)10 | No binding |
| 4 | (CUG)10 | 66 ± 2 |
| 5 | (CUG)10 | 70 ± 1 |
| 10 | (CUG)10 | 40.4 ± 0.3 |
| 11 | (CUG)10 | 57 ± 1 |
| 11 | (CCUG)10 | 73.6 ± 0.8 |
Values are a composite fit of at least two complete titrations ± 1 SD on the fit.
Analysis of binding kinetics
While equilibrium binding constants are essential for understanding binding selectivity, they do not provide a complete picture of the recognition process. In the ligand–protein binding field, an emerging consensus holds that kinetic constants, and in particular the off-rate, are key determinants of selectivity (42–44). Similar observations have been reported for DNA-binding compounds (45–48), but to date there is little information addressing this hypothesis in the context of RNA binding (and no information with regard to how this affects compound behavior in a cellular environment or in vivo, where compounds must compete with other cellular constituents for target RNAs).
For SPR-derived kinetic constants, we observe that on-rates generally do not vary substantially for individual compounds across a series of RNAs. In contrast, off rates strongly reflect differences in selectivity, consistent with the above hypothesis. These data are perhaps easier to conceptualize in terms of dissociative half-life, or t1/2, for the interaction with target sequences (Table 2). Comparisons among compounds and target sequences based on dissociative half-life are a useful addition to commonly employed metrics such as the selectivity index, and may have some advantages since the former is an absolute measurement while the latter is defined based on a changeable set of RNAs. For the compounds studied, 11 displays the largest differences in t1/2, consistent with its highest selectivity for CUG repeats: an ∼25-fold difference between (CUG)10 and (CCUG)10, and an ∼47-fold difference between (CUG)10 and the HIV-1 FSS.
Table 2.
Residence time (t1/2, s) for compound binding to RNA
| Compound | (CUG)2 | (CUG)4 | (CUG)10 | (CCUG)10 | (CAG)10 | Duplex | HIV-1 FSS |
|---|---|---|---|---|---|---|---|
| 4 | 757 | 1650 | 548 | 745 | 462 | 801 | 67 |
| 5 | 888 | 704 | 483 | 231 | 330 | 180 | 177 |
| 6 | ND | ND | 20 | 2 | 36 | 17 | 27 |
| 7 | ND | ND | 15 | 1 | 30 | ND | 35 |
| 8 | ND | ND | 230 | 253 | 227 | 256 | ND |
| 9 | ND | ND | 257 | 256 | 173 | 374 | 347 |
| 10 | ND | ND | 193 | 130 | ND | ND | ND |
| 11 | 109 | 806 | 1272 | 50 | 56 | NB | 27 |
| Neomycin | ND | ND | 14 | 4 | ND | ND | ND |
‘NB’ = no binding; ‘ND’ = not determined.
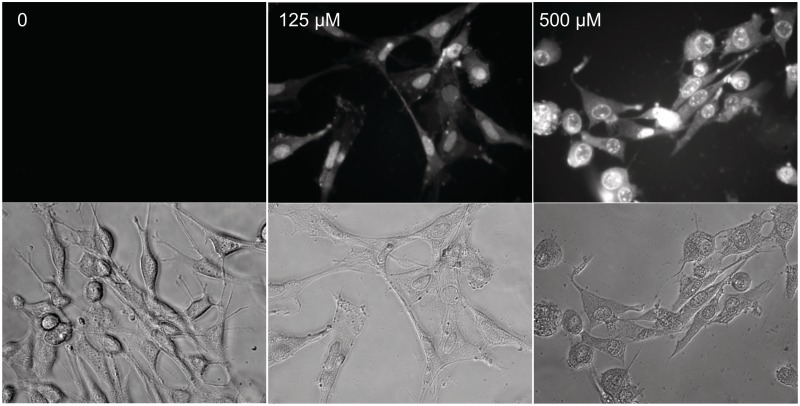
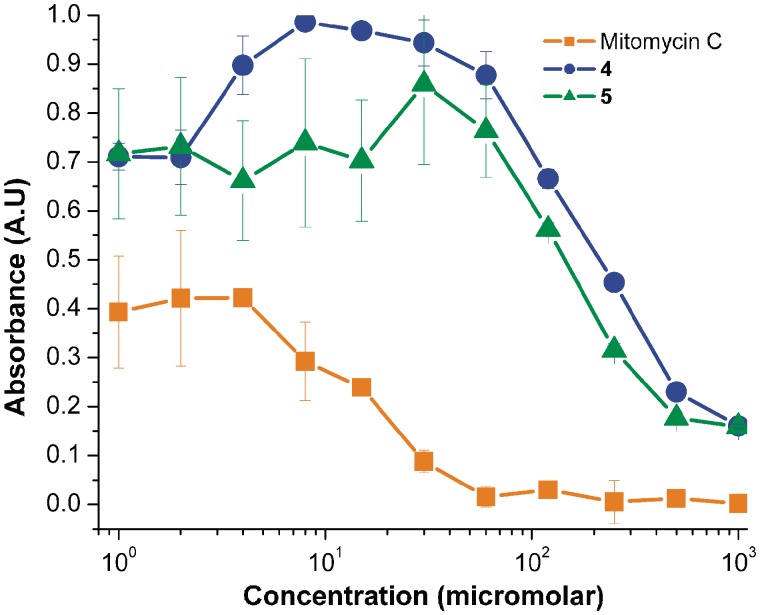
