Abstract
Cortical interneurons originate in the ganglionic eminences of the subpallium and migrate into the cortex in well-defined tangential streams. At the start of corticogenesis, two streams of migrating neurons are evident: a superficial one at the level of the preplate (PPL), and a deeper one at the level of the intermediate zone (IZ). Currently, little is known about the signalling mechanisms that regulate interneuron migration, and almost nothing is known about the molecules that may be involved in their choice of migratory stream. Here, we performed a microarray analysis, comparing the changes in gene expression between cells migrating in the PPL and those migrating in the IZ at embryonic day 13.5. This analysis identified genes, many of them novel, that were upregulated in one of the two streams. Moreover, polymerase chain reaction, in situ hybridization experiments and immunohistochemistry showed the expression of these genes in interneurons migrating within the PPL or IZ, suggesting that they play a role in their migration and choice of stream.
Keywords: developing cerebral cortex, interneurons, microarray, migration
Introduction
Interneurons, which constitute a morphologically, neurochemically and functionally diverse group of cortical cell types, are essential modulators of neuronal activity in the cerebral cortex. Abundant evidence indicates that alterations in the number, distribution and function of these GABA-releasing inhibitory neurons in humans may lead to neurological and psychiatric disorders (Benes & Berretta, 2001; Cossart et al., 2005; Gant et al., 2009). Thus, the mechanisms that control interneuron development have received considerable attention in recent years.
Tracing, fate-mapping and loss of function analyses in rodents have shown that the vast majority of cortical interneurons arise from the medial ganglionic eminence (MGE) and caudal ganglionic eminence (Tamamaki et al., 1997; Lavdas et al., 1999; Nery et al., 2002; Xu et al., 2004) and from the embryonic preoptic area (Gelman et al., 2009). These studies have also traced in detail the three tortuous migratory paths that interneurons follow from their origins in the subpallium to the cortex (reviewed by Corbin et al., 2001; Marín & Rubenstein, 2003; Métin et al., 2006). Specifically, an early cohort [embryonic day (E)12 in the mouse] first reaches the cortex and migrates at the level of the preplate (PPL). Slightly later in development (E13–15), a second and more prominent cohort migrates predominantly through the intermediate zone (IZ). At the later stages of corticogenesis and after the formation of the cortical plate (CP), three distinct tangential migratory streams are in evidence in the developing cortex, located in the marginal zone (MZ), lower IZ/subventricular zone (SVZ), and subplate (SP) (Lavdas et al., 1999; Anderson et al., 2001; Métin et al., 2006). Intricate molecular mechanisms are at play in the subpallium, repelling interneurons from their origins and sorting them into striatal or cortical types. The molecules involved in these processes include the Slit proteins and their Robo receptors, as well as the class 3 semaphorin family and their neuropilin and plexin receptors (Marín et al., 2001; Andrews et al., 2006; Barber et al., 2009; Hernández-Miranda et al., 2011). Migrating interneurons destined for the cortex utilize attractive cues, prominent among them neuregulin1/ErbB4 (Flames et al., 2004), to move round the corticostriatal notch and enter the cortical mantle (reviewed by Hernández-Miranda et al., 2010).
A number of factors have been identified as regulators of tangential (and radial) interneuron migration, including neurotrophic factors (brain-derived neurotrophic factor, neurotrophin 4, and glial cell-derived neurotrophic factor) (Polleux et al., 2002; Pozas & Ibáñez, 2005) and the chemokine CXCL12 (Stumm et al., 2003; Liapi et al., 2008). CXCL12 is produced by meningeal cells, Cajal–Retzius (CR) cells in the MZ, and cells in the IZ/SVZ, consistent with a role for this molecule in the intracortical guidance of cortical interneurons (Borrell & Marín, 2006; Stumm et al., 2007). In mice deficient in this chemokine, or its receptor CXCR4, interneurons alter their tangential migratory routes and invade the CP prematurely (Stumm et al., 2003; Tiveron et al., 2006; Li et al., 2008; Lopez-Bendito et al., 2008).
But what are the factors that determine the choice of stream by migrating interneurons as they enter the cortex? We reasoned that there exist molecular/genetic differences between interneurons that underlie their choice of one of the three tangential pathways. Here, we sought to identify genes involved in migratory stream specification by comparing the gene expression profiles of cells in the PPL with those of cells migrating through the IZ in early corticogenesis. We performed laser capture microdissection (LCM) on the cortices of glutamic acid decarboxylase-67 (GAD-67)–green fluorescent protein (GFP) transgenic mice (Tamamaki et al., 2003) in order to isolate interneuron-enriched populations of cells from the two zones. Subsequent microarray analysis revealed a large number of genes that are differentially expressed in the two cell populations, including those encoding cell surface proteins and regulators of intracellular signalling pathways. We focused on genes that are specifically upregulated in each stream, and examined their expression profiles by in situ hybridization. Our results support a role for a number of mostly novel genes in the migration of interneurons along specific routes.
Materials and methods
Animals
All experimental procedures were performed in accordance with the UK Animals (Scientific Procedures) Act 1986 and institutional guidelines. GAD67–GFP (Δneo) mice (Tamamaki et al., 2003) were maintained on a C57/BL6J background. The day on which the vaginal plug was found was considered to be E0.5. Mice of both sexes were used in all experiments.
LCM
Embryonic brains (E13.5) were dissected in RNase-free phosphate-buffered saline (PBS), placed in cryostat moulds, and frozen in Tissue-Tek OCT (Sakura Finetek Europe, Zoeterwoude, The Netherlands). Forebrains were sectioned, allowed to adhere to LCM membrane-mounted slides (Zeiss MicroImaging, Jena, Germany), and stored at −80 °C until use. For LCM, slides were individually thawed for 30 s, fixed in cold methanol for 1 min, and rinsed rapidly in PBS. Slides were dehydrated through 70–100% ethanol, and allowed to dry thoroughly (30 s–1 min). GAD67–GFP-positive cells were excised from the PPL and IZ within 15 min of drying, by use of a Zeiss Palm Microbeam system (Zeiss MicroImaging). GAD67–GFP-positive cells were allowed to adhere to capture tube lids (Zeiss MicroImaging). Tubes were placed on dry ice and kept at −80 °C until RNA extraction.
RNA extraction and microarray analysis
Total RNA from cortical PPL-derived and IZ-derived cells was extracted immediately after collection with the Qiagen RNeasy Miniplus kit (Qiagen, Chatsworth, CA, USA). RNA was sent to the Wolfson Institute for Biomedical Research (UCL Genomics, London, UK) for cDNA production, hybridization, and scanning. The quality of the RNA was assessed with an Agilent bioanalyser nanochip (Agilent, Palo Alto, CA, USA). All RNA had 18S and 28S rRNA bands. RNA (100 ng per chip) was converted to single-strand, sense-strand cDNA with the Affymetrix Sense target labelling protocol and the Mouse Gene 1.0ST Array kit (Affymetrix, High Wycombe, UK). After fragmenting and end-labelling, the cDNA was hybridized to Mouse Gene Gene-1_0-st-v1 Arrays (Affymetrix) at 45 °C for 16 h according to the manufacturer’s instructions. The arrays were then washed and stained on the Fluidics station 450 with the hybridization, wash and stain kits, and scanned on the GeneChip Scanner 3000. Analysis of microarray data was performed at the Bloomsbury Centre for Bioinformatics, Department of Computer Science (UCL). Raw data were summarized and normalized with the rma algorithm (Irizarry et al., 2003) implemented in the Affymetrix Expression Console software. limma (Linear Models for Microarray Analysis) (Smyth, 2004) was used to identify differentially expressed genes. limma applies a modified t-test to each probe set that uses an empirical Bayes approach for estimating sample variances. The moderated t-statistic calculated by limma is more robust than the ordinary t-statistic with small sample sizes. The P-values were corrected for multiple testing with the Benjamini–Hochberg correction, and a corrected P-value threshold of 0.01, together with a fold cut-off of > 2, was used to select differentially expressed genes.
