Abstract
Prion diseases or transmissible spongiform encephalopathies (TSEs) are infectious neurodegenerative disorders leading to death. These include Cresutzfeldt-Jakob disease (CJD), familial, sporadic and variant CJD and kuru in humans; and animal TSEs include scrapie in sheep, bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease (CWD) of mule deer and elk, and transmissible mink encephalopathy. All these TSEs share common pathological features such as accumulation of mis-folded prion proteins in the central nervous system leading to cellular dysfunction and cell death. It is important to characterize the molecular pathways and events leading to prion induced neurodegeneration. Here we discuss the impact of the functional genomics approaches including microarrays, subtractive hybridization and microRNA profiling in elucidating transcriptional cascades at different stages of disease. Many of these transcriptional changes have been observed in multiple neurodegenerative diseases which may aid in identification of biomarkers for disease. A comprehensive characterization of expression profiles implicated in neurodegenerative disorders will undoubtedly advance our understanding on neuropathology and dysfunction during prion disease and other neurodegenerative disorders. We also present an outlook on the future work which may focus on analysis of structural genetic variation, genome and transcriptome sequencing using next generation sequencing with an integrated approach on animal and human TSE related studies.
Keywords: Prion, TSEs, gene expression, functional candiate genes, PRNP, microarray.
1. INTRODUCTION
Prion diseases, or transmissible spongiform encephalopathies (TSEs), are invariably fatal neurodegenerative disorders affecting both humans and animals. Human prion diseases include Kuru, Creutzfeldt-Jakob disease (CJD) for about 85% of the cases, familial CJD in 10-15% of the cases and variant CJD which is limited to the UK and France [1, 2]. Animal TSEs (Table 1) include scrapie in sheep, bovine spongiform encephalopathy (BSE) in cattle, chronic wastings disease (CWD) in cervids, transmissible mink encephalopathy in mink, and feline spongiform encephalopathy (FSE) in cats, and exotic ungulate spongiform encephalopathy (EUE) of captive wild ruminants [3]. Animals affected by TSEs increase the risk of transmission to humans as well as seriously decline the farm animal production [4]. For example, since the BSE epidemic in 1986, slaughtering of millions of cattle and import ban on the beef have resulted in substantial losses. Prion diseases occur as sporadic, genetic, and transmissible disease, however, sporadic and heritable forms of the diseases are more frequent.
Table 1.
Transmissible Spongiform Encephalopathies (Prion Diseases) in Humans and Animals
| Type of Disease | Pathogenesis Mechanism / Mode of Transmission | |
|---|---|---|
| Human Diseases | ||
| Kuru | Infectious | Through Cannibalism |
| Sporadic CJD | Unknown | Spontaneous Conversion to PrPC to PrPSc Due to Spontaneous Mutation |
| Familial CJD | Genetic | PRNP Mutations |
| Latrogenic CJD | Infectious | Infection from Prion-Contaminated Material |
| Variant CJD | Infectious | Consumption of Infected Animals |
| Animal Diseases | ||
| Scrapie (Sheep and Goats) | Infectious | Exposure to Infected Sheep |
| Transmissible Mink Encephalopathy | Infectious | Infection from Prion Contaminated Feed |
| Bovine Spongiform Encephalopathy (BSE) | Infectious | Infected Meat and Bone Meal |
| Chronic Wasting Disease (CWD) | Infectious | Contaminated Pasture |
| Exotic Ungulate Spongiform Encephalopathy (EUE) | Infectious | Infection from Prion Contaminated Material |
| Feline Spongiform Encephalopathy (FSE) | Infectious | Infection from Prion Contaminated Feed |
Ingestion of contaminated biological material via food resulted in the acquired forms of diseases e.g., Kuru was the first known human TSE due to ritualistic cannibalism [5]. BSE is another massive common-source epidemic caused by meat and bone meal fed primarily to dairy cows [6, 7] and more recently vCJD in human has been associated with exposure to the BSE agents [1]. On the other hand, mutations in the normal prion protein encoded by the PRioN Protein (PRNP) gene, are linked to genetically inherited prion diseases including Gerstmann-Strausler-Sheinker (GSS) disease, fatal familial insomnia (FFI) and genetically associated CJD [2, 4].
2. PATHOGENESIS OF PRION DISEASE
In mammals, the conversion of the normal, cellular isoform of the prion protein (PrPC) to the disease-causing isoform (PrPSc) is the key process underlying prion diseases. PrPC and PrPSc have distinct conformations with PrPC has extensive α-helical content whereas PrPSc is rich in β-sheet structure with less α-helical content [8, 9], has the propensity to aggregate and has the resistance to proteolysis [10]. Prion diseases in animals can be characterized by PrPSc replication and accumulation, spongiform vacuolation and astrocytic gliosis, synaptic degeneration leading to neurodegeneration and lethality [11] while these symptoms are common in many neurodegenerative disorders [5, 12]. Approximately 45% of the PrPC protein is α-helical with two very short stretches of β-sheets and its conversion to PrPSc results in protein with ~30% α-helix and 45% β-sheet. Other ancillary proteins are also involved in this conversion process and PrPc appears to bind to PrPSc to form an intermediate complex [13]. The major route of transmission for many TSEs is through oral infection after which prions spread through the gut to secondary immune tissues and then to the brain [5]. Furthermore, prions have been detected in different lymphoid organs including the spleen, lymph nodes, Peyer’s patches and tonsils [14-16].
Prion protein (PrP) is encoded by a chromosomal gene, PRNP, with a single exon for the PrP open reading frame although the gene itself comprises of two to three exons [17-20]. The other exons of the PRNP gene contain untranslated sequences including the promoter and termination sites with multiple copies of GC-rich repeats in the PrP promoter [21]. Comparison of a total of 937 PRNP sequences from 83 species suggested a striking degree of conservation among the mammalian sequences [22]. However, variations in PrP sequences exist both between species and between individuals within species greatly affecting susceptibility to prion infection [23]. Increasing evidence suggests that other genes in addition to the PRNP genes also contribute to the genetic susceptibility of acquired TSEs, thus there is a need to improve our understanding of the molecular mechanisms underlying prion disease pathogenesis. Genome-wide studies in cattle [24-28], sheep [29, 30], mice [31, 32], and humans [33] have identified genomic regions and positional candidate genes, other than the prion gene, involved in TSE pathogenesis. This review highlights the recent advances in the field of prion diseases in human, mouse models and ruminant species to understand the complexities of molecular pathways using high throughput functional genomics technologies.
3. HIGH THROUGPUT GENE EXPRESSION STUDIES ASSOCIATED WITH HUMAN PRION DISEASES
The PRNP gene determines both susceptibility and phenotypes of prion diseases in humans, with point mutations leading to a specific pathological phenotype [34, 35]. A genome-wide study in patients with various forms of prion diseases (variant, sporadic, iatrogenic CJD and kuru patients) confirmed that the risk of developing prion diseases is strongly associated with the polymorphic codon 129 of the PRNP gene [33]. Presence of single nucleotide polymorphisms (SNPs) in the intron of PRNP, retinoic acid receptor-β protein (RARB), and SCG10/stathmin-like 2, a neuronal growth-associated protein (STMN2) increased the risk of prion diseases. Retinoic acid is known to regulate the expression of the prion protein in cell culture [36], and SCG10 modulates microtubule stability in neuronal cells, which, in turn, might potentially modulate prion neurotoxicity [37] indicating the involvement of these genes in the pathogenesis of prion disease.
