Abstract
Background & objectives:
Tuberculosis is (TB) responsible for high morbidity and mortality worldwide. Cytokines play a major role in defense against Mycobacterium tuberculosis infection. Polymorphisms in the genes encoding the various pro- and anti-inflammatory cytokines have been associated with tuberculosis susceptibility. In this study we examined association of 25 sequence polymorphisms in six candidate cytokine genes namely IFNG, TNFB, IL4, IL1RA, IL1B and IL12 and their related haplotypes with risk of developing pulmonary tuberculosis (PTB) among north Indians.
Methods:
Pulmonary TB (n=110) patients and 215 healthy controls (HC) from north India were genotyped. Purified multiplex PCR products were subjected to mass spectrometry using Sequenom MassARRAY platform to generate the genotypes in a population-based case-control study.
Results:
Using multiple corrections, significant overall risk against PTB was observed at seven loci which included variants in IFNG at rs1861493 and rs1861494; IL1RA at rs4252019, IL4 variant rs2070874, IL12 variants rs3212220, rs2853694 and TNFB variant rs1041981. Analysis of gene structure revealed two haplotype blocks formed by IFNG variants rs1861493 and rs1861494. The TA haplotype was significantly over-represented (P=0.011) in the cases showing a two-fold risk in the current population (Odds ratio=1.59 CI=1.101 to 2.297) and TNFB variants at rs2229094 and rs1041981 contributed to two haplotypes which were in strong linkage disequilibrium (LD) with AT haplotype showing a three-fold risk (P=0.0011, Odds ratio=3, CI=0.1939 to 0.7445) of developing PTB in north Indians.
Interpretation & conclusions:
Our study showed six novel associations of cytokine gene variants with susceptibility to PTB in north Indians. Variants of IFNG and TNFB emerged as factors imposing a significant risk of developing PTB in north Indians apart from risk indicated by IL1RA, IL4 and IL12.
Keywords: Cytokine gene variant, haplotype, Mycobacterium tuberculosis, pulmonary tuberculosis, single nucleotide polymorphisms
Tuberculosis (TB) causes significant morbidity and mortality throughout the world1. The vast majority of individuals infected with Mycobacterium tuberculosis (up to 95%) remain healthy, probably because of mounting an effective immune response against M. tuberculosis. In 1949, Haldane proposed that the maintenance of multiple genes that confer relative susceptibilities on the host to infectious diseases would be favoured by evolution. In support of this hypothesis, certain populations appear to be at risk for both increased susceptibility to infection2 and progressive clinical disease due to mycobacteria3. Several case-control studies have identified association between TB and candidate genes potentially involved in immune response to TB4,5. A growing body of evidence supports a role of host genetic components in the development of tuberculosis. The observation of familial clustering of disease with higher concordance of tuberculosis disease in monozygotic versus dizygotic twins6, the ethnic clustering of tuberculosis disease with a higher prevalence of tuberculosis in individuals of recent African descent2, as well as the demonstration of both common polymorphisms and rare mutations which confer susceptibility to mycobacterial species in humans7 point significantly in this direction. These studies suggest that unique environment and natural selective factors may be responsible for the development of ethnic-specific host genetic factors associated with TB.
The first step in innate host defense is cellular uptake of M. tuberculosis, which involves different cellular receptors and humoral factors. The subsequent inflammatory response is regulated by the production of pro- and anti-inflammatory cytokines and chemokines. Interferon-gamma (IFN-γ one of the most important cytokines involved in macrophage activation, stimulating anti-tumour and anti-microbicidal activities as well as expression of MHC-II8,9. Interleukin-4 (IL-4), an anti-inflammatory cytokine has been implicated to downregulate IFN-γ, and thus has a deleterious effect on TB patients10. It also promotes the induction of Th2 cells11. IL-12, a heterodimeric pro-inflammatory cytokine produced by activated macrophages, monocytes, β-lymphocytes and dendritic cells is the principal Th1 response inducing cytokine11. This cytokine is important for sustaining a sufficient number of memory/effector Th1 cells to mediate long-term protection to intracellular pathogen. Like tumour necrosis factor-alpha (TNF-α), IL-1β is mainly produced by monocytes, macrophages, and dendritic cells12. In tuberculosis patients, IL-1β is expressed in excess13 and at the site of disease14. Implicated mainly in tuberculosis pleurisy, a usually self-resolving type of primary tuberculosis, one may hypothesize that an increased IL-1β /IL-1Rα ratio protects against a more severe form of tuberculosis.