Fluorescence-activated cell sorting (FACS)
Timed pregnant dams were killed at E13.5 and E15.5. Embryonic brains were dissected in cold artificial cerebrospinal fluid. The forebrain was isolated, and the meninges, olfactory bulb and septum were removed. The cortex and ganglionic eminence (GE) were separated, and the (presumptive) hippocampus was separated from the cortex. Cortex and GE cells were dissociated by incubation in 0.05% trypsin with 100 μg/mL DNase I in Neurobasal medium (Invitrogen, Paisley, UK) at 37 °C for 15 min. Trypsinization was quenched by addition of neurobasal medium containing 10% heat-inactivated fetal bovine serum (Invitrogen) at 37 °C for 5 min. Cells were washed three times in Neurobasal medium (without fetal bovine serum) to remove serum for FACS. Cells were resuspended in Neurobasal medium without phenol red (Invitrogen) containing l-glutamine (Invitrogen) and B-27 supplement (1: 50; Invitrogen). Dissociated cells from 8 to 10 embryos were pooled for each FACS. FACS was performed by the Wolfson Scientific Support Services (UCL) with a MoFlo Sorter (Dako, Copenhagen, Denmark). A non-green embryo was used as a control for fluorescence. Cells were excited with a 488-nm argon laser and detected with a 530/40 (FL1) bandpass filter. A cell purity of 95–98.5% was obtained for each sorting.
Quantitative polymerase chain reaction (qPCR) validation
For validation of the differentially expressed genes, qPCR was performed on 20 genes. Embryonic dissection, LCM and RNA extraction were performed as previously described, and RNA was treated with DNase I (Amplification grade; Invitrogen) to remove any remaining trace amounts of DNA. cDNA was generated with 20 ng of RNA by use of the Qiagen Whole Transcriptome Amplification Kit (Qiagen), as described in the manufacturer’s protocol. Primers for qPCR were designed by SigmaGenosys (Sigma-Aldrich, Poole, UK), and were as shown in Supporting Information Table S1. The qPCR reaction was performed with SYBR Green reagent (Sigma, Poole, UK) on a Chromo4 PTC-200 Real-Time PCR Detector system (Bio-Rad, Hercules, CA, USA). Polymerase chain reaction (PCR) conditions were 94 °C for 2 min, followed by 40 three-step cycles of 94 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin were used for endogenous reference gene controls. Each primer set amplified a single PCR product of predicted size as determined by melt-curve analysis following PCR and by agarose gel electrophoresis, and had approximately equal amplification efficiencies when validated with a serial dilution of representative cDNA. Each qPCR was performed in triplicate, and relative quantification was determined according to the ΔΔc(t) method (Livak & Schmittgen, 2001).
In situ hybridization
In situ hybridization was performed as described previously (Faux et al., 2010). Briefly, embryonic brains were dissected in PBS and fixed in 4% paraformaldehyde in PBS for 4 h at 4 °C, and this was followed by cryoprotection in 30% diethyl pyrocarbonate-treated sucrose in PBS overnight at 4 °C. Brains were frozen in Tissue-Tek OCT (Sakura Finetek) and sectioned with a cryostat (20 μm; Bright Instruments, Huntingdon, UK). Sections were dried at room temperature for 2 h before overnight incubation at 65 °C in hybridization buffer [1 × diethyl pyrocarbonate-treated ‘salts’ (200 mm NaCl, 5 mm EDTA, 10 mm Tris, pH 7.5, 5 mm NaH2PO4.2H2O, 5 mm Na2HPO4); 50% deionized formamide (Ambion, Austin, TX, USA); 0.1 mg/mL RNase-free yeast tRNA (Invitrogen); 1 × Denhardts (RNase/DNase-free; Invitrogen); 10% dextran sulphate (Sigma)] containing 100–500 ng/mL digoxigenin-labelled RNA probes. Probes were generated by linearization of plasmids with appropriate enzymes and reverse transcription PCR to obtain antisense probes. A number of probes were obtained from the Max-Planck Institute of Biophysical Chemistry, Göttingen, Germany (Supporting Information Table S2). Other probes (Lhx6 and Reelin) were a kind gift from N. Kessaris (Wolfson Institute, UCL, UK). Following hybridization, sections were washed three times in wash solution (50% formamide, 1 × SSC, 0.1% Tween-20) at 65 °C and twice at room temperature in 1 × MABT (20 mm maleic acid, 30 mm NaCl, 0.1% Tween-20) before being incubated in blocking solution [2% blocking reagent (Roche Applied Science, Burgess Hill, UK), 10% normal goat serum (Vector, Burlingame, CA, USA) in MABT] and then overnight in alkaline phosphatase-conjugated anti-digoxigenin antibody (1: 1500; Roche Applied Science). Nitro Blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate diluted 1: 1000 in MABT with 5% poly(vinyl alcohol) was used for colorimetric detection at 37 °C for 8–20 h. Fast Red (Roche Applied Science) was used for fluorescent colour detection of probes by incubation in 100 mm Tris (pH 8.0) and 400 mm NaCl containing Fast Red for approximately 2 h at 37 °C. Fluorescent in situ hybridization was followed by immunohistochemical detection of GFP as described below. Sections were mounted with Glycergel Mounting Medium (Dako). Photographs were taken with a Leica DM microscope and a Leica DC 500 digital camera. All images were processed with Photoshop CS2 software (Adobe, San Jose, CA, USA).
Immunohistochemistry
Embryonic brains and cryosections were prepared as previously described. Sections and dissociated cortices were blocked for 1 h in PBS containing 5% normal goat serum, and then incubated in rabbit polyclonal anti-Cnr1 (1: 100; Sigma-Aldrich) and rabbit polyclonal anti-Dab1 (1: 100; Sigma-Aldrich) at room temperature overnight. They were then washed in PBS and incubated in biotinylated goat anti-rabbit (1: 200; Vector Laboratories) for 2 h. Antibody staining was enhanced with a tyramide signal amplification system (Perkin Elmer, Boston, MA, USA), according to the manufacturer’s instructions. Sections were washed and incubated with 4′,6-diamidino-2-phenylindole (1: 20 000; Sigma-Aldrich). Images were collected with an SP2 Leica confocal microscope. Sequential images were subsequently reconstructed with Metamorph imaging software (Universal Imaging Corporation, West Chester, PA, USA).
Results
Isolation of PPL and IZ cells by LCM
Examination of the forebrains of GAD67–GFP transgenic mice during corticogenesis revealed cells undergoing tangential migration (Fig. 1A), as previously described (Tamamaki et al., 2003; Métin et al., 2006). At E13.5, GFP-positive (interneurons) cells were observed primarily in the PPL and IZ (Fig. 1B). Using LCM on coronally cut sections at this age, we isolated GAD67–GFP-enriched populations of cells from these two zones (Fig. 1C and D).
Figure 1.
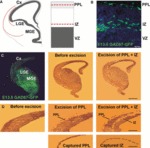
Tangential migration of interneurons into the cerebral cortex. (A) Schematic diagrams depicting the streams of migrating interneurons at E13.5. Red lines indicate the laminar positions of the PPL and IZ streams. (B) Coronal section through the cortex of an E13.5 GAD67–GFP transgenic mouse showing abundant migrating cells in the PPL and IZ streams. (C and D) Intact section through the forebrain of one hemisphere of an E13.5 GAD67–GFP transgenic mouse and after excision and capture of the PPL and IZ with a laser-capture microscope. Scale bars: (A) 100 μm; (C and D) 500 μm. Cx, cerebral cortex; LGE, lateral ganglionic eminence; VZ, ventricular zone.
Microarray analysis and validation
In order to identify genes that may be involved in the choice of migratory stream by cortical interneurons, we compared gene expression in the PPL with that in cells isolated from the IZ by performing microarray analysis. Genes were considered to be differentially expressed if a greater than two-fold change in expression was found, together with a corrected P-value threshold of 0.05. The number of genes upregulated in PPL cells at E13.5 was 113, and that in IZ cells was 69. In order to examine more closely the overall changes in expression, genes were classified into six categories according to their molecular function (Fig. 2).
