Preface
Hypoxia inducible factors (HIFs) are broadly expressed in human cancers, and HIF1α and HIF2α were previously suspected of promoting tumor progression through largely overlapping functions. However, this relatively simple model has now been challenged in light of recent data from genome-wide analyses of human tumors, genetically engineered mouse models of cancer, and systems biology approaches that reveal unique and sometimes opposing HIFa activities in both normal physiology and disease. These effects are mediated in part through regulation of unique target genes, as well as direct and indirect interactions with important oncoproteins and tumor suppressors, including MYC and p53. As HIF inhibitors are currently under clinical evaluation as cancer therapeutics, a more thorough understanding of unique roles performed by HIF1α and HIF2α in human neoplasia is warranted. This Review summarizes our rapidly changing understanding of shared and independent HIF1α and HIF2α activities in tumor growth and progression, and the implications for using selective HIF inhibitors as cancer therapeutics.
Introduction
Oxygen (O2) levels are known to vary widely across sub-domains of solid tumors, due to rapid cell division and aberrant tumor angiogenesis and blood flow. Although extended exposure to complete O2 deprivation (anoxia) can result in necrosis, viable hypoxic cancer cells often surround necrotic zones. Tumor hypoxia has long been associated with increased malignancy, poor prognosis and resistance to radiotherapy and chemotherapy (reviewed in1,2), prompting intensive research into cellular responses to O2 deprivation. Particular interest has been focused on the mechanisms by wh ich hypoxic tumor cells alter their transcriptional profiles to modulate glycolysis, proliferation, survival and invasion to persist under conditions of hypoxic stress3.
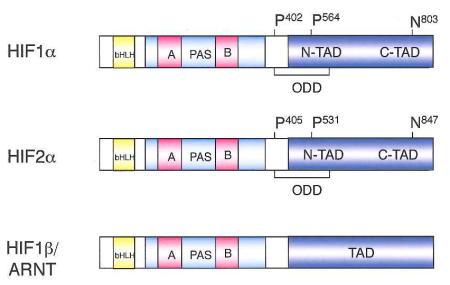
The Hypoxia Inducible Factor (HIF) transcription factors mediate the primary transcriptional responses to hypoxic stress in normal and transformed cells. HIFs are heterodimeric complexes composed of bHLH-PAS proteins including an O2-Iabile alpha subunit (HIF1α, HIF2α, or HIF3α) and a stable beta subunit (HIF1β, also known as ARNT), which together bind hypoxia responsive elements (HREs) containing a conserved RCGTG core sequence (see Box 1). Hypoxic HIF activity is controlled primarily through post-translational modification and stabilization of HIF1α and HIF2α subunits, so that HIFa protein levels and overall HIF transcriptional activity increase as cells become more hypoxic. The central molecular mechanisms underlying the O2-lability of HIFa subunits were first elaborated in 2001 by multiple groups, and are the subject of several recent reviews4,5 (Box 1). Briefly, HIFa subunits are modified by HIF-specific prolyl-hydroxylases (PHDs) in the presence of O2, leading to normoxic proteasomal degradation mediated in part by the Von Hippel Lindau tumor suppressor protein (pVHL) (Box 1). It is also important to note that elevated oncogenic signaling in cancer cells can induce HIFa expression through O2-independent mechanisms including increased transcription and/or translation of HIFα mRNAB6.
Box 1. O2-dependent HIF regulation.
Using molecular O2 and 2-oxoglutarate as substrates, HIF prolyl hydroxlase (PHD) enzymes4 hydroxylate two specific proline residues that reside in the 02-dependent degradation domain (ODD) of HIF-a proteins. These hydroxylation events occur on P402 and P564 in HIF1α, and P405 and P531 in HIF2α, respectively, and are required for the Von Hippel-Lindau (pVHL) tumor suppressor protein, the recognition component of an E3-ubiquitin ligase, to bind and degrade HIFα subunits under normoxic conditions. Hypoxia inhibits PHD activity through a number of mechanisms, including substrate limitation (reviewed in4), resulting in HIFα stabilization, heterodimerization with HIF1β/ARNT, and increased HIF transcriptional activity. Hypoxic conditions also inhibit a second hydroxylation of a conserved HIFα C-terminal asparagine residue by the FIH hydroxylase, an event that blocks the interaction between HIFα and the transcriptional co-activators p300/CBp149-151. Thus, whereas PHD-mediated hydroxylation destabilizes HIFα subunits, FIH-mediated hydroxylation inhibits their transcriptional activity.
HIF1α was first described by Semenza and colleagues in 1995, and was shown to playa central role in mediating O2-dependent transcriptional responses7. The identification of HIF2α by independent groups in 1997 (initially called endothelial PAS protein 1 (EPAS1)8, HIF-related factor (HRF)9, HIF1α-like factor (HLF)10, and member of PAS family 2 (MOP2)11) indicated that HIF regulation was more complex. Whereas HIF1α appears to be expressed in nearly all cell types, RNA in situ hybridization on mouse embryos revealed that HIF2α expression is more restricted, and particularly abundant in blood vessels. This observation led to the hypothesis that the primary role of HIF2α is to modulate vascular endothelial cell (Ee) function, an idea supported in part by the close correlation of HIF2α and VEGF mRNA expression patterns8. A more complex view emerged as HIF2α protein expression was identified in multiple cell types in hypoxic rat kidney, lung, and colonic epithelia, as well as hepatocytes, macrophages, muscle cells and astrocytes12, indicating that both HIF1α and HIF2α are co-expressed in a large number of cell types.
The majority of HIF transcriptional responses have been attributed to HIF1α and HIF2α; however, a third HIFα subunit (HIF3α) has also been described13. HIF3α mRNA is differentially spliced to produce multiple isoforms that either promote or inhibit the activity of other HIF complexes, although little is yet known about the impact of HIF3α on hypoxic tumor progression14-17. Similarly, a second ARNT protein (ARNT2) has been identified18 and shown to regulate neuronal development19 and exhibit overlapping activity with ARNT20; however, its activity in human cancer cells has not been studied in depth21. Although it will be important to determine whether (and how) HIF3α and ARNT2 affect HIF-mediated responses in cancers, the available evidence suggests that HIF1α and HIF2α account for the vast majority of HIF-dependent effects on tumor growth and progression described to date.
Elevated expression of HIF1α and HIF2α protein has been observed in a broad array of human cancer cell types, and associated with poor prognosis in many cases (Table 1). Particular attention has been focused on renal clear cell carcinomas (RCCs), approximately 90% of which lose function of the Von Hippel-Landau tumor suppressor protein (pVHL), which binds prolyl-hydroxylated HIFα subunits and targets them for ubiquitin-mediated proteolysis22 (Box 1). pVHL-deficient RCC cell lines consequently cannot degrade HIFα subunits in an 02-dependent manner, and have been used extensively to investigate the roles of HIF1α and HIF2α in tumor growth.
Table 1.
Correlation between HIFa protein expression and poor prognosis in human cancers*
| Cancer type | HIF1α | HIF2α | References |
|---|---|---|---|
| Astrocytoma | + | + | 161,162 |
| Bladder | + | ND | 163 |
| Breast | + | + | 164,165 |
| Cervical | + | +a | 166,167 |
| Colorectal | + | + | 168 |
| Gastric | + | NC | 169, 170 |
| Gastric | + | ND | 171 |
| GIST | + | ND | 172 |
| Glioblastoma | ND | + | 162 |
| Glioma | NCb | +b | 69 |
| Head/neck | + | + | 173,174 |
| Hepatocellular | ND | + | 175 |
| Lung (NSCLC) | + | + | 176 |
| Lung (NSCLC) | + | ND | 177 |
| Lung (NSCLC) | NC | + | 178 |
| Melanoma | + | + | 179 |
| Neuroblastoma | FP | + | 180 |
| Ovarian | + | ND | 181 |
| Ovarian | + | +d | 182 |
| Pancreatic | + | ND | 183,184 |
| Prostate | + | +d | 185 |
| Renal | FP | ND | 186 |
| Renal | + | ND | 187 |
NC, no correlation; ND, not determined; FP, correlation between HIFα expression and favorable prognosis
HIF2α expression in macrophages
Correlation using mRNA levels
Correlation of HIF1α expression with favorable prognosis
Correlation with cytoplasmic HIF2α expression
The observations summarized in Table 1 have led to the general view that elevated HIFα protein expression in tumor cells, whether induced by hypoxia or aberrant oncogenic signaling, actively drives tumor growth and progression by regulating the expression of critical target genes. Disparate correlations have been observed in some tumor types; for example, HIF1α expression has been associated with both better and worse prognosis in separate analyses of renal and non-small cell lung cancers (see Table 1). The basis of these apparent discrepancies is not understood, but may reflect the consequences of HIF activity in different cancer subtypes, or at different stages of tumor progression. In some tumors, including gastric cancers and glioma, only one HIFα subunit is correlated with prognosis, suggesting it plays a particularly important role or predominant role in these tumor cell types. Interestingly, multiple recent studies have also revealed unexpected tumor suppressive activities of HIF1α and HIF2α in specific contexts23-26. Although initially viewed as having largely overlapping functions, there is now mounting evidence that HIF1α and HIF2α can promote highly divergent – even opposing – outcomes when expressed in the same cell type. It appears that HIF1α and HIF2α mediate these disparate responses partly through independent regulation of distinct target genes, but also through direct and indirect interactions with complexes containing important oncoproteins and tumor suppressors.
Direct regulation of gene expression by HIF1α and HIF2α
Numerous early studies revealed that either HIF1α or HIF2α could regulate the expression of many hypoxically induced genes, but that each HIFα isoform also had unique targets (Table 2)27,28. By swapping protein domains between HIF1α and HIF2α, several groups demonstrated that this transcriptional specificity resided in the N-terminal activation domain (N-TAO), suggesting that differential interactions with transcriptional co-factors likely determine differential gene activation29,30. Recently, multiple groups have used chromatin immunoprecipitation coupled to tiled microarrays (ChiP-chip) to assess HIFα binding across the genome31-35. These analyses confirmed the RCGTG core binding sequence, and revealed no additional sequences absolutely required for HIF binding32,35.
Table 2.