Different approaches including human genome wide association studies (GWAS), mouse mapping and differential expression studies have suggested the association of Shadoo (Sho, shadow of prion protein)(SPRN) and E3 ubiquitin ligase (HECTD2) with risk of sporadic and variant CJD [38-40]. Shadoo is a neuronally expressed glycoprotein of unknown function. Due to a conserved physiological activity between PrPC and Sho, it might be acting on similar signaling pathways in human prion pathobiology [41]. By homology, HECTD2 is an E3 ubiquitin ligase and is involved in the ubiquitinylation of proteins for targeted degradation by the proteasome [40]. The ubiquitin-proteosome system has also been implicated in prion diseases and a number of other neurodegenerative disorders [40].
In another study [42], microarray analysis of the frontal cortex of 15 patients with sporadic CJD revealed the involvement of 79 up-regulated (e.g., metallothionein-1 and 2) and 275 down-regulated genes (e.g., Synaptosomal-associated protein 25 (SNAP25) modulating synaptic function and plasticity) compared to healthy controls. Metallothionein-1 and -2 are cysteine-rich intracellular proteins with a high capacity to bind to zinc and copper ions and are known to be mainly present in the cytoplasm of astrocytes of the cerebral cortex and white matter [43]. Altered expression of genes involved in metal ion binding activity would suggest an aberrant ion homeostasis in CJD [42]. SNAP-25 is involved in modulating synaptic function and plasticity and the synaptic impairment is a major pathological mechanism of CJD [44]. Further characterization of these genes is crucial to understanding the molecular basis of pathological process of prion diseases.
4. IDENTIFICATION OF CANDIDATE GENES THROUGH FUNCTIONAL GENOMIC STUDIES IN ANIMAL PROTEIN DISEASES
It has been well documented that besides PRNP, other genes are also involved in the pathogenesis of prion diseases [45]. Large scale gene expression profiling of infected vs. control animals to identify differentially expressed (DE) genes may help in understanding novel genes and pathways that are switched on and off at various time points during the pathogenesis of prion diseases. Different functional genomics technologies have been used to detected differential gene expression including cDNA libraries [46], mRNA differential display [47], suppression subtractive hybridization [48], microarrays [49-54] and more recently next generation sequencing [55]. These studies in experimental animal models or from clinically-infected animals have revealed multiple genes and signaling pathways that may be involved in TSE pathogenesis.
4.1. Prion Related Gene Expression Changes in Animal Models
Due to the availability of limited material from prion-infected samples at pre-clinical and clinical stages for humans and other animals; and due to the similarities between the mouse and human genome, the mouse models have been used in several prion pathogenesis studies. These models have provided the numbers and replication required for designing large scale studies to identify the genes of interest at different time points throughout disease pathogenesis [56]. Furthermore, mouse models can also be used for testing the candidate genes from over-expressing transgenics to knockouts, to determine their effect on prion pathology.
In brain tissues from experimental scrapie-infection animal models (Table 2 provides a list of high throughput studies conducted in mouse models), differentially expressed (DE) genes have been identified at different stages of prion diseases using microarray analysis [47, 57-60]. The global analysis of the overall transcriptional response in pre-clinical and clinical central nervous system tissue of mice in response to prion infection revealed that many of the individual genes identified had an association with the neurodegenerative process in prion disease as well as Alzheimer's disease (AD) [57]. The differential expression of genes involved in cellular stress (oxidative stress and ER stress), activated ER and mitochondrial apoptotic pathways, protease inhibitors, calcium binding proteins, endosome/lysosome function, immunity, synapse function, metal ion binding, and activated cholesterol biosynthesis, have been shown in the scrapie-infected hippocampus [58] and brain [59, 60]. Furthermore, all these studies have indicated similarities between gene expression patterns found in brains affected by AD and aging, respectively.
Table 2.
High Throughput Gene Expression Studies in Different Tissues from Prion-infected Mouse Model Leading to the Identification of Major Functional Candidate Genes / Markers / Pathways
| Study | Tissue | Technology Used | Functional Candidate Genes / Markers / Pathways |
|---|---|---|---|
| Dandoy-Dron et al., 1998 [47] | Brain from scrapie-infected mouse | mRNA differential display | Cathepsin S, the C1q B-chain of complement, apolipoprotein D, scrapie-responsive gene (ScRG-1) and ScRG-2 |
| Booth et al., 2004 [57] | Brain from scrapie-infected mouse | cDNA microarray | 158 differentially expressed (DE) genes Functional groups - secreted extracellular proteins, lysosomal proteases, defense and immune response-related proteins, signal transduction genes and cell growth, biogenesis-related genes |
| Brown et al., 2005 [58] | Hippocampal tissue from scrapie-infected mouse | Affymetrix high-density oligonucleotide probe arrays | 78 DE genes- sterol-C4-methyl oxidase and sterol-C5-desaturase (cholesterol biosynthesis), complement component C1qβ, short coiled-coil protein, signal recognition particle 9, THUMP domain-containing 1, neurofilament-L |
| Riemer et al., 2004 [49] | Cortex, medulla from scrapie-infected mouse | Mouse Genome U74Av2 arrays | 114 DE genes- proteinase inhibitor 2 (SPI-2); α-2-macroglobulin, lipocalin 24, CCAAT/ enhancer-binding protein delta (C/EBPdelta) |
| Xiang et al., 2004 [59] | Brain from scrapie-infected mouse | Affymetrix Mouse Expression Arrays | 121 DE genes - several members of the cathepsin family, protease inhibitors, S100 calcium binding proteins |
| Skinner et al., 2006 [60] | Brain from scrapie-infected mouse | cDNA microarrays | 400 DE genes - chemokine (C-X3-C) receptor 1, CD9 antigen, ATPase Na+/K+ transporting beta 1 polypeptide, cathepsin B, glial fibrillary acidic protein, and apolipoprotein E |
| Sorensen et al., 2008 [51] | Brain from multiple scrapie infected mouse | cDNA microarrays | 349 prion-related genes (PRGs)- transforming growth factor (TGF)-beta 1; extracellular signal-regulated kinase-mitogen-activated protein kinase (ERK/MAPK) signaling; transcription regulators, Early Growth responsive protein (EGR1) and CAMP responsive element binding protein 1 (CREB1) |
| Kim et al., 2008 [61] | Brain and spleen from multiple scrapie infected mouse | Affymetrix microarray | 67 DE genes - prolactin (Prl), Gh, and pro-opiomelanocortin-alpha (Pomc1); ATPase Na+/K+ transporting beta 1 polypeptide (Atp1b1); Acidic leucine-rich nuclear phosphoprotein 32 family member A (ANP32A) |
| Hwang et al., 2009 [62] | Eight distinct mouse strain-prion strain combinations | Microarray Subtractive analysis |
A core of 333 genes central to prion disease |
| Huzarewich et al., 2011 [63] | Spleen from scrapie-infected mouse enriched for dendritic cells and macrophages | Mouse genome 4x44K version microarray | 1753 DE genes- Leucine-rich proteoglycan decorin (DCN); osteoglycin (OGN), proline arginine-rich and leucine-rich repeat protein (PRELP), and chondroadherin (CHAD) |
| Tortosa et al., 2011 [64] | Transgenic mice overexpressing bovine cellular prion protein (PrPc) | Mouse Genome 430 2.0 arrays | 87 DE genes- Neuronal PAS domain protein 3 (Npas3, a transcription factor involved in the neuronal signaling]) and ribonucleotide reductase M2 B (Rrm2b, related to DNA) |
The differential expression of the genes involved in immunity, protein folding, ubiquitin/proteosome or in the endosome/lysosome system was observed in the brain and the spleen in scrapie-infected mice prior to the onset of clinical symptoms [61]. Altered expression of four genes, ATPase, Na+/K+ transporting, beta 1 polypeptide (Atp1b1), Growth hormone (Gh), acidic leucine-rich nuclear phosphoprotein 32 family member A (Anp32a) and Granulin (Grn) at the very early time of post-infection provided insights into the mechanism of pathogenesis [61]. Hwang et al. [62] performed a detailed characterization using two prion strains, different incubation times, and mice from six different genetic backgrounds and identified a core of 333 genes central to prion disease that were differentially expressed in all five of the combinations involving mice with normal levels of prion protein. Transcriptional analysis of follicular dendritic cells and macrophage enriched splenic cells revealed the genes related to iron metabolism and homeostasis as the major pathways [63]. Genome wide expression study in mice inoculated with BSE homogenate indicated changes in two main biological processes, neural cell metabolism and defense mechanisms [64]. Some of the genes identified in these studies may serve as markers for prion disease diagnosis which could be putative candidates for drug therapies.