Figure 2.
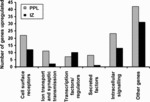
Numbers of genes upregulated in the PPL (grey bars) and IZ (black bars) migratory streams at E13.5. Genes were classified into the categories listed according to their molecular function.
As an initial validation of our microarray data, we examined changes in expression of Reelin and Dact1, genes that are known to be strongly expressed in the PPL (Ogawa et al., 1995; Faux et al., 2010; respectively), Robo2, which is highly expressed in the IZ (Andrews et al., 2007), and Lhx6, which is expressed by all cortical interneurons and has been shown to play a crucial role in their migration (Alifragis et al., 2004). As expected, Reelin and Dact1 were both expressed at higher levels in the PPL than in the IZ (Supporting Information Tables S6 and S7, respectively), whereas the opposite was the case for Robo2 expression (Supporting Information Table S9). No significant changes were observed in the levels of expression of Lhx6 or in specific interneuron subtype markers such as calbindin, calretinin, and somatostatin (data not shown).
Genes with higher expression levels in the PPL are listed in Supporting Information Tables S3–S8, and those with higher expression levels in the IZ are shown in Supporting Information Tables S9–S14. qPCR, carried out on a set of eight genes that showed higher expression levels in the PPL and on four that showed higher expression levels in the IZ at E13.5, was subsequently used to further validate the observed changes in expression (Tables 1 and 2). In this analysis, all genes were found to have fold changes in the same direction as the microarray. Interestingly, the fold changes observed by qPCR were much higher than those found with microarray, in agreement with previous observations (Faux et al., 2010). Thus, our microarray analysis, together with the qPCR, identified a number of genes with upregulated expression in specific streams of migrating cortical interneurons.
Table 1.
qPCR comparing expression profiles of cell surface receptor genes that are upregulated in PPL cells as compared with IZ cells
| Upregulated in PPL | Upregulated in IZ | |||
|---|---|---|---|---|
| Gene | Microarray | qPCR | Microarray | qPCR |
| Cdh8 | ++ | +++ | ||
| Cnr1 | + | +++ | ||
| Dact1 | + | ++ | ||
| Epha3 | + | +++ | ||
| Flrt2 | + | +++ | ||
| Lhx6 | NC | NC | NC | NC |
| Mc4r | + | +++ | ||
| Nelf | + | +++ | ||
| Neuritin | + | +++ | ||
| Ptpro | + | +++ | ||
| Reelin | + | +++ | ||
| Robo2 | + | +++ | ||
NC, no change; +, 2–5-fold greater; ++, 5–15-fold greater; +++, > 15-fold greater. Reference genes are in bold.
Table 2.
qPCR comparing expression profiles of intracellular signalling genes that are upregulated in PPL cells as compared with IZ cells
| Upregulated in PPL | Upregulated in IZ | |||
|---|---|---|---|---|
| Gene | Microarray | qPCR | Microarray | qPCR |
| Cdc42ep3 | + | +++ | ||
| Dab1 | + | +++ | ||
| Dact1 | + | ++ | ||
| Lhx6 | NC | NC | NC | NC |
| Plcb1 | + | +++ | ||
| Rasgef1b | + | +++ | ||
| Reelin | + | +++ | ||
| Robo2 | + | +++ | ||
NC, no change; +, 2–5-fold greater; ++, 5–15-fold greater; +++, > 15-fold greater. Reference genes are in bold.
Cell surface receptors – new candidate genes for choice of migratory stream
Cell surface molecules are involved in essential developmental processes, such as migration, neurite outgrowth, and synapse formation. We present genes thought to be involved in cellular interactions that are enriched within the PPL (Supporting Information Table S3) and IZ (Supporting Information Table S9) interneuron streams at E13.5. With the exception of Cnr1 (Morozov et al., 2009), EphA4 (Rudolph et al., 2010), Nrp1 (Marín et al., 2001), Robo1 (Andrews et al., 2006), Robo2 (Andrews et al., 2008), and Sstr2 (Beneyto et al., 2011), these receptor genes have not previously been implicated in cortical interneuron development. However, two recent microarray studies that examined the differential expression of genes between interneurons and non-interneurons (presumptive pyramidal cells) in the cortex, and between cortical interneurons and GE cells, identified a number of cell surface receptor genes that are expressed in the cortical interneuron population (Batista-Brito et al., 2008; Faux et al., 2010). Some of these genes, including Alcam, Cdh8, Cdh10, Csmd3, Fat3, Flrt2, Islr2, Mdga2, Nrn1, Nelf, Pcdh11x, Plxnc1, Ptpro, and Ptprz1, were also identified here, and appear to be differentially expressed in the two migratory streams analysed (Supporting Information Tables S3 and S9). We subsequently performed in situ hybridization at E13.5 and E15.5, the early phase and mid-phase of tangential interneuron migration, for a selection of these genes to further confirm their expression in the PPL/MZ or IZ/SVZ.
Several receptor genes demonstrated strong specific expression in the PPL, but not in the IZ, at E13.5 (Fig. 3, upper panels). These included the G-protein-coupled cannabinoid 1 receptor gene (Cnr1) (Fig. 3C and C′), Flrt2, which encodes a glycosylated membrane protein that acts as a regulator of fibroblast growth factor signalling (Fig. 3E and E′), the nasal embryonic luteinizing hormone-releasing hormone factor gene (Nelf) (Fig. 3G and G′), and the protein tyrosine phosphotase receptor type O gene (Ptpro) (Fig. 3I and I′. At E15.5, all four genes showed increased expression in the MZ and CP, but not in the IZ/SVZ (Fig. 3D, D′, F, F′, H, H′, J, and J′), confirming previous cortical expression studies on Cnr1 (Berrendero et al., 1998) and Nelf (Kramer & Wray, 2001). We also observed weak expression of Flrt2, Nelf and Ptpro in the mantle zone of the MGE (Fig. 3E, F, G, H, I, and J), suggesting that they may have other developmental function(s) in addition to a specific role in tangential interneuron migration within the cortical PPL/MZ.
Figure 3.
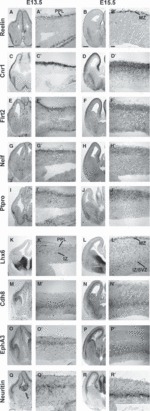
Expression of receptor genes in the interneuron migratory streams in the developing forebrain as seen by in situ hybridization at E13.5 and E15.5. A higher-magnification image of the cortex is shown beside each low-magnification panel of the forebrain. The upper panels show the expression of receptor genes in the PPL at E13.5 and MZ at E15.5. The lower panels show the expression of receptor genes predominantly in the IZ at E13.5 and E15.5. (A–B′) The expression of Reelin was used as an internal control, as it is known that it is expressed exclusively in cells (presumptive CR cells) in the PPL at E13.5 and in the MZ at E15.5. (C–J′) Expression of receptor genes Cnr1 (C–D′), Flrt2 (E–F′), Nelf (G–H′) and Ptpro (I–J) was localized predominantly within the PPL at E13.5 and within the MZ at E15.5. (K–L′) The interneuron marker Lhx6 was also used as an internal control, as it is known to be expressed in both the PPL and IZ at E13.5 (K–K′), and more widely, but predominantly in the MZ and IZ/SVZ, at E15.5 (L–L′). (M–R′) Expression of the receptor genes Cdh8 (M–N′), EphA3 (O–P′) and Neuritin (Q–R′) within the IZ at E13.5 and within the IZ/SVZ at E15.5. Scale bar in A–B′: 200 μm.