Representative shared and unique target genes regulated by HIF1α and HIF2α
| Gene | Function | HIF1α | HIF2α | Cell type |
|---|---|---|---|---|
| GLUT1 | Glucose transport | + | + | RCC27, mouse ES36, 37 |
| ADRP | Lipid metabolism | + | + | RCC27 |
| CAXII | pH homeostasis | + | + | RCC27 |
| FILAG | Cytoskeletal structure | + | + | RCC27 |
| IL-6 | Immune cytokine | + | + | RCC27 |
| ADM1 | Angiogenesis | + | + | RCC27 |
| VEGF | Angiogenesis | + | + | RCC, Hep3B27-29 |
| VEGF | Angiogenesis | + | − | Mouse EC51, mouseES36,37 |
| BNIP3 | Autophagy, apoptosis | + | − | RCC28 |
| HK1 | Glycolysis | + | − | mouse ES36, 37 |
| HK2 | Glycolysis | + | − | RCC27, mouse ES36, 37 |
| PFK | Glycolysis | + | − | RCC27, mouse ES36,37 |
| ALDA | Glycolysis | + | − | RCC27, mouse ES36, 37 |
| PGK1 | Glycolysis | + | − | RCC27, mouse ES36, 37 |
| LDHA | Glycolysis | + | − | RCC27, mouse ES36, 37 |
| INOS | NO production | + | − | Macrophages49 |
| ARG | Inhibitor of NO production | − | + | Macrophages49 |
| EPO | Erythropoiesis | − | + | Kidney41, 42, 65, liver188 |
| OCT4 | Stem cell identity | − | + | Mouse ES157 |
| SCGB3A1 | Secretoglobin 3A1 | − | + | NSCLC26 |
| TGFα | Growth Factor | − | + | RCC28, 189 |
| CCND1 | Cell cycle progression | − | + | RCC28 |
| DLL4 | NOTCH signaling, EC branching | − | + | Mouse ECS53 |
| ANG2 | Blood vessel remodeling | − | + | Mouse ECS53 |
Direct comparison of HIF1α and HIF2α binding in MCF7 breast cancer cells demonstrated that although some sites bind HIF1α exclusively, many others bind both HIFα subunits with equal affinity, despite the fact that HIF2α contributes to the hypoxic expression of relatively few genes in these cells33. Subsequent analysis using high-resolution ChiP-seq techniques revealed that HIFs bind to approximately 500 high-affinity sites across the genome, many of which are located at great distances (>100 kbp) from the genes they regulate35. Perhaps not surprisingly, HIF1α and HIF2α were shown to bind preferentially to specific genes each is known to preferentially regulate (Table 2): for example, a significantly higher level of HIF1α binding was associated with glycolytic pathway genes, whereas relatively greater HIF2α binding was observed at the Oct4 locus. Strikingly, however, significant levels of both proteins were detected at essentially all HIF binding sites, further implicating differential interactions with specific co-factors, perhaps mediated by distinct posttranslational modifications, in controlling target gene specificity35. Interestingly, it appears that HIFs are recruited to genes already expressed in norm oxic cells (as revealed by DNAsel hypersensitivity), and are therefore unlikely to direct hypoxic changes in chromatin structure of target genes32,35. The spectrum of HIF target genes may therefore be determined largely by underlying cell type-specific patterns of chromatin structure, a speculation supported by the limited concordance (40-60%) of HIF binding sites detected in MCF7 and RCC cells35. Intriguingly, several reports indicate that HIF1α binds and regulates the expression of multiple Jumonji-domain containing histone demethylases (JMJHDs), which may contribute directly to changes in hypoxic target gene expression31,34.
Assessing HIF1α and HIF2α function in tumor models
Multiple xenograft tumor models (Table 3) support the contention that HIF1α and HIF2α promote tumor progression by regulating both shared and unique target genes. As global deletion of the mouse Hif1α gene results in lethality at E9.536,37, and HIF2α deficiency causes embryonic and perinatal lethality38-40, or severe developmental abnormalities41 conditional alleles37, 42 were required to investigate the specific effects of HIFα deficiency in autochthonous mouse tumor models. Only a small number of studies have been reported to date (Table 3), but these have demonstrated independent roles for HIF1α and HIF2α in different cancers, as well as stromal cell types, at various stages of tumor growth and progression. For example, HIF1α deletion in a mouse mammary tumor virus (MMTV) promoter-driven Polyoma middle T cancer model reduced overall tumor burden and pulmonary metastasis, even when equivalent tumor burdens were allowed to occur in control mice43 (Table 3).
Table 3. Mouse models testing altered expression of HIFα proteins in tumour growth and progression.
| Tumour or cell type | HIFlα status | HIF2α status | Phenotypes | Refs |
|---|---|---|---|---|
| Xenograft tumours | ||||
| Teratoma | Loss-of-function knockout | Wild-type | Reduced growth and angiogenesis | 37,190 |
| Teratoma | Wild-type | Loss-of-function knockout | Increased growth | 25 |
| Fibrosarcoma | Loss-of-funct ion knockout | Wild-type | Reduced growth | 191 |
| RCC | Gain offunction | Wild-type | Reduced growth | 23,28 |
| RCC | Wild-type | Gain offunction | Increased growth | 28,192 |
| Autochthonous tumours | ||||
| MMlV-PyMT mammary tumours |
Conditional knockout | Wild-type | Reduced metastasis | 43 |
| KRAS-driven NSCLC | Conditional knockout | Wild-type | No effect | 26 |
| KRAS-driven NSCLC | Wild-type | Conditional knockout | Increased tumour burden and progression | 26 |
| p53-driven thymic lymphoma |
Heterozygous germline knockout |
Wild-type | Decreased tumour incidence | 156 |
| Tumour-associated stromal cells | ||||
| Tumour-associated macrophages |
Conditional knockout | Wild-type | Reduced NO production. increased T cell-mediated tumour immunosurveillance and reduced autochthonous mammary tumour growth |
48,49 |
| Tumour-associated macrophages |
Wild-type | Conditional knockout | Decreased macrophage infiltration into autochthonous liver and colon tumours and decreased tumour growth |
50 |
| Vascular ECs | Conditional knockout | Wild-type | Decreased xenograft tumour angiogenesis and growth | 51 |
| Vascular ECs | Wild-type | Conditional knockout | Non-productive angiogenic sprouting and impaired vessel remodelling |
53 |
EC, endothelial cell: MMTV, mouse mammary tumour virus; NO, nitric oxide; NSCLC, non-small-cell lung cancer; PyMT, polyoma middle T antigen; RCC, renal cell carcinoma.
In a direct comparison of HIF1α and HIF2α function in a KRAS-driven lung tumor model, HIF1α deletion had surprisingly little effect on tumor burden and progression, whereas loss of HIF2α actually increased tumor growth and progression26. This latter effect correlates to HIF2α-driven expression of the Scgb3a1 gene, which encodes the putative tumor suppressor secretoglobin 3a1 protein44. Surprisingly, overexpression of a stabilized HIF2α protein in the identical KRAS lung tumor model also promoted tumor angiogenesis and invasion by increasing expression of vascular endothelial growth factor (VEGF) and SNAIL45, respectively. The observation that either HIF2α overexpression or deletion can promote tumor growth in the same tumor context, albeit by different mechanisms, suggests that effective targeting of HIFα subunits in cancer treatment may be complicated. Growth of pVHL-deficient mouse liver hemangiomas was similarly shown to be specifically dependent on HIF2α, but not HIF1α46
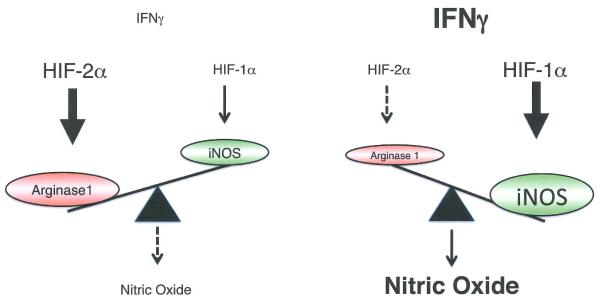
HIF1α and HIF2α deficiency in tumor-associated stromal cells also revealed isoform-specific effects on cancer progression (Figure 1). Initial gene expression studies revealed independent effects of HIF1α and HIF2α in primary human macrophages, as well as cultured murine macrophages47. HIF1α deletion in macrophages reduced overall tumor growth in a murine breast cancer model by reducing inducible nitric oxide synthase (iN OS) expression and consequent production of NO, which inhibits T cell responses in vitro and immune surveillance in vivo48. Intriguingly, HIF2α drives macrophage expression of arginase149, which catabolizes and thereby reduces pools of L-arginine, from which NO is produced. The two HIFα isoforms therefore appear to oppose one another to properly regulate overall macrophage NO levels. Interestingly, deletion of HIF2α (but not HIF1α) in mouse macrophages also significantly inhibits the expression of cytokine and chemokine receptors, including the macrophage colony-stimulating factor 1 receptor (M-CSFR, also known as CSF1 R) and CXCR450, thereby limiting macrophage migration into autochthonous liver and colorectal cancers and reducing overall tumor burden.
Figure 1. HIF1α and HIF2α exhibit antagonistic functions in nitric oxide (NO) production.
Under low IFNy conditions, HIF2α is more abundant and induces arginase1 expression, resulting in NO production. Under high IFNy conditions, HIF2α is diminished and HIF1α dominates so that iNOS can utilize arginine for NO generation. These physiologically antagonistic functions allow the HIFα subunits to coordinately regulate NO production in a cytokine-induced and transcription-dependent fashion.
Loss of either HIF1α or HIF2α in mouse vascular endothelial cells (ECs) reduces tumor expansion in xenograft models, although through different mechanisms. EC-specific HIF1α deletion reduces VEGFR2 receptor expression, thereby inhibiting VEGF signaling and EC proliferation, survival and expansion in hypoxic tumor zones51. In contrast, loss of HIF2α function in ECs reduced expression of ephrin A152, delta-like ligand 4 (DLL4) and angiopoietin 2 (ANG2)53, which correlated with unproductive sprouting and aberrant vessel remodeling and xenograft tumor growth. Collectively, these results reveal complex roles for HIF1α and HIF2α in distinct tumor and stromal cell types, although it will be important to test their function in additional tumor models.
Differential regulation of HIFα isoform expression
What molecular mechanisms contribute to the differential regulation of HIF1α and HIF2α? Control of HIF activity has been traditionally attributed to O2-dependent posttranslational stabilization of HIFα subunits; however, recent data indicate that control of HIF1α and HIF2α expression can be selectively regulated at the level of transcription, translation, and protein stability (summarized in Figure 2A).
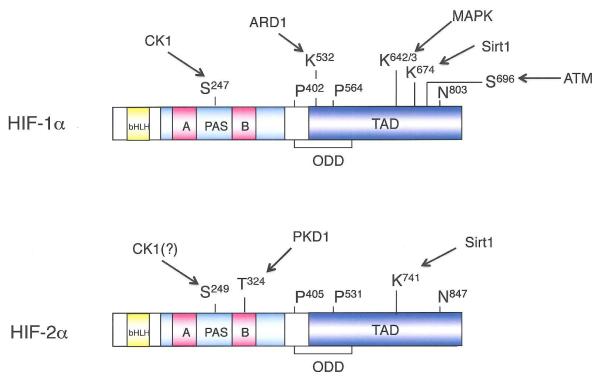
Figure 2. HIF1α and HIF2α are post-translationally modified, and differentially regulated by multiple mechanisms.