4.2. Prion Related Gene Expression in Ruminants
Gene expression profiling studies in natural target ruminant species (cattle, sheep, elk/deer) infected by natural route (oral infection) are important for understanding the pathogenesis of the prion diseases. In cattle, orally infected with BSE agent (12 and 45 months post-infection), 101 DE genes in Peyer’s patch tissues [52] and 176 DE genes in medulla [53, 54] have been identified. These genes are mainly associated with the synapse function (e.g., tachykinin, synuclein, neuropeptide Y, cocaine, amphetamine-responsive transcript, and synaptosomal-associated protein 25 kDa); calcium ion regulation (e.g., parvalbumin, visinin-like, and cadherin); immune and inflammatory response (major histocompatibility complex (MHC) class II), and apoptosis (cholinergic receptor).
Another study [65] investigated the effect of prion pathogenesis on gene expression in cattle using microarray and 114 genes related to immune response, apoptosis, cell adhesion, stress response, and transcription were found to be differentially regulated (Table 3). Due to inherent limitations of microarrays including sequence-specific probe hybridization, background and cross-hybridization of related genes, digital gene expression (DGE) tag profiling using next generation sequencing was used to compare the transcriptomic profiles of medulla tissues from cattle infected with BSE [55]. This study identified 190 DE transcripts from different pathways including neuroactive ligand–receptor interaction, regulation of the actin cytoskeleton, focal adhesion, SNARE interactions in vesicular transport, T-cell receptor signaling, calcium signaling, TGF-beta signaling, and MAPK signaling. Inaddition, the Tag profiling was successful in identifying additional pathways like ErbB signaling, the T cell receptor, the Wnt signaling, antigen processing, cytokine-cytokine receptor interaction, Gap junction, and the PPAR signaling as compared to the previous microarray studies on BSE-infected medulla tissues [53, 55]. The common DE genes detected in all these studies [52-55, 65] on cattle were: S100 calcium binding and Calmodulin; Prolactin-related protein; GTPase, IMAP family member, Histocompatibility complex, class II, Metallopeptidase and Myosin, Glutathione S transferase A, Aldo-Keto reductase family and Nuclear receptor subfamily group H.
Table 3.
High Throughput Gene Expression Studies in Prion-infected Different Tissues from the Ruminants Leading to the Identification of Major Functional Candidate Genes / Markers / Pathways
| Study | Animal/Tissue | Technology Used | Functional Candidate Genes / Markers / Pathways |
|---|---|---|---|
| Khaniya et al., 2009 [52] | BSE-infected cattle Peyer’s patch | Microarray | 90 DE genes- Major histocompatibility complex (MHC) class II, MHC class II DQ alpha, leukocyte-derived arginine aminopeptidase (L-RAP) |
| Tang et al., 2009 [65] | BSE-infected cattle brains | Microarray | 114 DE genes- immune response, apoptosis, cell adhesion, stress response, and transcription- S100 calcium binding and Calmodulin; Prolactin-related protein; GTPase, IMAP family member, Histocompatibility complex, class II, Metallopeptidase; Myosin; Glutathione S transferase A; Aldo-Keto reductase family; Nuclear receptor subfamily group H |
| Tang et al., 2010 [66] | BSE-infected cattle brains | Microarray | 230 DE genes- immune response, apoptosis, cell adhesion, ER stress related response and transcription; ubiquitin-proteasome pathway; autophagy-lysosome system-glucose-regulated protein 94 (Grp94/gp96); glucose-regulated protein 170 (Grp170/Orp150); Inositol 1,4,5-triphosphate receptor (IP3, ER calcium-depletion stress); reticulon 1, 3 and 4 (ER stress induced apoptosis) |
| Almeida et al., 2011a, b [53, 54] | BSE-infected cattle caudal medulla tissues | Microarray | 176 DE genes- extracellular matrix (ECM) receptor, cell adhesion, neuroactive ligand–receptor interaction, SNARE interactions in vesicular transport, and MAPK signalling pathway synapse pathway - tachykinin, synuclein, neuropeptide Y, cocaine, amphetamine-responsive transcript, and synaptosomal-associated protein 25 kDa (SNAP25); calcium ion regulation (parvalbumin, visinin-like, and cadherin) |
| Basu et al., 2011 [55] | BSE-infected cattle caudal medulla tissues | Tag profiling Solexa sequencing | 190 DE genes- neuroactive ligand-receptor interaction, regulation of the actin cytoskeleton, focal adhesion, SNARE interactions in vesicular transport, T-cell receptor signaling pathway, Calcium signaling pathway, TGF-beta signaling pathway, MAPK signaling pathway SNAP25, vesicle-associated membrane protein 1, MHC class II, DQ alpha 5, thrombospondin 1, guanine nucleotide exchange factor isoforms |
| Panelli et al., 2011 [67] | Bovine amyloidotic spongiform encephalopathy infected cattle white blood cells | Microarray | 56 DE genes- T- and B-cell development and activation, inflammatory responses |
| Filali et al., 2011 [68] | Scrapie-infected sheep caudal medulla tissues | 4x44K microarray | 350 DE genes- immune response, ion transport, cell adhesion, and transcription- calpain 6, galanin 1 and pancreatitis associated protein 1; three downregulated (collagen 1 α2, collagen 3 α2) and melatonin receptor 1b (MTNR1B) |
| Gossner et al., 2011 [69] | Scrapie-infected sheep lymph nodes and spleen | Microarray | 52 DE genes in lymph nodes and 37 DE genes in spleen Up-regulation of genes related to apoptosis and repression of genes linked to inflammation and oxidative stress |
| Basu et al., 2012 [70] | Brain, midbrain, thalamus, spleen, RPLN and tonsil of CWD-infected elk | Microarray using Bovine-specific oligos | 329 DE genes in the brain, 249 DE genes in the spleen, 30 DE genes in the retropharyngeal lymph node (RPLN) and 55 DE genes in the tonsil - neuronal signaling and synapse function, calcium signaling, apoptosis and cell death and immune cell trafficking and inflammatory response |
Up-regulation of three chaperones including endoplasmic reticulum (ER) chaperones, Grp94 and Grp170 has strongly suggested the presence of ER stress and the activation of the unfolded protein response (UPR) in BSE-infected cattle [66]. The patterns of gene expression in white blood cells following oral infection of cattle with Bovine amyloidotic spongiform encephalopathy (BASE) has also been investigated and significant change in expression of genes linked to T- and B-cell development and activation, and to inflammatory responses was observed [67]. High-density whole genome association study in 143 BSE case and 173 control animals elucidated chromosomal regions and positional candidate genes associated with disease [28]. These positional candidate genes include hypothetical gene LOC521010, similar to FK506 binding protein 2 encoding a protein from immunophilin protein family with a role in protein folding, leucine-rich repeats and immunoglobulin-like domains 2, LRIG2, a protein known to be involved in protein-protein interactions, and Repulsive guidance molecule family member A, a glycosyl phosphatidylinositol -anchored glycoprotein, with a role as an axon guidance protein in the central nervous system [28].