Prior to analyzing the ability of selected compounds to interfere with CUGexp mediated effects in model cell lines, it was crucial to first establish that they are capable of crossing the cell membrane, and are non-toxic at experimentally relevant concentrations. Cell permeability was assessed in human fibroblasts, and in mouse C2C12 myoblasts. In both cases, penetration of the cell membrane was readily observed because of the fluorescence of the benzo[g]quinoline chromophore; compounds also appear to localize preferentially in the nucleus (Figure 3). Since the sequestration of MBNL1 by pathogenic CUGexp RNA occurs entirely in the nucleus, this apparent selective localization even in non CUGexp-containing cell lines is particularly notable. Of course, the mechanism of compound entry is not revealed by this analysis. Interestingly, benzo[g]quinoline 2 showed only a modest ability to enter cells, and ‘scrambled’ compound 9 was significantly less able to enter cells than 4, 5 or 11 (Supplementary Figures S7–S10). Toxicity of 4 and 5 to mouse myoblasts was measured by MTT assay (49). No significant toxicity was observed at concentrations up to 100 µM. In contrast, mitomycin C, a commonly employed DNA-targeted cancer chemotherapeutic agent, showed significant toxicity at all concentrations tested (Figure 4). Similar results were obtained in preliminary experiments with human fibroblasts (Supplementary Figure S11). Benzo[g]quinoline 2 was also tested, and found to have no toxicity in fibroblasts at concentrations up to 500 µM (Supplementary Figure S10), an expected result given its limited ability to cross the cell membrane.
Figure 3.
Compounds readily penetrate cell membranes and localize in the nucleus. Representative bright field (lower) and fluorescence (upper) images are shown for compound 5 in mouse myoblasts.
Figure 4.
MTT assays indicate low toxicity of 4 and 5 in mouse myoblasts. The significantly higher toxicity of mitomycin C is shown for comparison. Error bars indicate single SDs of three replicates of each concentration.
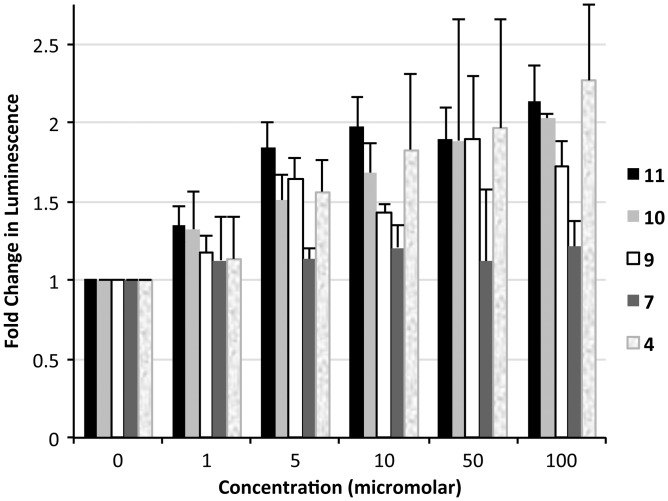
We then evaluated the ability of selected compounds to inhibit the nuclear retention of CUGexp RNA, a phenomenon that depends on formation of RNA–MBNL1 complexes, in mouse myoblasts. An engineered cell line (C5-14) expressing a luciferase-encoding mRNA with 800 CUG repeats in the 3′-UTR was used. Formation of (CUG)800 RNA–MBNL1 complexes in the C5-14 cell line suppresses translation of this mRNA, and therefore suppresses the cellular level of luciferase. Inhibition of protein complexation to the (CUG)800 RNA allows translation to occur, restoring luciferase expression. A second cell line (C1-S) carrying an analogous luciferase construct lacking the CUG repeats in the 3′-UTR was used as a positive control for luciferase expression. After treatment with a morpholino antisense oligonucleotide (CAG-25) complementary to the CUG repeat RNA, C5-14 cells showed an increase in luciferase expression consistent with direct interaction with the (CUG)800 mRNA in the nucleus (J. Hoskins et al., manuscript in preparation). The increase in luciferase activity is accompanied by a disruption of CUG foci, as seen by fluorescence in-situ hybridization (FISH). We hypothesized that binding of our compounds to the (CUG)800 repeat in C5-14 cells would likewise promote release of the luciferase transcripts from the nucleus, resulting in increased luciferase expression and activity. In contrast, luciferase activity in positive control (C1-S) cells should not be affected by the presence of CUG RNA binding compounds. Indeed, incubation of various concentrations of 4, 10, or 11 (0–100 µM) with C5-14 myoblasts resulted in concentration-dependent increases in luciferase activity, consistent with our hypothesis (Figure 5). Compound 7 (lacking selectivity to CUGexp RNA) had no effect on luciferase activity when incubated with C5-14 cells under the same conditions. No statistically significant change in luciferase signal was observed for C1-S cells (which lack CUG repeats) treated with compounds 10 or 11; however, we observed a concentration-dependent decrease in luciferase activity when these cells were incubated with 4. Since the WST-1 viability results showed no observable toxicity of 4 to the C1-S cells at the assay concentrations, we speculated that this effect could result from an off-target effect of compound 4 causing a global decrease in protein synthesis. The concentration of total protein in cell lysates of both C5-14 and C1-S cells, as measured by Bradford assay (Supplementary Figures S12 and S13), indeed showed a decreasing level of protein expression with increasing concentration of 4. The fact that this effect is not observed for 10 and 11 likely results from their greater selectivity for CUG repeats. Thus, CUGexp binding compounds are capable of releasing the sequestered mRNA transcripts into the cytoplasm for translation.
Figure 5.
Compounds are able to promote expression of a (CUG)800-containing luciferase construct in mouse myoblasts. Firefly luciferase activity is plotted as a ratio of the normalized luminescence from cells containing (CUG)800 in the 3′-UTR of the luciferase mRNA (C5–14) to normalized luminescence of cells containing no CUG repeats (C1-S). Error bars indicate 1 SD of luminescence from the average of three replicate wells.
Compounds partially restore splicing in a mouse model of DM1
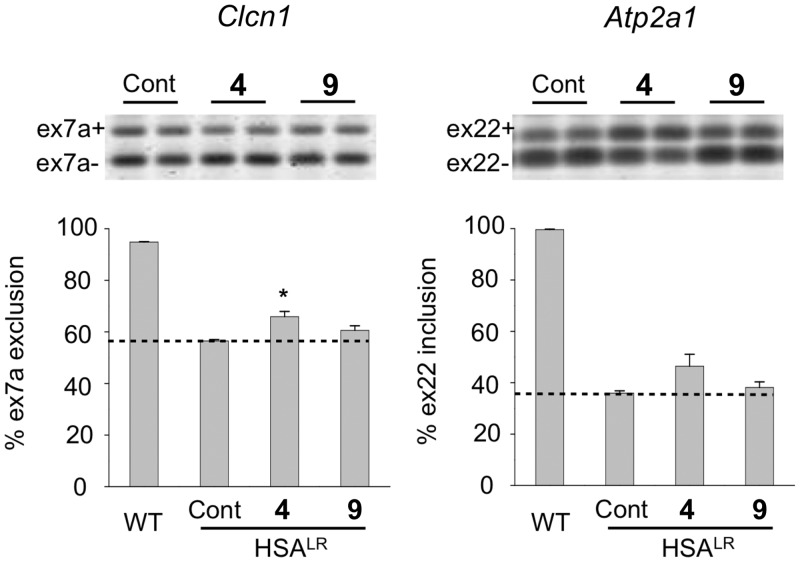
Of course, a critical test of a compound's performance is its ability to function in vivo. DM1 has several well-studied mouse models (31). We examined the activity of compounds 4, 9 and 11 in the HSALR mouse model, in which transgenic mice carry a long CTG repeat inserted into the human skeletal actin (HSA) gene in skeletal muscle (50). These mice exhibit several DM1-like phenotypic characteristics. They also display misregulated alternative splicing of genes, including Clcn1 and Atp2a1, which are physiological targets of MBNL1 regulation. Not all compounds were tested due both to the complexity and cost of mouse studies, and due to the limited availability of compound 10.
