Abstract
Purpose
Non-invasive PET imaging with radiolabeled RGD peptides for αvβ3 integrin targeting has become an important tool for tumor diagnosis and treatment monitoring in both pre-clinical and clinical studies. To better understand the molecular process and tracer pharmacokinetics, we introduced kinetic modeling in the investigation of 18F-labeled RGD peptide monomer 18F-FP-c(RGDyK) (denoted as 18F-FPRGD) and dimer 18F-FP-PEG3-E[c(RGDyK)]2 (denoted as 18F-FPPRGD2).
Procedures
MDA-MB-435 tumor-bearing mice underwent 60 min dynamic PET scans following the injection of either 18F-FPRGD or 18F-FPPRGD2. Blocking studies with pre-injection of a blocking mass dose were performed for both monomeric and dimeric RGD groups. 18F-FPRAD (RAD) was used as a negative control. Kinetic parameters (K1, k2, k3, k4) of a three-compartment model were fitted to the dynamic data to allow quantitative comparisons between the monomeric and dimeric RGD peptides.
Results
Dimeric RGD peptide tracer showed significantly higher binding potential (BpND = k3/k4, 5.87 ± 0.31) than that of the monomeric analog (2.75 ± 0.48, p = 0.0022, n = 4/group). The BpND values showed a significantly greater ratio (dimer/monomer ~2.1) than the difference in %ID/g uptake measured from static images (dimer/monomer ~1.5, p = 0.0045). Significant decrease in BpND was found in the blocked groups compared with the unblocked ones (dimer p = 0.00024, monomer p = 0.005, n = 4/group). Similarly, the RAD control group showed the lowest BpND value among all the test groups, as the RAD peptide does not bind to integrin αvβ3. Volume of distribution (VT = K1/k2(1+k3/k4)) could be separated into non-specific (VND = K1/k2) and specific (VS = K1k3/(k2k4)) components. Specific distribution volume (VS) was the dominant component of VT in the unblocked groups and decreased in the blocked groups. Unblocked RGD dimer also showed higher VS than that of the monomer (dimer VS = 2.38 ± 0.15, monomer VS = 0.90 ± 0.17, p = 0.0013, n = 4/group), well correlated with BpND calculations. Little difference in VND was found among all groups. Moreover, parametric maps allowed quantitative analysis at voxel level and provided higher tumor-to-background contrast for BpND maps than the static images. Tumor heterogeneity in kinetic parameters was found in parametric images, which couldn’t be clearly identified in static intensity images.
Conclusions
The pharmacokinetics of both monomeric and dimeric RGD peptide tracers was compared, and the RGD dimers showed significantly higher binding affinity than the monomeric analogs. Kinetic parameters were demonstrated to be valuable for separating specific and non-specific binding and may allow more sensitive and detailed quantification than simple standard uptake value (SUV) analysis.
Keywords: Positron Emission Tomography, kinetic modeling, quantitative analysis, RGD peptide, Integrin
Introduction
The expression of integrin αvβ3 on sprouting capillary cells and its interaction with specific matrix ligands play a key role in human tumor-induced angiogenesis and metastasis [1–6]. Non-invasive PET imaging of integrin αvβ3 has become an important tool for tumor diagnosis and treatment monitoring in both pre-clinical and clinical studies [4, 5, 7–11]. Suitably labeled RGD peptides with prominent binding affinity and high binding selectivity for integrin receptors are increasingly used to target and monitor integrin expression level, especially αvβ3 in tumor regions [12–16]. Among them, 18F-labeled cyclic RGDyK peptide dimer with mini-pegylation has favorable properties for PET imaging [12]. This radiotracer has been used in clinical trial and showed potentials in biomedical and clinical imaging [17]. It has been proven that the dimeric RGD peptides have better receptor-binding characteristics than those of the monomeric analogs [18]. In our previous studies of static images, 18F-FB-E[c(RGDyK)]2 (18F-FRGD2) showed more than 1.5 times as much tumor uptake in the same animal model as compared with the monomeric tracer 18F-FB-c(RGDyK) (18F-FRGD) [19, 20].
Compared with static images, dynamic PET imaging followed by kinetic modeling offers several advantages. First of all, it quantitatively measures the transport rates and provides means to measure the metabolic or specific binding rates of the tracer. It also facilitates the separation of specific signal from non-specific signal and can be used to discern specific binding in tissue [21]. Parametric mapping emphasizes the spatial distribution of the specific signal at the voxel level, and allows interpretation of physiological function, pharmacokinetics, as well as the behavior of target molecule [22]. This quantitative information may provide a more sensitive measure of early tumor response to treatment, compared with the semi-quantitative values (e.g. %ID/g) extracted from static images.
With 18F-galacto-RGD peptide, Beer et al. [23] used one- and two-compartment models to perform the pharmacokinetic analysis on patient data. Ferl et al. [24] conducted pharmacokinetic analysis of 64Cu-DOTA-RGD in preclinical models and demonstrated that a 2-tissue compartment, 4-parameter model with internalization is more appropriate to describe RGD tracer compared with the 1-tissue compartment (2-parameter) model and a 2-tissue compartment irreversible (3-parameter) model. In our previous study, we utilized the Logan graphical analysis with reference tissue model to fit the dynamic time activity curves (TACs) for 18F-labeled RGD tracers [19]. Although these studies have implied that the RGD kinetics agree with a reversible three-compartment model, it is still not clear how the kinetic parameters reflect the tracer binding affinity and whether the parametric map can provide more sensitive information. Therefore, an appropriate compartment model and a blocking study should be applied to accurately characterize the kinetics of RGD peptide tracers with different receptor binding characteristics. Furthermore, appropriate kinetic parameters and parametric images need to be employed for quantitative analysis and comparison between different RGD peptides.
Herein we used a reversible three-compartment model to analyze dynamic PET data of 18F-labeled RGD peptide monomer 18F-FP-c(RGDyK) (denoted as 18F-FPRGD) and dimer 18F-FP-PEG3-E[c(RGDyK)]2 (denoted as 18F-FPPRGD2). To validate the accuracy of the model, blocking studies with unlabeled peptide were performed. Studies with 18F-FP-c(RADyK) (18F-FPRAD), a peptide of very similar structure, but with negligible affinity to integrin receptors, were also conducted as a control. Tissue uptake of 18F-FPRAD was used to estimate the non-specific uptake of the RGD peptides. We also estimated specific binding and non-specific binding by calculating “macro” parameters such as binding potential (BpND) and volumes of distribution (VT). Furthermore, parametric maps were obtained by Logan graphical analysis with reference tissue at voxel level for quantitative comparison between monomeric and dimeric RGD tracers.