Recently, transcriptomics analysis in the caudal medulla oblongata of scrapie-symptomatic sheep using a CVI custom designed 4x44K microarray platform identified 148 genes (Table 3) that are primarily associated with the immune response, ion transport, cell adhesion, and transcription [68]. Transcriptome analysis of scrapie-infected lymph nodes and spleen tissues from sheep have revealed the repression of genes linked to inflammation and oxidative stress, and the up-regulation of genes related to apoptosis [69]. We performed first comprehensive microarray analysis of high-throughput gene expression associated with CWD disease using bovine specific oligos. The differential expression of a set of key genes from different pathways from multiple organs of CWD infected elk include major regulatory and signaling networks, neuronal signaling, synapse function in neurological disease, calcium signaling, apoptosis and cell death, and immune cell trafficking and inflammatory response [70].
In this review, we have summarized the emerging role of functional genomic technologies in identifying genetic changes which could be used as potentially diagnostic biomarkers for risk determination and as general indicators of disease progression. Most of the expression profiling studies performed so far are in different models after clinical symptoms appear, however, prion diseases and other neurodegenerative disorders have a long pre-symptomic phase [71]. Additional studies will be necessary to investigate differential gene expression through full transcriptomic analysis from animals at both pre-clinical and clinical time points to identify new genes, proteins and pathways involved in the pathogenesis of TSEs in order to select gene(s) which could be used as a pre-clinical diagnostic marker.
5. PRION DISEASES AND OTHER NEURODEGENERATIVE DISORDERS
Mis-folding of the prion protein and its progressive accumulation into aggregates is the common feature of many major neurodegenerative diseases, like AD, Parkinson's disease (PD), fronto-temporal dementia, Huntington's disease (HD) which is shared by TSEs or prion disorders like CJD [72, 73]. In all these neurological diseases, there is continued synthesis of mutant, aggregation-prone proteins such as tau (in Taupathies), α-synuclein (in PD), polyglutamine-containing proteins (in HD), and amyloid-β (in AD) for many decades before neuropathology symptoms are evident [13]. Furthermore, these proteins or protein aggregates indicate ‘prion-like’ phenomenon of CJD and BSE and have been shown to spread to other cells and brain regions resulting in the progressive deterioration [74, 75]. Prion diseases and AD share some common morphological and pathophysiological features including the copper binding properties of the amyloidogenic proteins, oxidative stress and the evidence of free radical damage, the formation of amyloid plaques and neurofibrillary pathology [13, 76]. Histological and biochemical studies [77, 78] have recently suggested that Alzheimer’s disease and TSE pathologies synergistically interact with the possibility of a direct interaction between the proteins leading to cross-seeding and increased pathogenesis with the possibility of molecular cross talk between both diseases. This interaction may disrupt interaction between PrPc and a co-receptor impairing the neuron’s signal-transduction pathways leading to amyloid β oligomerization outside the cell and intracellular accumulation of tau protein leading to synaptic plasticity, axonal damage and neuronal death [77-79].
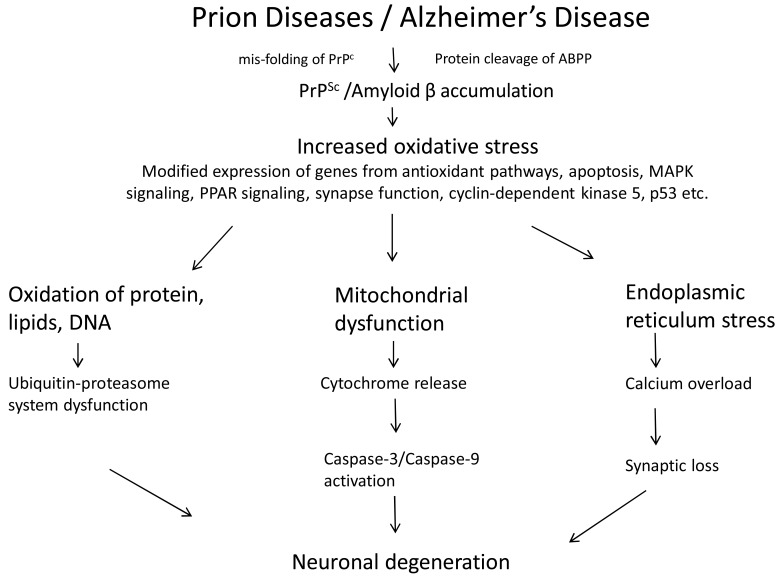
In addition to the common histopathological features, oxidative stress Fig. (1) is an important factor that has been implicated in the pathogenesis of a number of neurodegenerative disorders [58]. The misfolding of proteins or the development of plaques is followed by loss of synapses and possible abnormalities in mitochondrial function. Increased sensitivity to oxidative stress in neurodegenerative diseases is through alteration of anti-oxidant enzyme functions, and increased lipid peroxidation resulting in impairment of anti-oxidant response [80]. There is increasing evidence that secondary mitochondrial dysfunction by inhibition of several key enzymes in the mitochondrial energy production pathway or through the triggering of caspase-dependent apoptosis occurs in a number of major neurodegenerative diseases [81]. In addition, oxidative stress and apoptosis in prion diseases results in endoplasmic reticulum stress through the disruption of calcium homeostasis in the cell causing calcium overload in the cytoplasm leading to synaptic loss and neuronal death [82]. Other studies have also revealed that the overall level of oxidative damage to proteins, lipids and DNA may result in ubiquitin-proteasome dysfunction leading to neuronal degeneration [83]. Since the molecular pathways involved in different neurodegenerative diseases are similar, further studies to explore these molecular interactions, using high throughput functional genomics technologies combined with new bioinformatics capabilities, will provide a new conceptual framework for the pathogenesis of different neurodegenerative diseases and will have important therapeutic implications.
