Abstract
Protein-coding genes, guiding differentiation of ES cells into neural cells, have extensively been studied in the past. However, for the class of ncRNAs only the involvement of some specific microRNAs (miRNAs) has been described. Thus, to characterize the entire small non-coding RNA (ncRNA) transcriptome, involved in the differentiation of mouse ES cells into neural cells, we have generated three specialized ribonucleo-protein particle (RNP)-derived cDNA libraries, i.e. from pluripotent ES cells, neural progenitors and differentiated neural cells, respectively. By high-throughput sequencing and transcriptional profiling we identified several novel miRNAs to be involved in ES cell differentiation, as well as seven small nucleolar RNAs. In addition, expression of 7SL, 7SK and vault-2 RNAs was significantly up-regulated during ES cell differentiation. About half of ncRNA sequences from the three cDNA libraries mapped to intergenic or intragenic regions, designated as interRNAs and intraRNAs, respectively. Thereby, novel ncRNA candidates exhibited a predominant size of 18–30 nt, thus resembling miRNA species, but, with few exceptions, lacking canonical miRNA features. Additionally, these novel intraRNAs and interRNAs were not only found to be differentially expressed in stem-cell derivatives, but also in primary cultures of hippocampal neurons and astrocytes, strengthening their potential function in neural ES cell differentiation.
INTRODUCTION
In recent years, the number of proposed non-coding RNA (ncRNA) transcripts has dramatically been rising. For example, in the human genome there is an estimated number of up to 450 000 ncRNA transcripts predicted (1). In agreement, a recent study designated as ENCODE project (2), which focused on 1% of the human genome in high resolution, revealed that up to 90% of the human genome might be transcribed, with only 1.5% of RNA transcripts encoding for proteins. Thus, it has been proposed that the remaining 88.5% of RNA transcripts might serve as a source for regulatory ncRNAs (3). However, it is currently still unclear which of the 450 000 predicted ncRNA candidates, encoded on the human genome, are functional and which ones represent spurious transcription products or degradation intermediates (4). Therefore, it is important to clearly identify the functional portion of the ncRNA transcriptome in model organisms. Several features might be employed to filter out and preselect functional, regulatory ncRNAs from a background of spurious transcription/degradation intermediates such as (i) analysis of differential expression of ncRNAs during cell differentiation and development, (ii) ncRNA expression in disease or (iii) ncRNA expression during development. In addition, since most functional ncRNAs are known to bind to proteins forming ribonucleo-protein particles (RNPs), isolation by RNPs might increase the likelihood for identifying functional ncRNAs (5).
For embryonic stem (ES) cell maintenance and pluripotency, non-coding RNAs have recently emerged as important regulators of gene expression (6–8). Up till now, specific microRNAs, a class of small regulatory ncRNAs, sized 21–24 nt (9,10), have been investigated in neural development during ES cell differentiation. In particular, expression of the ES cell specific miR-290 cluster, harboring miRNAs-290, −291, −293, −294 and −295, respectively, has been shown to be significantly down-regulated upon differentiation (11), while regulating de novo methylation in ES cells by repressing the transcriptional repressor Rlb2 (12,13). Inhibition of mir-145 in human ES cells has been shown to reduce their capacity for differentiation (14), while maturation repression of the pre-let7 miRNA precursor by the Lin28 protein is a mechanism blocking commitment to neural fate (15). In addition, microRNA-array analysis in ES cells versus differentiated cells reveals specific microRNA expression signatures (16,17), thus implying transcriptome changes during differentiation related to microRNA function. Notably, lack of expression of pre-microRNA-processing proteins Dicer (18,19) and DGCR8 (20) was shown to result in severe differentiation defects.
In order to identify the complete set of small ncRNAs involved in neural differentiation of mouse ES cells in vitro, we have generated specialized, RNP-derived cDNA libraries, as previously described (21,22), for three differentiation stages. Transcriptional profiling by high-throughput sequencing revealed the presence of numerous differentially expressed known and novel ncRNAs, which predominantly locate to intergenic and intronic regions of the mouse genome. The majority of the novel ncRNAs exhibited a sequence length bias of 18–30 nt. For selected, newly identified ncRNA transcripts differential expression in primary hippocampal neurons and astrocytes was confirmed by real-time PCR.
MATERIALS AND METHODS
RNP library generation
RNP libraries were generated as previously described (21,22). Briefly, cells were lyzed and cell extracts were size-fractionated on 10–30% glycerol gradients. Subsequently, glycerol gradients were fractionated and phenol–chloroform precipitated. The extracted RNA was 3′-C-tailed using poly (A) polymerase from yeast (Epicenter, Madison, USA). A 19-mer 5′-adaptor (–GTCAGCAATCCCTAACCAG, bold and underlined are ribonucleotides) was ligated by T4 RNA ligase (Fermentas, St. Leon-Rot, Germany) to the C-tailed RNA. The RNAs were reverse transcribed by using an anchor primer (5′-AGGAGCCATCGTATGTCGGGGGGGGH) and amplified by PCR at 53°C annealing, for 25 cycles using the following primers: 5′-libPCR GTCAGCAATCCCTAACGAG, 3′-libPCR AGGAGCCATCGTATGTCG. The cDNA was PAGE purified and size selected from 20 to 400 bp. cDNA was cloned into the pGEM-T vector, (Promega, Mannheim, Germany) for diagnostic Sanger sequencing before high-thoughput sequencing. For Solexa sequencing, additional bar-coded forward primers were added to cDNAs by PCR:
5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATACGTCAGCAATCCCTAACGAG-3′ for the ES library,
5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCATCGTCAGCAATCCCTAACGAG for the NP library 5′AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGACAGTCAGCAATCCCTAACGAG for the N/G library. The following reverse primer was employed for all libraries: 5′-CAAGCAGAAGACGGCATACGAGCTCTTCCGATCTAGGAGCCATCGTATGTCG 3′. The eluted cDNAs were analyzed by high-throughput sequencing employing the Solexa (Illumina) platform. Sequencing was performed with the Genome Analyzer GAII at FASTERIS SA, Plan-les-Quates, (Switzerland). Reads were generated as single reads with a maximum of 76 bp in length.
ES cell cultures
The mouse ES cell line used was ES–E14TG2a (passage 10–14) (23). ES cells were cultured as previously described (24), with some modifications, on a feeder layer of mouse embryonic fibroblasts; feeder cells were inactivated with 10 μg/ml mitomycin C for 2.5 h. ES cell culture medium (ESCM) consisted on Knockout Dulbecco’s Modified Eagle’s medium with Knockout serum replacement (15%), 2 mM Glutamax, 0.1 mM MEM non-essential amino acids, 0.5 mM 2-mercaptoethanol (all from Gibco Invitrogen Corporation, Paisley, UK) supplemented with 1000 U/ml leukaemia inhibitory factor (LIF) (Chemicon, Temecula, CA). Cell cultures were maintained in a humidified atmosphere with 5% CO2, at 37°C. Medium was changed every day. ES cell colonies were passaged with 0.25% Trypsin–EDTA (Gibco) at a 1:3–1:5 split ratio every other day.
Neural differentiation cultures
Neural differentiation of ES cells occurred in discrete steps, which include neural induction, neural proliferation and neural specification, modified from previously published protocols (25–27).
