Abstract
Background & objectives:
mRNA is more rapidly destroyed in cells than rRNA or genomic DNA, an assay targeting bacterial mRNA would provide a better guide to mycobacterial viability than amplification tests directed at DNA or rRNA targets. This study was carried out to standardize reverse transcriptase PCR (RT-PCR) targeting 85B gene for the rapid detection of viable Mycobacterium tuberculosis from sputum specimens of suspected TB patients at Chennai, South India and to detect MDR-TB circulating in this population.
Methods:
Sputum samples from clinically suspected tuberculosis patients (n=301) and 78 controls were included in the study. The sputum samples were collected in sterile diethyl pyrocarbonate (DEPC) treated containers and transported in ice to the laboratory within 2 h to prevent degradation of RNA. RT-PCR targeting 85B gene, mycobacterial culture and phenotypic drug susceptibility testing for the first line drugs streptomycin (S), isoniazid (H), rifampicin (R), ethambutol (E) and pyrazinamide (Z) were performed by BACTEC microMGIT culture system for all the sputum specimens.
Results:
All the 78 controls were negative for culture and RT-PCR. Among the 301 sputum specimens from patients, 231 (76.8%) were RT-PCR positive and 70 (23.2%) were negative. There were 166 M. tuberculosis isolates, of which 11 (2.9%) were MDR-TB, 33 (8.7%) were polyresistant, 31 (8.2%) were monoresistant and 91 (30.2%) were sensitive to all five first line anti-tuberculous drugs by phenotypic drug susceptibility testing. Monoresistance was higher with Z [20 (20.8%)], followed by S [6 (3%)].
Interpretation & conclusions:
RT-PCR targeting 85B gene of M. tuberculosis was a specific, rapid, reliable technique to detect the M. tuberculosis directly from sputum specimens. Our results showed that 2.9 per cent of M. tuberculosis isolates in the study population of Chennai were MDR.
Keywords: Drug susceptibility, MDR-TB, Mycobacterium tuberculosis, RT-PCR, sputum
Tuberculosis continues to be a leading cause of mortality and morbidity worldwide1. The need for rapid diagnostic tests has led to the development of molecular methods for the detection of Mycobacterium tuberculosis. Polymerase chain reaction (PCR) has proven to be a useful technique in the diagnosis of tuberculosis infection. The problems with any PCR method is the risk of obtaining false-positive results due to contamination of clinical specimens with M. tuberculosis DNA product from the PCR laboratory, and the inability of the PCR method to detect a difference between viable and nonviable organisms2. The average half-life of bacterial mRNA is three min3. Because mRNA is more rapidly destroyed in cells than rRNA or genomic DNA, an assay targeting bacterial mRNA would provide a better guide to mycobacterial viability than amplification tests directed at DNA or rRNA targets. The target for amplification of mRNA, 85B gene is one of three homologous proteins that are part of the 85 antigen complex of mycobacteria. This target was selected because the 85B antigen constitutes up to 41 per cent of the total mycobacterial protein in log-phase culture supernatants4. It is reasonable to expect similarly high levels of 85B mRNA expression. The 85 antigen complex is present in all mycobacteria, and there is considerable evidence that the complex contains both species-specific and shared epitopes4. Thus, this antigen would provide a target, which is universally present.
Drug resistant tuberculosis has been reported since the early days of introduction of anti-tubercular chemotherapy, but recently multi-drug resistant tuberculosis (MDR-TB) has been an area of growing concern, and is posing threat to global efforts of tuberculosis control. Management of MDR-TB is difficult, expensive, challenging and quite often leads to treatment failure. Diagnosis is confirmed by drug susceptibility testing from reliable and reputed laboratories under constant quality control.
Thus, the present study was focused on the standardization and application of RT-PCR targeting 85B gene for the detection of M. tuberculosis from sputum specimens collected from clinically suspected cases of TB and to detect mutations responsible for drug resistance in this gene in clinical isolates of M. tuberculosis obtained from TB patients at Chennai, south India.
Material & Methods
Collection of sputum specimen: A total of 379 sputum samples from clinically suspected tuberculosis patients (301) and controls (78) attending the OPD of Institute of Thoracic Medicine, Chetpet, Chennai, were included in the study. The sputum samples were collected in sterile diethyl pyrocarbonate (DEPC) (Sigma Aldrich, India) treated containers and transported in ice to the laboratory within 2 h to prevent degradation of RNA after getting the patients consent. This study was approved by the Institutional Research and Ethics committee of Vision Research Foundation (VRF), Chennai.