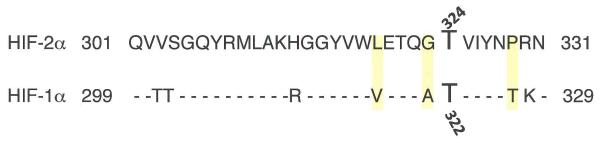
(A) Multiple mechanisms differentially regulate HIF1α and HIF2α at the levels of transcription or mRNA stability (red), mRNA translation (green), and protein stability (blue). In most cases, these regulatory events have opposite effects on HIF1α and HIF2α expression, or appear to be specific for only one HIFα isoform. See text for details. (NE), no effect. (8) Summary of phosphorylations, acetylations, and hydroxylations of the two HIFα subunits by CK1, ARD1, PHDs, FIH, MAPK, SIRT1, PKD1, and ATM. It should be noted that ARD1 acetylates HIF1α, while SIRT1 deacetylates both HIF1α and HIF2α. (C) Sequence alignment of HIF2α residues 301-331 with a similar region of HIF1α; shaded residues are unique to HIF2α and allow the selective phosphorylation of HIF2α T324 by PKD1.
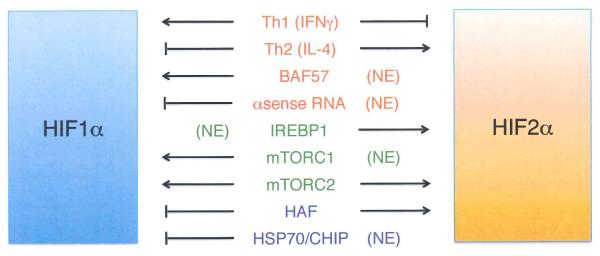
Differential transcription
Surprisingly, relatively little is known about the transcriptional regulation of the Hif1α and Epas1 (encoding HIF2α) genes. Nuclear factor-KB (NF-KB) regulates the transcription of the Hif1α gene54-57. Moreover, Th1 cytokines stimulate this NF-κB-HIF1α pathway to activate a range of HIF1α target genes, whereas Th2 cytokines interleukin-4 (IL-4) and IL-1 0 differentially activate Epas1 expression49, although the precise mechanisms involved are not clear. Expression of the Hif1α locus, in contrast to Epas1, is also regulated by the SWIISNF chromatin remodeling protein BAF5758 Additional investigation into differential Hif1α and Epas1 transcription is certainly warranted.
Differential mRNA translation
It is well established that elevated HIFα mRNA translation rates increase HIFα protein levels and activity, particularly in cells with activated PI3K1AKT/mTOR signaling, a common feature of cancer cells (reviewed in6). Intriguingly, HIF1α expression in RCC cell lines appears to be regulated by both mTORC1 and mTORC2 kinase complexes, whereas HIF2α expression is mTORC2-dependent and mTORC1-independent59 Other forms of differential translation control have been reported for HIFα proteins60, 61. For example, the iron response element binding protein 1 (IREBP1) was shown to bind a canonical iron response element (IRE) in the HIF2α 5′ UTR, thereby inhibiting translation61. This effect appears to be specific for HIF2α, as IREBP1 fails to bind the HIF1α transcript or regulate its translation, despite the presence of a near-consensus IRE in the HIF1α 5′ UTR62 This regulation is also consistent with the identification of HIF2α as the primary regulator of erythropoiesis and cellular iron metabolism in vivo42, 63-60.
Differential stability
As HIF1α and HIF2α protein levels are both modulated in a similar way by PHD-pVHL-dependent mechanisms (Box 1), the observation that HIF1α and HIF2α proteins accumulate at different O2 levels in specific cell types came as a surprise. Pahlman and colleagues first demonstrated that HIF2α protein is stabilized at moderate (2-5% O2) levels, whereas HIF1α accumulates only at lower (0-2% O2) levels in HeLa and neuroblastoma cells67, 68 (similar results were later reported for glioma cells69). Hypoxic neuroblastoma68 and lung adenocarcinoma cells60 maintain elevated HIF2α protein levels during long-term hypoxic culture (48 hours); in contrast, HIF1α levels increase acutely upon hypoxic exposure, but then decline after several hours. The HIF-mediated expression of antisense transcripts from the Hif1 a (but not Epas1) locus, results in Hif1α mRNA destabilization and may explain the gradual and specific reduction of HIF1α protein60
Two HIF1α specific E3 ubiquitin ligases have been described recently that may also contribute to the differential stability of HIF1α and HIF2α. HIF-associated factor (HAF) binds and destabilizes HIF1α under normoxic and hypoxic conditions in a pVHL-independent, proteasome-dependent manner, but has no effect on HIF2α levels70. Instead, HAF binds HIF2α at a distinct C-terminal region and promotes HIF2α transcriptional activity, effectively switching cells from a HIF1α to a HIF2α transcriptional program71. In addition, heat shock protein 70 (HSP70) and carboxyl terminus of Hsc70-interaction protein (CHIP), a recently identified E3-ubiquitin ligase, were shown to bind and degrade HIF1α (but not HIF2α) under conditions of prolonged hypoxia in cultured cells, whereas rapid reoxygenation destabilized both HIF1α and HIF2α proteins in a PHD-pVHL-dependent manner72 Precisely how these novel ubiquitylation events are regulated, either by hypoxia or other stimuli, and how they affect HIF activity in cancer progression is not yet known.
Posttranslational modifications and differential HIFα activity
The regulation of HIFα subunits by posttranslational proline and asparagine hydroxylation, catalyzed by PHD and factor inhibiting HIF (FIH, also known as HIF1AN) enzymes, respectively (Box 1), has been extensively reviewed elsewhere4, 63, 73-70. Interestingly, specific PHD enzymes exhibit biased activity toward HIF1α and HIF2α; for example, PHD3 preferentially hydroxylates HIF2α in multiple celilines76. Peet and colleagues have also shown that FIH preferentially hydroxylates HIF1α in certain cell lines, owing to the identity of the amino acid immediately C-terminal to the hydroxylated asparagine (valine in HIF1α, alanine in HIF2α)77. These results suggest that differential N-hydroxylation might regulate HIF1α and HIF2α activity, although the largely HIF-independent neurological phenotypes of FIH-deficient mice78 indicate that other factors are likely involved.
In addition to hydroxylation, both HIF1α and HIF2α are subject to an array of distinct, O2-independent posttranslational modifications, and growing evidence indicates that at least some of these are specific for either HIF1α or HIF2α, and may promote their differential activity (Figure 2B), These include:
Phosphorylation
Early work showed that both HIF1α and HIF2α are phosphorylated79, 80, and recent work suggests that isoform-specific phosphorylation may impact tumor progression, Specifically, Huang and colleagues demonstrated that HIF1α represses Myc-dependent expression of the DNA damage repair protein nibrin (NBS1) by displacing the SP1 transcription factor from the MYC transcriptional complex81, HIF2α, in contrast, is inhibited from interacting with SP1 through phosphorylation on T324 by protein kinase D1 (PKD1), a modification dependent on a neighboring proline residue unique to HIF2α (Figure 2C), When a proline residue was introduced into the corresponding position in HIF1α, PKD1 also phosphorylated the modified HIF1α protein, which consequently lost the ability to displace SP1 from MYC81, Other specific phosphorylation events catalyzed by MAPK82, casein kinase 1 (CK1)83 and ataxia telangectasia mutated (ATM)84 have been demonstrated to modulate HIF1α activity, although it is not yet known whether these also occur in HIF2α, It will be important to determine the degree to which these various phosphorylation events distinguish HIF1α or HIF2α activation, and whether they represent another mechanism of parallel regulation in cancer cells.
Acetylation
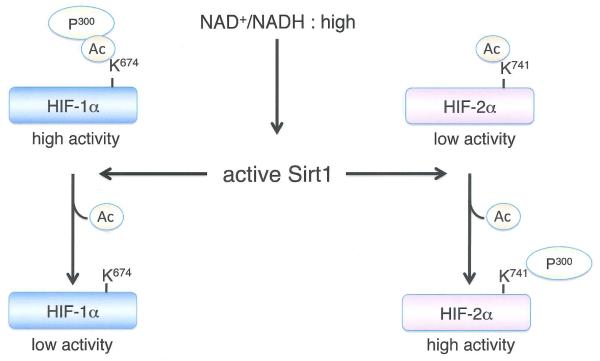
HIFα activity is also modulated by multiple sirtuins, a family of redox-sensitive, NAD+-dependent deacetylases and/or ADP-ribosyltransferases. Mammalian cells express a family of sirtuins (SIRT1-7) that regulate complex changes in gene expression, metabolism, and cellular redox status, and have also been implicated in controlling longevity, although this idea remains highly controversial85. SIRT1 forms a complex with HIF2α and deacetylates conserved lysine residues in the N-TAD, which enhances HIF2α transcriptional activity in vitro and in vivo86 SIRT1 was also reported to deacetylate lysine residues in HIF1α, which resulted in HIF1α transcriptional repression87 although this effect was not universally observed86 (Figure 3).
Figure 3. Differential regulation of HIF1α and HIF2α by SIRT1.
(A) High levels of NAD+ inactivate SIRT1, resulting in decreased HIF1α transcriptional activity and enhanced HIF2α stimulation of target genes like erythropoietin. (8) Distinct effects of HIF1α and HIF2α on MYC complex formation and promoter occupancy. Hypoxic cells exclusively expressing HIF1α exhibit decreased MYC activity due to diminished association with MAX and SP1, as well as reduced MYC stability. HIF1α also induces MXI1 expression, which inhibits MYC target gene expression (see text for details). Cells expressing HIF2α exhibit increased MYC complex formation and target gene activation, although the mechanisms involved are not fully understood.
The apparently opposing effects of SIRT1 on HIF1α and HIF2α could skew cells toward either HIF1α or HIF2α transcriptional programs in response to changing metabolic activity in hypoxic tumors. Park and colleagues87 proposed a positive feedback mechanism in which HIF1α promotes glycolysis, reducing NAD+/NADH ratios under hypoxia and inhibiting SIRT1, thereby further augmenting HIF1α activity. Presumably, inhibiting SIRT1 under these conditions would also decrease HIF2α activity, although the relative sensitivity of endogenous HIF1α and HIF2α proteins to SIRT1-mediated effects over a range of O2 levels is not yet clear, and the kinetics of these responses may differ. It would be interesting to determine whether deacetylation by SIRT1 contributes to the high relative abundance of HIF2α at intermediate O2 levels. There appear to be yet more wrinkles in this story, as both HIF1α and HIF2α were shown to bind the SIRT1 gene promoter and induce its expression under hypoxia88, and AKT activity can induce both HIF1α and SIRT1 expression by downregulating miR199a-5p expression89.