Neural induction medium (NIM) consisted of Dulbecco’s Modified Eagle Medium DMEM/F12 with Glutamax (Gibco) containing 25 μg/ml insulin, 100 μg/ml transferrin, 5 ng/ml sodium selenite, 2.5 μg/ml fibronectin (all from Sigma-Aldrich St. Louis, MO, USA) and 0.1 mM MEM nonessential amino acids solution.
Neural proliferation medium (NPM) consisted of DMEM/F12 with Glutamax, 1% N2 supplement and 0.1 mM MEM nonessential amino acids, supplemented with Fibroblast Growth Factor 2 (FGF2) and Epidermal Growth Factor (EGF) (10 ng/ml each).
Neuronal differentiation medium (NDM) medium consisted of Neurobasal medium with 2% (vol/vol) B27 supplement without vitamin A (Gibco), 2 mM glutamax, 1 μM cAMP, 200 μM ascorbic acid and 20 µg/ml laminin (all from Sigma). Media were changed every 48 h.
ES cells, at Day 0 of neural differentiation (D0), were separated from feeder cells, dissociated with Accutase (Sigma), and cultured as cell suspension (2 × 105 cells/ml) in ESCM without LIF in non-adhesive bacterial-grade dish. Cells that became floating cellular aggregates were cultured in the same medium for 2 days. The adherent colony culture was initiated by plating cellular aggregates on plastic or glass surface coated with polyornithine (Sigma, 15 μg/ml for plastic and 50 μg/ml for glass, 1 h RT) and laminin (20 μg/ml ON 4°C), and further cultured in NIM. Attached aggregates flattened over 1–2 days and columnar primitive neuroepithelial (NE) cells developed and formed neural rosettes at approximately D6. The neural tube-like rosettes formed during neural induction were used as a selection criterion; neural colonies were mechanically separated, collected and dissociated with Accutase, and cultured as cell suspension or in adherent culture onto polyornithine/laminin-coated plates in NPM (2 × 105 cells/ml) for 4 days, for the proliferation of the neural progenitor cells. For neural differentiation/specification, neural progenitors were dissociated with Accutase and plated onto polyornithine/laminin-coated plates or coverslips, followed by culturing in NDM.
Colonies and cellular morphologies were monitored by phase contrast microscopy. ES or neural colonies were mechanically detached or split under a stereomicroscope. Three independent experiments were performed. Samples for RNP libraries generation, RNA extraction or immunocytochemistry were taken at the following time-points: D0 (stage designated as ES), D10 (stage designated as NP) and D20 (stage designated as N/G).
Initial mapping and analysis of the cDNA libraries
Prior to analysis, the pool of sequencing reads has been divided into three libraries according to sequence barcodes: ATAC for ES library, CATC for NP library and GACA for N/G library. The analysis of the individual libraries has been performed employing the APART pipeline (automated pipeline for annontation of RNA transcripts) (28). All APART modules were used. For removal of adapter sequences, we employed a 20-nt window of the adaptor sequences with three allowed mismatches and included removal of the C-tail, located between the insert and 3′-adaptor. For downstream analysis, only reads with a minimal length of 18 nt by which both adaptors had been detected, were used. For sequence reads, containing only a partial 3′-adaptor sequence, at least six terminal bases were required to match the C-tail, or the beginning of the 3′-adaptor sequence. Next, sequences were aligned to the mouse genome (mm9), allowing for a single mismatch. Assembly and annotation was performed employing the APART pipeline, employing default settings.
Transcriptional profiling of three cDNA libraries from ES cells
For differential expression analysis the non-clustered list of contigs generated by APART, has been employed. The identification of overlapping contigs between libraries has been performed through BEDTools package (29) with 50% overlap threshold. Contigs appearing in at least two libraries have been selected for subsequent differential expression analysis. Next, the contig redundancy caused by allowing for multiple read matches has been removed by using in-house perl scripts employing APART clustering data. Normalization of the read number has been carried out utilizing the package edgeR (30) from Bioconductor (31). In order to test for differences in expression edgeR and several in-house scripts have been used.
miRNA prediction
For identification of novel miRNAs, present in our dataset, the software miRDeep2 (32) was employed, in particular scripts mapper.pl and miRDeep2.pl (33). Mapping was performed by using the default parameter set and the FASTQ files from each library, except that the adaptor sequences were already removed prior to analysis. The miRDeep2 script was then applied to find potential miRNAs in the data set based on the suggested parameters. Results were compared to data assembled in the differential expression analysis by using the package rtracklayer (34) of the Bioconductor project.
Real-time PCR
Total RNA was isolated from mouse ES cells, primary hippocampal neurons and primary astrocytes with TRI Reagent (Sigma-Aldrich, Vienna, Austria) according to the manufacturer’s protocol. An amount of 500 ng of total RNA were poly-A tailed and reverse transcribed to cDNA using the microRNA first strand synthesis kit (Agilent Technologies, Böblingen, Germany) technologies, following the manufacturer’s protocol.
The cDNA was used as template for the real-time PCR on ES cells, primary hippocampal neurons and astrocytes. The universal reverse primer provided with the kit was used together with the following forward primers in 5′–3′ orientation:
| mmu_interRNA30: | CATCCCACTTCTGACACCACAAA |
| mmu-interRNA32: | GCCTACCAAACCCTGTCAA |
| mmu-intraRNA16: | CCATAAGGTAGGCATTGCA |
| mmu-interRNA6: | GAACGTGAGCTGGGTTTAGACCGTC |
| mmu-interRNA36: | GATCCCACTTCTGACACCA |
| mmu-interRNA26: | GGGGAATCTGACTGTCTA |
| mmu-intraRNA11: | CAAGATAAGATTTCCCCG |
| mmu-intraRNA140: | AGTCCCTGCCCTTTGTACA |
| mmu-interRNA143: | TCTAAATTTTCCACCTTTTTCAGTTT |
| mmu-interRNA105: | GCGTTGGTGGTATAGTGGTGAGCATAGCT |
| mmu-interRNA140: | GGGCCGGCGGCGGCGGCG |
| mmu-interRNA35: | ATCCCACTTCTGACACCA |
| mmu-interRNA68: | TCCTCGTTAGTATAGTGGTTAGTA |
Primers were ordered from Sigma-Aldrich. Real-time PCR was performed using Power SYBR® Green PCR Master Mix (Applied Biosystems, Darmstadt, Germany). Reactions were performed at 60°C annealing for 1 min and for 40 cycles. Normalization was performed with U6.
RESULTS AND DISCUSSION
Generation of three specialized RNP-derived cDNA libraries from mouse ES cell neural differentiation
In order to identify novel ncRNAs that regulate ES cell neural differentiation we generated three RNP-derived cDNA libraries from three stages of neural differentiation of mouse ES cells: (i) pluripotent stem cells (designated as ES), (ii) neural progenitor cells (designated as NP) and (iii) a differentiated heterogeneous population of neurons and glial cells (designated as N/G), as previously reported (21,22). Briefly, mouse ES cells (expressing the pluripotent markers Nanog and Oct4) were differentiated into NP (expressing the neural markers nestin and Sox1), which subsequently spontaneously differentiated to neurons (expressiong Tau) and glial cells (expressing Gfap and Osp) by withdrawing growth factors FGF2 and EGF (see ‘Materials and Methods’ section and Supplementary Figure S1A).
Real-time PCR analysis of marker genes revealed high levels of Oct4 and Nanog in ES cells, while their levels were significantly down-regulated at the NP and not detected at the N/G stage. The neural marker genes Nestin and Sox1, were significantly up-regulated at NP and N/G stages, while the neuronal (Tau), astrocytic (Gfap) and oligodendrocytic (Osp) gene expression was highly up-regulated in the last stage (as compared to ES stage; see Supplementary Figure S1B).