Fig. (1).
Involvement of oxidative stress in prion diseases and other neurodegenerative diseases leading to mitochondrial dysfuntion, endoplasmic reticulum stress through altered gene functions.
6. FUTURE DIRECTIONS
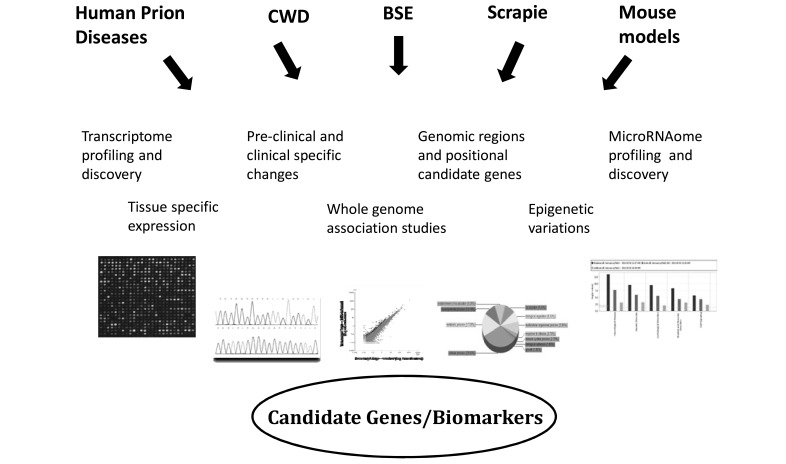
Until now, most of the functional genomics studies have employed microarrays and genome wide association studies to identify potential candidate genes or pathways involved in prion disease in different models or organisms. With the recent advances in next generation sequencing, exome sequencing, large scale data integration and data mining, it should be possible to investigate other key regulators including miRNA, and transcription factors playing a role in the pathogenesis mechanisms [84]. Some preliminary studies are beginning to establish the role of miRNA in prion diseases. The first comprehensive microarray analysis of miRNA expression in the brains of mice infected with mouse-adapted scrapie reported the de-regulation of 15 miRNAs controlling a number of genes and signaling pathways during prion induced neurodegeneration [85]. Another study provided evidence of up-regulation of miRNA-146a in sporadic Creutzfeldt-Jakob disease (sCJD) and Gerstmann-Straussler-Scheinker syndrome (GSS) suggesting its role in innate immune response and antiviral immunity [86]. Forthcoming directions of research will focus on a co-ordinated approach to link transcriptome and microRNAome data with extensive computational analysis, next generation sequencing studies spanning the entire genome or exome, analysis of structural genetic variation (copy number variations through deletions, duplications, and inversions) and genome wide association analysis. All these studies Fig. (2) will provide new insights of the regulatory networks involved in prion pathogenesis of human and animal prion diseases. Prions are also emerging as ‘extreme case of epigenetic inheritance’ and prion based mechanisms may influence genetic information [87]. Understanding of the epigenetic information in addition to the genetic information is becoming important for the preventive diagnosis and treatment of diseases [88]. The availability of next generation sequencing technologies will empower the study of epigenetic variations including DNA methylation, histone modifications, and chromatin accessibility which may be involved in prion associated neurodegenerative disorders. Furthermore, studies using functional proteomics and new physiology techniques such as laser capture microdissection on subpopulations of cells in pre- and post-symptomics phases of prions and other neurodegenerative diseases will complement the genomics studies to determine selective biomarkers.
Fig. (2).
Understanding of complex prion diseases: Identification of genes and genetic markers through functional genomics.
ACKNOWLEDGEMENT
Declared none.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Johnson RT. Prion diseases. Lancet Neurol. 2005;4:635–642. doi: 10.1016/S1474-4422(05)70192-7. [DOI] [PubMed] [Google Scholar]
- 2.Aguzzi A, Heikenwalder M. Pathogenesis of prion diseases: current status and future outlook. Nat. Rev. Microbiol. 2006;4:765–775. doi: 10.1038/nrmicro1492. [DOI] [PubMed] [Google Scholar]
- 3.Imran M, Mahmood S. An overview of animal prion diseases. Virology J. 2011;8:493–501. doi: 10.1186/1743-422X-8-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huzarewich RL, Siemens AG, Booth SA. Applications of ‘omics’ to prion biomarker discovery. J. Biomedicine Biotech. 2010. doi:10.1155/2010/613504. [DOI] [PMC free article] [PubMed]
- 5.Aguzzi A, Heikenwalder M. Prion diseases: cannibals and garbage piles. Nature. 2003;423:127–129. doi: 10.1038/423127a. [DOI] [PubMed] [Google Scholar]
- 6.Wilesmith JW, Ryan JB, Atkinson MJ. Bovine spongiform encephalopathy: epidemiological studies on the origin. Vet. Res. 1991;128:99–203. doi: 10.1136/vr.128.9.199. [DOI] [PubMed] [Google Scholar]
- 7.Nathanson N, Wilesmith J, Griot C. Bovine spongiform encephalopathy (BSE): causes and consequences of a common source epidemic. Am. J. Epidemiol. 1997;145:959–969. doi: 10.1093/oxfordjournals.aje.a009064. [DOI] [PubMed] [Google Scholar]
- 8.Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 9.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biljan I, Ilc G, Giachin G, Raspadori A, Zhukov I, Plavec J, Legname G. Toward the Molecular Basis of Inherited Prion Diseases: NMR Structure of the Human Prion Protein with V210I Mutation. J. Mol. Biol. 2011;412:660–673. doi: 10.1016/j.jmb.2011.07.067. [DOI] [PubMed] [Google Scholar]
- 11.Prusiner SB. Prions. Proc. Natl. Acad. Sci. USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguzzi A, Polymenidou M. Mammalian prion biology. One century of evolving concepts. Cell. 2004;116:313–327. doi: 10.1016/s0092-8674(03)01031-6. [DOI] [PubMed] [Google Scholar]
- 13.Fraser H, Dickinson AG. Studies of the lymphoreticular system in the pathogenesis of scrapie: the role of spleen and thymus. J. Comp. Pathol. 1978;88:563–573. doi: 10.1016/0021-9975(78)90010-5. [DOI] [PubMed] [Google Scholar]
- 14.Kimberlin RH, Walker CA. Pathogenesis of mouse scrapie: dynamics of agent replication in spleen, spinal cord and brain after infection by different routes. J. Comp. Pathol. 1979;89:551–562. doi: 10.1016/0021-9975(79)90046-x. [DOI] [PubMed] [Google Scholar]
- 15.Mabbott N A, MacPherson GG. Prions and their lethal journey to the brain. Nature Rev. Microbiol. 2006;4:201–211. doi: 10.1038/nrmicro1346. [DOI] [PubMed] [Google Scholar]
- 16.Basler K, Oesch B, Scott M, Westaway D, Walchli M, Groth F, McKinley MP, Prusiner SB, Weissmann C. Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell. 1986;46:417–428. doi: 10.1016/0092-8674(86)90662-8. [DOI] [PubMed] [Google Scholar]
- 17.Westaway D, Goodman PA, Mirenda AA, McKinley MP, Carlson GA, Prusiner SB. Distinct prion protein in short and long scrapie incubtation period mice. Cell. 1987;51:651–662. doi: 10.1016/0092-8674(87)90134-6. [DOI] [PubMed] [Google Scholar]