The 301 sputum specimens included in the study were categorized based on clinical diagnosis (chest radiograph) and direct smear by the clinician into category I: chest radiograph and direct smear positive for acid fast bacilli (AFB) (n=149), category II: chest radiograph positive and direct smear negative for AFB (n=115), category III: patients with prior anti-tuberculous treatment (ATT) (n=37).
The 78 control sputum specimens were negative by chest radiograph and direct smear for AFB.
RNA extraction and cDNA conversion: RNA was extracted from the sputum specimen by standard TRIzol protocol5 with few modifications. Briefly, 500 μl of TRI reagent (trizol) (Applied Biosystems, USA) was added to 200 μl of sputum specimen (the mucous portion of the sputum specimen was used) and mixed well by vortexing to dissolve the sputum in Trizol reagent. To this mixture, 200 μl of chloroform was added and mixed well again, incubated at room temperature for 15 min, followed by centrifugation at 8,000 g for 15 min in a cooling centrifuge (Remi, India).The aqueous layer was transferred to a new sterile (1.5 ml) vial and 500 μl of isopropanol was added, mixed well using a sterile Pasteur pipette and centrifuged at 8,000 g for 10 min in cooling centrifuge. The isopropanol was decanted and 1 ml of 75 per cent ethanol was added to the pellet, mixed well and centrifuged at 8,000 g for 5 min in a cooling centrifuge. The 75 per cent ethanol was decanted and the pellet was air-dried. After drying, 30 μl of sterile DEPC treated water was added and mixed well.
The extracted RNA was converted to cDNA using cDNA conversion kit (Applied Biosystems, USA)6. The Single-tube nested reverse transcriptase PCR (STN RT-PCR) targeting 85B gene was performed using PCR machine (Eppendorf Mastercycler, Germany).
Protocols followed for optimization of RT-PCR: After extraction of RNA, it was run in an agarose gel incorporated with ethidium bromide to rule out the contamination with DNA. If smearing was seen, the RNA extraction protocol was repeated. Also Turbo DNase treatment was done to remove the contaminating DNA (Applied Biosystems, USA). The concentration of RNA was quantitated using spectrophotometer and if the concentration was between 30 and 50 μg/μl, it was used for performing RT-PCR. The specificity of RT-PCR was checked, using the other species of mycobacterium.
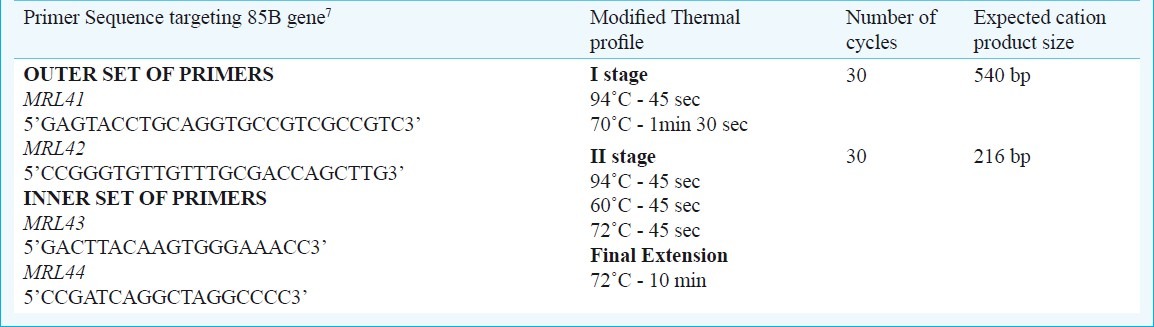
STN RT-PCR: The 216 bp region of 85B gene was amplified using two sets of primers7.
The first and second stages of amplification reactions in a 50 μl reaction volume contained 200 μM (8 μl) of each dNTPs [dATP, dTTP, dGTP, dCTP (Bangalore Genei)], 1 μM of outer and inner set of primers (1 μl each), 5 μl of 10 × buffer [10 mM Tris - HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2], 3 units (1 μl) of AmpliTaq Gold DNA polymerase (Applied Biosystems, USA) and 5 μl of 50 μg/μl of cDNA and 27 μl of sterile milliQ water. The primer sequence and modified thermal profile followed (the final extension was reduced to 10 min instead of 30 min as mentioned by Jou et al7) are given in Table I.
Table I.