From each of the three stages, specialized RNP libraries encoding small ncRNAs were generated (see ‘Materials and Methods’ section). Subsequently, cDNA libraries were analyzed by high-throughput sequencing employing the Solexa platform and ∼26 Mio. sequence reads for the three libraries were obtained [sequences have been deposited in the Sequence Read Archive (NCBI) with the accession number: SRP008250].
Transcriptional profiling of three cDNA libraries from ES cell neural differentiation
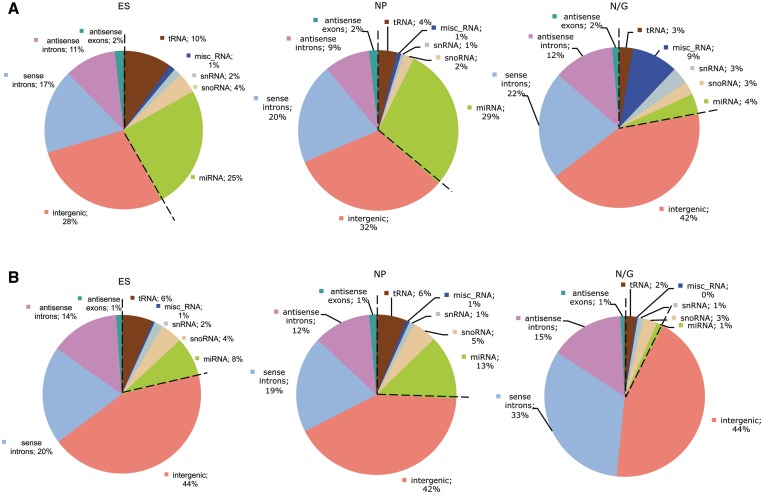
In order to determine differentially expressed RNA transcripts within the ES, NP and N/G stages, we bioinformatically analyzed sequence reads from the respective cDNA libraries by APART (automated pipeline for annotation of RNA transcripts), a bioinformatical algorithm recently developed in our lab (28). Thereby, differential expression analysis was based on barcoding of cDNAs from the three libraries prior to cloning and sequencing. Subsequently, two bioinformatical analyses were performed by employing APART: (i) we first annotated all sequence reads to known or unknown ncRNA species, e.g. miRNAs, snoRNAs, tRNAs, intraRNAs (i.e. intragenic RNAs) or interRNAs (intergenic RNAs; Figure 1A); (ii) we next grouped all identical sequences into ‘contigs’ and determined the distribution of these unique RNA transcripts in our libraries (Figure 1B). By this approach, 706 contigs with expression changes >2-fold between any of the three cDNA libraries and a mean expression value of at least 28 were identified (Supplementary File S1). Only those sequences were included which appeared at least in two of the three cDNA libraries in order to assess expression differences.
Figure 1.
Distribution of ncRNA sequences between the three cDNA libraries. (A) Distribution based on sequence reads. (B) Distribution of contigs. The most abundant known ncRNAs are shown in addition to reads mapping in intergenic and intronic regions (i.e. intergenic, sense/antisense intron). ES = embryonic stem cells, NP = neural progenitors, N/G = neurons and glial cells.
In this analysis, miRNAs represented the majority of differentially expressed known ncRNA sequences: while in the N/G library, miRNAs accounted for only 4% of all sequences, in ES or NP libraries 25–29% of sequences corresponded to miRNAs (Figure 1A). This is due to significantly fewer miRNA species being present at the N/G stage, as demonstrated by the number of unique contigs (1% versus 8–13%; Figure 1B), which hints to a more prominent participation of miRNAs in regulation of gene expression during the early stages of ES cell differentiation. Interestingly, a 9-fold increase in the abundance of sequences from the class of miscellaneous ncRNAs is observed in total sequence reads at the N/G stage, mainly due to an increase of 7SK and 7SL ncRNA expression (Figure 1A; see below).
The majority of novel ncRNA transcripts in all three cDNA libraries mapped to either intergenic or intronic regions of the mouse genome, which were designated as interRNAs (i.e. intergenic RNAs) or intraRNAs (i.e. intragenic RNAs such as intronically encoded RNAs), respectively (Figure 1B). Sequence reads representing intraRNAs or interRNAs showed a rather similar distribution in all cell stages (60% in ES and NP stages or 75% in the N/G stage, respectively). We have previously shown that intergenic regions between and intragenic regions within protein-coding genes harbour the majority of functional known ncRNAs (21), in particular snoRNAs and miRNAs, suggesting the presence of novel representatives of these ncRNA classes also in our cDNA libraries (see below).
From intraRNA species, about one-third of sequences mapped to introns in antisense orientation while the remaining sequences derived from the sense orientation of introns (Figure 1B). In addition, a small number of sequences were annotated to exons of mRNAs, in antisense orientation (Figure 1B). Previously, by analysis of a mouse brain cDNA library encoding small ncRNAs we have presented evidence that ncRNAs mapping in antisense orientation to introns or exon/intron boundaries might be involved in regulation of alternative splicing of respective genes (21).
Validation of ES cell neural differentiation by transcriptional profiling of miRNAs
To validate the transcriptional profiling analysis, we investigated expression of known miRNA species, previously described to be involved in neural differentiation. Indeed, we verified expression of stem cell specific microRNAs in the ES library, in particular abundant expression of the mir-290 (11) and the miR-17-92 clusters (35), as well as miRNAs miR-199, miR-106, miR-299 and miR-214 (Figure 2 and Supplementary File S2), as previously reported (11,36). In addition, we observed expression of these miRNA species being significantly down-regulated upon ES cell differentiation in the NP and N/G population of cells (see above and Figure 2), in agreement with earlier studies (35,36).
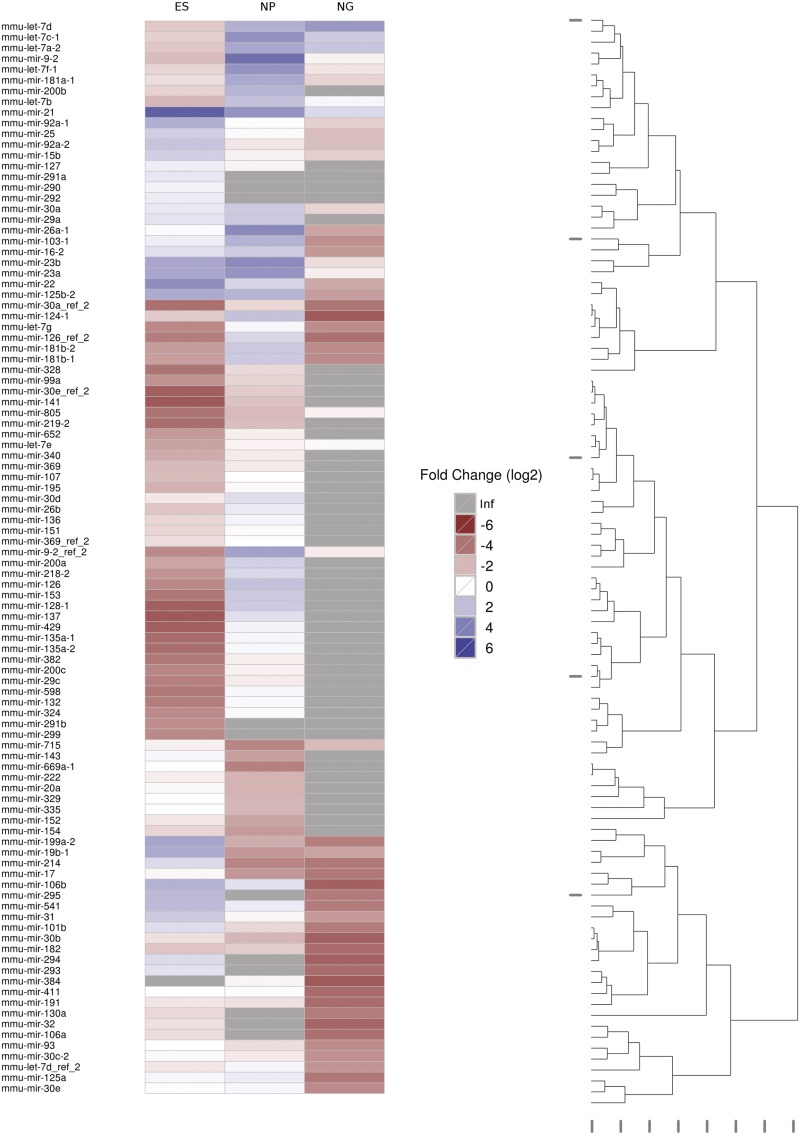
Figure 2.