Specific expression of several receptor genes was evident in the IZ at E13.5 and E15.5 (Fig. 3, lower panels). These included the calcium-dependent cell adhesion glycoprotein cadherin-8 gene (Cdh8) (Fig. 3M–N′) (Takeichi, 1988), the ephrin-A tyrosine kinase receptor A3 gene (EphA3) (Fig. 3O–P′), and the glycosylphosphatidylinositol-linked neuritin receptor gene [(Neuritin, also known as Cpg15) (Fig. 3Q–R′)]. At E13.5, all three genes appeared to be specifically expressed in the IZ interneuron stream. At E15.5, they were expressed in the IZ/SVZ interneuron stream, but Cdh8 and EphA3 showed expanded expression within the IZ, confirming previous studies that localized the products of these genes in thalamocortical and commissural fibres (Korematsu & Redies, 1997; Kudo et al., 2005). Furthermore, we observed varying levels of expression for Cdh8 and EphA3 in different sites of the subpallium (Fig. 3M, N, O, and P), in agreement with earlier reports (Korematsu & Redies, 1997; Kudo et al., 2005).
Cell signalling pathways – new candidate genes for choice of migratory stream
In addition to cell surface receptor genes that can directly affect cell migration and axon guidance events, our microarray analysis also identified various genes that modulate cell signalling pathways, which in turn can influence cell migration. In Supporting Information Tables S7 and S13, we list genes involved in intracellular signalling pathways whose expression is enriched within the PPL (Supporting Information Table S7) and IZ (Supporting Information Table S13). With the exception of Dab1 (Hammond et al., 2006), Dact1 (Faux et al., 2010), Prkra (Deng et al., 2009), and Syngap1 (Muhia et al., 2010), all other genes have not previously been implicated in cortical interneuron development. However, recent microarray studies identified a number of signalling genes that are expressed in cortical interneuron populations (Batista-Brito et al., 2008; Faux et al., 2010). These included Bub1, Cdc42ep3, Dab1, Dcn, Elmod1, Gng5, Plk2, Rasgef1b and Unc5d, which were confirmed here and appear to be expressed differentially in the two streams (Supporting Information Tables S7 and S13).
Using in situ hybridization, we examined the expression of several signalling genes that showed significant levels of expression in either the PPL/MZ (Fig. 4, upper panels) or the IZ (Fig. 4, lower panels). For example, Dab1 showed specific expression in the PPL, but not in the IZ, at E13.5 (Fig. 4C and C′) and, at E15.5, it showed stronger expression in the MZ and CP, as well as in the SP interneuron stream (Fig. 4D and D′). Three signalling genes that showed specific expression in the IZ interneuron stream at E13.5 included the CDC42 effector protein (Rho GTPase binding) 3 gene (Cdc42ep3) (Fig. 4G and G′), the phosphoinositide-specific phospholipase Cβ1 gene (Plcb1) (Fig. 4I and I′), and the gene encoding RasGEF1b, a guanine-nucleotide exchange factor (Rasgef1b) (Fig. 4K and K′). At E15.5, both Cdc42ep3 and Rasgef1b expression appeared to be limited to the IZ/SVZ interneuron stream (Fig. 4H, H, respectively), whereas Plcb1 showed an expanded expression band around the IZ (Fig. 4J and J′), in agreement with previous findings (Watanabe et al., 1998).
Figure 4.
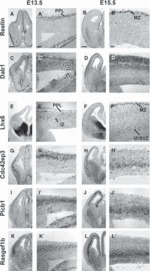
Expression of cell signalling genes in the interneuron migratory streams in the developing forebrain as seen by in situ hybridization at E13.5 and E15.5. A higher-magnification image of the cortex is shown beside each low-magnification panel of the forebrain. The upper panels show the expression of receptor genes in the PPL at E13.5 and in the MZ at E15.5. The lower panels show the expression of receptor genes predominantly in the IZ at E13.5 and E15.5. (A–B′) The expression of Reelin was used as an internal control, as it is known that it is expressed exclusively in cells (presumptive CR cells) in the PPL at E13.5 and in the MZ at E15.5. (C–D′) Expression of Dab1 is observed only within the PPL at E13.5 and in the MZ and SP (after the splitting of the CP) at E15.5. (E–F′) The interneuron marker Lhx6 was also used as an internal control, as it is known to be expressed in both the PPL and IZ at E13.5 (E–E′), and more widely, but predominantly in the MZ and IZ/SVZ, at E15.5 (F–F′). (G–L′) Expression of the cell signalling genes Cdc42ep3 (G–H′), Plcb1 (I–J′) and Rasgef1b (K–L′) was observed predominantly within the IZ at E13.5, and within the IZ/SVZ at E15.5. Scale bar in A–B′: 200 μm.
Although PPL and IZ cell populations isolated by LCM contain predominantly interneurons, they are likely to also contain other cell types. Thus, the PPL population undoubtedly also included CR cells, and the IZ samples were likely to also contain pyramidal neurons migrating through this zone en route to the CP. To confirm whether all genes identified in each of the two migratory streams are indeed expressed in interneurons, we first assessed their expression by PCR in FACS populations of GAD67–GFP-positive and GFP-negative cells derived from the cortex and GAD67–GFP-positive GE cells at E13.5 and E15.5 (Fig. 5). With the single exception of Mc4r at E13.5, all other genes were shown to be expressed in both cortical and GE GAD67–GFP-positive populations at both ages.
Figure 5.
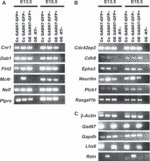
Agarose gel electophoresis of qPCR products. qPCR was performed on cortex-derived and GE-derived GAD67–GFP-positive and GAD67–GFP-negative cells at E13.5 and E15.5. The genes examined are listed next to the gel bands. (A) Genes shown to be upregulated in the cortical PPL/MZ. Nearly all genes were expressed in the three samples at both ages. The only exception was Mc4r, which was found to be expressed only in GAD67–GFP-negative cells (presumptive pyramidal neuron progenitors) in the cortex at E13.5. (B) Genes shown to be upregulated in the IZ. (C) Control genes. Cx, cortex; -RT, without reverse transcriptase (negative control).
To further confirm the expression of some of these genes in interneurons within specific tangential migratory streams, we performed double-labelling experiments. For example, we carried out immunofluorescence investigations for Cnr1 and Dab1 (red) on sections taken from GAD67–GFP brains at E15.5 (Fig. 6A–F). Co-localization between GFP and either Cnr1 or Dab1 (yellow) was observed in the MZ, but not in the IZ/SVZ (Fig. 6C and F), suggesting that some interneurons in the MZ express Cnr1 and Dab1. This was confirmed further by Cnr1 and Dab1 immunohistochemistry on GAD67–GFP-positive dissociated cortical cell cultures at E15.5 (Fig. 6G–L).
Figure 6.
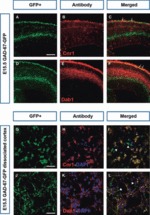
Expression of Cnr1 and Dab1 in interneurons. (A–F) Coronal sections from GAD67–GFP-positive mice at E15.5 were immunostained with anti-Cnr1 (B) and anti-Dab1 (E) (red). Co-localization (yellow; arrows) of Cnr1 and Dab1 with GFP is evident in some neurons in the MZ in the merged panels (C and F). (G–L) Dissociated cortical cell cultures prepared from E15.5 GAD67–GFP-positive cortices were immunostained with anti-Cnr1 (H) and anti-Dab1 (K). GFP single-positive cells are indicated by arrowheads, and Cnr1/GFP or Dab1/GFP double-positive cells are indicated by arrows in the merged panels (I and L). Scale bars: (A) 200 μm; (G) 15 μm.
Taken together, our findings have identified a number of genes whose expression is upregulated in specific interneuron populations, either within the PPL/MZ or within the IZ/SVZ, during corticogenesis. These genes may be potentially important for the migration of these cells and their choice of migratory stream.