Other sirtuins have also been shown to regulate HIFα activity. Mostoslavsky and colleagues identified SIRT6 as a HIF1α repressor, and showed that SIRT6 deficiency increased HIF1α-dependent glucose uptake and glycolytic activity at the expense of mitochondrial respiration90. Although the precise mechanisms regulating interactions between SIRT6 and HIF1α are not yet clear, SIRT6 deficiency increases both HIF1α synthesis and stability, suggesting that the effects of SIRT6 may be at least partly indirect. In addition, the mitochondrial SIRT3 deacetylase indirectly regulates HIFα stabilization by suppressing the formation of mitochondrial reactive oxygen species (ROS)91 which, in turn, promote HIF1α stabilization92-94. For this reason, SIRT3-deficient cells display HIF1α-dependent increases in glucose transport, glycolysis and proliferation95,96. The implications of these findings for tumor progression have not been explored in depth, but are likely to be both complex and important.
It is possible that other acetylation and deacetylation events regulate HIF activity: for example, the mouse arrest defective-1 (mARD1) protein was reported to destabilize HIF1α by acetylating K53297, an event apparently reversed by recruitment of HDAC1 to HIF1α by metastasis-associated protein 1 (MTA1)98 Other researchers, in contrast, observed neither interaction between mARD1 and HIF1α, nor any effects of hypoxia on mARD1 activity, and the importance of this regulatory event remains in dispute99. Finally, a growing number of reports indicate that HIFα proteins are subject to numerous other posttranslational modifications, including sumoylation, S-nitrosylation, and neddylation100-106 although whether any of these differentially regulate HIF1α and HIF2α is as yet unknown.
HIFs, oncogenes, and tumor suppressors - balancing HIF1α and HIF2α
Although HIF1α and HIF2α clearly influence tumor progression by directly regulating unique and shared target genes (Table 2), recent evidence indicates that these HIFα proteins also affect tumor progression by exerting distinct, often opposing effects on critical oncoproteins and tumor suppressors including MYC, p53, and mTOR.
HIFα and c-Myc
In many cell types, hypoxia suppresses proliferation. Koshiji et al. were the first to demonstrate that acute HIF1α stabilization at 1% O2 produces cell cycle arrest by inhibiting the protooncoprotein MYC107, a bHLH/leucine zipper (bHLH/LZ) transcription factor that is overexpressed in >40% of human cancers. MYC controls the G1/S cell cycle transition by forming heterodimers with the related protein MAX, binding conserved E-box sequences (CTCGAG), and promoting expression of genes encoding cyclin D2 (CCND2), E2F1, and ornithine decarboxylase 1 (ODC1), for example. MYC simultaneously inhibits the expression of CDKN1A and CDKN1B genes encoding cyclin-dependent kinase inhibitors (CKls) p21 and p27, respectively108, in part by displacing the SP1 protein from the transcription factor MIZ1. MYC also promotes proliferation by inducing the expression of essentially all glycolytic enzymes and enhancing protein synthesis, thereby increasing cell growth.
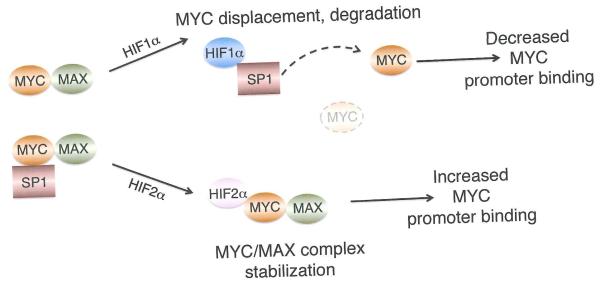
Under hypoxic conditions, HIF1α binds to SP1, displacing MYC from multiple target genes including CDKN1A, MSH2, MSH6, and NBS181,109. (Figure 3B). Gordan et al. subsequently showed that HIF1α rapidly disrupted the association of MYC with MAX and MIZ1, thus reducing MYC promoter occupancy at the CDKN1A, CDKN1B, CDKN2B (which encodes p15), ODC1, CCND2, and E2F1 genes110 A more chronic adaptation results from HIF1α mediated induction of MXI1, which interacts with MAX at E-boxes to inhibit the expression of ODC1 and peroxisome proliferator-activated receptor-y coactovator 1β(PGC-1β)111, 112, suppressing mitochondrial biogenesis and function. Moreover, HIF1α promotes MYC degradation under chronic hypoxia111,112. Through these multiple mechanisms, HIF1α effectively limits MYC-dependent anabolic metabolism, protein synthesis and cell division, an important hypoxic adaptation. Intriguingly, HIF1α also drives expression of the glycolytic pathway genes, permitting hypoxic cells to inhibit MYC driven macromolecular synthesis whilst producing ATP from glycolysis.
Surprisingly, transformed cells expressing HIF2α exclusively exhibit enhanced MYC activity, with more rapid entry into S phase of the cell cycle, increased CCND2, E2F1, and ODC1 gene expression, and elevated MYC promoter occupancy110 Moreover, HIF2α promotes cell cycle progression in hypoxic cells via transcriptional effects on both MYC activated (CCND2, E2F1) and repressed (p21, p27) target genes, and interactions with MAX, SP1, and MIZ1. This impact on MYC likely contributes to HIF2α-mediated neoplastic progression of renal clear cell carcinoma (RCC) tumorigenesis following loss of the VHL tumor suppressor113. Of note, RCC cells exclusively expressing HIF2α also displayed reduced genomic instability, correlating with increased MYC-dependent expression of genes encoding DNA repair proteins (including BRCA 1, BARD1, XRCC2, BUB1, and CENPE)113. These resu lts reveal a critical collaborative role for HIF2α and MYC in promoting genomic integrity and resistance to replication stress.
How do HIF1α and HIF2α exert these opposing roles on MYC? Multiple mechanisms appear to be involved: for example, HIF1α binds to SP1 via the PAS-B domain, whereas HIF2α fails to do so because it is phosphorylated by PKD1, blocking its ability to interact with SP181. In contrast, HIF2α forms a complex with MAX, causing a dose-dependent stabilization of MYC-MAX and MYC-MAX-SP1 complexes, resulting in increased MYC-MAX binding at CCND2, E2F1, p21, and p27110. These effects occur rapidly and can be detected after only 1-2 hours at 0.5% O2, suggesting they are independent of HIF2α transcriptional activity, which peaks at approximately 16 hours at 0.5% O2. A specific role for MXI1 in this differential regulation is currently unclear, as both HIF1α and HIF2α appear to contribute to MXI1 expression in VHL-deficient RCC cells112. How the “competition” between HIF1α and HIF2α is moderated in a given cell type, in terms of their respective influence on MYC activity, is equally mysterious at present.
The relative expression levels of MYC and HIFα proteins also play an important role in regulating tumor cell proliferation and metabolism. Many cancer cells exhibit subtle alterations in MYC levels as a consequence of elevated oncogenic signaling, whereas other cells express MYC at very high levels due to chromosomal amplifications, translocations, and mutations within MYC coding exons108. It appears that high levels of MYC sequester and tightly bind MAX, thereby relieving potential inhibition by HIF1α114. For example, most genes induced by ectopic MYC expression were not transcriptionally repressed by hypoxia in a B-cell tumor model. The picture is more complex, however, as HIF1α can actually cooperate with MYC to induce the expression of specific target genes, including those encoding the glycolytic enzyme hexokinase 2 (HK2), pyruvate dehydrogenase kinase 1 (PDK1), and VEGFA114. Similarly, high levels of NMYC override HIF1α inhibition of cell cycle progression while cooperating with HIF1α to promote phosphoglycerate kinase 1 (PGK1), (HK2), and lactate dehydrogenase A (LDHA) expression in neuroblastomas with MYCN gene amplification115. In summary, when MYC family members are highly overexpressed, they not only overcome the inhibitory effects of HIF1α, but MYC and HIF1α collaborate to favor glycolysis and continued proliferation under decreased O2 availability. In contrast, tumors with lower MYC levels are susceptible to HIF1α inhibition, explaining the anti-tumorigenic effects of HIF1α in certain cancers such as RCC116.
HIF1α, HIF2α, and p53
Low O2 and other stresses associated with tumor growth (such as growth factor withdrawal, nutrient deprivation, and acidosis) activate p53, a critical tumor suppressor that is mutated or silenced in a majority of human cancers117. While it is maintained at low levels in normal cells by MDM2-mediated degradation, p53 is posttranslationally modified and stabilized in response to numerous stimuli, including abnormal proliferation signals, osmotic stress, DNA damage, and hypoxia118. p53 forms homotetramers that bind and regulate numerous genes involved in metabolism, DNA repair, cell cycle arrest, and cell death, thereby coordinating cellular responses to microenvironmental stress117
HIF1α and HIF2α display opposing effects on the p53 pathway. Numerous studies have shown that p53 accumu lation occurs within hypoxic regions of solid tumors, and correlates with cells undergoing apoptosis, although this may only occur when also accompan ied by acidosis and nutrient deprivation119. An et al. orig inally suggested that transcriptionally active wild type p53 is stabilized through a physical association with HIF1α120 Sanchez-Puig further reported that the HIF1α ODD and N-terminal TAD domains bind to p53 tetramers under physiological conditions121; however, subsequent reports suggested that MDM2 mediates the interaction between p53 and HIF1α by acting as a bridge between the two transcription factors122. Whereas HIF1α fails to bind p53 in vitro, it directly binds MDM2, suppressing MDM2-dependent ubiquitylation of p53 in vivo and p53 nuclear export. Surprisingly, MDM2 overexpression actually promotes p53 accumulation and target gene stimu lation when HIF1α is activated in hypoxic cells122. Furthermore, HIF1α appears to enhance p53 activation by ionizing radiation (IR), resulting in increased p53 phosphorylation and p53-mediated apoptosis123 IR significantly increases HIF1α activity in tumors due to increased reactive oxygen and nitrogen species, and rad iation combined with hypoxia lead to increased p53 phosphorylation in a HIF1α dependent manner, by a mechanism that rema ins unclear.
It should also be noted that the relationship between HIF1α and p53 provides a potential negative feedback loop for HIF1α activity. Ravi et al. have suggested that p53 can induce HIF1α turnover, by promoting its MDM2-mediated ubiquitylation and proteasomal degradation124. Therefore, p53 loss in colon cancer cells enhanced HIF1α levels and augmented VEGFA expression and tumor angiogenesis, suggesting that inactivating p53 mutations can contribute to the “angiogenic switch” during colorectal tumorigenesis.