The 100 most differentially expressed miRNAs. Expression values were normalized against the mean expression value (257.6). Fold changes are depicted in a log2 scale. Inf: infinite fold-change because expression was not detectable in the respective library. The dendrogram to the right indicates the Euclidean distance of the expression values.
We were also able to verify expression of miRNAs miR-21, miR-15b, miR-669, miR-329, miR-335, miR-16, miR-411 and miR-541 in mouse ES cells (11,35) and, in addition, we observed that their expression was significantly down-regulated in the later stages of ES cell differentiation (i.e. NP, N/G stages; Figure 2). Notably, we find, as reported, the stem-cell-specific miR-302 (16,37–41) exclusively expressed in ES cells while completely absent at the NP and N/G stage (Supplementary File S2).
Conversely, expression of neural-specific microRNAs mir-9 and miR-124, as well as the miRNA let-7 family, which have been reported to modulate stem cell derived neurogenesis (42), was found to be significantly up-regulated at the NP stage in our analysis (Figure 2). For human ES cells, several microRNAs have been reported to be up-regulated upon spontaneous differentiation, such as mir-181a, mir-181b-2, mir-26a, mir-23b, mir-137 (39), consistent with their mouse homologues from this study.
Interestingly, we also identified microRNAs exhibiting expression patterns differing from previous studies. Thereby, expression of microRNAs miR-29a (16,36), miR-135a (39), miR-141 (40,43), miR-340 (41), miR-200c (16,37,43), miR-328 (36) and miR-30e (40) has been reported to be up-regulated in ES cells. In contrast, we find an increase in their expression at the NP stage (Figure 2). As for miR-135a, miR-141, miR-340, and miR-200c this is probably due to the fact that previous studies were carried out in human ES cells (16,39–41), which derive from a different developmental stage than mouse ES cells. In addition, expression analyses carried out in mouse ES cells (36) were only extended to embryoid bodies (EBs) and not to neural progenitor cells, which might explain the observed up-regulation of expression of miR-29a and miR-328 in NPs with respect to ES cells in our study.
Finally, we have identified expression of specific miRNAs, not previously being reported in ES cells, such as miR-715. Also, we observed microRNAs whose expression has been reported in mouse ES cells (35), without further analysis on their differential expression, to be significantly up-regulated in NPs and NGs (Figure 2). From these, miR-200a, miR-200b, miR-218-2, miR-153, miR-103-1, miR-26b, miR-30d, miR-324, miR-429 and miR-382, were found to be the highest expressed at the NP stage, while miR-805 and miR-674 were the highest expressed at the N/G stage (Figure 2 and Supplementary File S3). We also observed expression of miR-598 being up-regulated at the NP stage while miR-877, miR-682, miR-678 and miR-217 are expressed at the N/G stage, only. Taken together, our data demonstrate that the regulation of gene expression during neural differentiation by a large number of miRNAs might be even more complex than previously anticipated.
Differentially expressed known ncRNAs from the three stages of ES cell neural differentiation
In addition to miRNAs, also several C/D box snoRNAs (small nucleolar RNAs) were found among the most prominent differentially expressed known ncRNA species (Figure 3), designated as SNORD12, 29, 31, 35, 74, 101, 104 and 115 (44). The majority of snoRNAs has been reported to guide covalent modifications of ribosomal RNAs (rRNAs) or small nuclear RNAs (snRNAs), respectively (45,46). Thereby, most snoRNAs, with few exceptions, are encoded within intronic sequences of protein-coding genes (45,47,48). Up till now, two classes of snoRNAs have been described, i.e. C/D box snoRNAs and H/ACA box snoRNAs. Unlike canonical snoRNAs, so-called ‘orphan’ snoRNAs lack any complementarity to rRNA or snRNA targets and might target other RNA molecules such as mRNAs (45,46,49).
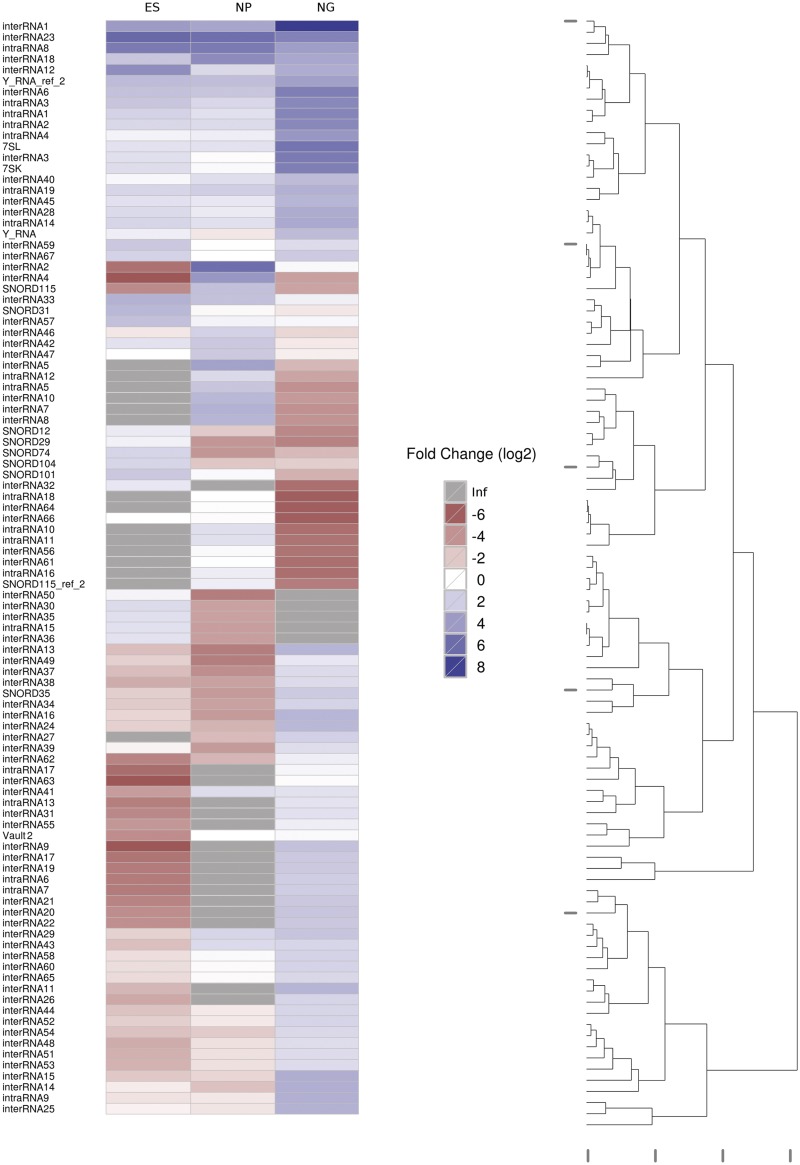
Figure 3.