- 18.Hsiao K, Baker HF, Crow TJ, Poulter M, Owen F, Terwilliger JD, Westaway D, Ott J, Prusiner SB. Linkage of a prion protein missense variant to Gerstmann-Sträussler syndrome. Nature. 1989;338:342–345. doi: 10.1038/338342a0. [DOI] [PubMed] [Google Scholar]
- 19.Gabriel J-M, Oesch B, Kretzschmar H, Scott M, Prusiner SB. Molecular cloning of a candidate chicken prion protein. Proc. Natl. Acad. Sci. 1992;89:9097–9101. doi: 10.1073/pnas.89.19.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguzzi A, Calella AM. Prions: protein aggregation and infectious diseases. Physiol. Rev. 2009;89:1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- 21.Mcknight S, Tjian R. Transriptional selectivity of viral genes in mammalian cells. Cell. 1986;46:795–805. doi: 10.1016/0092-8674(86)90061-9. [DOI] [PubMed] [Google Scholar]
- 22.Rongyan Z, Xianglong L, Lanhui L, Xiangyun L, Fujun F. Evolution and differentiation of the prion protein gene (PRNP) among species. J. Heredity. 2008;99:647–652. doi: 10.1093/jhered/esn073. [DOI] [PubMed] [Google Scholar]
- 23.Colby DW, Prusiner SB. Prions. Cold Spring Harb. Perspect. Biol. 2011;3:a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernández-Sánchez J, Waddington D, Wiener P, Haley CS, Williams JL. Genome-wide search for markers associated with bovine spongiform encephalopathy. Mamm. Genome. 2002;13:164–168. doi: 10.1007/BF02684022. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, de Koning DJ, Hernández-Sánchez J, Haley CS, Williams JL, Wiener P. Mapping of multiple quantitative trait loci affecting bovine spongiform encephalopathy. Genetics. 2004;167:1863–1872. doi: 10.1534/genetics.104.026401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murdoch BM, Clawson ML, Laegreid WW, Stothard P, Settles M, McKay S, Prasad A, Wang Z, Moore SS, Williams JL. A 2cM genome-wide scan of European Holstein cattle affected by classical BSE. BMC Genet. 2010;29:11–20. doi: 10.1186/1471-2156-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murdoch BM, Clawson ML, Yue S, Basu U, McKay S, Settles M, Capoferri R, Laegreid WW, Williams JL, Moore SS. PRNP haplotype associated with classical BSE incidence in European Holstein cattle. PLoS ONE. 2010;5:pii: e12786. doi: 10.1371/journal.pone.0012786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murdoch BM, Murdoch GK, Settles M, McKay S, Williams JL, Moore SS. Genome-wide can identifies loci associated with classical BSE occurrence. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0026819. e26819.doi: 10.1371/journal.pone. 0026819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez L, Arranz JJ, San Primitivo F. Identification of a new leucine haplotype (ALQ) at codon 154 in the ovine prion protein gene in Spanish sheep. J Anim. Sci. 2006;2:259–265. doi: 10.2527/2006.842259x. [DOI] [PubMed] [Google Scholar]
- 30.Moreno CR, Cosseddu GM, Schibler L, Roig A, Moazami-Goudarzi K, Andreoletti O, Eychenne F, Lajous D, Schelcher F, Cribiu EP, Laurent P, Vaiman D, Elsen JM. Identification of new quantitative trait loci (other than the PRNP, gene) modulating the scrapie incubation period in sheep. Genetics. 2008;179:723–726. doi: 10.1534/genetics.108.088146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno CR, Lantier F, Lantier I, Sarradin P, Elsen JM. Detection of new quantitative trait loci for susceptibility to transmissible spongiform encephalopathies in mice. Genetics. 2003;165:2085–2091. doi: 10.1093/genetics/165.4.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lloyd SE, Uphill JB, Targonski PV, Fisher EM, Collinge J. Identification of genetic loci affecting mouse-adapted bovine spongiform encephalopathy incubation time in mice. Neurogenetics. 2002;4:77–81. doi: 10.1007/s10048-002-0133-9. [DOI] [PubMed] [Google Scholar]
- 33.Mead S, Poulter M, Uphill J, Beck J, Whitfield J, Webb TE, Campbell T, Adamson G, Deriziotis P, Tabrizi SJ, Hummerich H, Verzilli C, Alpers MP, Whittaker JC, Collinge J. Genetic risk factors for variant Creutzfeldt-Jakob disease: a genome-wide association study. Lancet Neurol. 2009;8:57–66. doi: 10.1016/S1474-4422(08)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gambetti P, Kong Q, Zou W, Parchi P, Chen SG. Sporadic and familial CJD: classification and characterisation. Br. Med. Bull. 2003;66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 35.Pocchiari M, Poleggi A, Principe S, Graziano S, Cardone F. Genomic and post-genomic analyses of human prion diseases. Genome Med. 2009;1:63–71. doi: 10.1186/gm63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rybner C, Hillion J, Sahravui T, Lanotte M, Botti J. All-trans retinoic acid downregulates prion protein expression independently of granulocyte maturation. Leukemia. 2002;16:940–948. doi: 10.1038/sj.leu.2402443. [DOI] [PubMed] [Google Scholar]
- 37.Kristiansen M, Messenger MJ, Klöhn PC, Brandner S, Wadsworth JD, Collinge J, Tabrizi SJ. Disease-related prion protein forms aggresomes in neuronal cells leading to caspase activation and apoptosis. J. Biol. Chem. 2005;280:38851–38861. doi: 10.1074/jbc.M506600200. [DOI] [PubMed] [Google Scholar]
- 38.Beck JA, Campbell TA, Adamson G, Poulter M, Uphill JB, Molou E, Collinge J, Mead S. Association of a null allele of SPRN with variant Creutzfeldt-Jakob disease. J. Med. Genet. 2008;45:813–817. doi: 10.1136/jmg.2008.061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lloyd SE, Maytham EG, Pota H, Grizenkova J, Molou E, Uphill J, Hummerich H, Whitfield J, Alpers MP, Mead S, Collinge J. HECTD2 is associated with susceptibility to mouse and human prion disease. PLoS Genet. 2009;5:e1000383. doi: 10.1371/journal.pgen.1000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lloyd S, Mead S, Collinge J. Genetics of prion disease. Top Curr. Chem. 2011;305:1–22. doi: 10.1007/128_2011_157. [DOI] [PubMed] [Google Scholar]