Discussion
Since the discovery in the late 1990s that cortical interneurons have their origins in the subpallium, at least in rodents, numerous studies have traced in detail their long and tortuous migratory paths (reviewed in Marín & Rubenstein, 2003; Métin et al., 2006; Hernández-Miranda et al., 2010). These cells move round the corticostriatal notch, enter the neocortex, and migrate initially along well-defined tangential streams in the PPL/MZ, IZ, and SP, before moving radially to populate the CP. To date, only a few signalling and guidance molecules have been found that directly control their migration, and almost nothing is known about their choice of migratory stream. In the present study, we utilized a whole genome analysis of mRNAs expressed in the PPL and IZ at an early stage of corticogenesis. We focused our analysis on groups of genes (cell surface receptors and intracellular signalling) that are known to play a role in neuronal migration, both in the cortex and in other areas of the developing brain, as well as on genes that are thought to be involved in migration-related events such as neurite outgrowth and guidance, cell adhesion, and intracellular signalling. This analysis identified a number of genes that are differentially upregulated in the two streams and may be involved in the choice of pathway by migrating cortical interneurons.
Cell surface receptors
Many of the cell surface genes found to be upregulated in cortical interneurons have previously been shown to play a role in cell migration, either directly or indirectly, by regulating specific events that are essential for this process, such as neurite outgrowth and branching. One of the cell surface receptor genes identified here, Cnr1, appeared to be specifically upregulated in the PPL/MZ. Cnr1 is one of the most widely expressed G-protein-coupled receptors in the mammalian brain (Herkenham et al., 1991), and has been shown previously to be expressed in interneurons (Katona et al., 1999). In addition, it has been identified in pyramidal cells (Berrendero et al., 1998), in a subclass of hippocampal GABAergic interneurons containing the neuropeptide cholecystokinin (Katona et al., 1999), and in neuronal elements of the striatum (Rodriguez et al., 2001), a finding confirmed in our present study. Schizophrenia subjects show a decrease in Cnr1 mRNA and protein levels in the prefrontal cortex (Eggan et al., 2008), as well as diminished parvalbumin mRNA in the same region (Hashimoto et al., 2003). In an attempt to investigate the role of Cnr1 signalling, a recent analysis of Cnr1 null mice showed decreased parvalbumin immunoreactivity in the cortex and striatum (Fitzgerald et al., 2011), highlighting its importance in interneuron development. Recent evidence has also pointed to a role for Cnr1 in neuronal migration, as loss of function in a neural stem cell line and rostral migratory stream decreased migration and, conversely, direct activation resulted in increased migration (Oudin et al., 2011).
Our analysis also identified cell surface molecules, such as Nrp1, Robo1 and Robo2, that have previously been shown to be expressed in cortical interneurons and play an important role in their development (Marín et al., 2001; Andrews et al., 2006, 2008). Furthermore, it revealed the expression of genes encoding cell surface proteins, including Cdh8, Nelf, Pcdh19, Plxnd1, Sema5a, Sorl1, and Vcan, which have not previously been shown to be expressed in interneurons, but are known to play key roles in cell migration and neuronal differentiation in the developing brain. Thus, Cdh8 is expressed in the IZ/SVZ, but not specifically in interneurons within this zone (Korematsu & Redies, 1997), and recent studies have suggested that it regulates mossy fiber fasciculation and targeting (Bekirov et al., 2008). Nelf plays a role in the outgrowth of olfactory axons and migration of gonadotropin-releasing hormone neurons (Kramer & Wray, 2000; Xu et al., 2010), whereas Neuritin functions to coordinately regulate the growth of dendritic and axonal arbours and to promote synaptic maturation (Nedivi et al., 1998; Cantallops et al., 2000; Javaherian & Cline, 2005). Plxnd1 has been shown to control migration of thymocytes (Choi et al., 2008), and Sema5a acts as an axon guidance cue for axial motor neurons (Hilario et al., 2009); like Vcan, it has been reported to induce neuronal differentiation and promote neurite outgrowth (Wu et al., 2004). Whether these genes have similar functions in interneuron migration and development requires further experimental assessment.
Intracellular signalling pathways
In addition to cell surface receptor genes that can directly affect cell migration and axon guidance events, our microarray analysis also identified various genes that modulate cell signalling pathways, which, in turn, can influence developmental events such as migration, neurite outgrowth, and synaptic function. Included in this group of genes is Dab1, which encodes an intracellular adaptor that is expressed in cells that respond to Reelin (Howell et al., 1997). As such, it is an essential component of the Reelin signalling pathway, and is required for correct radial migration of cortical pyramidal neurons (Howell et al., 1997; Franco et al., 2011). It has been reported (Rice et al., 1998; Trommsdorff et al., 1999), and confirmed in our present in situ hybridization experiments, that Dab1 is also expressed in the MGE, where interneurons commence their migration towards the cortex. The importance of Reelin function in cortical interneuron development has been demonstrated in organotypic slice culture and in vivo experiments (Morante-Oria et al., 2003). Subsequent studies of Dab1−/− mice showed abnormal cortical interneuron layering (Pla et al., 2006). Furthermore, pulse labelling experiments indicated that early-born (E12.5) interneurons in Dab1 mutant brains show layer inversion, whereas the lamination of late-born interneurons is unaffected (Hammond et al., 2006), suggesting that expression of Dab1 in early-born interneurons is required for correct cortical layering.
In the present study, we have also identified intracellular signalling molecules known to play a role in developmental events, including neuronal migration, that have not previously been shown to be expressed in interneurons. A number of the genes encoding these molecules, including Gpr12, Plcb1, and Syngap1, have been implicated in neuronal process development (Tanaka et al., 2007; Spires et al., 2005; Carlisle et al., 2008), and Rgs6 has been implicated in neuronal differentiation (Liu et al., 2002). Like Sorbs2 and Prkce, they have been shown to have a key role in cell migration (Roignot et al., 2010; Solecki et al., 2004). Whether these genes fulfil such roles in cortical interneuron development remains to be determined.
Our PCR analysis of FACS populations of GAD67–GFP-positive and GAD67–GFP-negative cells derived from the cortex indicated that all but one of the genes found to be upregulated in the PPL and IZ are expressed in interneurons, but the majority are also expressed in non-interneurons. It is possible that the expression of genes identified in cellular elements surrounding migrating interneurons contributes to a permissive environment for their migration. For example, Robo receptors are expressed on axons coursing through the IZ and in interneurons (Andrews et al., 2006, 2008), and Robo homophilic–heterophilic interactions have been shown to be important for neurite outgrowth (Hivert et al., 2002). Thus, Robo receptors may be required for interneuron migration along IZ axons, similar to what has been postulated for the neural adhesion molecule TAG-1 (Denaxa et al., 2001). Similarly, Reelin, which is predominantly expressed by CR cells in the PPL/MZ and is known to act as a chemoattractant for migrating neurons (Ogawa et al., 1995), may also contribute to a permissive environment that promotes the migration of interneurons. In support of this possibility is our finding that interneurons express the Reelin signalling molecule Dab1.
Interestingly, although a significant proportion of the genes found to be upregulated in the PPL and IZ migratory streams have so far not been associated with interneuron development, several have been linked to human neurological disorders. Thus, mutations in Nelf have been identified in patients with normosmic hypogonadotropic hypogonadism and Kallmann syndrome (Xu et al., 2010, 2011). Similarly, Nrp1 (Supporting Information Table S9), which shows a similar expression pattern to Nelf within gonadotropin-releasing hormone neurons, but is also expressed by cortical interneurons (Marín et al., 2001), has been suggested to play a role in the aetiology of hypogonadotropic hypogonadism (Cariboni et al., 2011). Mutations in Cdh8 and Plcb1 are associated with susceptibility to autism and the development of early-onset epilepsy, respectively (Pagnamenta et al., 2011; Kurian et al., 2010). Furthermore, alterations in mRNA levels of Cnr1, CDC42 and neuritin have been linked to the development of schizophrenia (Eggan et al., 2008; Hill et al., 2006; Chandler et al., 2010), and alterations in the level of Sorl1 mRNA have been linked to the development of Alzheimer’s disease (Kölsch et al., 2009). The observations that many of the genes identified here are associated with neurological disorders suggests that disruption of cortical interneuron development may contribute to the underlying aetiology of these disorders. Thus, future studies should aim at elucidating the function of these genes in cortical interneuron migration and development.