The 100 most differentially expressed novel ncRNA candidates. Further legend analogous to Figure 2.
We show that snoRNAs SNORD12, 29, 31, 74, 101 and 104 are abundantly expressed in ES cells, while their expression significantly decreases upon ES cell differentiation by ∼10- to 30-fold (Figure 3). In contrast, expression of SNORD35 and 115 is low in ES cells while their expression is significantly up-regulated in NP cells or N/G cells (Figure 3). Interestingly, SNORD31, 101 and 115 have no validated rRNA targets (49,50) and might be involved in regulation of other RNA species such as mRNAs (see below). In addition, unlike canonical snoRNAs, SNORD12, 29, 31, 74, 104 and 115 are reported to be encoded within intronic sequences of non-protein coding transcripts or hypothetical open reading frames, which might not code for proteins (44,49,51,52); up till now, the role of these non-protein-coding snoRNA host gene transcripts has been elusive; this implies the possibility that, in addition to intronically encoded snoRNAs, also their respective host gene transcripts might be involved in the regulation of neural differentiation.
Interestingly, earlier reports have implicated SNORD115 (also designated as HBII-52) to be involved in brain development and disease (49,53). Thereby, SNORD115 has been proposed to target the brain-specific serotonin receptor 2C mRNA (49), thus regulating alternative splicing and/or editing of the serotonin receptor 2C pre-mRNA (54,55). We observed an ∼100-fold increase in SNORD115 expression between mouse ES cells and differentiating cells, which was confirmed by northern blotting (Supplementary Figure S3). At the N/G stage, expression of SNORD115 is reduced by ∼50-fold in comparison to the NP stage. However, the N/G library mainly consists of astrocytes and oligodendrocytes with fewer neurons (see above); since SNORD115 has previously been shown to be expressed exclusively in neurons, this might explain the lower abundance at the N/G stage.
Previously, it has been demonstrated that SNORD115 maps to the Prader–Willi Syndrome (PWS) locus on chromosome 15 (49). PWS is a neurodevelopmental disease with patients showing severe obesity and varying degrees of mental retardation (56). Notably, two snoRNAs from that locus have directly been implicated in the etiology of the disease, namely SNORD115 and SNORD116 (also designated as HBII-85), respectively (57–60). It is thus interesting to note that expression of SNORD115 is significantly up-regulated at a very early stage of development, i.e. already during neural differentiation.
In addition, expression of three other known ncRNAs, i.e. 7SL, 7SK and vault-2 RNA, respectively, is up-regulated at the N/G stage compared to NP and ES cells (Figure 3). Thereby, 7SL RNA is an abundant ncRNA species, which serves as an integral part of the signal recognition particle (SRP) (61,62). The SRP has been shown to promote the insertion of proteins into the cellular membrane or to regulate protein secretion (63). These two processes might especially be required at the N/G stage where neural cells interconnect to form neural networks guided by receptors on the surface of cells, such as serotonin receptor 2C (see above); concomitantly, an increase in protein secretion might be required by secretion of neurotransmitter peptides.
Previously, 7SK RNA has been reported to negatively regulate RNA Polymerase II transcription by inactivating the positive transcription elongation factor b (P-TEFb) as well as affecting the function of the chromatin regulator HMGA1 (64,65). By this mechanism, an important transcriptional regulatory role of 7SK RNA in HMGA1-dependent cell differentiation regulation has been described (64). Hence, 7SK RNA might also be involved in the regulation of the transition from mouse ES cells into neural/glial population of cells.
Lastly, among differentially expressed known ncRNAs we have identified expression of vault-2 ncRNA (66), an ncRNA component of the vault RNP, to be upregulated at NP and N/G stages, respectively (Figure 3). Recently, by a subtractive hybridization approach we have observed expression of vault RNAs 1–3 to be highly up-regulated upon EBV (Epstein–Barr virus) infection of human B cells (67,68).
Size distribution of novel ncRNAs from the three stages of ES cell neural differentiation
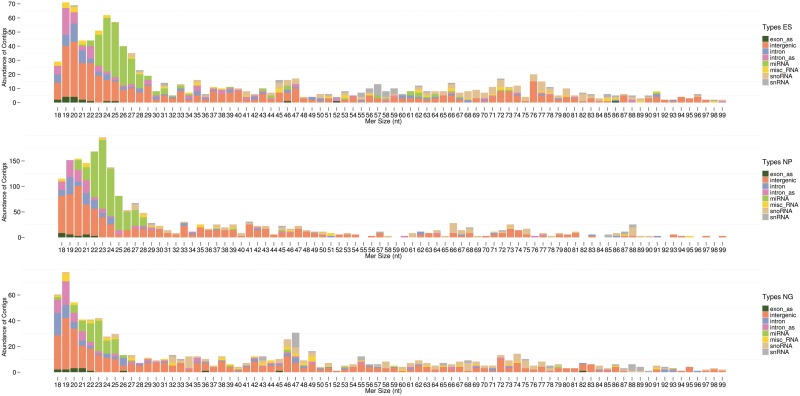
Analysis of the length distribution of novel and known ncRNA candidates from the three cDNA libraries showed a bias towards 18–30 nt sized ncRNA species in all three cDNA libraries (Figure 4). Thereby, in the NP library the 18–30 nt peak is ∼2-fold larger compared to ES and N/G libraries. A second, smaller peak of ncRNA sequences appears at RNAs sized 45–47 nt which increases at the N/G stage; a third potential peak, sized 71–78 nt, is also observed (Figure 4).
Figure 4.
Length distribution and abundance of contigs from the three RNP libraries. NcRNAs were clustered in unique contigs; the consensus sequence was employed to determine the absolute abundance versus the length of respective ncRNAs (contigs exhibiting a size >99 nt are rare and are therefore not shown).
The fraction of 18–30 nt sized ncRNA candidates is comprised of miRNA species in addition to inter- and intraRNAs, i.e. ncRNAs located in intergenic or intronic regions (Figure 4). Although we tried to sub-classify the latter RNA species, including motif finding algorithms provided by the MEME suite (69) and searching for structural conservation employing the Vienna RNA package (70) and RNAz (71), as of now, we could not identify any common motifs. In addition, a correlation of features such as RNA size did not result in satisfying findings. However, due to their size and high abundance, similar to the abundance of miRNAs, some intra- and interRNAs might represent, yet undiscovered, novel microRNAs. Therefore, we employed the miRDeep algorithm (32,33) to search for novel microRNAs in our dataset. Thereby, we investigated folding of the pre-miRNAs by mfold, as well as the conservation of seed sequences and the abundance of the miR* sequences in the cDNA libraries. In some cases, novel identified microRNA candidates were predicted to derive from snoRNA sequences. Since it has previously been reported that some snoRNAs can indeed serve as precursor RNAs for miRNAs (72–74), we included these candidates in our analysis. Based on above criteria, we characterized 17 novel microRNA candidates (Supplementary File S4). Thereby, the highest scoring miRNA candidate was mmu-interRNA26, whose expression was also validated by a mouse ESTs (expressed sequence tags) database screen; mmu-interRNA26 is abundantly expressed in the N/G library, unlike most other miRNAs (Figure 3), as well as in primary hippocampal neurons and astrocytes (see below).