- 41.Sakthivelua V, Seidelb RP, Winklhofera KF, Tatzelt J. Conserved stress-protective activity between prion protein and Shadoo. J. Biol. Chem. 2011;286:8901–8908. doi: 10.1074/jbc.M110.185470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang W, Windl O, Westner IM, Neumann M, Zerr I, Lederer RM, Kretzschmar HA. Cerebral gene expression profiles in sporadic Creutzfeldt-Jakob disease. Ann. Neurol. 2005;58:242–257. doi: 10.1002/ana.20551. [DOI] [PubMed] [Google Scholar]
- 43.Kawashima T, Doh-ura K, Torisu M, Uchida Y, Furuta A, Iwaki T. Differential expression of metallothioneins in human prion diseases. Dement. Geriatr. Cogn. Disord. 2000;11:251–262. doi: 10.1159/000017247. [DOI] [PubMed] [Google Scholar]
- 44.Ferrer I, Rivera R, Blanco R, Marti E. Expression of proteins linked to exocytosis and neurotransmission in patients with Creutzfeldt-Jakob disease. Neurobiol. Dis. 1999;6:92–100. doi: 10.1006/nbdi.1998.0226. [DOI] [PubMed] [Google Scholar]
- 45.Diaz C, Vitezica ZG, Rupp R, Andreoletti O, Elsen JM. Polygenic variation and transmission factors involved in the resistance/susceptibility to scrapie in a Romanov flock. J. Gen. Virol. 2005;86:849–857. doi: 10.1099/vir.0.80412-0. [DOI] [PubMed] [Google Scholar]
- 46.Diedrich JF, Bendheim PE, Kim YS, Carp RI, Haase AT. Scrapie-associated prion protein accumulates in astrocytes during scrapie infection. Proc. Natl. Acad. Sci. 1991;88:375–379. doi: 10.1073/pnas.88.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dandoy-Dron F, Guillo F, Benboudjema L, Deslys JP, Lasmezas C, Dormont D, Tovey MG, Dron M. Gene expression in scrapie. Cloning of a new scrapie-responsive gene and the identification of increased levels of seven other mRNA transcripts. J. Biol. Chem. 1998;273:7691–7697. doi: 10.1074/jbc.273.13.7691. [DOI] [PubMed] [Google Scholar]
- 48.Kopacek J, Sakaguchi S, Shigematsu K, Nishida N, Atarashi R, Nakaoke R, Moriuchi R, Niwa M, Katamine S. Upregulation of the genes encoding lysosomal hydrolases, a perforin-like protein, and peroxidases in the brains of mice affected with an experimental prion disease. J. Virol. 2000;74:411–417. doi: 10.1128/jvi.74.1.411-417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riemer C, Neidhold S, Burwinkel M, Schwarz A, Schultz J, Kratzschmar J, Monning U, Baier M. Gene expression profiling of scrapie-infected brain tissue. Biochem. Biophys. Res. Commun. 2004;323:556–564. doi: 10.1016/j.bbrc.2004.08.124. [DOI] [PubMed] [Google Scholar]
- 50.Greenwood AD, Horsch M, Stengel A, Vorberg I, Lutzny G, Maas E, Schädler S, Erfle V, Beckers J, Schätzl H, Leib-Mösch C. Cell line dependent RNA expression profiles of prion-infected mouse neuronal cells. J. Mol. Biol. 2005;349:487–500. doi: 10.1016/j.jmb.2005.03.076. [DOI] [PubMed] [Google Scholar]
- 51.Sorensen G, Medina S, Parchaliuk D, Phillipson C, Robertson C, Booth SA. Comprehensive transcriptional profiling of prion infection in mouse models reveals networks of responsive genes. BMC Genom. 2008;9:114–127. doi: 10.1186/1471-2164-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khaniya B, Almeida L, Basu U, Taniguchi M, Williams JL, Barreda DR, Moore SS, Guan LL. Microarray analysis of differentially expressed genes from Peyer’s patches of cattle orally challenged with bovine spongiform encephalopathy. J. Toxicol. Environ. Health A. 2009;72:1008–1013. doi: 10.1080/15287390903084199. [DOI] [PubMed] [Google Scholar]
- 53.Almeida LM, Basu U, Khaniya B, Taniguchi M, Williams JL, Moore SS, Guan LL. Gene expression in the medulla following oral infection of cattle with bovine spongiform encephalopathy. J. Toxicol. Environ. Health A. 2011;74:110–126. doi: 10.1080/15287394.2011.529061. [DOI] [PubMed] [Google Scholar]
- 54.Almeida LM, Basu U, Williams JL, Moore SS, Guan LL. Microarray analysis in caudal medulla of cattle orally challenged with bovine spongiform encephalopathy. Genet. Mol. Res. 2011;10 doi: 10.4238/2011.October.25.5. DOI: http://dx.doi.org/10.4238 . [DOI] [PubMed] [Google Scholar]
- 55. Basu U, Almeida L, Olson NE, Meng Y, Williams JL, Moore SS, Guan LL. Transcriptome analysis of the medulla tissue from cattle in response to bovine spongiform encephalopathy using digital gene expression tag profiling. J. Toxicol. Environ. Health A. 2011;74:127–137. doi: 10.1080/15287394.2011.529062. [DOI] [PubMed] [Google Scholar]
- 56.Lloyd S, Mead S, Collinge J. Genetics of prion diseases. Topics Curr. Chem. 2011;305:1–22. doi: 10.1007/128_2011_157. [DOI] [PubMed] [Google Scholar]
- 57.Booth S, Bowman C, Baumgartner R, Sorensen G, Robertson C, Coulthart M, Phillipson C, Somorjai RL. Identification of central nervous system genes involved in the host response to the scrapie agent during preclinical and clinical infection. J. Gen. Virol. 2004;85:3459–3471. doi: 10.1099/vir.0.80110-0. [DOI] [PubMed] [Google Scholar]
- 58.Brown AR, Rebus S, McKimmie CS, Robertson K, Williams A, Fazakerley JK. Gen expression profiling of the preclinical scrapie-infected hippocampus. Biochem. Biophys. Res. Commun. 2005;334:86–95. doi: 10.1016/j.bbrc.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 59.Xiang W, Windl O, Wunsch G, Dugas M, Kohlmann A, Dierkes N, Westner IM, Kretzschmar HA. Identification of differentially expressed genes in scrapie-infected mouse brains by using global gene expression technology. J. Virol. 2004;78:11051–11060. doi: 10.1128/JVI.78.20.11051-11060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skinner PJ, Abbassi H, Chesebro B, Race RE, Reilly C, Hasse AT. Gene expression alterations in brains of mice infected with three strains of scrapie. BMC Genomics. 2006;7:114–127. doi: 10.1186/1471-2164-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim HO, Snyder GP, Blazey TM, Race RE, Chesebro B, Skinner PJ. Prion disease induced alterations in gene expression in spleen and brain prior to clinical symptoms. Adv Appl. Bioinform. Chem. 2008;1:29–50. doi: 10.2147/aabc.s3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang D, Lee IY, Yoo H, Gehlenborg N, Cho J, Petritis B, Baxter D, Pitstick R, Young R, Spicer D, Price ND, Hohmann JG, DeArmond SJ, Carlson GA, Hood LE. A systems approach to prion disease. Mol. Systems Biol. 2009;5:252–255. doi: 10.1038/msb.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huzarewich RL, Medina S, Robertson C, Parchaliuk D, Booth SA. Transcriptional modulation in a leukocyte-depleted splenic cell population during prion disease. J. Toxicol. Environ. Health A. 2011;74:1504–1520. doi: 10.1080/15287394.2011.618979. [DOI] [PubMed] [Google Scholar]