Details of primer sequence, thermal profile and amplified product size
The analytical sensitivity of the PCR was tested by serial ten fold dilutions of RNA and converting each dilution to cDNA followed by amplification. The specificity of the primers was tested by amplifying the DNA extracted from the following mycobacterial species: M. tuberculosis H37Rv and H37Ra, M. bovis, M. intracellulare (ATCC 1403), M. kansasii (ATCC 1201), M. xenopi (ATCC 1432), M. gordonae, M. fortuitum (ATCC 1529), M. chelonae (ATCC 1524), M. abscessus (laboratory isolate), M. smegmatis (ATCC 607), M. phlei, M. thermoresistible, M. flavescens obtained from National Institute for Tuberculosis Research (ICMR), Chennai, India.
Detection of amplified products: The amplified product was subjected to electrophoresis on 2 per cent agarose gel incorporated with 0.5 μg/ml ethidium bromide for visualization by UV transilluminator (Vilber Lourmet, France).
Processing of sputum specimen for mycobacterial culture by BACTEC MicroMGIT culture system: Mycobacterial culture was performed by BACTEC MicroMGIT culture system, USA after standard decontamination procedures using NALC-NaOH. The cultures were incubated at 37°C for 42 days, before reporting the culture as negative.
Phenotypic drug susceptibility testing: After the isolation of M. tuberculosis isolates, phenotypic drug susceptibility testing was performed for streptomycin (S), isoniazid (H), rifampicin (R), ethambutol (E) and pyrazinamide (Z) by BACTEC MicroMGIT culture system following the manufacturer's instruction. Each vial of lyophilized drugs (streptomycin, isoniazid, rifampicin and ethambutol) was reconstituted with 4 ml of sterile distilled water and aliquoted in 200 μl amounts and stored at -20°C up to 6 months. The final concentrations used were 4.0, 0.2, 2.0 and 5.0 μg/ml, respectively. Lyophilized powder of pyrazinamide was reconstituted with 2.5 ml of sterile distilled water and aliquoted in 200 μl amounts and stored at -20°C up to 6 months. The final concentration of PZA was 100 μg/ml.
Results
The sensitivity of RT-PCR was 107fg/μl and it was specific for the detection of M. tuberculosis. Of the 379 sputum specimens (301 test specimens + 78 controls), none of the 78 specimens from controls were positive for RT-PCR for the detection of M. tuberculosis. Of the 301 sputum specimens from clinically suspected tuberculosis patients, 231 (76.8%) were RT-PCR positive and 70 (23.2%) were negative.
Of the 301 sputum samples from TB patients, 202 (67.2%) had concordant results (149 culture and RT-PCR positive, 53 culture and RT-PCR negative) while 99 (32.8%) were discordant [82 (27.2 %) culture negative and RT-PCR positive, 17 (5.6%) culture positive and RT-PCR negative]. All the 82 patients who were culture negative and RT-PCR positive were symptomatic and had positive clinical association with X-ray lesions.
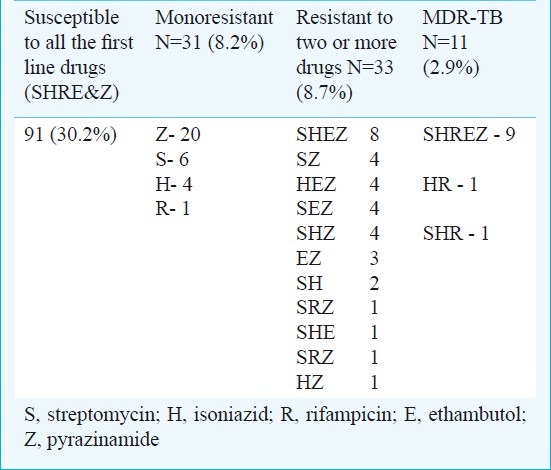
Of the 166 M. tuberculosis isolates, 11 (2.9%) were MDR-TB, 33 (8.7%) were polyresistant, 31 (8.2%) were monoresistant and 91 (30.2%) were sensitive to all five first line anti-tuberculous drugs by phenotypic drug susceptibility testing (Table II). Among 11 MDR patients (male 7, female 4), 7 were from newly diagnosed patients (primary resistance) and 4 were from previously treated patients (acquired resistance). The mean age of the patients was 34.7 yr (range 19-60), six patients were between 19-30 yr and five were between 40-60 yr. Of the 31 monoresistant isolates, 20 were resistant to pyrazinamide, six to streptomycin, four to isoniazid and one to rifampicin (Table II). Monoresistance was higher with Z (12%), followed by S (3.6%) and H (2.4%).
Table II.
Results of first line drug susceptibility testing of 166 M. tuberculosis isolates by BACTEC method
Discussion
Molecular methods in the diagnosis of tuberculosis have made a major impact in the early diagnosis of tuberculosis. Polymerase chain reaction methods targeting IS61108–11, MPB6412–14 and 65KDA11 have potential to detect M. tuberculosis specific DNA sequences. The major drawback in PCR technique is that it detects the genome of both the live and dead M. tuberculosis from clinical specimen whereas the RT-PCR targeting the mRNA of M. tuberculosis detects only the live organism in the clinical specimen.