The remaining inter- and intraRNAs in the size range of 18–30 nt scored very low as potential miRNAs suggesting that these might represent a distinct class of entirely novel neural ncRNA candidates. Interestingly, we note that sequences of numerous inter- and intraRNAs are included in reported ESTs (Supplementary File S1), thus validating their expression. Several inter-and intraRNAs were found also to cover binding sites for transcription factor p300 and the transcriptional repressor CTCF, which are reported to regulate gene expression in ES cells and in the cortex and cerebellum of the mouse brain. We further identified a number of inter-and intraRNAs overlapping regions of chromatin acetylation, which has been reported in total brain and cerebellum (Supplementary File S1). Taken together, these results indicate that most inter- and intraRNAs are actively transcribed in the brain, while at least some of these might be involved in regulating brain function and development on a transcriptional level.
Processing analysis of known and novel ncRNA candidates by APART
In Eukarya, many functional known ncRNA species are processed from larger RNA transcripts (75). In particular, rRNA, miRNA-, snoRNA- or tRNA-precursor transcripts are processed by distinct processing pathways, which include site-specific exo- and endonucleases. By employing APART we have identified a large number of small stable RNA species, potentially derived from larger precursor transcripts (for selected examples see Supplementary Figure S2). Detection of RNA processing by APART is achieved by scanning of a contig-coverage plot to search for significant changes (28). APART considers a position as a putative processing site when the coverage shift between 2 nt is larger than one-third of the maximum coverage of the contig (Supplementary Figure S2).
For proof of principle we investigated miRNA processing by the APART algorithm for the class of miRNAs, which are reported to be processed by Drosha/Pasha and Dicer from larger primary and precursor miRNA transcripts (76,77). Indeed, we were able to identify this processing event for miRNAs mmu-let-7c-1 as well as for mmi-mir-21 (Supplementary Figure S2). In addition, in particular for the class of 18–30 nt sized ncRNA candidates, we were able to identify in many cases potential precursor transcripts (Figure 5) which might strengthen the functionality of these novel ncRNA candidates.
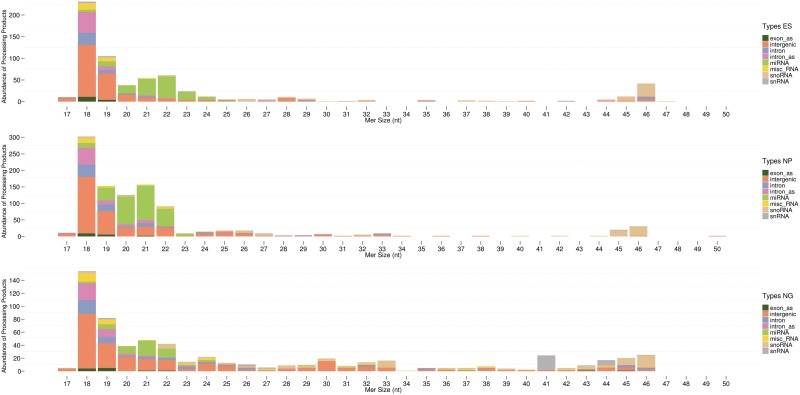
Figure 5.
Length distribution and abundance of processed contigs from the three RNP libraries. NcRNAs were clustered in unique contigs; the consensus sequence was employed to determine the absolute abundance of the processed ncRNAs, as assessed by APART, versus the length of the respective processed ncRNAs (processed contigs exhibiting a size >50 nt are rare and are therefore not shown).
Novel ncRNA candidates are expressed in primary hippocampal neurons and astrocytes
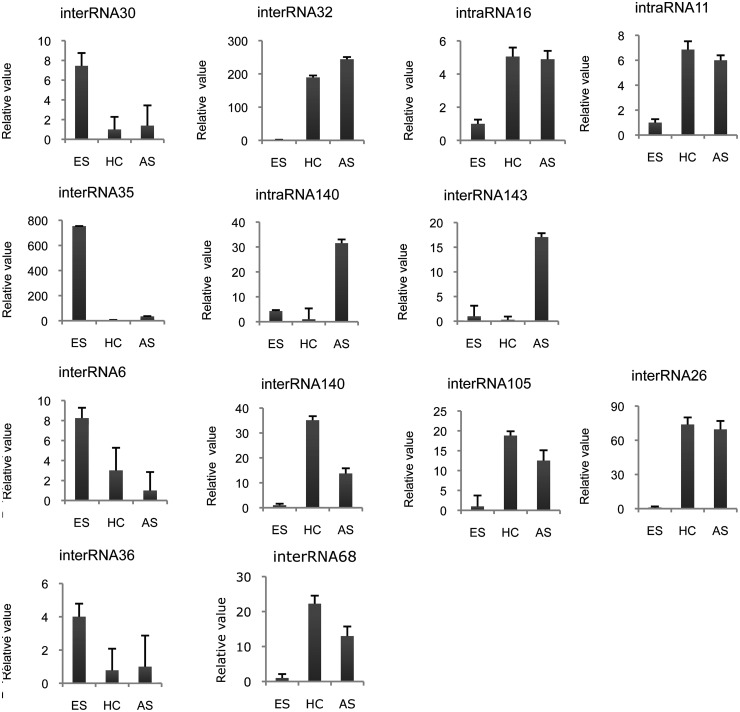
Due to the heterogeneity of the N/G stage, comprised of neurons and glial cells (astrocytes and oligodendrocytes) (Supplementary Figure S1), we additionally analyzed in which type of neural cells selected inter- and intraRNAs are expressed. To that aim, we analyzed primary cell cultures of neuronal and glial cells for the presence of selected novel ncRNA candidates by real-time PCR. Based on the library profiling by protein markers (see above), we employed astrocytes (AS) as well as primary hippocampal neurons (HC) (see Supplementary Methods section) to investigate ncRNA expression.
Indeed, we observed neural-specific as well as ES cell-specific expression of selected candidates (Figure 6). For example, interRNA32 and interRNA26 were highly expressed in HC and AS but showed very low expression in ES cells; thereby, up-regulation of expression was observed between 70- and 200-fold (Figure 6). Other ncRNAs, such as interRNA30, interRNA35 and interRNA36, which exhibited a reduced expression level during differentiation (i.e. in the NP and N/G stages), also show reduced expression in HC and AS cell cultures comprised mainly of a single cell type (Figure 6). Interestingly, interRNA143 and intraRNA140 exhibited a highly AS-specific expression when compared to HC and ES cells, thereby being potentially involved in AS differentiation. Conversely, interRNA105, interRNA140 and interRNA68 are higher expressed in HC than in AS cells, with interRNA140 being ∼3-fold higher expressed in HC compared to AS (Figure 6).
Figure 6.
Expression analysis of selected inter- and intraRNAs in primary hippocampal neurons and astrocytes by real-time PCR. Expression values were normalized against U6.