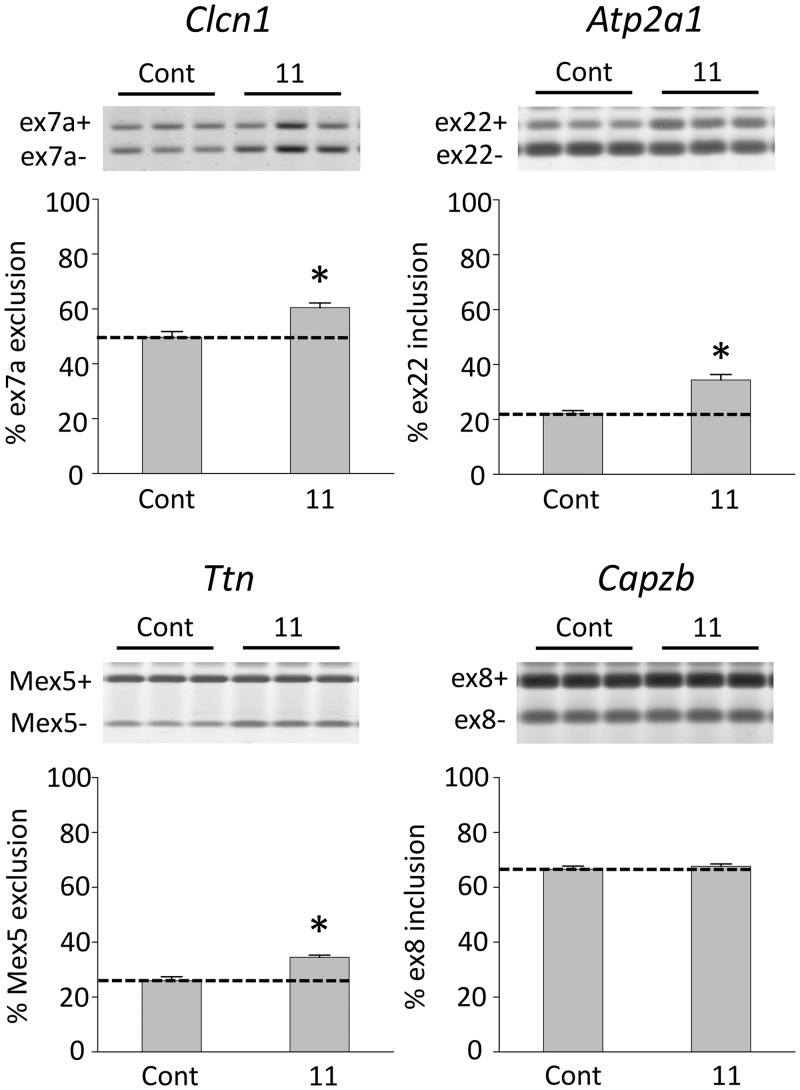
To examine effects on MBNL1-regulated splicing events we administered compounds 4, 9 or 11 to HSALR transgenic mice by daily intraperitoneal injection of 40 mg/kg for 5 days. In the initial experiment, analysis of splicing in hind limb (quadriceps) muscle showed that compound 4 produced a modest but statistically significant (P = 0.0379) improvement of Clcn1 splicing relative to saline-injected controls, whereas compound 9 did not (Figure 6). Statistically significant improvements in both Clcn1 and Atp2a1 (P = 0.0419 and P = 0.0223, respectively; Supplementary Figure S14) were observed following treatment with compound 11. To confirm these results, we carried out a further study in which five age-matched mice were treated with compound 11 via interperitoneal injection at 40 mg/kg once daily for 5 days, and compared with five age-matched controls. Once again, 11 provided a statistically significant improvement in Atp2a1 and Clcn1 splicing. Ttn, another transcript dependent on MBNL1-mediated splicing, also showed improvement (Figure 7). Splicing of Capzb, a developmentally regulated exon whose splicing does not depend on MBNL1, was not affected.