In contrast to HIF1α, HIF2α does not bind MDM2125, and appears to inhibit p53 indirectly by multiple mechanisms. Bertout et al. demonstrated that elevated HIF2α expression inhibits p53 phosphorylation and stabilization in ReG cell lines, whereas knocking down HIF2α expression increases p53 transcriptional activity and target gene expression125. Furthermore, HIF2α deficient cells exhibit elevated ATM activity and DNA double strand break formation, as well as increased levels of ROS after IR. HIF2α has been reported to regulate antioxidants such as superoxide dismutase 1 (SOD1), SOD2, glutathione peroxidase 1, and catalase in developing embryos and neonates41. However, in RGG cells, HIF2α instead decreases ROS accumulation by regulating the expression of distinct antioxidant enzymes (heme oxygenase 1, ceruloplasmin, glutathione peroxidase 8, and peroxiredoxin 3). Importantly, HIF2α expression in RCC tumor samples correlates with decreased p53 phosphorylation and target gene expression, and may contribute to radioresistance in HIF2α expressing RCCs125.
In parallel studies, Roberts et al. showed that HIF2α also suppresses p53 expression and function via indirect effects on MDM2126. AKT-mediated phosphorylation of MDM2 promotes its nuclear localization and enhanced p53 degradation, and represents an important pro-survival effect of AKT. AKT activation occurs downstream of growth factor receptors like EGFR and platelet-derived growth factor receptor (PDGFR), which are stimulated by transforming growth factor-α (TGF-α) and PDGFβ, specific transcriptional targets of HIF2α in RCC cells. Thus, HIF2α overexpression in VHL-deficient RCC can inhibit p53 through a growth factor receptor-AKT-MDM2 pathway, in addition to maintaining redox homeostasis. In aggregate, these findings suggest that HIF2α likely contributes to RCC tumor cell survival during both radiation and chemotherapy by multiple mechanisms.
HIFs regulate mTOR
Cell division requires high levels of protein synthesis and anabolic metabolism, which is regulated by the serine/threonine kinase mTOR in response to nutrient and growth factor availability. mTORC1 promotes ribosome biogenesis, mRNA translation, and nutrient import, while inhibiting autophagy127. Elevated mTORC1 activity is observed in the majority of human tumors, due to activation of upstream oncogenes (PI3K, AKT) and/or loss of tumor suppressors (PTEN, LKB1)128. In particular, the tuberous sclerosis proteins TSC1 and TSC2 together inhibit mTORC1 activity to limit cell growth under conditions of environmental stress, including reduced growth factor, glucose, amino acid, and O2 levels127.
Hypoxia suppresses mTORC1 through multiple mechanisms. For example, decreased ATP levels in severely hypoxic cells activate AMP-activated kinase (AMPK)129, which phosphorylates TSC2 (as well as the mTORC1-associated factor RAPTOR) to inhibit mTORC1 activity. In addition, HIF1α (but not HIF2α) induces expression of the DDIT4 gene130, which encodes REDD1, a protein that represses mTORC1 by promoting the release of sequestered TSC2 from 14-3-3 proteins131. Finally, the hypoxia-inducible proautophagic protein BNIP3 binds and inhibits RAS homolog enriched in brain (RHEB), resulting in decreased mTORC1 activity132. HIF1α dependent inhibition of mTORC1 may benefit cells by reducing ATP-intensive protein synthesis, while increasing autophagy, under conditions of hypoxic stress.
Growing evidence suggests that HIF2α may, in contrast, stimulate mTORC1 to promote cellular proliferation in O2-deprived cells. The focal adhesion kinase (FAK) family interacting protein of 200 kd (FIP200) gene has been identified as a HIF2α target through microarray studies27 and FIP200 has been proposed to interact with TSC1, thereby disrupting TSC1ITSC2 complexes and promoting mTORC1 activation133. In addition, FIP200 may promote TSC1 degradation by the ubiquitin-proteosome pathway134. HIF2α could also selectively enhance mTORC1 activity by positive effects on growth factor signaling, as HIF2α induces the expression of TGF-α, PDGF-β, and IGF-1, leading to AKT and mTORC1 activation in renal cancer cells126. Although additional work is clearly needed to further elucidate the molecular mechanisms by which HIF2α promotes mTORC1 functions, these results reveal another example in which HIF1α and HIF2α antagonize one another to balance hypoxic responses in key growth regulatory pathways.
HIFα and growth control
Why would the two HIFα subunits result in opposite effects on the c-Myc, p53, and mTORC1 pathways? The inhibitory activity of HIF1α towards these growth regulatory systems represents important energy conservation mechanisms in light of decreased ATP production during periods of O2 limitation, which are likely to be compounded by decreased availability of nutrients (glucose, amino acids, lipids) and growth factors in hypoxic subdomains of solid tumors. In contrast, the “pro-growth” effects of HIF2α may contribute to the ability of endothelial cells to proliferate during neoangiogenesis in ischemic tissues. It is interesting that HIF2α accumulates at higher O2 levels than HIF1α, which may allow its selective activation in blood vessels. In addition, the ability of HIF2α to promote cell growth in RCCs may explain why HIF1α expression is often silenced in these tumors. Selective135, as well as genome-wide136, sequence and copy number analyses have identified truncating Hif1α mutations in a small percentage of RCCs, as well as Hif1α heterozygosity in others137, supporting the hypothesis that inhibition of HIF1α function is a selective advantage for some RCCs. It is also tempting to speculate that the recent identification of Epas1 single nucleotide polymorphisms (SNPs) as a predisposing factor for RCC development136 could reveal genetic alterations that increase or expand HIF2α function.
Therapy
As HIF complexes are instrumental in cancer cell adaptation to hypoxic tumor microenvironments, the ability to selectively inhibit HIF activity would appear to be of clinical benefit1, 139. Historically, DNA-binding proteins have been difficult to target, but a large collection of compounds have been reported to inhibit HIF transcriptional activity, either directly or indirectly. For example, compounds including topoisomerase inhibitors (camptothecan, topotecan)140 and DNA intercalators (ech inomycin, daunorubicin, doxorubicin)141 block HIF heterodimerization and transcriptional activation, and interfere with xenograft tumor growth in a HIF-dependent manner. These observations are particularly interesting, given their frequent use as sequence non-specific DNA damaging agents in chemotherapy. Oncogenic signal transduction pathways also promote mTORC1-dependent HIF1α mRNA translation; consequently, receptor tyrosine kinase inhibitors (Herceptin, Gleevec, erlotinib, gefitinib) and mTOR inhibitors (rapamycin, temsirolimus, everolimus)142,143 are thought to reduce tumor angiogenesis, and possibly other hypoxic responses, by indirectly reducing HIFα protein synthesis. Other drugs have been shown to increase HIFα degradation, including HDAC inhibitors144,145, and compounds that disrupt HIFα binding to HSP90 (geldanamycin)146, 147. These studies indicate that HIF activity is susceptible to inhibition using a variety of drugs already approved for cancer treatment; however, the extent to which these drugs limit the growth and progression by specifically inhibiting HIF activity in autochthonous tumors is as yet unknown, and needs to be investigated.
Given the disparate effects of HIF1α and HIF2α on tumor growth and progression described in this review, it will also be critical to determine whether potential HIF inhibitors affect both HIFα subunits equally. There may be situations where se lective inhibition of either HIFα protein wou ld be especially beneficial; for example, inhibiting HIF1α may be particularly advantageous for highly glycolytic hypoxic tumors, whereas inhibiting only HIF2α is likely to be useful in treating RCCs. Intriguingly, IIiopouios and colleagues identified a series of small compounds that interfere with HIF2α mRNA translation by enhancing IREBP1 binding to the iron response element found in the 5′ UTR of HIF2α, but not HIF1α62 Selective agents of this kind could be particularly useful in cancers that express both HIF1α and HIF2α, where they have distinct roles. Furthermore, Dewhirst et al have shown that rad iation induces HIF1α and VEGFA, protecting endothelial cells from radiation-mediated apoptosis148. Treatment of mice harboring tumors with the HIF inhibitor YC-1 enhanced vessel destruction and slowed tumor growth; in another study148, the HIF inhibitor PX-478 reduced VEGFA expression, rendering xenografts more sensitive to ionizing radiation. As stated previously, endothelial VEGFA expression appears to be regulated primarily by HIF1α, suggesting that its selective inhibition would be beneficial.
Finally, HIF inhibition may be advantageous only up to a certain point. As discussed earlier, HIF2α overexpression, as well as HIF2α deletion, increases the growth of KRAS driven murine lung tumors, although by different mechanisms. Too much HIF2α increases VEGF and SNAIL expression, promoting angiogenesis and tumor invasion, whereas complete loss of HIF2α reduces the expression of the tumor suppressor Scgb3a1, a direct HIF2α target gene. These data suggest that successful HIF inhibition in cancer treatment may involve a narrower therapeutic window than initially envisioned. Although currently in early stages, the prospect of pharmacological HIF inhibition for cancer treatment, whether targeting HIF1α and HIF2α together, or either subunit individually, is an exciting one.
Box 2. HIFs in normal and cancer stem cells.
Stem cells reside in complex microenvironments or niches, and multiple studies revealed that O2 levels influence the ability of stem and/or progenitor cells to remain quiescent or undergo differentiation, depending on cell type152. Again, HIF1α and HIF2α exhibit distinct roles in stem cell regulation. HIF1α appears to playa dominant role in modulating152. WNT-β-catenin signaling in hypoxic ES cells and isolated neural stem cells (NSCs) of the embryonic mesencephalon and adult hippocampus153. WNT-β-catenin activity is closely associated with low O2 regions in the subgranular zone of the hippocampus, an important NSC niche, and Hif1α deletion impairs WNT-dependent processes, such as NSC proliferation, differentiation, and neuronal maturation. It should be noted that the opposite resu lt has been reported for colon cancer cells, where HIF1α inhibits WNT-β-catenin activity154, indicating that the interaction between HIF1α and WNT in stem cells is functionally distinct from more differentiated cells, including neoplastic cells. The basis for this difference is currently unknown. HIF1α has also been proposed to increase the intracellular stability of activated NOTCH1 and to promote NOTCH target gene activation of myogenic and neural precursor cells155. This has been extended to thymic lymphomas in p53 mutant mice where HIF1α promotes NOTCH1 activation and target gene expression156. However, the data on neuroblastoma stem cells suggest that both HIF1α and HIF2α can augment NOTCH pathway signaling.
In contrast, HIF2α (but not HIF1α) regulates the POU transcription factor OCT4 (also known as POU5F1)157. OCT4 is essential for maintaining an undifferentiated cell fate in embryonic stem (ES) cells, the embryonic epiblast, and primordial germ cells (PGCs). Finally, HIF2α is selectively expressed in CD133+ glioblastoma “stem” cells, whereas HIF1α is detected in both tumorigenic (i.e. stem) and non-tumorigenic populations, suggesting HIF2α has a unique role in the CD133+ fraction69. Similarly, human neuroblastomas exhibit small numbers of tumor initiating/stem cells expressing neural crest markers (ID2, NOTCH1, HES1, and Vimentin) and HIF2α158. Upon HIF2α inhibition, these cells undergo early sympathetic neuronal differentiation, and express markers such as HASH1 (also known asASCL1), ISL1, and SCG10 (also known as STMN2). It is noteworthy that the CD133+ glioblastoma and putative neuroblastoma tumor initiating/stem cells express high levels of HIF2α, although they reside in peri-endothelial niches159. While the extent of O2 saturation with in these capillaries is unknown, the data are consistent with the idea that HIF2α accumulates at higher levels of O2 than HIF1α. Alternatively, HIFα expression in distinct cancer cell subpopulations may be controlled by non-hypoxic stimuli, such as aberrant metabolism160.