TNF-β or lymhotoxin-alpha (LTα) is considered to be a proinflammatory cytokine and it is shown that secreted LTα is essential for the control of an intracellular bacterial infection15. Recently Allie et al16 suggested that LTα might not have a critical role in host defense to acute mycobacterial infection, independent of TNF, but certainly a contribution of LTα in the control of chronic M. tuberculosis infection is observed17.
Association studies from north India probing multiple loci across the spectrum of candidate cytokine genes are scanty. The present study, therefore, was aimed to bring in focus certain unexplored polymorphisms in the context of tuberculosis susceptibility in north Indian population. The role and importance of genetic background in tuberculosis has now become univocal with ethnicity playing a crucial role. Probing new loci relating to tuberculosis susceptibility could suggest novel approach in pharmacogenomics and therapy to combat this pathogen. Also it could provide an insight into predicting individual's genetic proneness to tuberculosis and of being future diagnostic tool for preventive therapy against tuberculosis.
Material & Methods
Study population: PTB patients above 18 yr of age (n=110) were enrolled randomly in the study between 2010-11 from Rajan Babu Institute of Pulmonary Medicine and Tuberculosis (RBIPMT), Kingsway Camp, New Delhi (India). The study was carried out in Department of Microbiology, V.P. Chest Institute, University of Delhi, Delhi. Enrolled patients were category I cases, clinically and radiologically (chest X-ray) diagnosed for pulmonary tuberculosis and confirmed by sputum microscopy and culture for Mycobacterium following the guidelines of Revised National TB Control Programme (RNCTP), Ministry of Health and Family Welfare, Government of India (http://www.tbcindia.nic.in). All patients were given free anti-tuberculosis drugs under DOTS (Directly Observed Treatment, short course) regimen of the Government of India. The mean age of PTB cases was 31.89 ± 2.6 yr while the ratio of male : female was 47:53.
Patients having any immunosuppressive presentation such as diabetes mellitus or HIV co-infection which are considered to be risk factors for tuberculosis development, and patients suspected to have extra-pulmonary tuberculosis along with pulmonary tuberculosis were excluded from the study. Structured questionnaires were used to document all other relevant information such as age, sex, ethnicity, socio-economic status, BCG vaccinations, and previous family history of tuberculosis. The healthy control (HC) group consisted of 215 randomly chosen nonconsanguineous BCG vaccinated students and laboratory personnel from the various departments of the University of Delhi who were willing to participate in the study with no signs, symptoms or history of previous mycobacterial infection. For HC mean age was 29.31 ± .82 yr and the ratio of male : female was 43:57.
Analysis of population stratification: Serious effort was made to avoid any false-positives arising as a result of population stratification. The self reported ethnicity of each subject and his/her parents was carefully considered. In addition, the genotype data were subjected to EIGENSTRAT principal component analysis for population stratification correction as illustrated by Price et al18.
All individuals were briefed about the study and a signed informed consent was obtained from the patient or his or her guardians before sample collection. The study was approved by the ethics committee of Vallabhbhai Patel Chest Institute, University of Delhi, India.
DNA extraction: Three ml of venous blood was collected in BD vacutainers containing ethylene diamine tetra acetic acid (EDTA) as anticoagulant and kept frozen until use. Genomic DNA was extracted from frozen whole blood using QiaAMP DNA kit (Qiagen, Germany). Extracted DNA was quantified by spectrophotometery, checked for purity and stored at -20°C until further analyses.
SNP selection and genotyping: Six candidate cytokine genes namely IFNG, TNFB, IL4, IL1RA, IL1B and IL12B, were selected owing to their suggested role in tuberculosis pathogenesis. All single nucleotide polymorphisms (SNPs) selected for genotyping were accessed from the public dbSNP (http://www.ncbi.nih.gov) and the HapMap (http://www.hapmap.org/). Most of the selected SNPs are from the intronic regions of the corresponding genes. We reasoned that not only the changes in the promoter but also of other unexplored regions of the gene may hamper its normal functioning leading to disease. The parameters taken into account while SNP selection were the frequency of <0.01 in dbSNP, reported allele frequency of at least 20 per cent in two world populations (from Hapmap), average spacing 1 kb but in closely spaced minor allele frequency was carefully considered. In addition, reported heterozygosity was considered in an effort to minimize selection of homozygous loci.