Abundance versus differential expression of ncRNA candidates
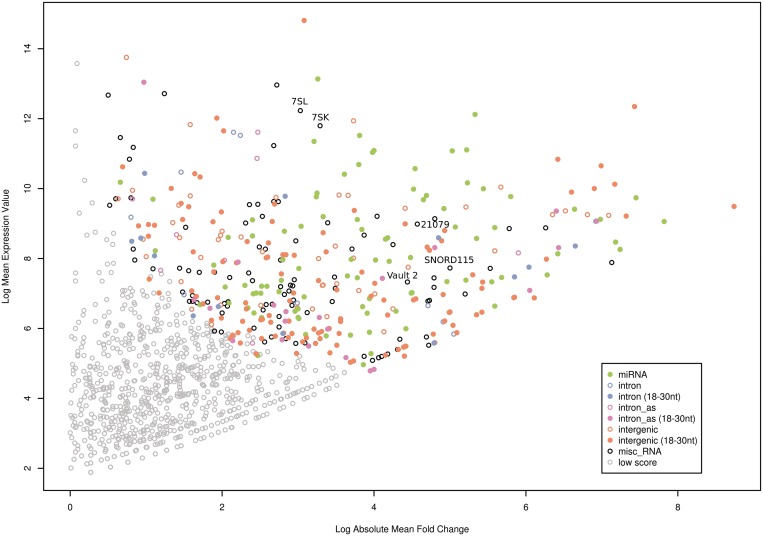
To further increase the likelihood for the identification of novel functional ncRNA candidates, we assessed their relative abundance in the libraries and compared it to their differential expression within the three differentiation stages of ES cells. The differentially expressed candidates were assigned scores, calculated by their mean expression value versus their absolute mean fold change in all three libraries. We rationalized that there should be a correlation between the abundance of a ncRNA, its differential expression, and the likelihood for representing a functional ncRNA species. This does not rule out, that lowly expressed ncRNAs with an even distribution between the three ES cell stages might not be functional. However, differential expression is one of the key indications of regulatory function of the gene products. Moreover, for future functional analysis such as the identification of protein binding partners, a robust, abundant and differential expression would be desirable.
The results of the abundance/differential expression plot show that ncRNA candidates with the highest scores (i.e. highest abundance/highest differential expression) are predominantly interRNAs or miRNAs, sized 18–30 nt next to miRNAs (Figure 7, upper right corner). We also identified other known ncRNAs such as 7SL RNA and 7SK RNA, vault-2 RNA and snoRNAs, in particular SNORD115 (see above), which are among the top-ranked candidates. The potentially novel miRNA candidate mmu-interRNA26 is indicated in Figure 7 (designated as 21 079, corresponding to the ID number indicated in Supplementary File S4). Differential expression of selected ncRNA candidates from this analysis was also verified by real-time PCR (Supplementary Figure S4). Further functional analysis of selected ncRNA candidates will thus focus on highest-ranked novel and known ncRNAs from our screen.
Figure 7.
Plot of the mean fold-change versus the mean expression of ncRNAs expressed in all three libraries. The ncRNAs are assigned scores (see Supplementary Material section) based on their mean expression value (y-axis) versus their absolute mean fold change (x-axis). Expression values and fold changes represent the mean in all three libraries. Fold changes are shown in a log2 scale. The grey dots (low score) refer to candidates having a mean expression value of <50 and a fold-change of <2.
CONCLUSION
In this study, we defined the small ncRNA transcriptome involved in regulation of mouse ES cell differentiation into neural cells in mouse by a deep-sequencing approach. In general, high-throughput deep-sequencing approaches have the potential to identify a large number of potential ncRNA candidates, however, these studies usually lack functional evidence of novel RNA transcripts. Functional analysis of potential ncRNA candidates, however, is highly time consuming, with currently no high-throughput methods available. Hence, it is important to preselect or enrich for functional ncRNA candidates in deep-sequencing screens for further analysis. In order to increase the likelihood for identification of such functional ncRNA species, we applied three novel selection filters: (i) cDNA libraries were generated from RNPs rather than from protein-devoid RNAs, since most functional eukaryal RNAs are known to form RNA–protein complexes; (ii) we analyzed differential expression of novel ncRNA candidates by generation of three RNP libraries from three stages of ES cell differentiation taking into consideration that many functional ncRNAs are regulated during development; (iii) since most functional ncRNAs have been shown to be processed from larger precursor RNAs, we also analyzed potential RNA processing products.
By these analyses, we identified several known ncRNAs from two abundant RNA classes, i.e. miRNAs and snoRNAs, respectively, which were differentially expressed. In addition, expression of three other known ncRNAs, i.e. 7SL, 7SK and vault-2 RNA, respectively, was shown to be significantly up-regulated during neural differentiation.
While regulation of gene expression by 7SK RNA is exerted on the transcriptional level, miRNAs and some selected snoRNAs have been reported to regulate gene expression on the post-transcriptional level; finally, 7SL RNA was shown to act on a post-translational level. Hence, regulation of gene expression during neural differentiation might be exerted by these ncRNAs on transcriptional, translational and post-translational levels.
The majority of differentially expressed ncRNAs were represented by novel ncRNA candidates and mapped to intergenic and intronic regions, designated as interRNAs or intraRNA, respectively. These RNA species predominantly exhibited sizes between 18 and 30 nt. Thereby, 17 of these interRNAs and intraRNAs might represent novel members from the class of miRNAs, as assessed by the miRDeep algorithm. However, the majority of interRNAs or intraRNAs could not be assigned as miRNAs and thus might represent novel candidates for regulatory ncRNAs in neural differentiation. For future analyses, it will be interesting to study the effects of known and novel ncRNAs on ES cell differentiation by overexpression or inactivation of selected ncRNA candidates.
ACCESSION NUMBERS
Sequence Read Archive (NCBI) accession number: SRP008250.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Files 1–4, Supplementary Figures 1–4, Supplementary Methods and Supplementary References [78–80].