MATERIALS AND METHODS
Preparation of 4-nitrophenyl 18F-2-fluoropropionate
4-Nitrophenyl 18F-2-fluoropropionate (18F-NPFP) was prepared on a GE TRACERLab FX F-N module according to a published procedure [25] with some modifications. Briefly, 5 mg of ethyl 2-bromopropionate in 0.5 mL of acetonitrile was reacted with anhydrous 18F-fluoride containing 15.0 mg of K-222 and 3.5 mg of potassium carbonate to form ethyl 18F-2-fluoropropionate. The radioactive ester was hydrolyzed to the corresponding carboxylic acid with 0.2 N KOH and then converted to 4-nitrophenyl 18F-2-fluoropropionate (18F-NPFP) with 20 mg of bis-4-nitropenyl carbonate (BNPC) in acetonitrile. The final product was purified with HPLC on a semi-prep Phenomenex Luna C18 column running at 5 mL/min with 40% acetonitrile/water containing 0.1% trifluoroacetic acid. The desired product was collected and trapped on a Waters Sep-Pak Plus C18 cartridge and eluted with 1 mL of methylene chloride.
Preparation of 18F-FPRGD
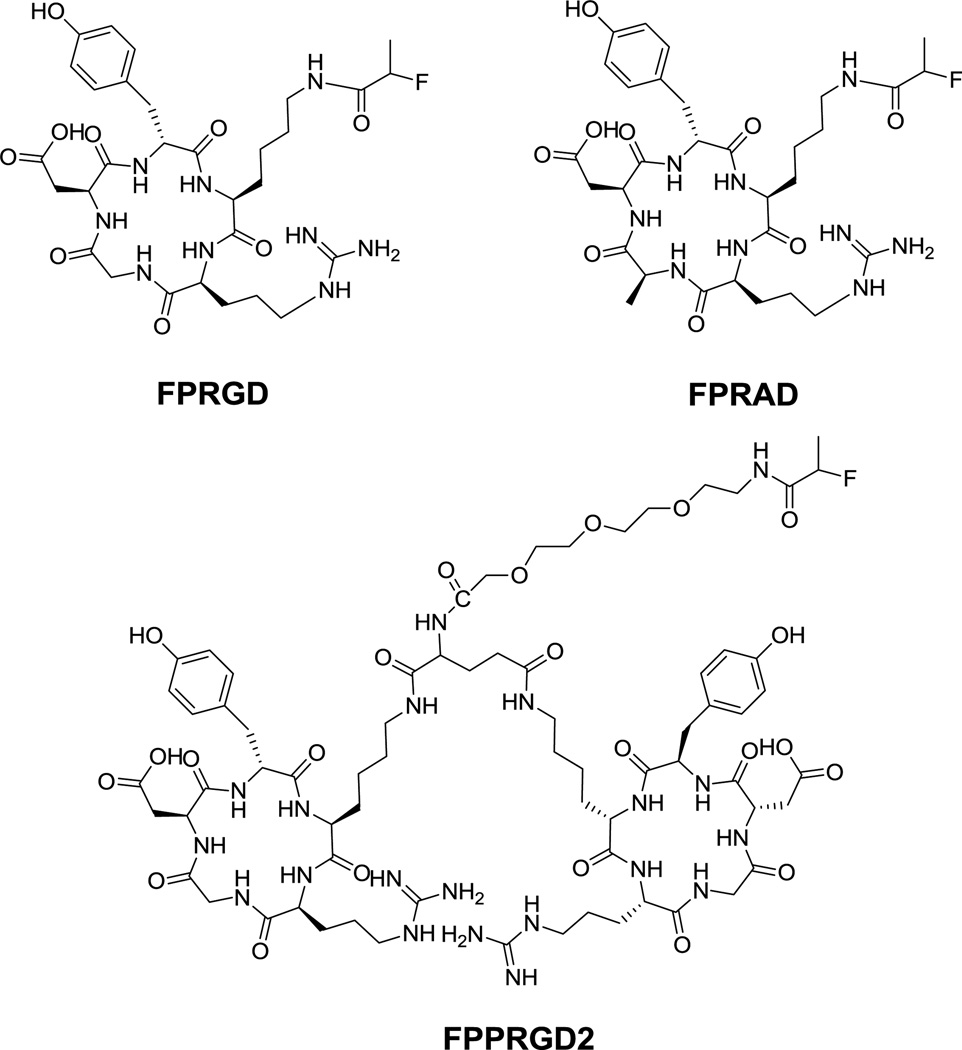
Methylene chloride was removed from the 18F-NPFP solution with argon flow at room temperature and 1.0 mg of c(RGDyK) in 0.1 mL of dimethyl sulfoxide containing 20 µL of diisopropylethylamine was added and heated at 100 °C for 10 min. The reaction mixture was cooled, diluted with 0.7 mL of water containing 25 µL of acetic acid, and injected onto a semi-prep HPLC column (Vydac C18) running a linear gradient starting from 5% A (0.1% TFA in acetonitrile) and 95% B (0.1% in water) for 2 min and increasing A to 65% at 32 min at 5 mL/min. The radioactive peak at retention time of 13.3 min was collected, diluted with 10 mL water, and the product trapped on a Varian Bond Elut C18 column (100 mg). The radioactivity trapped on the C18 column was eluted with 0.3 mL of 1 mM HCl ethanol solution and the eluate evaporated with argon flow. The residue was re-dissolved in normal saline for use in animal experiments. 18F-FPRAD and 18F-FPPRGD2 (Fig. 1) were prepared with similar procedure with HPLC retention time of 15.3 min and 13.3 min, respectively.
Fig. 1.
Schematic structures of RGD monomer FP-c(RGD)yK (FPRGD), dimer FP-PEG3-E[c(RGDyK)]2 (FPPRGD2), and FP-c(RADyK) (FPRAD).
Tumor model
The MDA-MB-435 tumor model, which expresses medium level of integrin αvβ3, was chosen for dynamic PET imaging [26]. The MDA-MB-435 cell line was purchased from the American Type Culture Collection (ATCC). The cells were grown in Leibovitz’s L-15 medium supplemented with 10% (v/v) fetal bovine serum at 37°C under 100% air atmosphere. The tumor model was established by injection of 5×106 cells into the left mammary fat pad of each female athymic nude mouse at 5–6 weeks of age (Harlan Laboratories). Tumor growth was monitored by caliper measurements three times a week after the tumors are palpable. The mice were used for PET imaging when the tumor volume reached about 300 mm3 (about 3 weeks after inoculation). The tumor volume was determined as the formula: V=a × (b2)/2, where a and b are the length and width of each tumor, respectively, in mm. All procedures in this animal study were conducted under a protocol approved by the NIH Clinical Center Animal Care and Use Committee (ACUC). Moreover, all mice were maintained in a specific pathogen-free facility in accordance with the requirements of the ACUC.