The primary objective of this study was to optimize RT-PCR, which could detect live M. tuberculosis directly from clinical specimens. Desjardin et al15 developed RT-PCR assay directed towards the more stable and abundant 16S rRNA and found it to be 10-fold less sensitive than the 85B gene. Also it was not sensitive enough to detect only the live M. tuberculosis as they could detect the presence of 16S ribosomal RNA in the sputum of patients after anti-tuberculosis treatment15. In the present study, the modified procedure followed with STN RT-PCR resulted in 100 per cent specificity and 69.1 per cent sensitivity. Jou et al7 showed a sensitivity of 100 per cent while Negi et al11 showed a sensitivity of 94.67 per cent. In a real time reverse transcriptase PCR study by Mdivani et al16 a diagnostic sensitivity of 85.2 and specificity of 88.6 per cent were reported.
The reason for culture positive and RT-PCR negative results may be delay in processing of the specimen or improper separation of sputum for RT-PCR. The RT-PCR positive but culture negative result could be attributed to various factors affecting the multiplication of M. tuberculosis including the processing of specimens, inoculation including the change in nutritional requirements of the bacteria. All the 82 RT-PCR positive but culture negative patients had X-ray lesions. The main constraint of performing RT-PCR as a routine laboratory technique is the stringent collection technique to be followed and the extraction of RNA within a few hours of collection of specimen.
With the increasing numbers of MDR-TB cases there is an urgent need to contain this scourge17. Globally, just under 30,000 cases of MDR tuberculosis were reported to the World Health Organization (WHO) in 2008 (7% of the estimated total), of whom less than one fifth were managed according to international guidelines1. The vast majorities of the remaining cases probably are not diagnosed or, if diagnosed, are mismanaged18–20. In a study conducted by Thomas et al21 in the Thiruvalluvar district in Tamil Nadu, south India of the 4467 patients registered, 2206 (49.5%) were culture positive and 81 (3.67%) were identified as MDR-TB. More number of patients need to be included in the study to have the exact incidence of MDR-TB in Chennai population.
To conclude, RT-PCR targeting 85B gene of M. tuberculosis is a rapid, reliable technique to detect the viable M. tuberculosis directly from sputum specimens and has a potential role in the early diagnosis of active tuberculosis.
Acknowledgment
The authors acknowledge the financial support by Chennai Willingdon Corporate Foundation, Chennai, for the research project.
References
- 1.Global tuberculosis control - surveillance, planning, financing. WHO Report 2008 WHO/HTM/TB/2008.393. Available from: http://www.who.int/tb/publications/global_report/2008/en/index.html .
- 2.Yoneda M, Fukui Y, Yamanouchi T. Extracellular proteins of tubercle bacilli. V. Distribution of alpha and beta antigens in various mycobacteria. Biken J. 1965;8:201–3. [PubMed] [Google Scholar]
- 3.Wiker HG, Harboe M, Lea TE. Purification and characterization of two protein antigens from the heterogeneous BCG85 complex in Mycobacterium bovis BCG. Int Arch Allergy Appl Immunol. 1986;81:298–306. doi: 10.1159/000234153. [DOI] [PubMed] [Google Scholar]
- 4.Salata RA, Sanson AJ, Malhotra IJ, Wiker HG, Harboe M, Phillips NB, et al. Purification and characterization of the 30,000 dalton native antigen of Mycobacterium tuberculosis and characterization of six monoclonal antibodies reactive with a major epitope of this antigen. J Lab Clin Med. 1991;118:589–98. [PubMed] [Google Scholar]
- 5.Desjardin LE, Perkins MD, Teixeira L, Perkins MD, Cave MD, Eisenach KD. Alkaline decontamination of sputum specimens adversely affects stability of mycobacterial mRNA. J Clin Microbiol. 1996;34:2435–9. doi: 10.1128/jcm.34.10.2435-2439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.High capacity cDNA Archive kit. Applied Biosystems. 2000 [Google Scholar]