Figure 6.
Compound 4 improves MBNL1-dependent splicing in vivo. Asterisk (*) indicates statistical significance (P = 0.0379); the dashed line is provided as a reference to splicing levels for HSALR mice (n = 3 for experimental, control and wild-type) in the absence of compound (control). The error bars indicate SEM.
Figure 7.
Compound 11 improves MBNL1-dependent splicing in vivo. Asterisks (*) indicate statistical significance (P < 0.005); the dashed line is provided as a reference to splicing levels for HSALR mice in the absence of compound (n = 5 for compound-treated and control). The level of an MBNL1-independent splicing event (Capzb) is unaffected by 11. Error bars indicate SEM.
DISCUSSION
Using a moderate-affinity ligand identified from a resin-bound dynamic combinatorial library as a starting point, we have successfully developed next-generation lead compounds able to selectively bind DM1 and DM2 RNA with high affinity. These compounds represent to our knowledge the first use of the benzo[g]quinoline moiety in an RNA-binding context, an important advance in that this substructure allows for direct visualization of compounds in cells via fluorescence. The strong selectivity of 11 for CUG repeats is particularly notable, as is its enhanced affinity for longer CUG repeat sequences. Selectivity for longer repeats is potentially highly advantageous: since isolated and short CUG repeat sequences are found throughout the transcriptome, binders must differentiate between these and longer repeats in order to avoid off-target effects in vivo.
As stated at the outset, our inclusion of the benzo[g]quinoline moiety in second-generation compounds was predicated on the assumption that binding of lead compounds to CUG repeats involved intercalation of the quinoline moieties. Therefore, the stoichiometries for 4 and 5 binding these repeats as determined by SPR were surprising. In each case, binding of 1 molecule of 4 or 5 per CUG via a mode in which both benzo[g]quinolines intercalate would require violation of the neighbor-exclusion rule (51). Further studies will be essential to fully understanding the structural parameters of binding by these two compounds, as well as the others described in this article.
Several of the molecules synthesized show low toxicity in human fibroblast and mouse myoblast cell lines, suggesting the high sequence selectivity displayed by these structures is transferable to a biological context. Compounds examined in this work are able to release the nuclear retention of a CUG repeat-containing transcript in a concentration-dependent manner in mouse myoblasts, and improve splicing of MBNL1-dependent targets in a mouse model of DM1. Thus, we have demonstrated for the first time that DCC can serve as a starting point from which to develop high-affinity sequence-selective RNA binding compounds with desirable biological activity in vitro (in cell culture) and in vivo (in mice). Activity in vivo was observed to correlate with in vitro potency. Interestingly, we also observed that mice treated with 4 exhibited significant acute toxicity effects while those treated with 11 did not. While the higher (CUGexp) sequence selectivity of 11 may contribute to its lower toxicity, further studies will be essential to understand these differences in detail. The amount of splicing improvement following treatment with 4 or 11 following daily interperitoneal injection at 40 mg/kg for 5 days is similar to that produced by pentamidine (8) at a dosage of 40 mg/kg once daily for 7 days, but we anticipate the greater selectivity of the compounds, we describe (particularly 11) may provide a more favorable pathway for future development.
In order to build on these initial successes, it will be necessary to increase the amount of compound-dependent splicing improvement observed in mouse models of DM1. It is possible that this goal can be at least partially accomplished by modification of the dosage and/or dosing schedule. Further improvement of RNA binding affinity, bioavailability and stability will also likely prove essential. Medicinal chemistry efforts based on compound 11 along those lines are currently in progress, as we continue to make strides toward the development of a viable anti-DM1 therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–44, Supplementary Figures 1–14 and Supplementary Methods.
FUNDING
National Institutes of Health (NINDS) [5R21NS071023, AR049077, U54NS48843]. Funding for open access charge: NIH – NINDS [5R21NS071023].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Matthew Baker and Donald Curran for assistance with preliminary studies on the synthesis of benzo[g]quinoline 2, and Prof. Brian R. McNaughton for helpful discussions. L.O.O. and B.L.M. designed all compounds. L.O.O. synthesized compounds 2–11, carried out all SPR and fluorescence binding assays, carried out all MTT and cell permeability experiments and completed all assays in mouse myoblasts (C1-S and C5-14 cells) in tandem with J.H. M.N. carried out all in vivo mouse studies. Data analysis was conducted by all authors and all authors contributed to writing the manuscript.