References
- 1.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 2.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–75. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–85. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–43. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 7.Wang GL, Jiang B-H, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-Ioop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 9.Flamme I, et al. HRF, a putative basic helix-Ioop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1 alpha and developmentally expressed in blood vessels. Mech. Dev. 1997;63:51–60. doi: 10.1016/s0925-4773(97)00674-6. [DOI] [PubMed] [Google Scholar]
- 10.Ema M, et al. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1 alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. USA. 1997;94:4273–8. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogenesch JB, et al. Characterization of a subset of the basic-helix-Ioop-helix-PAS superfam ily that interacts with components of the dioxin signaling pathway. J Bioi Chem. 1997;272:8581–93. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 12.Wiesener MS, et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. Faseb J. 2003;17:271–273. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- 13.Makino Y, Kanopka A, Wilson w.J., Tanaka H, Poellinger L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3alpha locus. J Bioi Chem. 2002;277:32405–8. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- 14.Heikkila M, Pasanen A, Kivirikko K1, Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3alpha variants in the hypoxia response. Cell Mol Life Sci. 2011 doi: 10.1007/s00018-011-0679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka T, Wiesener M, Bernhardt w., Eckardt KU, Warnecke C. The human HIF (hypoxia-inducible factor)-3alpha gene is a HIF-1 target gene and may modulate hypoxic gene induction. Biochem J. 2009;424:143–51. doi: 10.1042/BJ20090120. [DOI] [PubMed] [Google Scholar]
- 16.Maynard MA, et al. Multiple splice variants of the human HIF-3 alpha locus are targets of the von Hippel-Lindau E3 ubiquitin ligase complex. J Bioi Chem. 2003;278:11032–40. doi: 10.1074/jbc.M208681200. [DOI] [PubMed] [Google Scholar]
- 17.Maynard MA, et al. Dominant-negative HIF-3 alpha 4 suppresses VHL-null renal cell carcinoma progression. Cell Cycle. 2007;6:2810–6. doi: 10.4161/cc.6.22.4947. [DOI] [PubMed] [Google Scholar]
- 18.Hirose K, et al. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helixlPAS factor (Arnt2) with close sequence similarity to the Aryl hydrocarbon receptor nuclear translocator (Arnt) Mol. Cell. BioI. 1996;16:1706–1713. doi: 10.1128/mcb.16.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michaud JL, DeRossi C, May NR, Holdener BC, Fan C. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech. Dev. 2000;90:253–261. doi: 10.1016/s0925-4773(99)00328-7. [DOI] [PubMed] [Google Scholar]
- 20.Keith B, Adelman DM, Simon MC. Targeted mutation of the murine arylhydrocarbon receptor nuclear translocator 2 (Arnt2) gene reveals partial redundancy with Arnt. Proc. Natl. Acad. Sci. USA. 2001;98:6692–6697. doi: 10.1073/pnas.121494298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin XY, et al. siRNA-mediated knockdown of aryl hydrocarbon receptor nuclear translocator 2 affects hypoxia-inducible factor-1 regulatory signaling and metabolism in human breast cancer cells. FEBS Lett. 2011;585:3310–15. doi: 10.1016/j.febslet.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Kaelin WG., Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–73. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 23.Maranchie JK, et al. The contribution ofVHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–55. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 24.Kondo K, Kim w.y., Lechpammer M, Kaelin WG., Jr. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Bioi. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acker T, et al. Genetic evidence for a tumor suppressor role of HIF-2alpha. Cancer Cell. 2005;8:131–41. doi: 10.1016/j.ccr.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Mazumdar J, et al. HIF-2alpha deletion promotes Kras-driven lung tumor development. Proc Natl Acad Sci USA. 2010;107:14182–7. doi: 10.1073/pnas.1001296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu C-J, Wang L-Y, Chodosh LA, Keith B, Simon MC. Differential Roles of Hypoxia-Inducible Factor 1 alpha (HIF-1 alpha) and HIF-2alpha in Hypoxic Gene Regulation. Mol Bioi Cell. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raval RR, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Bioi. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu CJ, Sataur A, Wang L, Chen H, Simon MC. The N-Terminal Transactivation Domain Confers Target Gene Specificity of Hypoxia-inducible Factors HIF-1{alpha} and HIF-2{alpha} Mol Bioi Cell. 2007;18:4528–42. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau KW, Tian YM, Raval RR, Ratcliffe PJ, Pugh CW. Target gene selectivity of hypoxia-inducible factor-alpha in renal cancer cells is conveyed by post-DNA-binding mechanisms. Br J Cancer. 2007;96:1284–92. doi: 10.1038/sj.bjc.6603675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia X, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci USA. 2009;106:4260–5. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia X, Kung AL. Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Bioi. 2009;10:R113. doi: 10.1186/gb-2009-10-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mole DR, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1 alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Bioi Chern. 2009;284:16767–75. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krieg AJ, et al. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Bioi. 2010;30:344–53. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schodel J, et al. High-resolution genome-wide mapping of HIF-binding sites by ChiP-seq. Blood. 2011;117:e207–17. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer NV, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian H, Hammer RE, Matsumoto AM, Russell DW, McKnight SL. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Compernolle V, et al. Loss of HIF-2alpha and inh ibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat Med. 2002;8:702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- 40.Peng J, Zhang L, Drysdale L, Fong GH. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc. Natl. Acad. Sci. USA. 2000;97:8386–8391. doi: 10.1073/pnas.140087397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scortegagna M, et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1-1-mice. Nat Genet. 2003;35:331–40. doi: 10.1038/ng1266. [DOI] [PubMed] [Google Scholar]
- 42.Gruber M, et al. Acute postnatal ablation of Hif-2alpha results in anemia. Proc Natl Acad Sci USA. 2007;104:2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao D, Corle C, Seagroves TN, Johnson RS. Hypoxia-inducible factor-1alpha is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Res. 2007;67:563–72. doi: 10.1158/0008-5472.CAN-06-2701. [DOI] [PubMed] [Google Scholar]
- 44.Krop I, et al. HIN-1, an inhibitor of cell growth, invasion, and AKT activation. Cancer Res. 2005;65:9659–69. doi: 10.1158/0008-5472.CAN-05-1663. [DOI] [PubMed] [Google Scholar]
- 45.Kim WY, et al. HIF2alpha cooperates with RAS to promote lung tumorigenesis in mice. J Clin Invest. 2009;119:2160–70. doi: 10.1172/JCI38443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rankin EB, et al. Hypoxia-inducible factor-2 regulates vascular tumorigenesis in mice. Oncogene. 2008;27:5354–8. doi: 10.1038/onc.2008.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang HY, et al. Hypoxia-inducible factors 1 and 2 are important transcriptional effectors in primary macrophages experiencing hypoxia. Blood. 2009;114:844–59. doi: 10.1182/blood-2008-12-195941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doedens AL, et al. Macrophage expression of hypoxia-inducible factor-1 alpha suppresses T-cell function and promotes tumor progression. Cancer Res. 2010;70:7465–75. doi: 10.1158/0008-5472.CAN-10-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeda N, et al. Differential activation and antagonistic function of HIF-{alpha} isoforms in macrophages are essential for NO homeostasis. Genes Dev. 2010;24:491–501. doi: 10.1101/gad.1881410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imtiyaz HZ, et al. Hypoxia-inducible factor 2alpha regulates macrophage function in mouse models of acute and tumor inflammation. J Clin Invest. 2010;120:2699–714. doi: 10.1172/JCI39506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang N, et al. Loss of HIF-1alpha in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer Cell. 2004;6:485–95. doi: 10.1016/j.ccr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 52.Yamash ita T, et al. Hypoxia-inducible transcription factor-2alpha in endothelial cells regulates tumor neovascularization through activation of ephrin A1. J Bioi Chem. 2008;283:18926–36. doi: 10.1074/jbc.M709133200. [DOI] [PubMed] [Google Scholar]
- 53.Skuli N, et al. Endothelial deletion of hypoxia-inducible factor-2alpha (HIF-2alpha) alters vascular function and tumor angiogenesis. Blood. 2009;114:469–77. doi: 10.1182/blood-2008-12-193581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belaiba RS, et al. Hypoxia up-regulates hypoxia-inducible factor-1alpha transcription by involving phosphatidylinositol 3-kinase and nuclear factor kappaB in pulmonary artery smooth muscle cells. Mol Bioi Cell. 2007;18:4691–7. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frede S, Stockmann C, Freitag P, Fandrey J. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via p44/42 MAPK and NF-kappaB. Biochem J. 2006;396:517–27. doi: 10.1042/BJ20051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nizet V, Johnson RS. Interdependence of hypoxic and innate immune responses. Nat Rev Immunol. 2009;9:609–17. doi: 10.1038/nri2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rius J, et al. NF-kappaB links innate immun ity to the hypoxic response through transcriptional regulation of HIF-1 alpha. Nature. 2008;453:807–1. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kenneth NS, Mudie S, van Uden P, Rocha S. SWI/SNF regulates the cellular response to hypoxia. J Bioi Chem. 2009;284:4123–31. doi: 10.1074/jbc.M808491200. [DOI] [PubMed] [Google Scholar]
- 59.Toschi A, Lee E, Gad ir N, Ohh M, Foster DA. Differential dependence of hypoxia-inducible factors 1 alpha and 2 alpha on mTORC1 and mTORC2. J Bioi Chem. 2008;283:34495–9. doi: 10.1074/jbc.C800170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uchida T, et al. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1 alpha. J Bioi Chem. 2004;279:14871–8. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat Struct Mol Bioi. 2007;14:420–6. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 62.Zimmer M, et al. Small-molecule inhibitors of HIF-2a translation link its 5′UTR iron-responsive element to oxygen sensing. Mol Cell. 2008;32:838–48. doi: 10.1016/j.molcel.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee FS, Percy MJ. The HIF pathway and erythrocytosis. Annu Rev Patho/ 2011;6:165–92. doi: 10.1146/annurev-pathol-011110-130321. [DOI] [PubMed] [Google Scholar]