FUNDING
The Austrian Science Funds FWF (SFB F4411); the Austrian Ministry of Science and Research grant Gen-AU (D 110420-011-015); the PhD program W 1206-B18 (funding of K.S.) to A.H.; the Austrian Ministry of Science and Research grant Gen-AU (D110420-012-012) (funding of M. Z). Funding for open access charge: Austrian Ministry of Science and Research grant (SFB F4411).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Willingham AT, Gingeras TR. TUF love for “junk” DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattick JS, Taft RJ, Faulkner GJ. A global view of genomic information–moving beyond the gene and the master regulator. Trends Genet. 2009;26:21–28. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 4.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hüttenhofer A, Schattner P. The principles of guiding by RNA: chimeric RNA-protein enzymes. Nat. Rev. Genet. 2006;7:475–482. doi: 10.1038/nrg1855. [DOI] [PubMed] [Google Scholar]
- 6.Stadler BM, Ruohola-Baker H. Small RNAs: keeping stem cells in line. Cell. 2008;132:563–566. doi: 10.1016/j.cell.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallanna SK, Rizzino A. Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev. Biol. 2010;344:16–25. doi: 10.1016/j.ydbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 11.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 12.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 13.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 14.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 15.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell. Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 16.Lakshmipathy U, Love B, Goff LA, Jörnsten R, Graichen R, Hart RP, Chesnut JD. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2007;16:1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Ridzon D, Lee CT, Blake J, Sun Y, Strauss WM. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm. Genome. 2007;18:316–327. doi: 10.1007/s00335-007-9032-6. [DOI] [PubMed] [Google Scholar]
- 18.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rederstorff M, Bernhart SH, Tanzer A, Zywicki M, Perfler K, Lukasser M, Hofacker IL, Huttenhofer A. RNPomics: defining the ncRNA transcriptome by cDNA library generation from ribonucleo-protein particles. Nucleic Acids Res. 2010;38:e113. doi: 10.1093/nar/gkq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rederstorff M, Huttenhofer A. cDNA library generation from ribonucleoprotein particles. Nat. Protoc. 2011;6:166–174. doi: 10.1038/nprot.2010.186. [DOI] [PubMed] [Google Scholar]
- 23.Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 24.Shimizukawa R, Sakata A, Hirose M, Takahashi A, Iseki H, Liu Y, Kunita S, Sugiyama F, Yagami K. Establishment of a new embryonic stem cell line derived from C57BL/6 mouse expressing EGFP ubiquitously. Genesis. 2005;42:47–52. doi: 10.1002/gene.20122. [DOI] [PubMed] [Google Scholar]
- 25.Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat. Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- 26.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 27.Nat R, Salti A, Suciu L, Ström S, Dechant G. Pharmacological modulation of the hedgehog pathway differentially affects dorsal/ventral patterning in mouse and human embryonic stem cell models of telencephalic development. Stem Cells Dev. 2012 doi: 10.1089/scd.2011.0271. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Zywicki M, Bakowska-Zywicka K, Polacek N. Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res. 2012;40:4013–4024. doi: 10.1093/nar/gks020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 33.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence M, Gentleman R, Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tripathi R, Saini HK, Rad R, Abreu-Goodger C, van Dongen S, Enright AJ. Messenger RNA and microRNA profiling during early mouse EB formation. Gene Expr. Patterns. 2011;11:334–344. doi: 10.1016/j.gep.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Tzur G, Levy A, Meiri E, Barad O, Spector Y, Bentwich Z, Mizrahi L, Katzenellenbogen M, Ben-Shushan E, Reubinoff BE, et al. MicroRNA expression patterns and function in endodermal differentiation of human embryonic stem cells. PLoS One. 2008;3:e3726. doi: 10.1371/journal.pone.0003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, Fan JB, Loring JF. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 39.Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stadler B, Ivanovska I, Mehta K, Song S, Nelson A, Tan Y, Mathieu J, Darby C, Blau CA, Ware C, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J. Transl. Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bachellerie JP, Cavaillé J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 46.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 47.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 48.Kiss T. Biogenesis of small nuclear RNPs. J. Cell. Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 49.Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vitali P, Royo H, Seitz H, Bachellerie JP, Hüttenhofer A, Cavaille J. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 2003;31:6543–6551. doi: 10.1093/nar/gkg849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tycowski KT, Shu MD, Steitz JA. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 52.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Runte M, Hüttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 54.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 55.Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaillé J, Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butler MG. Prader-Willi syndrome: obesity due to genomic imprinting. Curr. Genomics. 2011;12:204–215. doi: 10.2174/138920211795677877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding F, Prints Y, Dhar MS, Johnson DK, Garnacho-Montero C, Nicholls RD, Francke U. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm. Genome. 2005;16:424–431. doi: 10.1007/s00335-005-2460-2. [DOI] [PubMed] [Google Scholar]
- 58.Skryabin BV, Gubar LV, Seeger B, Pfeiffer J, Handel S, Robeck T, Karpova E, Rozhdestvensky TS, Brosius J. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hogart A, Leung KN, Wang NJ, Wu DJ, Driscoll J, Vallero RO, Schanen NC, LaSalle JM. Chromosome 15q11-13 duplication syndrome brain reveals epigenetic alterations in gene expression not predicted from copy number. J. Med. Genet. 2009;46:86–93. doi: 10.1136/jmg.2008.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001;70:755–75. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 62.Wild K, Weichenrieder O, Strub K, Sinning I, Cusack S. Towards the structure of the mammalian signal recognition particle. Curr. Opin. Struct. Biol. 2002;12:72–81. doi: 10.1016/s0959-440x(02)00292-0. [DOI] [PubMed] [Google Scholar]
- 63.Lutcke H. Signal recognition particle (SRP), a ubiquitous initiator of protein translocation. Eur. J. Biochem. 1995;228:531–550. doi: 10.1111/j.1432-1033.1995.tb20293.x. [DOI] [PubMed] [Google Scholar]
- 64.Eilebrecht S, Benecke BJ, Benecke A. 7SK snRNA-mediated, gene-specific cooperativity of HMGA1 and P-TEFb. RNA Biol. 2011;8:1084–93. doi: 10.4161/rna.8.6.17015. [DOI] [PubMed] [Google Scholar]
- 65.Eilebrecht S, Brysbaert G, Wegert T, Urlaub H, Benecke B-J, Benecke A. 7SK small nuclear RNA directly affects HMGA1 function in transcription regulation. Nucleic Acids Res. 2011;39:2057–2072. doi: 10.1093/nar/gkq1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Zon A, Mossink MH, Scheper RJ, Sonneveld P, Wiemer EAC. The vault complex. Cell. Mol. Life Sci. 2003;60:1828–1837. doi: 10.1007/s00018-003-3030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mrázek J, Kreutmayer SB, Grässer FA, Polacek N, Hüttenhofer A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007;35:e73. doi: 10.1093/nar/gkm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nandy C, Mrazek J, Stoiber H, Grasser FA, Huttenhofer A, Polacek N. Epstein-barr virus-induced expression of a novel human vault RNA. J. Mol. Biol. 2009;388:776–784. doi: 10.1016/j.jmb.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 69.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hofacker IL. RNA secondary structure analysis using the Vienna RNA package. Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi1202s26. Chapter 12, Unit12 12. [DOI] [PubMed] [Google Scholar]
- 71.Gruber AR, Findeiβ S, Washietl S, Hofacker IL, Stadler PF. Rnaz 2.0: improved noncoding Rna detection. Pac. Symp. Biocomput. 2010;15:69–79. [PubMed] [Google Scholar]
- 72.Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 73.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tollervey D, Caceres JF. RNA processing marches on. Cell. 2000;103:703–709. doi: 10.1016/s0092-8674(00)00174-4. [DOI] [PubMed] [Google Scholar]
- 76.Conrad R, Barrier M, Ford LP. Role of miRNA and miRNA processing factors in development and disease. Birth Defects Res. C Embryo Today. 2006;78:107–117. doi: 10.1002/bdrc.20068. [DOI] [PubMed] [Google Scholar]
- 77.Lehrbach NJ, Miska EA. Regulation of pre-miRNA Processing. Adv. Exp. Med. Biol. 2011;700:67–75. doi: 10.1007/978-1-4419-7823-3_7. [DOI] [PubMed] [Google Scholar]
- 78.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Polleux F, Ghosh A. The slice overlay assay: a versatile tool to study the influence of extracellular signals on neuronal development. Sci. STKE. 2002 doi: 10.1126/stke.2002.136.pl9. 2002, pl9. [DOI] [PubMed] [Google Scholar]
- 80.Zeitelhofer M, Vessey JP, Xie Y, Tübing F, Thomas S, Kiebler M, Dahm R. High-efficiency transfection of mammalian neurons via nucleofection. Nat. Protoc. 2007;2:1692–1704. doi: 10.1038/nprot.2007.226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.