All SNPs were genotyped using the matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Sequenom Inc., USA). Assays for all SNPs were designed using SpectroDESIGNER software (Sequenom Inc., USA) All SNPs were genotyped using the iPLEX assays (www.sequenom.com/iplex). Briefly, as template, 5 ng of genomic DNA was used in a multiplex PCR reaction. The PCR product was further purified before the primer extension reaction to generate allele-specific base extension products. The base-extension products were detected in the MALDI-TOF mass spectrometer to determine genotypes.
Genetic and statistical analyses: Hardy-Weinberg equilibrium was calculated in both PTB cases and HC separately to ensure that the samples were within allelic population equilibrium by using Haploview v 4.2 (http://www.broad.mit.edu/mpg/haploview/). A stringent cut-off offered by the Haploview v 4.2 was used for further analysis (minimum genotype =75% and minimum minor allele frequency 0.0010). The samples and variations failing this test were not selected for further analysis. PLINK v 1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) was used to test for multiple comparison and P value after Bonferroni corrections was considered significant. Haplotype block generation was performed using the algorithm by Gabriel et al19 implemented in the Haploview software which was also used for initial association testing. The statistical significance of P value of haplotypes was assessed by permutation analysis (N=10,000) with Haploview v 4.2.
Genetic association testing was done using a 2 × 2 contingency table. Odds ratio, two tailed P value was calculated for alleles. 2 × 2 Computations were done using GraphPad Prism (version 5.00 for Windows, Graph Pad Software, San Diego California, USA; www.graphpad.com). Two-tailed P<0.05 was considered statistically significant.
Results
Table I shows the location and characteristics of the SNPs included in the study and Table II shows the associations after multiple corrections carried out using PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) which were found to be associated with susceptibility to PTB in north Indians in this study.
Table I.
Location and base-pair positions of single neucleotide polymorphisms (SNPs) of various cytokine genes passing the exclusion criteria and minor allele frequency (MAF) in controls
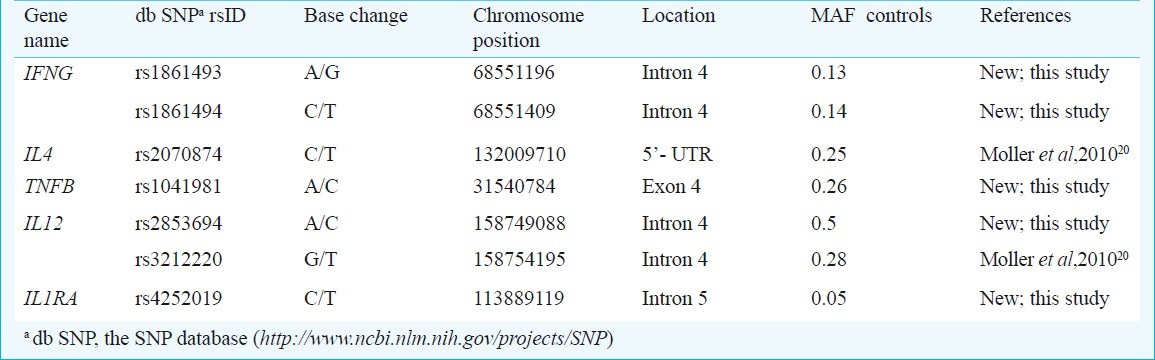
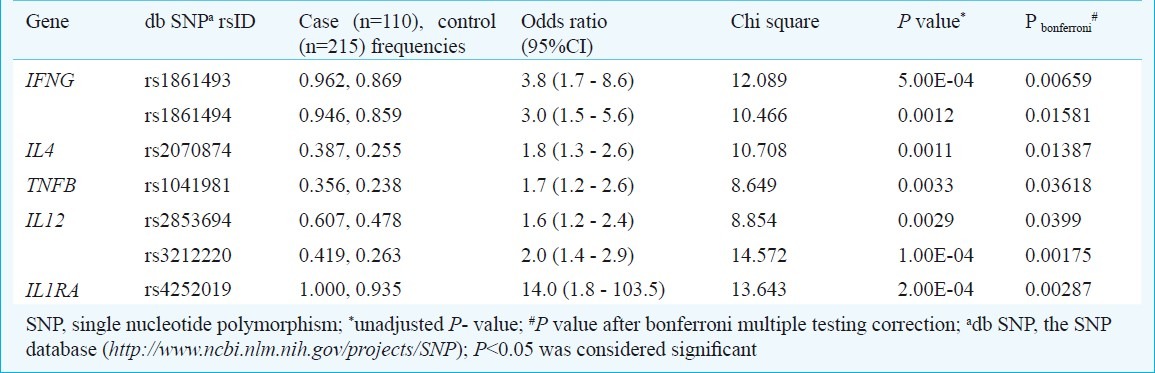
Table II.