REFERENCES
- 1.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 2.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov. Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 4.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osborne RJ, Thornton CA. RNA-dominant diseases. Hum. Mol. Genet. 2006;1B5:162–169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 6.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 7.Machuca-Tzili L, Brook D, Hilton-Jones D. Clinical and molecular aspects of the myotonic dystrophies: a review. Muscle Nerve. 2005;32:1–18. doi: 10.1002/mus.20301. [DOI] [PubMed] [Google Scholar]
- 8.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler T. Myotonic dystrophy: therapeutic strategies for the future. Neurotherapeutics. 2008;5:592–600. doi: 10.1016/j.nurt.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dervan PB, Bürli RW. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 1999;3:688–693. doi: 10.1016/s1367-5931(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 12.Corbett PT, Leclaire J, Vial L, West KR, Wietor JL, Sanders JK, Otto S. Dynamic combinatorial chemistry. Chem. Rev. 2006;106:3652–3711. doi: 10.1021/cr020452p. [DOI] [PubMed] [Google Scholar]
- 13.Gareiss PC, McNaughton BR, Sobczak K, Thornton CA, Miller BL. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA−MBNL1 interaction in vitro: discovery of lead compounds targeting myotonic dystrophy (DM1) J. Am. Chem. Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNaughton BR, Miller BL. Resin-bound dynamic combinatorial chemistry. Org. Lett. 2006;8:1803–1806. doi: 10.1021/ol060330+. [DOI] [PubMed] [Google Scholar]
- 15.García-López A, Llamusí B, Orzáez M, Pérez-Payá E, Artero RD. In vivo discovery of a peptide that prevents CUG-RNA hairpin formation and reverses RNA toxicity in myotonic dystrophy models. Proc. Natl Acad. Sci. USA. 2011;108:11866–11871. doi: 10.1073/pnas.1018213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc. Natl Acad. Sci. USA. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong CH, Fu Y, Ramisetty SR, Baranger AM, Zimmerman SC. Selective inhibition of MBNL1-CCUG interaction by small molecules toward potential therapeutic agents for myotonic dystrophy type 2 (DM2) Nucleic Acids Res. 2011;39:8881–8890. doi: 10.1093/nar/gkr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MM, Pushechnikov A, Disney MD. Rational and modular design of potent ligands targeting the RNA that causes myotonic dystrophy 2. ACS Chem. Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MM, Childs-Disney JL, Pushechnikov A, French JM, Sobczak K, Thornton CA, Disney MD. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: targeting the RNAs that cause myotonic muscular dystrophy. J. Am. Chem. Soc. 2009;131:17464–17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palde PB, Ofori LO, Gareiss PC, Lerea J, Miller BL. Strategies for recognition of stem-loop RNA structures by synthetic ligands: application to the HIV-1 frameshift stimulatory sequence. J. Med. Chem. 2010;53:6018–6027. doi: 10.1021/jm100231t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varvaresou A, Iakovou K. Molecular modeling study of intercalation complexes of tricyclic carboxamides with d(CCGGCGCCGG)2 and d(CGCGAATTCGCG)2. J. Mol. Model. 2011;17:2041–2050. doi: 10.1007/s00894-010-0891-5. [DOI] [PubMed] [Google Scholar]
- 22.Wainwright M. Acridine-a neglected antibacterial chromophore. J. Antimicrob. Chemother. 2001;47:1–13. doi: 10.1093/jac/47.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Horstmann MA, Hassenpflug WA, zur Stadt U, Escherich G, Janka G, Kabisch H. Amsacrine combined with etoposide and high-dose methylprednisolone as salvage therapy in acute lymphoblastic leukemia in children. Haematologica. 2005;90:1701–1703. [PubMed] [Google Scholar]
- 24.Petrov OE, Soprunova NY, Binh LN, Kozyreva NP, Bekhli AF. In vitro interaction of chick erythrocyte DNA with substituted 4-amino benzo[g]quinolines. Khim.-Farmat. Zhurnal. 1982;16:1304–1306. [Google Scholar]
- 25.Kienzle F. The reaction of phthalaldehydes with 3-nitropropionates A simple route to 3-nitro-2-naphthoic acids. Helv. Chim. Acta. 1980;63:2364–2369. [Google Scholar]