- 7.Jou NT, Yoshimori RB, Mason GR, Louie JS, Liebling MR. Single-tube, nested, reverse transcriptase PCR for detection of viable Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:1161–5. doi: 10.1128/jcm.35.5.1161-1165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JY, Lee LN, Chou CS, Huang CY, Wang SK, Lai HC, et al. Performance assessment of a nested-PCR assay (the RAPID BAP-MTB) and the BD ProbeTec ET system for detection of Mycobacterium tuberculosis in clinical specimens. J Clin Microbiol. 2004;42:4599–603. doi: 10.1128/JCM.42.10.4599-4603.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunisha P, Madhavan HN, Jayanthi U, Therese KL. Polymerase chain reaction using IS6110 primer to detect Mycobacterium tuberculosis in clinical samples. Indian J Pathol Microbiol. 2000;43:395–402. [PubMed] [Google Scholar]
- 10.Negi SS, Anand R, Pasha ST, Gupta S, Basir SF, Khare S, et al. Diagnostic potential of IS6110, 38kDa, 65 kDa and 85B sequence-based polymerase chain reaction in the diagnosis of Mycobacterium tuberculosis in clinical samples. Indian J Med Microbiol. 2007;25:43–9. doi: 10.4103/0255-0857.31061. [DOI] [PubMed] [Google Scholar]
- 11.Biswas J, Therese KL, Madhavan HN. Use of polymerase chain reaction in detection of Mycobacterium tuberculosis complex DNA from vitreous sample of Eales’ disease. Br J Ophthalmol. 1999;83:994. doi: 10.1136/bjo.83.8.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Therese KL, Jayanthi U, Madhavan HN. Application of nested polymerase chain reaction (nPCR) using MPB64 gene primers to detect Mycobacterium tuberculosis DNA in clinical specimens from extrapulmonary tuberculosis patients. Indian J Med Res. 2005;122:165–70. [PubMed] [Google Scholar]
- 13.Madhavan HN, Therese KL, Gunisha P, Jayanthi U, Biswas J. Polymerase chain reaction for detection of Mycobacterium tuberculosis in epiretinal membrane in Eales’ disease. Invest Ophthalmol Vis Sci. 2000;41:822–5. [PubMed] [Google Scholar]
- 14.Biswas J, Krishnakumar S, Rupauliha P, Misra S, Bharadwaj I, Therese KL. Detection of Mycobacterium tuberculosis by nested polymerase chain reaction in a case of subconjunctival tuberculosis. Cornea. 2002;21:123–5. doi: 10.1097/00003226-200201000-00027. [DOI] [PubMed] [Google Scholar]
- 15.Desjardin LE, Perkins MD, Wolski K, Haun S, Teixeira L, Chen Y, et al. Measurement of sputum Mycobacterium tuberculosis messenger RNA as a surrogate for response to chemotherapy. Am J Respir Crit Care Med. 1999;160:203–10. doi: 10.1164/ajrccm.160.1.9811006. [DOI] [PubMed] [Google Scholar]
- 16.Mdivani N, Li H, Akhalaia M, Gegia M, Goginashvili L, Kernodle DS, et al. Monitoring therapeutic efficacy by real-time detection of Mycobacterium tuberculosis mRNA in sputum. Clin Chem. 2009;55:1694–700. doi: 10.1373/clinchem.2009.124396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris KA, Jr, Mukundan U, Musser JM, Kreiswirth BN, Lalitha MK. Genetic diversity and evidence for acquired antimicrobial resistance in Mycobacterium tuberculosis at a large hospital in South India. Int J Infect Dis. 2000;4:140–7. doi: 10.1016/s1201-9712(00)90075-4. [DOI] [PubMed] [Google Scholar]
- 18.Tupasi TE, Gupta R, Quelapio MI, Orillaza RB, Mira NR, Mangubat NV, et al. Feasibility and cost-effectiveness of treating multidrug-resistant tuberculosis: a cohort study in the Philippines. PLoS Med. 2006;3:e352. doi: 10.1371/journal.pmed.0030352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orenstein EW, Basu S, Shah NS, Andrews JR, Friedland GH, Moll AP, et al. Treatment outcomes among patients with multidrug-resistant tuberculosis: systematic review and meta-analysis. Lancet Infect Dis. 2009;9:153–61. doi: 10.1016/S1473-3099(09)70041-6. [DOI] [PubMed] [Google Scholar]
- 20.Sotgiu G, Ferrara G, Matteelli A, Richardson MD, Centis R, Ruesch-Gerdes S, et al. Epidemiology and clinical management of XDR-TB: a systematic review by TBNET. Eur Respir J. 2009;33:871–81. doi: 10.1183/09031936.00168008. [DOI] [PubMed] [Google Scholar]
- 21.Thomas A, Ramachandran R, Rehaman F, Jaggarajamma K, Santha T, Selvakumar N, et al. Management of multi drug resistance tuberculosis in the field: Tuberculosis Research Centre experience. Indian J Tuberc. 2007;54:117–24. [PubMed] [Google Scholar]