Allelic associations in after adjustment for multiple testing
Population stratification correction: To access any underlying structure in the study population that could confound the apparent genetic association population stratification correction was carried out using Eigenstrat Principal Component analysis method as illustrated by Price et al18. The method models ancestry difference between cases and controls and any other compared group based on the supplied genotype data. Our cases and controls formed a homogenous group devoid of any stratification. According to Indian Genome Variation Consortium (IGVC)20 north Indians fall into Indo-European lineage. Our cases and controls matched with supplied marker data of Indo-European ancestry thereby ruling out completely any underlying structure in the population.
Allelic association of cytokine SNPs and the risk of pulmonary tuberculosis: Among the 25 studied SNPs, from six candidate cytokine genes the variants of IFNG, IL1RA, IL4, IL12 and TNFB were found to be associated with susceptibility to PTB in north Indians. All studied variants passing the exclusion criteria were in Hardy-Weinberg equilibrium in both cases and controls. Allelic association when probed in variants passing the exclusion criteria yielded six loci showing high risk for PTB susceptibility.
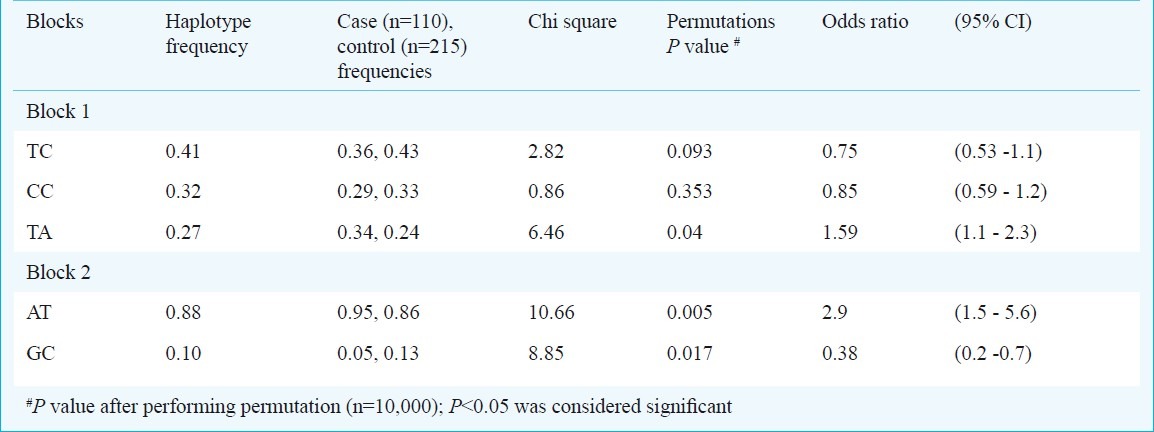
IFNG polymorphism and PTB susceptibility: After adjusting for multiple testing corrections the IFNG intronic variants at rs1861493 [χ2 =12.089, Pbonferroni = 0.006593, odds ratio (95%CI) =3.8 (1.7 - 8.6)] and rs1861494 (χ2 =10.466, Pbonferroni = 0.01581, odds ratio (95%CI) =3.0 (1.5 - 5.6)] showed a significant risk of developing pulmonary tuberculosis in north Indians with over-representation of the associated A and T alleles among PTB patients, respectively. Investigation of the gene structure and linkage disequilibrium pattern showed haplotypes formed by IFNG variants rs1861493 and rs1861494 which were in high linkage disequilibrium (LD) (Fig.). Three combinations of haplotype were seen namely TC, CC and TA, of which TA haplotype was over-represented in the cases and imposed a two-fold risk of developing pulmonary tuberculosis in north Indians (Table III).