Acknowledgments
We would like to thank Y. Yanagawa and K. Obata for providing the GAD67–GFP transgenic mice, N. Kessaris for a number of in situ probes, and Mary Rahman for technical support. Funding for the research was provided by grants from the Wellcome Trust (Programme Grants 074549 to J. G. Parnavelas, and 089775 to J. G. Parnavelas and W. D. Andrews).
Glossary
- CP
cortical plate
- CR
Cajal–Retzius
- E
embryonic day
- FACS
fluorescence-activated cell sorting
- GAD-67
glutamic acid decarboxylase-67
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GE
ganglionic eminence
- GFP
green fluorescent protein
- IZ
intermediate zone
- LCM
laser capture microdissection
- MGE
medial ganglionic eminence
- MZ
marginal zone
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PPL
preplate
- qPCR
quantitative polymerase chain reaction
- SP
subplate
- SVZ
subventricular zone
Supporting information
Additional supporting information can be found in the online version of this article
Table S1. qPCR primer details.
Table S2. Genepaint PCR in situ probe details.
Table S3. Cell surface receptors with upregulated expression in the cortical PPL.
Table S4. Ion transport and synaptic transmission genes with upregulated expression in the cortical PPL.
Table S5. Transcription factors/regulators with upregulated expression in the cortical PPL.
Table S6. Secreted factors with upregulated expression in the cortical PPL.
Table S7. Intracellular signalling molecules with upregulated expression in the cortical PPL.
Table S8. Other genes from different classes with upregulated expression in the cortical PPL.
Table S9. Cell surface receptors with upregulated expression in the cortical IZ.
Table S10. Ion transport and synaptic transmission genes with upregulated expression in the cortical IZ.
Table S11. Transcription factors/regulators with upregulated expression in the cortical IZ.
Table S12. Secreted factors with with upregulated expression in the cortical IZ.
Table S13. Intracellular signalling molecules with upregulated expression in the cortical IZ.
Table S14. Other genes from different classes with upregulated expression in the cortical IZ.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset by Wiley-Blackwell. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Alifragis P, Liapi A, Parnavelas JG. Lhx6 regulates the migration of cortical interneurons from the ventral telencephalon but does not specify their GABA phenotype. J. Neurosci. 2004;24:5643–5648. doi: 10.1523/JNEUROSCI.1245-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Marín O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- Andrews W, Liapi A, Plachez C, Camurri L, Zhang J, Mori S, Murakami F, Parnavelas JG, Sundaresan V, Richards LJ. Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development. 2006;133:2243–2252. doi: 10.1242/dev.02379. [DOI] [PubMed] [Google Scholar]
- Andrews WD, Barber M, Parnavelas JG. Slit–Robo interactions during cortical development. J. Anat. 2007;211:188–198. doi: 10.1111/j.1469-7580.2007.00750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews W, Barber M, Hernadez-Miranda LR, Xian J, Rakic S, Sundaresan V, Rabbitts TH, Pannell R, Rabbitts P, Thompson H, Erskine L, Murakami F, Parnavelas JG. The role of Slit–Robo signaling in the generation, migration and morphological differentiation of cortical interneurons. Dev. Biol. 2008;313:648–658. doi: 10.1016/j.ydbio.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Barber M, Di Meglio T, Andrews WD, Hernández-Miranda LR, Murakami F, Chédotal A, Parnavelas JG. The role of Robo3 in the development of cortical interneurons. Cereb. Cortex. 2009;19(Suppl 1):i22–i31. doi: 10.1093/cercor/bhp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R, Machold R, Klein C, Fishell G. Gene expression in cortical interneuron precursors is prescient of their mature function. Cereb. Cortex. 2008;18:2306–2317. doi: 10.1093/cercor/bhm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekirov IH, Nagy V, Svoronos A, Huntley GW, Benson DL. Cadherin-8 and N-cadherin differentially regulate pre- and postsynaptic development of the hippocampal mossy fiber pathway. Hippocampus. 2008;18:349–363. doi: 10.1002/hipo.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Berretta S. GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology. 2001;25:1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- Beneyto M, Morrism HM, Rovenskym KC, Lewis DA. Lamina- and cell-specific alterations in cortical somatostatin receptor 2 mRNA expression in schizophrenia. Neuropharmacology. 2011 doi: 10.1016/j.neuropharm.2010.12.029. doi: 10.1016/j.neuropharm.2010.12.029 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrendero F, García-Gil L, Hernández ML, Romero J, Cebeira M, de Miguel R, Ramos JA, Fernández-Ruiz JJ. Localization of mRNA expression and activation of signal transduction mechanisms for cannabinoid receptor in rat brain during fetal development. Development. 1998;125:3179–3188. doi: 10.1242/dev.125.16.3179. [DOI] [PubMed] [Google Scholar]
- Borrell V, Marín O. Meninges control tangential migration of hem-derived Cajal-Retzius cells via CXCL12/CXCR4 signaling. Nat. Neurosci. 2006;9:1284–1293. doi: 10.1038/nn1764. [DOI] [PubMed] [Google Scholar]
- Cantallops I, Haas K, Cline HT. Postsynaptic CPG15 promotes synaptic maturation and presynaptic axon arbor elaboration in vivo. Nat. Neurosci. 2000;3:1004–1011. doi: 10.1038/79823. [DOI] [PubMed] [Google Scholar]
- Cariboni A, Davidson K, Rakic S, Maggi R, Parnavelas JG, Ruhrberg C. Defective gonadotropin-releasing hormone neuron migration in mice lacking SEMA3A signalling through NRP1 and NRP2: implications for the aetiology of hypogonadotropic hypogonadism. Hum. Mol. Genet. 2011;20:336–344. doi: 10.1093/hmg/ddq468. [DOI] [PubMed] [Google Scholar]
- Carlisle HJ, Manzerra P, Marcora E, Kennedy MB. SynGAP regulates steady-state and activity-dependent phosphorylation of cofilin. J. Neurosci. 2008;28:13673–13683. doi: 10.1523/JNEUROSCI.4695-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler D, Dragović M, Cooper M, Badcock JC, Mullin BH, Faulkner D, Wilson SG, Hallmayer J, Howell S, Rock D, Palmer LJ, Kalaydjieva L, Jablensky A. Impact of neuritin 1 (NRN1) polymorphisms on fluid intelligence in schizophrenia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2010;153B:428–437. doi: 10.1002/ajmg.b.30996. [DOI] [PubMed] [Google Scholar]
- Choi YI, Duke-Cohan JS, Ahmed WB, Handley MA, Mann F, Epstein JA, Clayton LK, Reinherz EL. PlexinD1 glycoprotein controls migration of positively selected thymocytes into the medulla. Immunity. 2008;29:888–898. doi: 10.1016/j.immuni.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin JG, Nery S, Fishell G. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat. Neurosci. 2001;4(Suppl):1177–1182. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- Cossart R, Bernard C, Ben-Ari Y. Multiple facets of GABAergic neurons and synapses: multiple fates of GABA signalling in epilepsies. Trends Neurosci. 2005;28:108–115. doi: 10.1016/j.tins.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Denaxa M, Chan CH, Schachner M, Parnavelas JG, Karagogeos D. The adhesion molecule TAG-1 mediates the migration of cortical interneurons from the ganglionic eminence along the corticofugal fiber system. Development. 2001;128:4635–4644. doi: 10.1242/dev.128.22.4635. [DOI] [PubMed] [Google Scholar]