Dynamic PET imaging
Dynamic PET scans were performed using an Inveon microPET/CT scanner (Siemens Medical Solutions). The animals underwent 15-min CT scans followed by the dynamic PET scan using the same animal bed for ROI quantification. Each MDA-MB-435 tumor-bearing mouse was placed at the center of the field of view (FOV) focusing on the tumor location, where the highest detection sensitivity can be achieved. Sixty-min dynamic PET data acquisitions were performed following tail-vein injection of ~3.7 MBq (100 µCi) of radiotracer (18F-FPRGD, 18F-FPPRGD2, or 18F-FPRAD) under isoflurane anesthesia. Monomer RGD peptide c(RGDyK) was injected 10 min before scanning for the blocking studies. During the scanning, the body temperature of mice was maintained by a thermostat-controlled thermal heater. PET images were reconstructed with 2 iterations of 3-dimensional ordered-subsets expectation maximum (3D OSEM) with 14 subsets, followed by 18 iterations maximum a posteriori (MAP) algorithm with a smoothing parameter of 0.1. Frame rates were 10×30 s, 5×60 s, 5×120 s and 10×240 s.
ROI quantification and derivatives of time activity curves
In dynamic PET image analysis, regions of interest (ROIs) were drawn manually on individual tumor and correlative organs with Inveon Research Workplace (IRW) 3.0. The time–activity curves were derived by superimposing the ellipsoid volume of interest (VOI) to the target organs of each time frame of the entire 60-min dynamic image sequence. PET/CT fused images were acquired for accurate VOI quantification. The value of each time point represents the overall concentration of radioactivity in the tissue. The activity concentrations were determined by the mean pixel values within each VOI, which were converted to µCi/mL by using a calibration constant. Assuming the tissue density of ~1 g/mL, the VOI activity was converted to µCi/g and normalized as percent injected dose per gram (%ID/g).
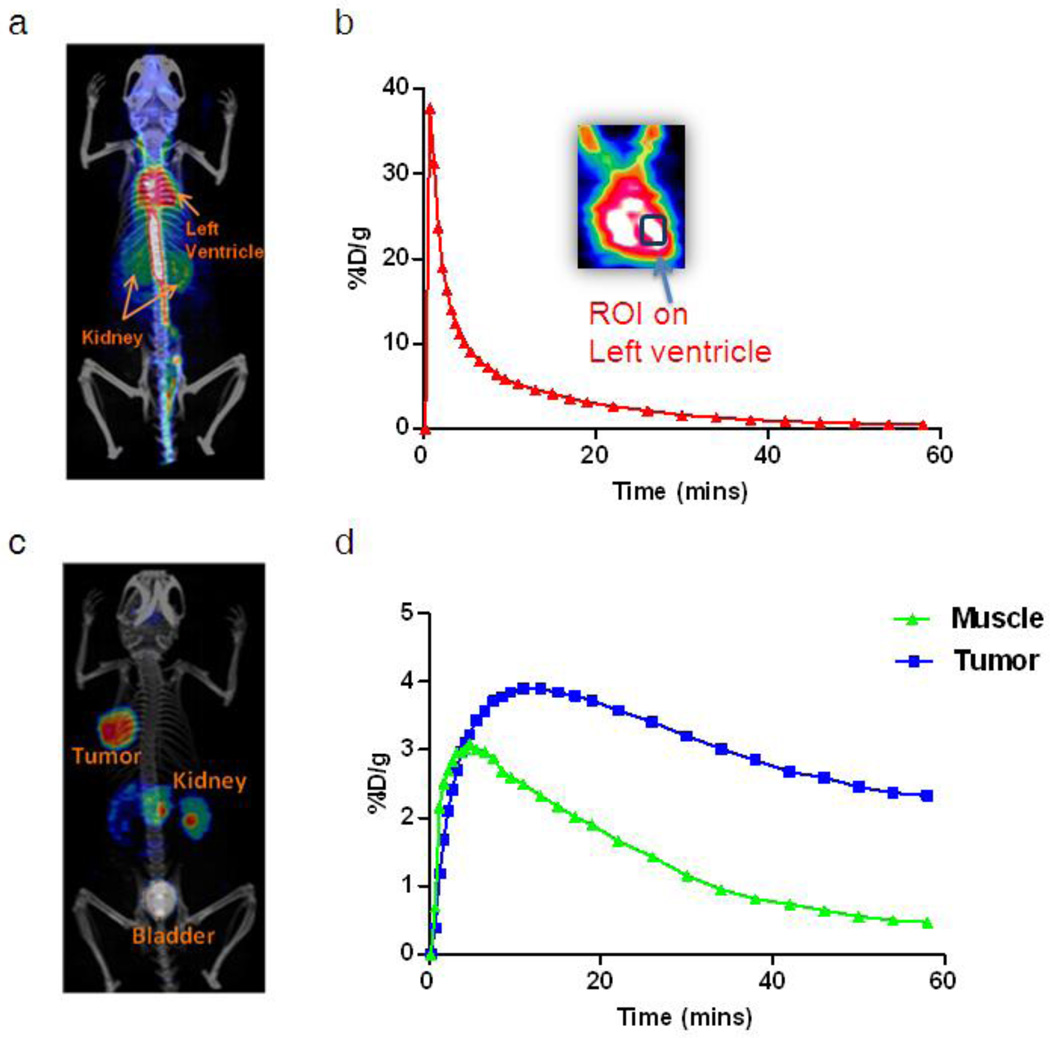
Heart could be identified clearly in the PET images. A representative 2D projection of PET image fused with CT image for left ventricle identification was shown in Fig. 2a. The corresponding blood input function (red curve) with zoomed coronal image at left ventricle was shown in Fig. 2b. The arterial blood input function was estimated by drawing a VOI in the region of left ventricle on reconstructed PET/CT images at the 0.5 min frame (the second frame of dynamic PET image series). It permits the extraction of the input function from the left ventricle while keeping the peak of tracer concentration in blood with good accuracy. Tumor and muscle VOI were determined in the last frame of the 60 min dynamic PET images (Fig. 2c). A region of muscle contralateral to the tumor was selected as the reference tissue (the region with the same blood input function but without specific binding). The corresponding time courses are shown in Fig. 2d. The impact of partial volume effect and spillover may slightly affect the accuracy of evaluation but was ignored in this study. Because of the high performance of the Inveon scanner, partial volume effect for the tumor region of interest analysis was also considered to be negligible.
Fig. 2.
a A representative PET/CT fusion 2D projection image at 0.5 min (the second frame of dynamic PET image series). b A blood time activity curve is extracted by drawing a region of interest over the left ventricle. c A representative PET/CT fusion 2D projection image at 60 min (the last frame of dynamic PET image series). d Time activity curves corresponding to the regions of tumor and muscle.
Kinetic modeling and parameter estimation
Kinetic analysis was performed by importing the TACs into a three-compartment model [27]. The three-compartment model describes RGD tracer kinetics in the tumor region, where Cp represents tracer concentration in the arterial blood plasma, Ct represents the free or non-specific binding component in the interstitial and intracellular space and Cm represents integrin specific component of the RGD tracer. The transport and binding rates of the tracer are: transfer from arterial plasma to tissue (K1 [mL/g/min]), clearance from tissue (k2 [1/min]), on-rate for specific binding (k3 [1/min]), and target dissociation rate (k4 [1/min]).