Fig.
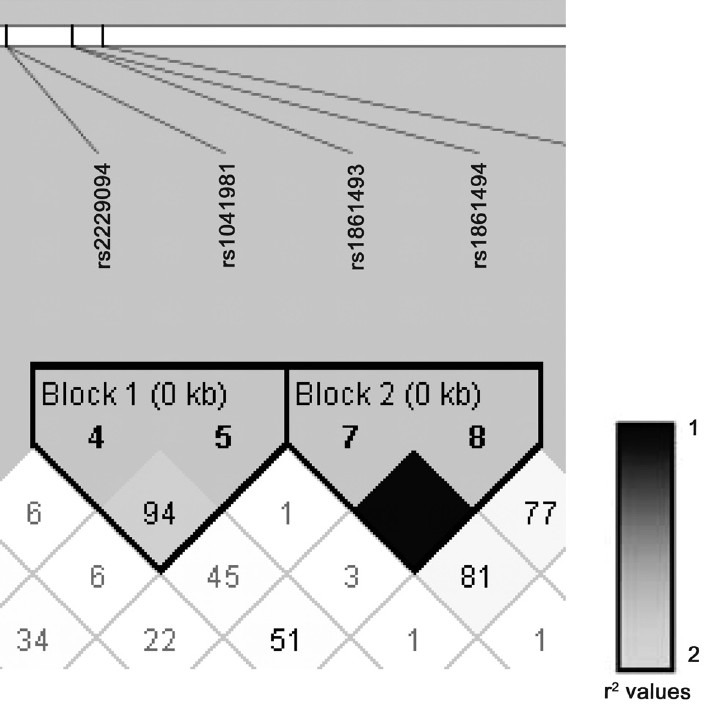
Linkage disequilibrium (LD) plot and haplotype structure of cytokine gene variants in PTB cases. D’ values are displayed within each diamond, missing values indicate D’ = 100%. Colour scheme gradient indicates r2 values. Length of each block, in kilobases (kb), is shown in brackets.
Table III.
Heplotype blocks and frequencies
IL4 polymorphism and PTB susceptibility: IL4 variant rs2070874 [x2=10.708, P bonferroni = 0.01387, odds ratio (95%CI) = 1.8 (1.3 - 2.6)] showed a two-fold risk by T allele in north Indians. The other studied IL-4 variant rs2243270 passing the exclusion criteria did not show any association towards susceptibility to pulmonary tuberculosis in this population.
IL1RA polymorphism and PTB susceptibility: The significantly associated locus of IL1RA included intronic variant at rs4252019 [χ2=13.643, P bonferroni = 0.00287, Odds ratio (95%CI) = 14.0 (1.8 - 103.5)] showing a 14-fold risk. Other variant such as rs315919 and rs380092 did not show any association towards susceptibility to pulmonary tuberculosis in this population.
IL12 polymorphism and PTB susceptibility: IL12 variants rs3212220 [χ2 = 14.572, Pbonferroni = 0.00175, Odds ratio (95%CI) = 2.0 (1.4 - 2.9)] and rs2853694 [χ2 =8.854, Pbonferroni = 0.0399, odds ratio (95%CI) = 1.6 (1.2 - 2.4)] showed a two-fold risk associated with T and A alleles, respectively.
IL1B polymorphism and PTB susceptibility: The selected IL1B variants did not show any direct influence on PTB susceptibility in north Indians
TNFB polymorphism and PTB susceptibility: TNFB variants at rs1041981 [χ2 =8.649, Pbonferroni = 0.03618, Odds ratio (95%CI) = 1.7 (1.2 - 2.6)] a synonymous change showed a two-fold risk of association for PTB in north Indians. Interestingly rs1041981 contributed to a haplotype block with rs2229094 confirming the importance of this locus in risk of developing PTB in north Indians. The two haplotypes observed were AT and GC of which AT was over-represented in PTB cases and imposed a three-fold risk of developing PTB in north Indians.
Discussion
The host genetic bias contributing to susceptibility and progression of pulmonary tuberculosis might involve interactions between multiple alleles located on different genes and chromosomes21. In order to overcome this drawback we planned selection of different cytokine gene and multiple loci to cover a wide spectrum of immune response associated cytokines.