- Deng P, Pang ZP, Lei Z, Xu ZC. Excitatory roles of protein kinase C in striatal cholinergic interneurons. J. Neurophysiol. 2009;102:2453–2461. doi: 10.1152/jn.00325.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan SM, Hashimoto T, Lewis DA. Reduced cortical cannabinoid 1 receptor messenger RNA and protein expression in schizophrenia. Arch. Gen. Psychiatry. 2008;65:772–784. doi: 10.1001/archpsyc.65.7.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux C, Rakic S, Andrews W, Yanagawa Y, Obata K, Parnavelas JG. Differential gene expression in migrating cortical interneurons during mouse forebrain development. J. Comp. Neurol. 2010;518:1232–1248. doi: 10.1002/cne.22271. [DOI] [PubMed] [Google Scholar]
- Fitzgerald ML, Lupica CR, Pickel VM. Decreased parvalbumin immunoreactivity in the cortex and striatum of mice lacking the CB1 receptor. Synapse. 2011;65:827–831. doi: 10.1002/syn.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Long JE, Garratt AN, Fischer TM, Gassmann M, Birchmeier C, Lai C, Rubenstein JL, Marín O. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44:251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Müller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gant JC, Thibault O, Blalock EM, Yang J, Bachstetter A, Kotick J, Schauwecker PE, Hauser KF, Smith GM, Mervis R, Li Y, Barnes GN. Decreased number of interneurons and increased seizures in neuropilin 2 deficient mice: implications for autism and epilepsy. Epilepsia. 2009;50:629–645. doi: 10.1111/j.1528-1167.2008.01725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman DM, Martini FJ, Nóbrega-Pereira S, Pierani A, Kessaris N, Marín O. The embryonic preoptic area is a novel source of cortical GABAergic interneurons. J. Neurosci. 2009;29:9380–9389. doi: 10.1523/JNEUROSCI.0604-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond V, So E, Gunnersen J, Valcanis H, Kalloniatis M, Tan SS. Layer positioning of late-born cortical interneurons is dependent on Reelin but not p35 signaling. J. Neurosci. 2006;26:1646–1655. doi: 10.1523/JNEUROSCI.3651-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, Sampson AR, Lewis DA. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J. Neurosci. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkenham M, Lynn AB, Johnson MR, Melvin LS, de Costa BR, Rice KC. Characterization and localization of cannabinoid receptors in rat brain: a quantitative in vitro autoradiographic study. J. Neurosci. 1991;11:563–583. doi: 10.1523/JNEUROSCI.11-02-00563.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Miranda LR, Parnavelas JG, Chiara F. Molecules and mechanisms involved in the generation and migration of cortical interneurons. A.S.N. Neuro. 2010;2:e00031. doi: 10.1042/AN20090053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Miranda LR, Cariboni A, Faux C, Ruhrberg C, Cho JH, Cloutier JF, Eickholt BJ, Parnavelas JG, Andrews WD. Robo1 regulates semaphorin signaling to guide the migration of cortical interneurons through the ventral forebrain. J. Neurosci. 2011;31:6174–6187. doi: 10.1523/JNEUROSCI.5464-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilario JD, Rodino-Klapac LR, Wang C, Beattie CE. Semaphorin 5A is a bifunctional axon guidance cue for axial motoneurons in vivo. Dev. Biol. 2009;326:190–200. doi: 10.1016/j.ydbio.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol. Psychiatry. 2006;11:557–566. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- Hivert B, Liu Z, Chuang CY, Doherty P, Sundaresan V. Robo1 and Robo2 are homophilic binding molecules that promote axonal growth. Mol. Cell. Neurosci. 2002;21:534–545. doi: 10.1006/mcne.2002.1193. [DOI] [PubMed] [Google Scholar]
- Howell BW, Hawkes R, Soriano P, Cooper JA. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Javaherian A, Cline HT. Coordinated motor neuron axon growth and neuromuscular synaptogenesis are promoted by CPG15 in vivo. Neuron. 2005;45:505–512. doi: 10.1016/j.neuron.2004.12.051. [DOI] [PubMed] [Google Scholar]
- Katona I, Sperlágh B, Sík A, Käfalvi A, Vizi ES, Mackie K, Freund TF. Presynaptically located CB1 cannabinoid receptors regulate GABA release from axon terminals of specific hippocampal interneurons. J. Neurosci. 1999;19:4544–4558. doi: 10.1523/JNEUROSCI.19-11-04544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölsch H, Jessen F, Wiltfang J, Lewczuk P, Dichgans M, Teipel SJ, Kornhuber J, Frölich L, Heuser I, Peters O, Wiese B, Kaduszkiewicz H, van den Bussche H, Hüll M, Kurz A, Rüther E, Henn FA, Maier W. Association of SORL1 gene variants with Alzheimer’s disease. Brain Res. 2009;1264:1–6. doi: 10.1016/j.brainres.2009.01.044. [DOI] [PubMed] [Google Scholar]
- Korematsu K, Redies C. Expression of cadherin-8 mRNA in the developing mouse central nervous system. J. Comp. Neurol. 1997;387:291–306. [PubMed] [Google Scholar]
- Kramer PR, Wray S. Novel gene expressed in nasal region influences outgrowth of olfactory axons and migration of luteinizing hormone-releasing hormone (LHRH) neurons. Genes Dev. 2000;14:1824–1834. [PMC free article] [PubMed] [Google Scholar]
- Kramer PR, Wray S. Nasal embryonic LHRH factor (NELF) expression within the CNS and PNS of the rodent. Brain Res. Gene Expr. Patterns. 2001;1:23–26. doi: 10.1016/s1567-133x(01)00004-7. [DOI] [PubMed] [Google Scholar]
- Kudo C, Ajioka I, Hirata Y, Nakajima K. Expression profiles of EphA3 at both the RNA and protein level in the developing mammalian forebrain. J. Comp. Neurol. 2005;487:255–269. doi: 10.1002/cne.20551. [DOI] [PubMed] [Google Scholar]
- Kurian MA, Meyer E, Vassallo G, Morgan NV, Prakash N, Pasha S, Hai NA, Shuib S, Rahman F, Wassmer E, Cross JH, O’Callaghan FJ, Osborne JP, Scheffer IE, Gissen P, Maher ER. Phospholipase C beta 1 deficiency is associated with early-onset epileptic encephalopathy. Brain. 2010;133:2964–2970. doi: 10.1093/brain/awq238. [DOI] [PubMed] [Google Scholar]
- Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J. Neurosci. 1999;19:7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Adesnik H, Li J, Long J, Nicoll RA, Rubenstein JL, Pleasure SJ. Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J. Neurosci. 2008;28:1085–1098. doi: 10.1523/JNEUROSCI.4602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liapi A, Pritchett J, Jones O, Fujii N, Parnavelas JG, Nadarajah B. Stromal-derived factor 1 signalling regulates radial and tangential migration in the developing cerebral cortex. Dev. Neurosci. 2008;30:117–131. doi: 10.1159/000109857. [DOI] [PubMed] [Google Scholar]
- Liu Z, Chatterjee TK, Fisher RA. RGS6 interacts with SCG10 and promotes neuronal differentiation. Role of the G gamma subunit-like (GGL) domain of RGS6. J. Biol. Chem. 2002;277:37832–37839. doi: 10.1074/jbc.M205908200. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez-Bendito G, Sanchez-Alcaniz JA, Pla R, Borrell V, Pico E, Valdeolmillos M, Marín O. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J. Neurosci. 2008;28:1613–1624. doi: 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O, Rubenstein JL. Cell migration in the forebrain. Annu. Rev. Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- Marín O, Yaron A, Bagri A, Tessier-Lavigne M, Rubenstein JL. Sorting of striatal and cortical interneurons regulated by semaphorin–neuropilin interactions. Science. 2001;293:872–875. doi: 10.1126/science.1061891. [DOI] [PubMed] [Google Scholar]