Model equations are illustrated as:
| Eq. (1) |
| Eq. (2) |
Values of K1–k4 were determined by fitting the models to the time-activity curves. The arterial blood input function determined by ROI quantification in the left ventricle was used as Cp in the model equation calculation. To minimize the sum of the residuals, the efficiency of fitting was assessed with the Akaike Information Criterion (AIC) [28].
Some combinations of parameters were also calculated, such as binding potential (BpND, equation 3) that reflects the binding affinity, and volume of distribution (VT) that reflects the tissue-to-plasma concentration ratio. VT can be regarded as the sum of specific and nonspecific distribution in the tissue. Parametric maps were generated by applying kinetic modeling at voxel level [24, 29].
BpND is defined as [30]:
| Eq. (3) |
VT can be divided into two elements according to the pharmacokinetics: specific volume of distribution (VS) and non-displaceable (VND), that is, nonspecific volume of distribution:
| Eq. (4) |
| Eq. (5) |
Total volume of distribution:
| Eq. (6) |
Where K1, k2, k3, and k4 were calculated by fitting the model to 60-min dynamic PET data [24].
Parametric map estimation
Voxel-wise parametric mapping was applied to whole body images by using the Logan graphical analysis. The parametric maps of volume of distribution (VT) were generated for each group by using blood input function. And the reference tissue model was applied for the parametric imaging of binding potential (BpND). The VT parametric map was calculated using equation 7 below.
| Eq. (7) |
The BpND map was calculated using equations 8, and the value of binding potential is BpND = DVR-1.
| Eq. (8) |
Statistical Analysis
Quantitative kinetic parameters determined from dynamic PET data were expressed as means ± SD. Differences between either blocked and unblocked or dimeric and monomeric RGD groups were evaluated using unpaired Student t test. P values less than 0.05 were considered statistically significant.
RESULTS
Time-activity curves
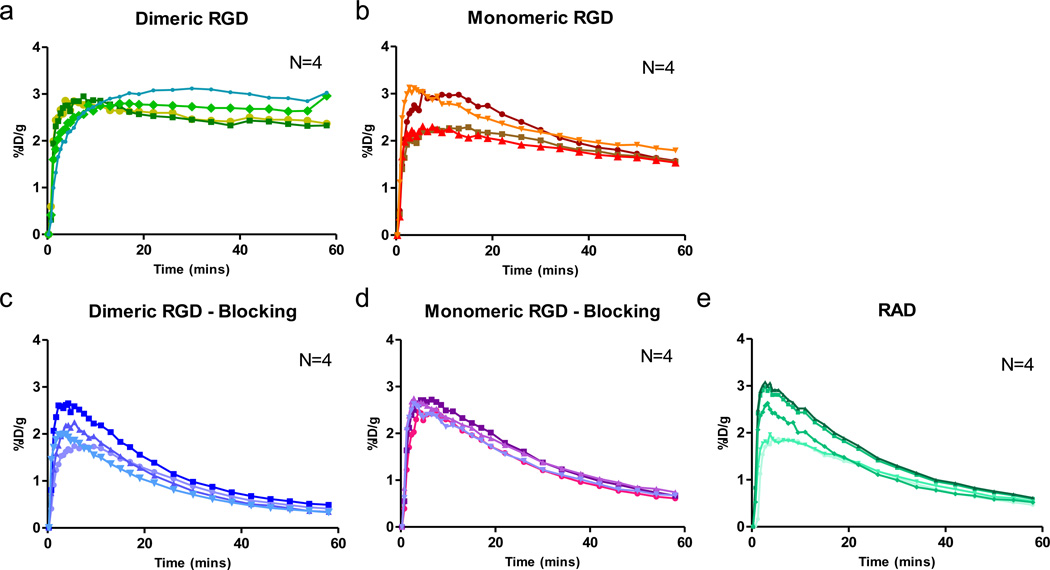
The time-activity curves of tumor uptake after administration of RGD dimer (18F-FPPRGD2), monomer (18F-FPRGD) or RAD (18F-FPRAD) and the corresponding blocking studies were analyzed and presented in Fig. 3. Without a blocking dose, high and rapid tumor tissue uptake was observed that reached a plateau after 30 min. The tumor uptake in the blocked group decreased continuously over time and was extremely low at late time points. Compared with the monomeric RGD, dimeric RGD showed much slower tumor washout. As a control, RAD peptide showed the most rapid tumor washout since there was no specific binding to integrin.
Fig. 3.
Tumor time-activity curves derived from 60-min dynamic PET scans for (a) unblocked group of mice after administration of dimeric RGD peptide tracer 18F-FPPRGD2, (b) unblocked group of monomeric RGD peptide tracer 18F-FPRGD, (c) blocked group of dimeric RGD, (d) blocked group of monomeric RGD, and (e) 18F-FPRAD control. (n = 4/group).
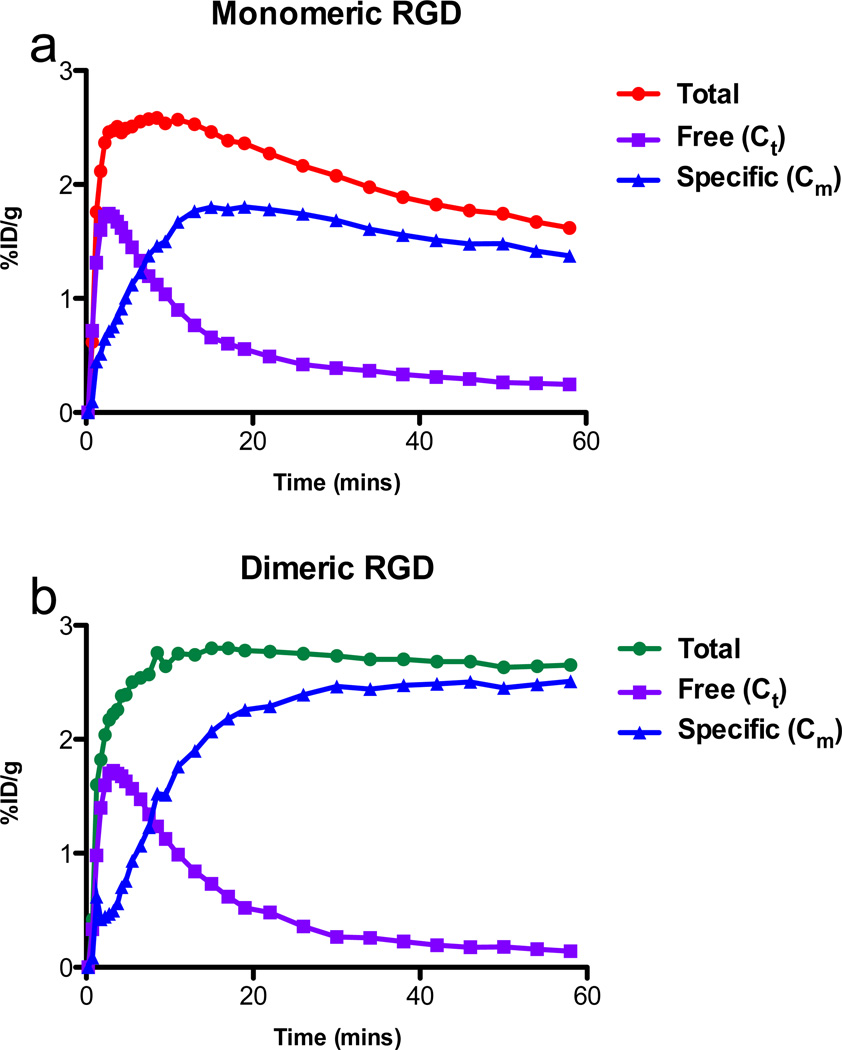
From the model fitting, non-specific (Ct) and specific binding (Cm) components were separated from the tumor uptakes of dimeric and monomeric RGD tracers, respectively (Fig. 4). The curves representing specific binding revealed that RGD dimer has increasing specific uptake in the tumor throughout the total time course of the experiment, and the rate of increase is higher than that of monomer. The specific bound component of dimeric RGD in the tumor region is almost twice as much as that of the monomer at 1-h. Both dimeric and monomeric groups showed similar non-specific (free) time curves with RAD.