Case-control studies involving carefully chosen locus across ethnicities are valiant means of identifying novel associations pertaining to disease susceptibility. Association that arises may be a result of the polymorphism in question being functional or it being in linkage disequilibrium with another functional allele or a result of confounding association due to population stratification. To overcome such false positives, we carefully considered the self reported ethnicity of the study groups and further checked for any genetic heterogeneity in our data by Eigenstrat principal component analysis illustrated by Price et al18 and found that the present data were free from any underlying population structure. Thus, this uniform data represent north Indian population for association analysis.
The IFN-γ being a crucial cytokine in immunopathogenesis of TB has been subject to several polymorphisms studies for pulmonary tuberculosis susceptibility. The locus probed here namely rs1861494 has not been studied in susceptibility to PTB but extensively studied in many other diseases such as leprosy22 and asthma23. Kumar et al24 found an association of this locus with susceptibility to asthma in Indians and could identify a haplotype. They also showed that alleles of rs1861494 A/G have differential affinity to bind to putative nuclear factor. In the present study, we found significant risk for the locus in susceptibility to PTB. The other probed locus rs1861493 has been studied in idiopathic inflammatory myopathy24 and asthma23 but not in pulmonary tuberculosis. We also identified a risk haplotype contributed by rs1861493 and rs1861494 emphasizing the importance of the above mentioned loci as risk factors for developing pulmonary tuberculosis in north Indians.
IL4 locus rs2070874 has been an important locus of investigation in various diseases including asthma and rheumatoid arthritis25. Its role in TB was reported not to be significant in Iranian pulmonary TB patients26 and recently in South Africans TB patients also the locus did not show any association27. In the present study this locus showed a two-fold risk in the north Indian population.
IL1RA locus rs4252019 has shown significant risk of development of pulmonary TB in north Indians. The variant rs4252019 has been shown to be associated with prostate cancer risk28 but not pulmonary tuberculosis. Interestingly, the variant showed a 14-fold risk of developing PTB in the population studied here and emerged as a major locus to look out for in further studies.
IL12 variants rs3212220 and rs2853694 showed a significant risk associated with development of PTB in north Indians. The variant rs321220 has been shown to contribute to a haplotype by Moller et al20. We have also predicted its importance in our previous study29. Based on the analysis of serum IL-12 level, we demonstrated that for IL12 variant rs3212220 TT genotype among active PTB cases showed significantly higher serum IL-12 level when compared to either GT or GG. The present study revealed T allele to be a risk allele in the present population. Similarly, rs2853694 a novel variant in the context of developing tuberculosis29 was predicted to be of importance and was validated in the present study. For rs2853694 among active PTB cases AA genotype showed a trend towards higher serum IL-12 level in contrast to a reverse trend observed in HC where AA accounted for low serum IL-1229. The present study showed A allele at rs2853694 to be a risk allele for the north Indian population in the context of PTB susceptibility. An interesting observation was that both the higher serum cytokine producers i.e. TT genotype for rs3212220 and AA genotype for rs2853694 emerged as respective risk alleles T and A for this population, indicating that overproduction of IL-12 by these individuals might be interfering with the cytokine homeostasis and thus affecting the immune function of the cytokine in these individuals making them prone to infection. Our observation was further supported by the work of Leandro et al30, who indicated that role of IL-12 as potent inducer of IFN-γ lied in its efficacy at low concentrations. In the present study it is observed that the PTB patients with IL12 risk allele genotypes are not efficient inducers of IFN-γ which in turn interferes with the protective immunity in these individuals, whereas a low profile of IL-12 in HC elicits an effective and optimal immune response rendering these individuals healthy.
TNFB though not usually considered for PTB association studies, was taken up in the current study because of its role in control of intracellular bacterial infection15. The variant rs1041981 emerged as a significant risk locus for PTB susceptibility in north Indians. The variant also contributed to a haplotype with rs2229094 and reinstated the role of TNFB polymorphisms in PTB.