- 64.Mastrogiannaki M, et al. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice. J Clin Invest. 2009;119:1159–66. doi: 10.1172/JCI38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rankin EB, et al. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest. 2007;117:1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morita M, et al. HLF/HIF-2alpha is a key factor in retinopathy of prematurity in association with erythropoietin. Embo J. 2003;22:1134–46. doi: 10.1093/emboj/cdg117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nilsson H, et al. HIF-2alpha expression in human fetal paraganglia and neuroblastoma: relation to sympathetic d ifferentiation, glucose deficiency, and hypoxia. Exp Cell Res. 2005;303:447–56. doi: 10.1016/j.yexcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 68.Holmquist-Mengelbier L, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–23. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 69.Li Z, et al. Hypoxia-inducible factors regu late tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–13. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koh MY, Darnay BG, Powis G. Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1 alpha, leading to its oxygen-independent degradation. Mol Cell Bioi. 2008;28:7081–95. doi: 10.1128/MCB.00773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koh MY, Lemos R, Jr., Liu X, Powis G. The Hypoxia-Associated Factor Switches Cells from HIF-1 {alpha}- to HIF-2{alpha}-Dependent Signaling Promoting Stem Cell Characteristics, Aggressive Tumor Growth and Invasion. Cancer Res. 2011;71:4015–27. doi: 10.1158/0008-5472.CAN-10-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo W, et al. Hsp70 and CHIP selectively mediate ubiqu itination and degradation of hypoxia-inducible factor (HIF)-1 alpha but Not HIF-2alpha. J Bioi Chem. 2010;285:3651–63. doi: 10.1074/jbc.M109.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jokilehto T, Jaakkola PM. The role of HIF prolyl hydroxylases in tumour growth. J Cell Mol Med. 2010;14:758–70. doi: 10.1111/j.1582-4934.2010.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chan DA, Giaccia AJ. PHD2 in tumour angiogenesis. BrJ Cancer. 2010;103:1–5. doi: 10.1038/sj.bjc.6605682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cockman ME, Webb JD, Ratcliffe PJ. FIH-dependent asparaginyl hydroxylation of ankyrin repeat domain-containing proteins. Ann N Y Acad Sci. 2009;1177:9–18. doi: 10.1111/j.1749-6632.2009.05042.x. [DOI] [PubMed] [Google Scholar]
- 76.Appelhoff RJ, et al. Differential function of the prolyl hydroxylases, PHD1, 2 and 3 in the regulation of Hypoxia inducible factor (HIF) J BioI Chem. 2004;279:38458–65. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 77.Bracken CP, et al. Cell-specific regulation of hypoxia-inducible factor (HIF)-1 alpha and HIF-2alpha stabilization and transactivation in a graded oxygen environment. J BioI Chem. 2006;281:22575–85. doi: 10.1074/jbc.M600288200. [DOI] [PubMed] [Google Scholar]
- 78.Zhang N, et al. The asparaginyl hydroxylase factor in hibiting HIF-1alpha is an essential regulator of metabolism. Cell Metab. 2010;11:364–78. doi: 10.1016/j.cmet.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J BioI Chem. 1999;274:32631–7. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- 80.Conrad P.w., Freeman TL, Beitner-Johnson D, Millhorn DE. EPAS1 trans-activation during hypoxia requires p42/p44 MAPK. J BioI Chem. 1999;274:33709–13. doi: 10.1074/jbc.274.47.33709. [DOI] [PubMed] [Google Scholar]
- 81.To KK, Sedelnikova OA, Samons M, Bonner WM, Huang LE. The phosphorylation status of PAS-B distinguishes HIF-1alpha from HIF-2alpha in NBS1 repression. Embo J. 2006;25:4784–94. doi: 10.1038/sj.emboj.7601369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mylonis I, et al. Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxia-inducible factor-1 alpha. J BioI Chem. 2006;281:33095–106. doi: 10.1074/jbc.M605058200. [DOI] [PubMed] [Google Scholar]
- 83.Kalousi A, et al. Casein kinase 1 regulates human hypoxia-inducible factor HIF-1. J Cell Sci. 2010;123:2976–86. doi: 10.1242/jcs.068122. [DOI] [PubMed] [Google Scholar]
- 84.Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1alpha. Mol Cell. 2010;40:509–20. doi: 10.1016/j.molcel.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Finkel T, Deng C.x., Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–91. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dioum EM, et al. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–93. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 87.Lim JH, et al. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1 alpha. Mol Cell. 2010;38:864–78. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 88.Chen R, Dioum EM, Hogg RT, Gerard RD, Garcia JA. Hypoxia increases sirtuin 1 expression in a hypoxia-inducible factor-dependent manner. J BioI Chem. 2011;286:13869–78. doi: 10.1074/jbc.M110.175414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sayed D, Abdellatif M. AKT-ing via microRNA. Cell Cycle. 2010;9:3213–7. doi: 10.4161/cc.9.16.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhong L, et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bell EL, Guarente L. The SirT3 Divining Rod Points to Oxidative Stress. Mol Cell. 2011;42:561–8. doi: 10.1016/j.molcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guzy RD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–8. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 93.Mansfield KD, et al. Cytochrome C is required for cellular oxygen sensing and hypoxic HIF activation. Cell Metabolism. 2005;1:393–9. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brunelle JK, et al. Oxygen sensing requ ires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–14. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 95.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–96. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Finley LW, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–28. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jeong JW, et al. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell. 2002;111:709–20. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- 98.Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1 alpha protein by recru iting histone deacetylase 1. Embo J. 2006;25:1231–41. doi: 10.1038/sj.emboj.7601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bilton R, et al. Arrest-defective-1 protein, an acetyltransferase, does not alter stability of hypoxia-inducible factor (HIF)-1 alpha and is not induced by hypoxia or HIF. J BioI Chem. 2005;280:31132–40. doi: 10.1074/jbc.M504482200. [DOI] [PubMed] [Google Scholar]
- 100.Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1 alpha during hypoxia. Cell. 2007;131:584–95. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Hagen M, Overmeer RM, Abolvardi SS, Vertegaal AC. RNF4 and VHL regulate the proteasomal degradation of SUMO-conjugated Hypoxia-Inducible Factor-2alpha. Nucleic Acids Res. 2010;38:1922–31. doi: 10.1093/nar/gkp1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bae SH, et al. Sumoylation increases HIF-1alpha stability and its transcriptional activity. Biochem Biophys Res Commun. 2004;324:394–400. doi: 10.1016/j.bbrc.2004.09.068. [DOI] [PubMed] [Google Scholar]
- 103.Carbia-Nagashima A, et al. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1 alpha during hypoxia. Cell. 2007;131:309–23. doi: 10.1016/j.cell.2007.07.044. [DOI] [PubMed] [Google Scholar]
- 104.Huang C, et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. Embo J. 2009;28:2748–62. doi: 10.1038/emboj.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li F, et al. Regulation of HIF-1 alpha stability through S-n itrosylation. Mol Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ryu JH, et al. Hypoxia-inducible factor alpha subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J Bioi Chem. 2011;286:6963–70. doi: 10.1074/jbc.M110.188706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koshiji M, et al. HIF-1alpha induces cell cycle arrest by functional ly counteracting Myc. Embo J. 2004;23:1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dang C.v., Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–6. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 109.Koshiji M, et al. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 110.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcri ptiona l activity. Cancer Cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Corn PG, et al. Mxi1 is induced by hypoxia in a HIF-1-dependent manner and protects cells from c-Myc-induced apoptosis. Cancer Bioi Ther. 2005;4:1285–94. doi: 10.4161/cbt.4.11.2299. [DOI] [PubMed] [Google Scholar]
- 112.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellu lar respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 113.Gordan JD, et al. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14:435–46. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim JW, Gao P, Liu YC, Semenza GL, Dang C.v. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Bioi. 2007;27:7381–93. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qing G, et al. Combinatorial regu lation of neuroblastoma tumor progression by N-Myc and hypoxia inducible factor HIF-1 alpha. Cancer Res. 2010;70:10351–61. doi: 10.1158/0008-5472.CAN-10-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kaelin WG., Jr. Kidney cancer: now available in a new flavor. Cancer Cell. 2008;14:423–4. doi: 10.1016/j.ccr.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 117.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 118.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–50. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 119.Pan Y, Oprysko PR, Asham AM, Koch CJ, Simon MC. p53 cannot be induced by hypoxia alone but responds to the hypoxic microenvironment. Oncogene. 2004 doi: 10.1038/sj.onc.1207657. [DOI] [PubMed] [Google Scholar]
- 120.An WG, et al. Stabilization of wild-type p53 by hypoxia-inducible factor 1 alpha. Nature. 1998;392:405–8. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 121.Sanchez-Puig N, Veprintsev DB, Fersht AR. Binding of natively unfolded HIF-1 alpha ODD domain to p53. Mol Cell. 2005;17:11–21. doi: 10.1016/j.molcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 122.Chen D, Li M, Luo J, Gu W. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J Bioi Chem. 2003;278:13595–8. doi: 10.1074/jbc.C200694200. [DOI] [PubMed] [Google Scholar]
- 123.Moeller BJ, et al. Pleiotropic effects of HIF-1 blockade on tumor radiosensitivity. Cancer Cell. 2005;8:99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 124.Ravi R, et al. Regu lation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 125.Bertout JA, et al. HIF2alpha inh ibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc Natl Acad Sci USA. 2009;106:14391–6. doi: 10.1073/pnas.0907357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roberts AM, et al. Suppression of hypoxia-inducible factor 2alpha restores p53 activity via Hdm2 and reverses chemoresistance of renal carcinoma cells. Cancer Res. 2009;69:9056–64. doi: 10.1158/0008-5472.CAN-09-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sengupta S, Peterson TR, Sabatin i DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–22. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008;27(Suppl 2):S43–51. doi: 10.1038/onc.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu L, et al. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–31. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1rrSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–51. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li Y, et al. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J BioI Chem. 2007;282:35803–13. doi: 10.1074/jbc.M705231200. [DOI] [PubMed] [Google Scholar]
- 133.Gan B, Melkoumian ZK, Wu X, Guan KL, Guan JL. Identification of FIP200 interaction with the TSC1-TSC2 complex and its role in regulation of cell size control. J Cell BioI. 2005;170:379–89. doi: 10.1083/jcb.200411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chano T, et al. Neuromuscular abundance of RB1CC1 contributes to the non-proliferating enlarged cell phenotype through both RB1 maintenance and TSC1 degradation. Int J Mol Med. 2006;18:425–32. [PubMed] [Google Scholar]