Fig. 4.
Mean time activity curves of RGD monomer (a) and dimer (b). Tumor total uptake, free or non-specific binding tracer separated by model fitting.
Binding potential and volumes of distribution
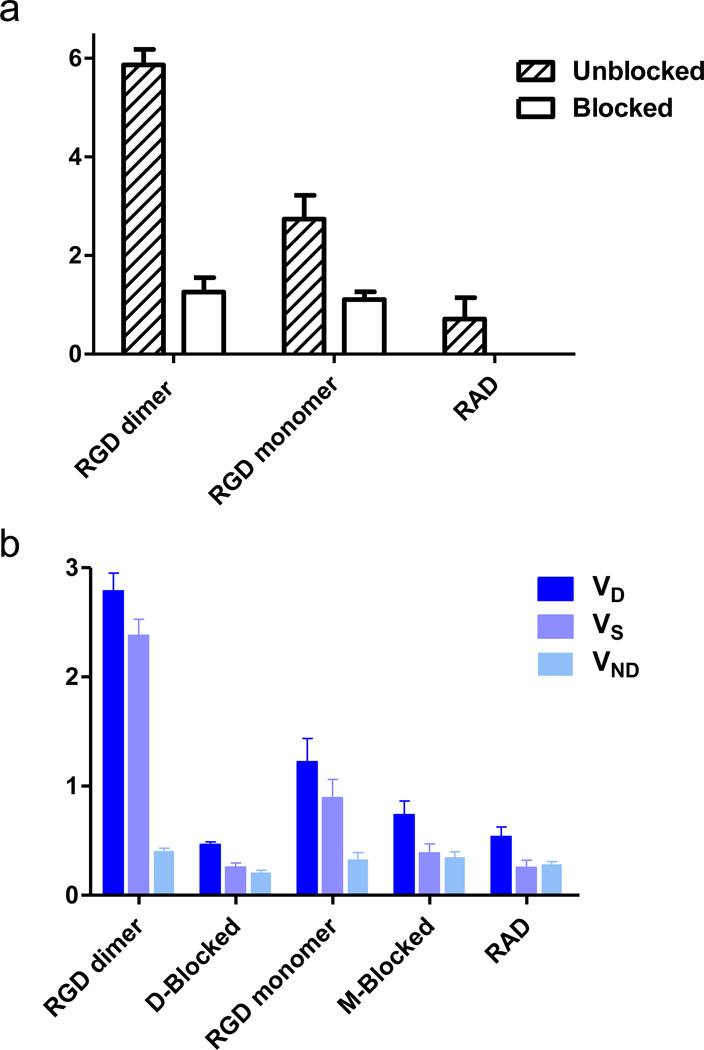
As shown in Fig. 5a, dimeric RGD showed significantly higher BpND (5.87 ± 0.31) than that of the monomeric analog (2.75 ± 0.048, p = 0.0022, n = 4/group). The dimer showed more than 2 times higher BpND value than the monomer, which is greater than the difference in %ID/g uptake (dimer is 1.5 times as much as monomer) measured from static images [31]. After blocking with an excess amount of unlabeled RGD peptide, a significant decrease in BpND was found in tumor-bearing mice administered either 18F-FPPRGD2 (~80% decreased, p < 0.001) or 18F-FPPRGD (~60% decreased, p < 0.01). Similar to the blocking studies, the RAD control group showed the lowest BpND (0.89 ± 0.19).
Fig. 5.
a Binding potential (BpND) of 18F-labeled RGD peptide tracers. b volumes of distribution (VT) of 18F-labeled RGD peptide tracers. The binding potential was calculated as BpND = k3/k4 reflecting the binding affinity, and the volume of distribution (VT = K1/k2(1+ k3/k4)) reflects the tissue-to-plasma concentration ratio. VT can be regarded as the sum of specific (VS = K1·k3/(k2·k4)) and nonspecific (VND = K1/k2) distribution.
By applying kinetic modeling, the total volume of distribution (VT) could be separated into non-displaceable (VND) and specific (VS) components to enable accurate assessment of the magnitude of specific binding of the two RGD compounds. Fig. 5b plots the mean ± SD of the total VT, specific VS and non-specific VND components for blocking and unblocking studies. The individual parameter values of K1, k2, k3 and k4 were summarized in Table 1. As shown, VS was the dominant component of the total distribution volume in the unblocked group and decreased in the blocked group.18F-FPPRGD2 also showed significantly higher VS than 18F-FPRGD (dimer VS = 2.38 ± 0.15, monomer VS = 0.90 ± 0.17, p = 0.0013, n = 4/group), similar to the dimer and monomer BpND calculations. Little difference in VND was found among different groups, suggesting comparable non-specific binding in tumor region.
TABLE 1.
Estimated Parameter Values for Compartmental Model Fitting
| Group | K1 | k2 | k3 | k4 | BpND | VT | VS | VND |
|---|---|---|---|---|---|---|---|---|
| RGD Dimer | ||||||||
| Unblock | 0.130±0.048 | 0.316±0.097 | 0.098±0.028 | 0.017±0.004 | 5.865±0.312 | 2.788±0.163 | 2.381±0.147 | 0.406±0.024 |
| Blocked | 0.082±0.031 | 0398±0.167 | 0.064±0.023 | 0.050±0.009 | 1.257±0.293 | 0.467±0.021 | 0.258±0.038 | 0.209±0.020 |
| RGD Monomer | ||||||||
| Unblock | 0.147±0.062 | 0.461±0.224 | 0.096±0.065 | 0.033±0.017 | 2.747±0.483 | 1.224±0.213 | 0.895±0.166 | 0.329±0.063 |
| Blocked | 0.276±0.256 | 0.735±0.587 | 0.051±0.016 | 0.046±0.012 | 1.110±0.155 | 0.739±0.125 | 0.389±0.083 | 0.350±0.048 |
| RAD | ||||||||
| 0.095±0.030 | 0.334±0.092 | 0.047±0.011 | 0.055±0.020 | 0.893±0.192 | 0.538±0.089 | 0.255±0.067 | 0.283±0.026 | |
Parametric mapping
As a graphical analysis method, Logan plot is robust for kinetic modeling, which is computationally appropriate for parametric mapping [32]. Moreover, it is applicable to ligands binding reversibly to receptors, such as cyclic RGD peptides to integrin αvβ3. By applying the Logan graphical analysis at the voxel level, we obtained the parametric maps for volumes of distribution (VT) and binding potential (BpND), respectively (Fig. 6).