Overall, five of the loci namely rs1861493 and rs1861494 (IFNG), rs4252019 (IL1RA) rs1041981 (TNFB) and rs2853694 (IL12) studied in patients of pulmonary tuberculosis showed a significant risk towards susceptibility to pulmonary tuberculosis in north Indians. We also report here the significant risk imposed by IL4 variant rs2070874 in the active PTB patients. Six new associations and three new associated haplotypes contributing to the spectrum of cytokine gene polymorphisms and risk of developing tuberculosis in general and north Indians in particular, were detected.
Acknowledgment
The authors thank all patients and volunteers for participating in this study. The support of the Medical Superintendent and staff at Rajan Babu Institute of Pulmonary Medicine and Tuberculosis (RBIPMT), Kingsway Camp, New Delhi (India) for the help in sample collection is acknowledged. Authors acknowledge the Council of Scientific and Industrial Research (CSIR), New Delhi, for financial support.The first author was the Junior Research Fellow (JRF) in the CSIR project.
References
- 1.Raviglione MC, Snider DE, Jr, Kochi A. Global epidemiology of tuberculosis: morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–6. [PubMed] [Google Scholar]
- 2.Stead WW, Senner JW, Reddick WT, Lofgren JP. Racial differences in susceptibility to infection by Mycobacterium tuberculosis. N Engl J Med. 1990;322:422–7. doi: 10.1056/NEJM199002153220702. [DOI] [PubMed] [Google Scholar]
- 3.Bellamy RJ, Hill AV. Host genetic susceptibility to human tuberculosis. Novartis Found Symp. 1998;217:3–13. doi: 10.1002/0470846526.ch2. [DOI] [PubMed] [Google Scholar]
- 4.Newport MJ, Huxley CM, Huston S, Hawrylowicz CM, Oostra BA, Williamson R, et al. A mutation in the interferon- γ-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941–5. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 5.Altare F, Durandy A, Lammas D, Emile JF, Lamhamedi S, Le Deist F, et al. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science. 1998;280:1432–5. doi: 10.1126/science.280.5368.1432. [DOI] [PubMed] [Google Scholar]
- 6.Comstock GW. Tuberculosis in twins: a re-analysis of the Prophit survey. Am Rev Respir Dis. 1978;117:621–4. doi: 10.1164/arrd.1978.117.4.621. [DOI] [PubMed] [Google Scholar]
- 7.Doffinger R, Dupuis S, Picard C, Fieschi C, Feinberg J, Barcenas-Morales G, et al. Inherited disorders of IL-12 and IFN gamma-mediated immunity: a molecular genetics update. Mol Immunol. 2002;38:903–9. doi: 10.1016/s0161-5890(02)00017-2. [DOI] [PubMed] [Google Scholar]
- 8.Pace JL, Russell SW, Torres BA, Johnson HM, Gray PW. Recombinant mouse γ-interferon induces the priming step in macrophage activation for tumor cell killing. J Immunol. 1983;130:2011–3. [PubMed] [Google Scholar]
- 9.Nathan CF, Murray HW, Wiebe ME, Rubin BY. Identification of interferon-γ as the lymphokine that activates human macrophage oxidative metabolism and anti-microbial activity. J Exp Med. 1983;58:670–89. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powrie F, Coffman RL. Inhibition of cell-mediated immunity by IL4 and IL10. Res Immunol. 1993;144:639–43. doi: 10.1016/s0923-2494(05)80019-7. [DOI] [PubMed] [Google Scholar]
- 11.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–93. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 12.Roach TI, Barton CH, Chatterjee D, Blackwell JM. Macrophage activation: lipoarabinomannan from avirulent and virulent strains of Mycobacterium tuberculosis differentially induces the early genes c-fos, KC, JE, and tumor necrosis factor-alpha. J Immunol. 1993;150:1886–96. [PubMed] [Google Scholar]
- 13.Schauf V, Rom WN, Smith KA, Sampaio EP, Meyn PA, Tramontana JM, et al. Cytokine gene activation and modified responsiveness to interleukin-2 in the blood of tuberculosis patients. J Infect Dis. 1993;168:1056–9. doi: 10.1093/infdis/168.4.1056. [DOI] [PubMed] [Google Scholar]