- Métin C, Baudoin JP, Rakić S, Parnavelas JG. Cell and molecular mechanisms involved in the migration of cortical interneurons. Eur. J. Neurosci. 2006;23:894–900. doi: 10.1111/j.1460-9568.2006.04630.x. [DOI] [PubMed] [Google Scholar]
- Morante-Oria J, Carleton A, Ortino B, Kremer EJ, Fairén A, Lledo PM. Subpallial origin of a population of projecting pioneer neurons during corticogenesis. Proc. Natl. Acad. Sci. USA. 2003;100:12468–12473. doi: 10.1073/pnas.1633692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov YM, Torii M, Rakic P. Origin, early commitment, migratory routes, and destination of cannabinoid type 1 receptor-containing interneurons. Cereb. Cortex. 2009;19(Suppl 1):i78–i89. doi: 10.1093/cercor/bhp028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhia M, Yee BK, Feldon J, Markopoulos F, Knuesel I. Disruption of hippocampus-regulated behavioural and cognitive processes by heterozygous constitutive deletion of SynGAP. Eur. J. Neurosci. 2010;31:529–543. doi: 10.1111/j.1460-9568.2010.07079.x. [DOI] [PubMed] [Google Scholar]
- Nedivi E, Wu GY, Cline HT. Promotion of dendritic growth by CPG15, an activity-induced signaling molecule. Science. 1998;281:1863–1866. doi: 10.1126/science.281.5384.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery S, Fishell G, Corbin JG. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat. Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Miyata T, Nakajima K, Yagyu K, Seike M, Ikenaka K, Yamamoto H, Mikoshiba K. The reeler gene-associated antigen on Cajal-Retzius neurons is a crucial molecule for laminar organization of cortical neurons. Neuron. 1995;14:899–912. doi: 10.1016/0896-6273(95)90329-1. [DOI] [PubMed] [Google Scholar]
- Oudin MJ, Gajendra S, Williams G, Hobbs C, Lalli G, Doherty P. Endocannabinoids regulate the migration of subventricular zone-derived neuroblasts in the postnatal brain. J. Neurosci. 2011;31:4000–4011. doi: 10.1523/JNEUROSCI.5483-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnamenta AT, Khan H, Walker S, Gerrelli D, Wing K, Bonaglia MC, Giorda R, Berney T, Mani E, Molteni M, Pinto D, Le Couteur A, Hallmayer J, Sutcliffe JS, Szatmari P, Paterson AD, Scherer SW, Vieland VJ, Monaco AP. Rare familial 16q21 microdeletions under a linkage peak implicate cadherin 8 (CDH8) in susceptibility to autism and learning disability. J. Med. Genet. 2011;48:48–54. doi: 10.1136/jmg.2010.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla R, Borrell V, Flames N, Marín O. Layer acquisition by cortical GABAergic interneurons is independent of Reelin signaling. J. Neurosci. 2006;26:6924–6934. doi: 10.1523/JNEUROSCI.0245-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F, Whitford KL, Dijkhuizen PA, Vitalis T, Ghosh A. Control of cortical interneuron migration by neurotrophins and PI3-kinase signaling. Development. 2002;129:3147–3160. doi: 10.1242/dev.129.13.3147. [DOI] [PubMed] [Google Scholar]
- Pozas E, Ibáñez CF. GDNF and GFRalpha1 promote differentiation and tangential migration of cortical GABAergic neurons. Neuron. 2005;45:701–713. doi: 10.1016/j.neuron.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Rice DS, Sheldon M, D’Arcangelo G, Nakajima K, Goldowitz D, Curran T. Disabled-1 acts downstream of Reelin in a signaling pathway that controls laminar organization in the mammalian brain. Development. 1998;125:3719–3729. doi: 10.1242/dev.125.18.3719. [DOI] [PubMed] [Google Scholar]
- Rodriguez JJ, Mackie K, Pickel VM. Ultrastructural localization of the CB1 cannabinoid receptor in mu-opioid receptor patches of the rat caudate putamen nucleus. J. Neurosci. 2001;21:823–833. doi: 10.1523/JNEUROSCI.21-03-00823.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignot J, Taïeb D, Suliman M, Dusetti NJ, Iovanna JL, Soubeyran P. CIP4 is a new ArgBP2 interacting protein that modulates the ArgBP2 mediated control of WAVE1 phosphorylation and cancer cell migration. Cancer Lett. 2010;288:116–123. doi: 10.1016/j.canlet.2009.06.030. [DOI] [PubMed] [Google Scholar]
- Rudolph J, Zimmer G, Steinecke A, Barchmann S, Bolz J. Ephrins guide migrating cortical interneurons in the basal telencephalon. Cell Adh. Migr. 2010;4:400–408. doi: 10.4161/cam.4.3.11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. No. 1, Article 3. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat. Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- Spires TL, Molnár Z, Kind PC, Cordery PM, Upton AL, Blakemore C, Hannan AJ. Activity-dependent regulation of synapse and dendritic spine morphology in developing barrel cortex requires phospholipase C-beta1 signalling. Cereb. Cortex. 2005;15:385–393. doi: 10.1093/cercor/bhh141. [DOI] [PubMed] [Google Scholar]
- Stumm RK, Zhou C, Ara T, Lazarini F, Dubois-Dalcq M, Nagasawa T, Höllt V, Schulz S. CXCR4 regulates interneuron migration in the developing neocortex. J. Neurosci. 2003;23:5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm R, Kolodziej A, Schulz S, Kohtz JD, Höllt V. Patterns of SDF-1alpha and SDF-1gamma mRNAs, migration pathways, and phenotypes of CXCR4-expressing neurons in the developing rat telencephalon. J. Comp. Neurol. 2007;502:382–399. doi: 10.1002/cne.21336. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherins: cell–cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- Tamamaki N, Fujimori KE, Takauji R. Origin and route of tangentially migrating neurons in the developing neocortical intermediate zone. J. Neurosci. 1997;17:8313–8323. doi: 10.1523/JNEUROSCI.17-21-08313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomiokam R, Miyazakim J, Obatam K, Kaneko T. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J. Comp. Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Ishii K, Kasai K, Yoon SO, Saeki Y. Neural expression of G protein-coupled receptors GPR3, GPR6, and GPR12 up-regulates cyclic AMP levels and promotes neurite outgrowth. J. Biol. Chem. 2007;282:10506–10515. doi: 10.1074/jbc.M700911200. [DOI] [PubMed] [Google Scholar]
- Tiveron MC, Rossel M, Moepps B, Zhang YL, Seidenfaden R, Favor J, König N, Cremer H. Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J. Neurosci. 2006;26:13273–13278. doi: 10.1523/JNEUROSCI.4162-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Nakamura M, Sato K, Kano M, Simon MI, Inoue Y. Patterns of expression for the mRNA corresponding to the four isoforms of phospholipase C beta in mouse brain. Eur. J. Neurosci. 1998;10:2016–2025. doi: 10.1046/j.1460-9568.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- Wu Y, Sheng W, Chen L, Dong H, Lee V, Lu F, Wong CS, Lu WY, Yang BB. Versican V1 isoform induces neuronal differentiation and promotes neurite outgrowth. Mol. Biol. Cell. 2004;15:2093–2104. doi: 10.1091/mbc.E03-09-0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J. Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Bhagavath B, Kim HG, Halvorson L, Podolsky RS, Chorich LP, Prasad P, Xiong WC, Cameron RS, Layman LC. NELF is a nuclear protein involved in hypothalamic GnRH neuronal migration. Mol. Cell. Endocrinol. 2010;319:47–55. doi: 10.1016/j.mce.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Kim HG, Bhagavath B, Cho SG, Lee JH, Ha K, Meliciani I, Wenzel W, Podolsky RH, Chorich LP, Stackhouse KA, Grove AM, Odom LN, Ozata M, Bick DP, Sherins RJ, Kim SH, Cameron RS, Layman LC. Nasal embryonic LHRH factor (NELF) mutations in patients with normosmic hypogonadotropic hypogonadism and Kallmann syndrome. Fertil. Steril. 2011;95:1613–1620. doi: 10.1016/j.fertnstert.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