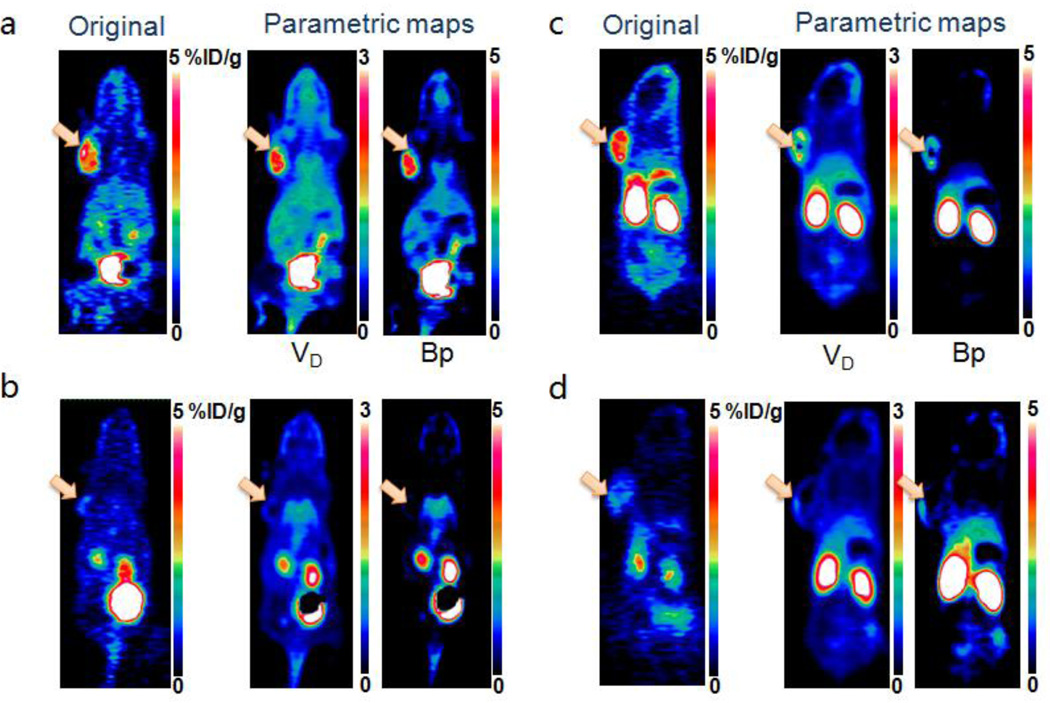
Fig. 6.
(a, b) Representative original static PET images at 60 min (left), parametric maps of volume of distribution (middle) and binding potential (right) for unblocked (a) and blocked (b) groups of mice after administration of dimeric RGD peptide tracer 18F-FPPRGD2. (c, d) Representative original static PET images at 60 min (left), parametric maps of volume of distribution (middle) and binding potential (right) for unblocking (c) and blocking (d) groups of mice after administration of monomeric RGD peptide tracer 18F-FPRGD. The arrows point to tumors.
In parametric maps, dimeric RGD showed significantly higher VT and BpND in the tumor region than the monomer, which is greater than the difference in %ID/g values determined from the original static images at 60 min with the same image scales. Extremely low binding potential in the tumor region in both blocked groups was found as compared with that in the unblocked groups. Parametric maps also provided higher tumor-to-muscle contrast ratio for BpND maps (70.8 ± 12.5) than the original static images at 1-h time point (5.29 ± 1.18, p = 0.035), e.g. in dimeric RGD unblocked group.
DISCUSSION
One main goal of molecular imaging is to visualize and quantify target expression level non-invasively by applying the molecular probe in a real-time manner. The well-developed PET image evaluation parameters with static images, such as standard uptake value (SUV) or %ID/g are widely used for molecular image quantification. However, besides the tracer binding affinity, the ability to convert tissue uptake into target concentration is unavoidably affected by other factors such as heterogeneity of blood supply, vascular permeability, and interstitial fluid pressure. Thus, there is an urgent need to delineate specific uptake from the non-specific accumulation of radioactive tracers in the tumor region, which will definitely facilitate tumor diagnosis and treatment monitoring.
By using dynamic PET imaging, the time-activity curves of various organs or tissue can be obtained and characterized. Dynamic PET imaging provides more information than a static image and may be useful in revealing the microenvironment of target tissue. In the kinetic analysis, the uptake of radiotracer into the tumor microenvironment can be modeled as different compartments, such as the homogeneous tracer concentration in plasma, tissue interstitial space or tumor cells. The tracer concentration in tumor tissue can be divided into two elements according to its molecular interaction in tissues, free and bound, which is also regarded as non-specific binding and specific binding. Usually, the tracer concentrations of three compartments in the compartment model are regarded as the amount of tracer trapped in tissue regions detected by the static PET image. By applying dynamic PET imaging and kinetic modeling, we can separate the specific and non-specific bindings. The kinetics of tracer uptake is described by the kc parameters representing the exchange rates between compartments. With appropriate model definition and dynamic curve fitting, kinetic modeling could quantitatively assess the tracer binding affinity for the receptors in vivo. In this study, by taking the intensively investigated RGD-integrin system as an example, we implemented a three compartment model to quantitatively estimate the kinetics of dimeric and monomeric RGD peptide tracers in an MDA-MB-435 tumor model.
According to the definition binding potential (BpND = k3/k4), the BpND value represents the specific binding affinity of tracer. The measurement of BpND could be affected by either tracer specific binding affinity or available receptor density. When comparing the BpND value of RGD dimer with monomer in the same xenograft, the available receptor density in each individual mouse could be regarded as the same, and then the resulting BpND only reflects the difference in the binding affinity of the tracers for integrin. According to the IC50 values in cell binding assay in our previous studies, the RGD dimer showed 3.6–3.8 fold higher binding affinity than the monomer [31, 33]. However, in static PET image quantification, the tumor uptake of the RGD dimer was only 1.5 times as much as that of the monomer. Comparing the binding potentials of dimeric and monomeric RGD peptides in Fig. 5a, significantly higher BpND of the dimeric RGD (5.87 ± 0.31) was found than that of the monomeric analog (2.75 ± 0.048). The RGD dimer showed more than a two-fold higher binding affinity than the monomer. Thus the BpND appears to be more sensitive than the differences illustrated in static images and much closer to the comparison of cell binding assay results. Thus, even a minor subtraction for non-specific binding provides greater sensitivity than a conventional %ID/g analysis, where the difference between the dimer and the monomer pertains only to the specific binding. On the other hand, the comparison between radioligand only and blocked studies for either RGD dimer or monomer group implies that the binding potential is an efficient indicator for available receptor density. Using the same RGD tracer, the blocking study resulted in significant decreases in binding potential.