- 64.Tortosa R, Castells X, Vidal E, Costa C, Ruiz de Villa Mdel C, Sánchez A, Barceló A, Torres JM, Pumarola M, Ariño J. Central nervous system gene expression changes in a transgenic mouse model for bovine spongiform encephalopathy. Vet. Res. 2011;42:109–123. doi: 10.1186/1297-9716-42-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang Y, Xiang W, Hawkins SAC, Kretzschmar HA, Windt O. Transcriptional changes in the brains of cattle orally infected with the bovine spongiform encephalopathy agent precede detection of infectivity. J. Virol. 2009;83:9464–9473. doi: 10.1128/JVI.00352-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Y, Xiang W, Terry L, Kretzschmar HA, Windl O. Transcriptional analysis implicates endoplasmic reticulum stress in Bovine Spongiform Encephalopathy. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0014207. e14207. doi:10.1371/journal.pone.0014207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panelli S, Strozzi F, Capoferri R, Barbieri I, artinelli N, Capucci L, Lombardi G, Williams JL. Analysis of gene expression in white blood cells of cattle orally challenged with Bovine amyloidotic spongiform encephalopathy. J. Toxicol. Environ. Health, Part A. 2011;74:96–102. doi: 10.1080/15287394.2011.529059. [DOI] [PubMed] [Google Scholar]
- 68.Filali H, Martin-Burriel I, Harders F, Varona L, Lyahyai J, Zaragoza P, Pumarola M, Badiola JJ, Bossers A, Bolea R. Gene expression profiling and association with prion-related lesions in the medulla oblongata of symptomatic natural scrapie animals. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0019909. e19909. doi: 10.1371/journal.pone.0019909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gossner A, Roupaka S, Foster J, Hunter N, Hopkins J. Transcriptional profiling of peripheral lymphoid tissue reveals genes and networks linked to SSBP/1 scrapie pathology in sheep. Vet. Microbiol. 2011;153:218–228. doi: 10.1016/j.vetmic.2011.05.036. [DOI] [PubMed] [Google Scholar]
- 70.Basu U, Almeida LM, Dudas S, Graham CE, Czub S, Moore SS, Guan LL. Gene expression alterations in rocky mountain elk infected with chronic wasting disease. Prion. 2012. [DOI] [PMC free article] [PubMed]
- 71.Benetti F, Gustincich S, Legname G. Gene expression profiling and therapeutic interventions in neurodegenerative diseases: a comprehensive study on potentiality and limits. Expert Opin. Drug Discov. 2012;7:245–258. doi: 10.1517/17460441.2012.659661. [DOI] [PubMed] [Google Scholar]
- 72.Chiti F, Dobson CM. Protein misfolding, functional amyloid and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 73.Jellinger KA. Interaction between pathogenic proteins in neurodegenerative disorders. J. Cell Mol. Med. 2011. DOI: 10.1111/j.1582-4934.2011.01507.x. [DOI] [PMC free article] [PubMed]
- 74.Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nature Rev. Neurosci. 2010;11:155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guest WC, Silverman M, Pokrishevsky E, O’Neil MA, Grad LI, Cashman NR. Generalixation of the prion hypothesis to other neurodegenerative diseases: An imperfect fit. J. Toxicol. Environ. Health, Part A. 2011;74:1433–1459. doi: 10.1080/15287394.2011.618967. [DOI] [PubMed] [Google Scholar]
- 76.Castellani RJ, Perry G, Smith MA. Prion diseases and Alzheimer’s disease, pathogenic overlap. Acta. Neurobiol. Exp. 2004;64:11–17. doi: 10.55782/ane-2004-1487. [DOI] [PubMed] [Google Scholar]
- 77.Cisse M, Mucke L. A prion protein connection. Nature. 2009;257:1090–1091. doi: 10.1038/4571090a. [DOI] [PubMed] [Google Scholar]
- 78.Morales R, Estrada LD, Diaz-Espinoxa R, Morales-Scheihing D, Jara MC, Castilla J, Soto C. Molecular cross talk between misfolded protein in animal models of Alzheimer’s and prion diseases. J. Neurosci. 2010;30:4528–4535. doi: 10.1523/JNEUROSCI.5924-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soto C. Transmissible protein: Expanding the prion heresy. Cell. 2012;149:968–977. doi: 10.1016/j.cell.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Milhavet O, Lehamnn S. Oxidative stress and the prion protein in transmissible spongiform encephalopathis. Brain Research Reviews. 2002;38:328–339. doi: 10.1016/s0165-0173(01)00150-3. [DOI] [PubMed] [Google Scholar]
- 81.Mancuso M, Siciliano G, Filosto M, Murri L. Mitochondrial dysfunction and Alzheimer’s disease: new developments. J. Alz. Dis. 2006;9:111–117. doi: 10.3233/jad-2006-9203. [DOI] [PubMed] [Google Scholar]
- 82.Soto C, Satani N. The intricate mechanisms of neurodegenration in prion diseases. Trends Mol Medicine. 2011;17:14–24. doi: 10.1016/j.molmed.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giasson BI, Lee VM-Y, Trojanowski JQ. The relationship between oxidative/nitrative stress and pathological inclusions in Alzheimer's and Parkinson's diseases. Free Radical Biol. Medicine. 2002;32:1264–1275. doi: 10.1016/s0891-5849(02)00804-3. [DOI] [PubMed] [Google Scholar]
- 84.Provost P. MicroRNAs as a molecular basis for mental retardation, Alzheimer's and prion diseases. Brain Res. 2010;1338:58–66. doi: 10.1016/j.brainres.2010.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lukiw WJ, Dua P, Pogue AI, Eicken C, Hill JM. Upregulation of micro RNA-146a (miRNA-146a), a marker for inflammatory neurodegeneration, in sporadic Creutzfeldt-Jakob disease (sCJD) and Gerstmann-Straussler-Scheinker (GSS) syndrome. J. Toxicol. Environ. Health A. 2011;74:1460–1468. doi: 10.1080/15287394.2011.618973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- 88.Chahwan R, Wontakal SN, Roa S. The multidimensional nature of epigenetic information and its role in disease. Discov. Med. 2011;11:233–243. [PubMed] [Google Scholar]