- 26.McNaughton BR, Miller BL. A mild and efficient one-step synthesis of quinolines. Org. Lett. 2003;5:4257–4259. doi: 10.1021/ol035333q. [DOI] [PubMed] [Google Scholar]
- 27.Myers AG, Gleason JL, Yoon T, Kung DW. Highly practical methodology for the synthesis of d- and l-α-amino acids, N-protected α-amino acids, and N-methyl-α-amino acids. J. Am. Chem. Soc. 1997;119:656–673. [Google Scholar]
- 28.Katsamba PS, Navratilova I, Calderon-Cacia M, Fan L, et al. Kinetic analysis of a high-affinity antibody/antigen interaction performed by multiple Biacore users. Anal. Biochem. 2006;352:208–221. doi: 10.1016/j.ab.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 29.Rich RL, Myszka DG. Grading the commercial optical biosensor literature-Class of 2008: “The Mighty Binders.”. J. Mol. Recognit. 2009;23:1–64. doi: 10.1002/jmr.1004. [DOI] [PubMed] [Google Scholar]
- 30.Rich RL, Papalia GA, Flynn PJ, Furneisen J, et al. A global benchmark study using affinity-based biosensors. Anal. Biochem. 2009;386:194–216. doi: 10.1016/j.ab.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes-Pereira M, Cooper TA, Gourdon G. Myotonic dystrophy mouse models: towards rational therapy development. Trends Mol. Med. 2011;17:506–517. doi: 10.1016/j.molmed.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Wilson WD. Quantitative analysis of small molecule-nucleic acid interactions with a biosensor surface and surface plasmon resonance detection. Methods Mol. Biol. 2010;613:1–23. doi: 10.1007/978-1-60327-418-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staple DW, Butcher SE. Solution structure of the HIV-1 frameshift inducing stem-loop RNA. Nucleic Acids Res. 2003;31:4326–4331. doi: 10.1093/nar/gkg654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munde M, et al. DNA minor groove induced dimerization of heterocyclic cations: compound structure, binding affinity, and specificity for a TTAA site. J. Mol. Biol. 2010;402:847–864. doi: 10.1016/j.jmb.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byers LD. Probe-dependent cooperativity patterns in Hill-plots. J. Chem. Educ. 1977;54:352. [Google Scholar]
- 37.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mooers BH, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc. Natl Acad. Sci. USA. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases. Nucleic Acids Res. 2010;38:8370–8376. doi: 10.1093/nar/gkq700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Structural insights into CUG repeats containing the “stretched U-U wobble”: implications for myotonic dystrophy. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu H, Tonge PJ. Drug-target residence time: critical information for lead optimization. Curr. Opin. Chem. Biol. 2010;14:467–474. doi: 10.1016/j.cbpa.2010.06.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang R, Monsma F. The importance of drug-target residence time. Curr. Opin. Drug Discov. Devel. 2009;12:488–496. [PubMed] [Google Scholar]
- 44.Copeland RA, Pompliano DL, Meek TD. Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 45.Feigon J, Denny WA, Leupin W, Kearns DR. Interactions of antitumor drugs with natural DNA: 1H NMR study of binding mode and kinetics. J. Med. Chem. 1984;27:450–465. doi: 10.1021/jm00370a007. [DOI] [PubMed] [Google Scholar]
- 46.Chaires JB, Dattagupta N, Crothers DM. Kinetics of the daunomycin-DNA interaction. Biochemistry. 1985;24:260–267. doi: 10.1021/bi00323a004. [DOI] [PubMed] [Google Scholar]
- 47.Phillips DR, Crothers DM. Kinetics and sequence specificity of drug-DNA interactions: an in vitro transcription assay. Biochemistry. 1986;25:7355–7362. doi: 10.1021/bi00371a017. [DOI] [PubMed] [Google Scholar]
- 48.Chen FM. Binding specificities of actinomycin D to non-self-complementary -XGCY-tetranucleotide sequences. Biochemistry. 1992;31:6223–6228. doi: 10.1021/bi00142a008. [DOI] [PubMed] [Google Scholar]
- 49.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 50.Mankodi A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 51.Crothers DM. Calculation of binding isotherms for heterogenous polymers. Biopolymers. 1968;6:575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.