Table 1 showed the values of transporting rates between the compartments for each tracer. It has been reported that dimeric and polymeric RGD peptides could enhance the receptor binding affinity through polyvalency effect, wherein the RGD sequence was locally increased [34]. The RGD dimer has comparable on-rate of specific binding (k3) but a two-fold lower off-rate (k4) compared with the monomer, which resulted in a higher binding potential of the RGD dimer. It is likely that the receptor binding of one RGD domain significantly enhances the local concentration of the other RGD domain within the dimer in the vicinity of the receptor, which may lead to an apparent slower rate of dissociation of the radiolabeled RGD dimer from integrin [19].
Furthermore, volume of distribution in the tumor region could be separated into specific and non-specific components and was demonstrated to be valuable for quantification. VT, the tissue to plasma concentration ratio, is the index of tracer present in the tumor tissue normalized by plasma tracer concentration when at equilibrium. VND (K1/k2) represents the non-specific binding and is determined by tracer influx and efflux rates between the tissue and plasma. VS (K1k3/(k2k4)) demonstrates the specific binding concentration and is affected by not only the specific binding affinity of the tracer, and the number of available binding sites but also the perfusion and clearance rates. As shown in Fig. 5b, the VS of dimeric RGD is about 2.6 times higher than the monomeric RGD, which is also significantly greater than the difference in %ID/g derived from static images. VND is about the same in both dimeric and monomeric groups, and is similar to that of RAD, which only shows non-specific binding in the tumor region. It is clear that the pre-injected blocking agent only reduced the specifically bound tracer signal in the PET images but not the perfusion component among all the groups. The kinetic parameters, especially the macro-parameters, are better indicators of tumor specific binding and enable more sensitive evaluation of tracer kinetics in tumors.
In previous studies [24, 35], the performances of two-compartment model, three-compartment reversible and irreversible were compared for receptor-ligand binding, e.g. RGD to integrin, to identify the most appropriate model. The discrimination process and evaluation was conducted by using Akaike information criterion (AIC). The three-compartment reversible model yielded the lowest AICs for fitting tumor time activity curves in unblocked RGD studies, which indicated that both 18F-labeled RGD dimer and monomer kinetics could be assessed by a reversible receptor binding model. Although the AIC analysis suggested that the two-compartment model (two parameters: K1 and k2) is sufficient to describe the blocked tracer and RAD kinetics, the three-compartment model appears to provide more accurate information. This observation is reasonable considering that the integrin receptors were incompletely blocked in the blocking experiments and RAD peptide may have very low affinity for integrin, resulting in low levels of bound tracers. In order to compare BpND = k3/k4 and VT = f(K1, k2, k3, k4) more objectively, we used three-compartment reversible model to estimate the kinetic parameters of the blocked and RAD studies.
The impact of spillover in regions constructed for extraction of arterial time activity curves is expected to be small, since the myocardium uptake of RGD is minimal. Direct arterial blood sampling is regarded as the gold standard of input function for kinetic modeling. Unfortunately, arterial blood sampling is technically quite challenging in small animal studies. To accurately assess the kinetics of RGD peptides, it may be more appropriate to apply curve fitting to the image derived input function which may require scaling by several blood samples at late time points.
In this study, we evaluated kinetic parameters for dimeric and monomeric RGD peptide tracers by using a three-compartment model and compared the binding potential and volumes of distribution derived from macro-parameters among all the text groups. The quantification of kinetic parameters may provide unique ways to assess the receptor density and specific concentration in the tumor region. To validate the correlation between the kinetic parameters and receptor density, different tumor models with varying receptor expression levels will be considered in our future studies. Furthermore, in our future studies the RAD data acquisition will be performed on the same animals to separate the RGD specific and non-specific binding concentration in the tumor region.
CONCLUSION
The pharmacokinetics of both monomeric and dimeric RGD peptide tracers were compared and the RGD dimer showed significantly higher binding affinity as assessed by the calculated binding potential (BpND) than the monomer analog in our in vivo study. Specific binding and non-specific binding uptake in the tumor region could be separated according to the macro kinetic parameters. Parametric maps of the macro-parameters at the voxel level modeling results in better tumor-to-muscle contrast and can potentially be used to assess tumor heterogeneity. Kinetic parameters may allow more sensitive and detailed quantification than simple SUV analysis for potential tumor diagnosis and therapy response monitoring applications.
ACKNOWLEDGMENTS
This work was supported in part, by the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), the International Cooperative Program of the National Science Foundation of China (NSFC) (81028009), and NSFC Grants (60972099, 61027006). NG was partially sponsored by the China Scholarship Council (CSC).
Footnotes
Conflict of Interest Disclosure. The authors declare that they have no conflict of interest.