- 135.Morris MR, et al. Mutation analysis of hypoxia-inducible factors HIF1A and HIF2A in renal cell carcinoma. Anticancer Res. 2009;29:4337–43. [PubMed] [Google Scholar]
- 136.Dalg liesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010 doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Beroukhim R, et al. Patterns of gene expression and copy-number alterations in von-hippellindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–81. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Purdue MP, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11 q13.3. Nat Genet. 2010;43:60–5. doi: 10.1038/ng.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Semenza GL. Defin ing the role of hypoxia-i nducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–34. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rapisarda A, et al. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 2002;62:4316–24. [PubMed] [Google Scholar]
- 141.Lee K, et al. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascu larization. Proc Natl Acad Sci USA. 2009;106:17910–5. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 142.Majumder PK, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 143.Thomas GV, et al. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–7. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- 144.Kong X, et al. Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1 alpha. Mol Cell Bioi. 2006;26:2019–28. doi: 10.1128/MCB.26.6.2019-2028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qian D.l., et al. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res. 2006;66:8814–21. doi: 10.1158/0008-5472.CAN-05-4598. [DOI] [PubMed] [Google Scholar]
- 146.Isaacs JS, et al. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J BioI Chem. 2002;277:29936–44. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- 147.Mabjeesh NJ, et al. Geldanamycin induces degradation of hypoxia-inducible factor 1 alpha protein via the proteosome pathway in prostate cancer cells. Cancer Res. 2002;62:2478–82. [PubMed] [Google Scholar]
- 148.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Rad iation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–41. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 149.Lando D, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regu lates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hewitson KS, et al. Hypoxia-inducible Factor (HIF) Asparagine Hydroxylase Is Identical to Factor Inhibiting HIF (FIH) and Is Related to the Cupin Structural Family. J BioI Chem. 2002;277:26351–5. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 152.Simon MC, Keith B. The role of oxygen availability in embryonic development and stem cell function. Nat Rev Mol Cell BioI. 2008;9:285–96. doi: 10.1038/nrm2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mazumdar J, et al. O2 regulates stem cells through Wntlbeta-catenin signalling. Nat Cell BioI. 2010;12:1007–13. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kaidi A, Williams AC, Paraskeva C. Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell BioI. 2007;9:210–217. doi: 10.1038/ncb1534. [DOI] [PubMed] [Google Scholar]
- 155.Gustafsson MV, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 156.Bertout JA, et al. Heterozygosity for hypoxia inducible factor 1 alpha decreases the incidence of thymic lymphomas in a p53 mutant mouse model. Cancer Res. 2009;69:3213–20. doi: 10.1158/0008-5472.CAN-08-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Covello KL, et al. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pietras A, et al. HIF-2alpha maintains an undifferentiated state in neural crest-like human neuroblastoma tumor-initiating cells. Proc Natl Acad Sci USA. 2009;106:16805–10. doi: 10.1073/pnas.0904606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gilbertson RJ, Rich JN. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7:733–6. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- 160.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–6. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Korkolopoulou P, et al. Hypoxia-inducible factor 1 alpha/vascular endothelial growth factor axis in astrocytomas. Associations with microvessel morphometry, proliferation and prognosis. Neuropathol Appl Neurobiol. 2004;30:267–78. doi: 10.1111/j.1365-2990.2003.00535.x. [DOI] [PubMed] [Google Scholar]
- 162.Scrideli CA, et al. Prognostic significance of co-overexpression of the EGFRIIGFBP-2/HIF-2A genes in astrocytomas. J Neuroonco/ 2007;83:233–9. doi: 10.1007/s11060-007-9328-0. [DOI] [PubMed] [Google Scholar]
- 163.Theodoropoulos VE, et al. Hypoxia-inducible factor 1 alpha expression correlates with angiogenesis and unfavorable prognosis in bladder cancer. Eur Uro/ 2004;46:200–8. doi: 10.1016/j.eururo.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 164.Yamamoto Y, et al. Hypoxia-inducible factor 1alpha is closely linked to an aggressive phenotype in breast cancer. Breast Cancer Res Treat. 2008;110:465–75. doi: 10.1007/s10549-007-9742-1. [DOI] [PubMed] [Google Scholar]
- 165.Helczynska K, et al. Hypoxia-Inducible Factor-2alpha correlates to distant recurrence and poor outcome in invasive breast cancer. Cancer Research. 2008;68:9212–9220. doi: 10.1158/0008-5472.CAN-08-1135. [DOI] [PubMed] [Google Scholar]
- 166.Birner P, et al. Overexpression of hypoxia-inducible factor 1 alpha is a marker for an unfavorable prognosis in early-stage invasive cervical cancer. Cancer Res. 2000;60:4693–6. [PubMed] [Google Scholar]
- 167.Kawanaka T, et al. Prognostic significance of HIF-2alpha expression on tumor infiltrating macrophages in patients with uterine cervical cancer undergoing radiotherapy. J Med Invest. 2008;55:78–86. doi: 10.2152/jmi.55.78. [DOI] [PubMed] [Google Scholar]
- 168.Yoshimura H, et al. Prognostic impact of hypoxia-inducible factors 1alpha and 2alpha in colorectal cancer patients: correlation with tumor angiogenesis and cyclooxygenase-2 expression. Clin Cancer Res. 2004;10:8554–60. doi: 10.1158/1078-0432.CCR-0946-03. [DOI] [PubMed] [Google Scholar]
- 169.Griffiths EA, et al. Hypoxia-inducible factor-1alpha expression in the gastric carcinogenesis sequence and its prognostic role in gastric and gastro-oesophageal adenocarcinomas. Br J Cancer. 2007;96:95–103. doi: 10.1038/sj.bjc.6603524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Griffiths EA, et al. Hypoxia-associated markers in gastric carcinogenesis and HIF-2alpha in gastric and gastro-oesophageal cancer prognosis. Br J Cancer. 2008;98:965–73. doi: 10.1038/sj.bjc.6604210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mizokami K, Kakeji Y, Oda S, Maehara Y. Relationsh ip of hypoxia-inducible factor 1alpha and p21WAF1/CIP1 expression to cell apoptosis and clinical outcome in patients with gastric cancer. World J Surg Oncol. 2006;4:94. doi: 10.1186/1477-7819-4-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Takahashi R, et al. Hypoxia-inducible factor-1alpha expression and angiogenesis in gastrointestinal stromal tumor of the stomach. Oncol Rep. 2003;10:797–802. [PubMed] [Google Scholar]
- 173.Koukourakis M1, et al. Hypoxia-inducible factor (HIF1A and HIF2A), angiogenesis, and chemoradiotherapy outcome of squamous cell head-and-neck cancer. Int J Radiat Oncol BioI Phys. 2002;53:1192–202. doi: 10.1016/s0360-3016(02)02848-1. [DOI] [PubMed] [Google Scholar]
- 174.Winter SC, et al. The relation between hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression with anemia and outcome in surgically treated head and neck cancer. Cancer. 2006;107:757–66. doi: 10.1002/cncr.21983. [DOI] [PubMed] [Google Scholar]
- 175.Bangoura G, et al. Prog nostic significance of HIF-2alpha/EPAS1 expression in hepatocellular carcinoma. World J Gastroentero/ 2007;13:3176–82. doi: 10.3748/wjg.v13.i23.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Giatromanolaki A, et al. Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br J Cancer. 2001;85:881–90. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kim SJ, et al. Expression of HIF-1alpha, CA IX, VEGF, and MMP-9 in surgically resected non-small cell lung cancer. Lung Cancer. 2005;49:325–35. doi: 10.1016/j.lungcan.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 178.Wu XH, Qian C, Yuan K. Correlations of hypoxia-inducible factor-1alpha/hypoxia-inducible factor-2alpha expression with ang iogenesis factors expression and prognosis in non-small cell lung cancer. Chin Med J (Engl) 2011;124:11–8. [PubMed] [Google Scholar]
- 179.Giatromanolaki A, et al. Hypoxia-inducible factors 1 alpha and 2alpha are related to vascular endothelial growth factor expression and a poorer prognosis in nodular malignant melanomas of the skin. Melanoma Res. 2003;13:493–501. doi: 10.1097/00008390-200310000-00008. [DOI] [PubMed] [Google Scholar]
- 180.Noguera R, et al. HIF-1alpha and HIF-2alpha are differentially regulated in vivo in neuroblastoma: high HIF-1 alpha correlates negatively to advanced clin ical stage and tumor vascu larization. Clin Cancer Res. 2009;15:7130–6. doi: 10.1158/1078-0432.CCR-09-0223. [DOI] [PubMed] [Google Scholar]
- 181.Daponte A, et al. Prog nostic significance of Hypoxia-Inducible Factor 1 alpha(HIF-1 alpha) expression in serous ovarian cancer: an immunohistochemical study. BMC Cancer. 2008;8:335. doi: 10.1186/1471-2407-8-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Osada R, et al. Expression of hypoxia-inducible factor 1 alpha, hypoxia-inducible factor 2alpha, and von Hippel-Lindau protein in epithelial ovarian neoplasms and allelic loss of von Hippel-Lindau gene: nuclear expression of hypoxia-inducible factor 1 alpha is an independent prognostic factor in ovarian carcinoma. Hum Patho/ 2007;38:1310–20. doi: 10.1016/j.humpath.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 183.Sun HC, et al. Expression of hypoxia-inducible factor-1 alpha and associated proteins in pancreatic ductal adenocarcinoma and their impact on prognosis. Int J Onco/ 2007;30:1359–67. [PubMed] [Google Scholar]
- 184.Shibaji T, et al. Prognostic significance of HIF-1 alpha overexpression in human pancreatic cancer. Anticancer Res. 2003;23:4721–7. [PubMed] [Google Scholar]
- 185.Nanni S, et al. Endothelial NOS, estrogen receptor beta, and HIFs cooperate in the activation of a prognostic transcriptional pattern in aggressive human prostate cancer. J Clin Invest. 2009;119:1093–108. doi: 10.1172/JCI35079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lidgren A, et al. The expression of hypoxia-inducible factor 1 alpha is a favorable independent prognostic factor in renal cell carcinoma. C/in Cancer Res. 2005;11:1129–35. [PubMed] [Google Scholar]
- 187.Klatte T, et al. Hypoxia-inducible factor 1 alpha in clear cell renal cell carcinoma. C/in Cancer Res. 2007;13:7388–93. doi: 10.1158/1078-0432.CCR-07-0411. [DOI] [PubMed] [Google Scholar]
- 188.Kapitsinou PP, et al. Hepatic HIF-2 regulates erythropoietic responses to hypoxia in renal anemia. B/ood. 2010;116:3039–48. doi: 10.1182/blood-2010-02-270322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gunaratnam L, et al. Hypoxia inducible factor activates the transforming growth factor-alpha/epidermal growth factor receptor growth stimulatory pathway in VHL(−/−) renal cell carcinoma cells. J Biol Chem. 2003;278:44966–74. doi: 10.1074/jbc.M305502200. [DOI] [PubMed] [Google Scholar]
- 190.Carmeliet P, et al. Role of HIF-1 alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–90. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 191.Ryan HE, et al. Hypoxia-inducible factor-1 alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–5. [PubMed] [Google Scholar]
- 192.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–46. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]