- 14.Law K, Weiden M, Harkin T, Tchou-Wong K, Chi C, Rom WN. Increased release of interleukin-1 beta, interleukin-6, and tumor necrosis factor-alpha by bronchoalveolar cells lavaged from involved sites in pulmonary tuberculosis. Am J Respir Crit Care Med. 1996;153:799–804. doi: 10.1164/ajrccm.153.2.8564135. [DOI] [PubMed] [Google Scholar]
- 15.Roach DR, Briscoe H, Saunders B, France MP, Riminton S, Britton WJ. Secreted lymphotoxin-a is essential for the control of an intracellular bacterial infection. J Exp Med. 2001;193:239–46. doi: 10.1084/jem.193.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allie N, Keeton R, Court N, Abel B, Fick L, Vasseur V, et al. Limited role for lymphotoxin α in the host immune response to Mycobacterium tuberculosis. J Immunol. 2010;185:4292–301. doi: 10.4049/jimmunol.1000650. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs M, Togbe D, Fremond C, Samarina A, Allie N, Botha T, et al. Tumor necrosis factor is critical to control tuberculosis infection. Microbes Infect. 2007;5:623–8. doi: 10.1016/j.micinf.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Price AL, Patterson J, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 20.Indian Genome Variation Consortium. Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet. 2008;87:3–20. doi: 10.1007/s12041-008-0002-x. [DOI] [PubMed] [Google Scholar]
- 21.Hill AV. The immunogenetics of human infectious diseases. Annu Rev Immunol. 1998;16:593–617. doi: 10.1146/annurev.immunol.16.1.593. [DOI] [PubMed] [Google Scholar]
- 22.Cardoso CC, Pereira AC, Brito-de-Souza VN, Dias-Baptista IM, Maniero VC, Venturini J, et al. IFNG +874 T>A single nucleotide polymorphism is associated with leprosy among Brazilians. Hum Genet. 2010;128:481–90. doi: 10.1007/s00439-010-0872-x. [DOI] [PubMed] [Google Scholar]
- 23.Kumar A, Ghosh B. A single nucleotide polymorphism (A --> G) in intron 3 of IFN gamma gene is associated with asthma. Genes Immun. 2008;9:294–301. doi: 10.1038/gene.2008.17. [DOI] [PubMed] [Google Scholar]
- 24.Chinoy H, Salway F, John S, Fertig N, Tait BD, Oddis CV, et al. Interferon-gamma and Interleukin-4 gene polymorphisms in UK Caucasian idiopathic inflammatory myopathy patients. Ann Rheum Dis. 2007;66:970–3. doi: 10.1136/ard.2006.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balsa A, Del Amo J, Blanco F, Caliz R, Silva L, Sanmarti R, et al. Prediction of functional impairment and remission in rheumatoid arthritis patients by biochemical variables and genetic polymorphisms. Rheumatology. 2010;49:458–66. doi: 10.1093/rheumatology/kep380. [DOI] [PubMed] [Google Scholar]
- 26.Amirzargar AA, Rezaei N, Jabbari H, Danesh AA, Khosravi F, Hajabdolbaghi M, et al. Cytokine single nucleotide polymorphisms in Iranian patients with pulmonary tuberculosis. Eur Cytokine Netw. 2006;17:84–9. [PubMed] [Google Scholar]
- 27.Möller M, Nebel A, Helden PDV, Schreiber S, Hoal EG. Analysis of eight genes modulating interferon gamma and human genetic susceptibility to tuberculosis: a case-control association study. BMC Infect Dis. 2010;10:154. doi: 10.1186/1471-2334-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tindall EA, Hayes VM, Petersen DC. Inflammatory genetic markers of prostate cancer risk. Cancers. 2010;2:1198–220. doi: 10.3390/cancers2021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abhimanyu, Mangangcha IR, Jha P, Arora K, Mukerji M, Banavaliker JN, et al. Differential serum cytokine levels are associated with cytokine gene polymorphisms in north Indians with active pulmonary tuberculosis. Infect Genet Evol. 2011;11:1015–22. doi: 10.1016/j.meegid.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 30.Leandro AC, Rocha MA, Cardoso CS, Bonecini-Almeida MG. Genetic polymorphisms in vitamin D receptor, vitamin D-binding protein, Toll-like receptor 2, nitric oxide synthase 2, and interferon-gamma genes and its association with susceptibility to tuberculosis. Braz J Med Biol Res. 2009;42:312–22. doi: 10.1590/s0100-879x2009000400002. [DOI] [PubMed] [Google Scholar]