REFERENCES
- 1.Beer AJ, Schwaiger M. Imaging of integrin alphavbeta3 expression. Cancer Metastasis Rev. 2008;27:631–644. doi: 10.1007/s10555-008-9158-3. [DOI] [PubMed] [Google Scholar]
- 2.Kimura RH, Cheng Z, Gambhir SS, et al. Engineered knottin peptides: a new class of agents for imaging integrin expression in living subjects. Cancer Res. 2009;69:2435–2442. doi: 10.1158/0008-5472.CAN-08-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 4.Haubner R, Wester HJ, Weber WA, et al. Noninvasive imaging of αvβ3 integrin expression using 18F-labeled RGD-containing glycopeptide and positron emission tomography. Cancer Res. 2001;61:1781–1785. [PubMed] [Google Scholar]
- 5.Niu G, Chen X. Why integrin as a primary target for imaging and therapy. Theranostics. 2011;1:30–47. doi: 10.7150/thno/v01p0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beer AJ, Kessler H, Wester HJ, et al. PET imaging of integrin αvβ3 expression. Theranostics. 2011;1:48–57. doi: 10.7150/thno/v01p0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X. Multimodality imaging of tumor integrin αvβ3 expression. Mini Rev Med Chem. 2006;6:227–234. doi: 10.2174/138955706775475975. [DOI] [PubMed] [Google Scholar]
- 8.Dumont RA, Deininger F, Haubner R, et al. Novel 64Cu- and 68Ga-labeled RGD conjugates show improved PET imaging of αvβ3 integrin expression and facile radiosynthesis. J Nucl Med. 2011;52:1276–1284. doi: 10.2967/jnumed.111.087700. [DOI] [PubMed] [Google Scholar]
- 9.Mittra ES, Goris ML, Iagaru AH, et al. Pilot pharmacokinetic and dosimetric studies of 18F-FPPRGD2: a PET radiopharmaceutical agent for imaging αvβ3 integrin levels. Radiology. 2011;260:182–191. doi: 10.1148/radiol.11101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin FT, Shen B, Liu S, et al. First experience with clinical-grade [18F]FPP(RGD)2: An automated multi-step radiosynthesis for clinical PET studies. Mol Imaging Biol. 2011 Mar 12; doi: 10.1007/s11307-011-0477-3. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battle MR, Goggi JL, Allen L, et al. Monitoring tumor response to antiangiogenic sunitinib therapy with 18F-fluciclatide, an 18F-labeled αvβ3-integrin and αvβ5-integrin imaging agent. J Nucl Med. 2011;52:424–430. doi: 10.2967/jnumed.110.077479. [DOI] [PubMed] [Google Scholar]
- 12.Liu S. Radiolabeled cyclic RGD peptides as integrin αvβ3-targeted radiotracers: maximizing binding affinity via bivalency. Bioconjug Chem. 2009;20:2199–2213. doi: 10.1021/bc900167c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schottelius M, Laufer B, Kessler H, et al. Ligands for mapping αvβ3-integrin expression in vivo. Acc Chem Res. 2009;42:969–980. doi: 10.1021/ar800243b. [DOI] [PubMed] [Google Scholar]
- 14.Cai W, Chen X. Preparation of peptide-conjugated quantum dots for tumor vasculature-targeted imaging. Nat Protoc. 2008;3:89–96. doi: 10.1038/nprot.2007.478. [DOI] [PubMed] [Google Scholar]
- 15.Jeong JM, Hong MK, Chang YS, et al. Preparation of a promising angiogenesis PET imaging agent: 68Ga-labeled c(RGDyK)-isothiocyanatobenzyl-1,4,7-triazacyclononane-1,4,7-triacetic acid and feasibility studies in mice. J Nucl Med. 2008;49:830–836. doi: 10.2967/jnumed.107.047423. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y, Zhang X, Xiong Z, et al. microPET imaging of glioma integrin αvβ3 expression using 64Cu-labeled tetrameric RGD peptide. J Nucl Med. 2005;46:1707–1718. [PubMed] [Google Scholar]
- 17.Sun X, Yan Y, Liu S, et al. 18F-FPPRGD2 and 18F-FDG PET of response to Abraxane therapy. J Nucl Med. 2011;52:140–146. doi: 10.2967/jnumed.110.080606. [DOI] [PubMed] [Google Scholar]
- 18.Liu S, Liu Z, Chen K, et al. 18F-labeled galacto and PEGylated RGD dimers for PET imaging of αvβ3 integrin expression. Mol Imaging Biol. 2010;12:530–538. doi: 10.1007/s11307-009-0284-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Xiong Z, Wu Y, et al. Quantitative PET imaging of tumor integrin alphavbeta3 expression with 18F-FRGD2. J Nucl Med. 2006;47:113–121. [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Liu S, Wang F, et al. Noninvasive imaging of tumor integrin expression using 18F-labeled RGD dimer peptide with PEG4 linkers. Eur J Nucl Med Mol Imaging. 2009;36:1296–1307. doi: 10.1007/s00259-009-1112-2. [DOI] [PubMed] [Google Scholar]
- 21.Shoghi KI. Quantitative small animal PET. Q J Nucl Med Mol Imaging. 2009;53:365–373. [PubMed] [Google Scholar]
- 22.Hong YT, Beech JS, Smith R, et al. Parametric mapping of [18F]fluoromisonidazole positron emission tomography using basis functions. J Cereb Blood Flow Metab. 2011;31:648–657. doi: 10.1038/jcbfm.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beer AJ, Haubner R, Goebel M, et al. Biodistribution and pharmacokinetics of the alphavbeta3-selective tracer 18F-galacto-RGD in cancer patients. J Nucl Med. 2005;46:1333–1341. [PubMed] [Google Scholar]
- 24.Ferl GZ, Dumont RA, Hildebrandt IJ, et al. Derivation of a compartmental model for quantifying 64Cu-DOTA-RGD kinetics in tumor-bearing mice. J Nucl Med. 2009;50:250–258. doi: 10.2967/jnumed.108.054049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang M, Gao H, Zhou Y, et al. 18F-labeled GRPR agonists and antagonists: A comparative study in prostate cancer imaging. Theranostics. 2011;1:220–229. doi: 10.7150/thno/v01p0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quan Q, Yang M, Gao H, et al. Imaging tumor endothelial marker 8 using an 18F-labeled peptide. Eur J Nucl Med Mol Imaging. 2011;38:1806–1815. doi: 10.1007/s00259-011-1871-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phelps ME, Huang SC, Hoffman EJ, et al. Tomographic measurement of local cerebral glucose metabolic rate in humans with 18F-2-fluoro-2-deoxy-D-glucose: validation of method. Ann Neurol. 1979;6:371–388. doi: 10.1002/ana.410060502. [DOI] [PubMed] [Google Scholar]
- 28.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr AC. 1974:716–723. [Google Scholar]
- 29.Watabe H, Ikoma Y, Kimura Y, et al. PET kinetic analysis--compartmental model. Ann Nucl Med. 2006;20:583–588. doi: 10.1007/BF02984655. [DOI] [PubMed] [Google Scholar]
- 30.Innis RB, Cunningham VJ, Delforge J, et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27:1533–1539. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- 31.Li ZB, Chen K, Chen X. 68Ga-labeled multimeric RGD peptides for microPET imaging of integrin αvβ3 expression. Eur J Nucl Med Mol Imaging. 2008;35:1100–1108. doi: 10.1007/s00259-007-0692-y. [DOI] [PubMed] [Google Scholar]
- 32.Tomasi G, Turkheimer F, Aboagye E. Importance of quantification for the analysis of PET data in oncology: Review of current methods and trends for the future. Mol Imaging Biol. 2011 Aug 13; doi: 10.1007/s11307-011-0514-2. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 33.Li ZB, Cai W, Cao Q, et al. 64Cu-labeled tetrameric and octameric RGD peptides for small-animal PET of tumor αvβ3 integrin expression. J Nucl Med. 2007;48:1162–1171. doi: 10.2967/jnumed.107.039859. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Tohme M, Park R, et al. Micro-PET imaging of αvβ3-integrin expression with 18F-labeled dimeric RGD peptide. Mol Imaging. 2004;3:96–104. doi: 10.1162/15353500200404109. [DOI] [PubMed] [Google Scholar]
- 35.Tomasi G, Kenny L, Mauri F, et al. Quantification of receptor-ligand binding with [18F]fluciclatide in metastatic breast cancer patients. Eur J Nucl Med Mol Imaging. 2011;38:2186–2197. doi: 10.1007/s00259-011-1907-9. [DOI] [PubMed] [Google Scholar]