Abstract
Global analysis of gene expression by DNA microarrays is nowadays a widely used tool, especially relevant for cancer research. It helps the understanding of complex biology of cancer tissue, allows identification of novel molecular markers, reveals previously unknown molecular subtypes of cancer that differ by clinical features like drug susceptibility or general prognosis. Our aim was to compare gene expression profiles in breast cancer that develop against a background of inherited predisposing mutations versus sporadic breast cancer. In this preliminary study we analysed seven hereditary, BRCA1 mutation-linked breast cancer tissues and seven sporadic cases that were carefully matched by histopathology and ER status. Additionally, we analysed 6 samples of normal breast tissue. We found that while the difference in gene expression profiles between tumour tissue and normal breast can be easily recognized by unsupervised algorithms, the difference between those two types of tumours is more discrete. However, by supervised methods of data analysis, we were able to select a set of genes that may differentiate between hereditary and sporadic tumours. The most significant difference concerns genes that code for proteins engaged in regulation of transcription, cellular metabolism, signalling, proliferation and cell death. Microarray results for chosen genes (TOB1, SEPHS2) were validated by real-time RT-PCR.
Keywords: hereditary breast cancer, DNA microarrays
Introduction
DNA microarrays have been recently widely employed in studies on breast cancer encompassing research on breast cancer cell lines and resected tumour tissues. Two directions of these studies seem to be especially spectacular and promising: the studies of the Norwegian/Stanford group that led to the recognition of several distinct molecular classes of breast cancers [1,2] and the studies of a group at the Netherlands Cancer Institute, which brought identification of a 70-gene prognostic profile for patients with node-negative breast cancer [3,4]. The results of those studies indicate that gene expression analysis by DNA microarrays may help the understanding of the molecular background underlying development and progression of breast cancer as well as providing a clinically useful tool for more personalized treatment [reviewed in: [5]]. It seems clear that a multi-gene approach will prove more useful and informative than currently used analysis of single markers.
Since the identification of two major predisposing genes, BRCA1 and BRCA2, and broad application of genetic testing, significant numbers of mutation carriers have been identified worldwide among breast cancer patients. This allowed further studies in order to estimate clinical features of those specific breast cancer cases. Some indications are accumulating that mutation-linked breast cancer may be a clinically distinct entity from the majority of malignant breast tumours. Among the characteristics of BRCA1 tumours are: earlier age of manifestation, high tumour grade, low oestrogen receptor content and elevated lymphocyte infiltration. In addition, these cases are often characterized by high proliferative activity, resulting in tumours with pushing margins and high mitotic index [6-12]. The data concerning survival in BRCA1 mutation carriers are confusing. There are intriguing observations that despite adverse prognostic indications, patients with BRCA1 mutations have survival rates similar to or even better than patients with sporadic breast cancer [[13-15], own unpublished data]. The long-term aim of our study is an attempt to elucidate the molecular basis underlying described discrepancies by comparing gene expression profiles of BRCA1-associated hereditary breast cancer and sporadic breast cancer cases. The first attempt to compare hereditary versus sporadic breast cancers by DNA microarray analysis was published by Hedenfalk et al., who used cDNA microarrays containing 6512 cDNA clones [16]. In our study we used HG U133 Plus 2.0 Gene Chip (Affymetrix), allowing detection of over 47,000 transcripts. We also attempted to make a more careful selection of tumour specimens, which were chosen exclusively from among ER(-) cases. Our group of tumours was also more uniform according to histopathology; only ductal carcinomas and medullary carcinomas, all grade 3, were analysed.
Materials and methods
Tissue samples
Frozen surgical specimens of breast cancer and adjacent normal breast tissue were obtained from the Pomeranian Medical Academy in Szczecin. Only tissues from patients without preceding chemotherapy were used for microarray experiments. For this initial study we chose seven cancer tissues from women with germline mutation in the BRCA1 gene and seven samples of sporadic breast cancer. Three cases had mutation C61G in exon 5, one at 4153delA in exon 11, and three harboured the 5382insC mutation in exon 20. Sporadic cases were obtained from women without a family history of breast/ovarian cancer, in which, additionally, the three most common BRCA1 mutations in Poland were excluded by genetic tests. Eight cases were diagnosed as grade 3 medullary or atypical medullary carcinoma, and 5 cases were grade 3 ductal carcinomas. All tumours were ER negative (immunohistochemistry on paraffin-embedded material). The percentage of cancer cells within tumour specimens was estimated by a pathologist; in the majority of samples it ranged from 70% to 90%, while in 3 samples it was approx. 50%. In addition, we analysed six samples of unchanged glandular tissue surrounding the tumour and obtained during mastectomy. The lack of tumour samples from BRCA2 mutation carriers in our study reflects the specificity of the mutational spectrum in BRCA genes in Central-Eastern Europe, where BRCA2 mutations are very rare (it was estimated by sequencing that BRCA2 mutations account for only about 5% of all BRCA1 and BRCA2 mutations found in Polish families with a strong familial history of breast/ovarian cancer [17]). In total we analysed 14 tumour samples and 6 normal breast samples.
RNA isolation
20-40 mg of frozen tissue was placed in a lysing solution (4M guanidine thiocyanate, 25 mM sodium citrate, 0.5% sodium N-laurylsarcosinate, 0.1M β-mercaptoethanol) and homogenized with Lysing Matrix D in a FastPrep instrument (QBioGene). Total RNA was extracted from the supernatant according to [18]. RNA cleanup and simultaneous on-column digestion of DNA traces with DNAse I (Qiagen) was done using the RNeasy Mini Kit (Qiagen), according to the manufacturer's instructions. RNA quantity was estimated with the ND-1000 Spectrophotometer (NanoDrop Technologies). RNA quality was controlled by microcapillary electrophoresis measurements in the Agilent 2100 Bioanalyzer using the RNA 6000 Nano LabChip Kit and analysed with RNA Integrity Number software (Agilent).
Oligonucleotide microarrays
We used HG U133 Plus 2.0 Gene Chip oligonucleotide arrays (Affymetrix). The hybridization target was prepared according to recommendations of microarray manufacturer. Briefly: 8 μg of total RNA was used for synthesis of double stranded cDNA, half volume of cDNA was used for synthesis of biotynylated cRNA with the BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics). Both cDNA and cRNA were purified with Gene Chip Sample Cleanup Module (Affymetrix). 16g of cRNA was fragmented and hybridized to the microarray for 16h at 45°C. After washing and staining microarrays were scanned with GeneChip Scanner 3000 (Affymetrix).
Statistical analysis of microarray data
Data were obtained using GCOS 1.2 software (Affymetrix). The preprocessing was performed by Robust Multi-Array Analysis (RMA). Hierarchical clustering, Principal Component Analysis (PCA) and supervised comparisons were carried out using GeneSpring 7.2 software (Silicon Genetics). For selection of genes differentially expressed between breast cancer and normal breast tissue we used the parametric Welch test. False Discovery Rate was estimated by Benjamini-Hochberg algorithm. For selection of genes differentially expressed between hereditary and sporadic breast cancer we used the Bioconductor limma package, based on linear models with empirical Bayesian approach. This method provides stable results even when the number of analysed arrays is small.
Quantitative RT-PCR
Quantitative RT-PCR analysis was performed using the ABI 7700 Sequence Detection System and dedicated software (Applied Biosystems). The reactions were performed using the MasterAmp Real-Time RT-PCR Kit (Epicentre), according to the manufacturer's recommendations. Primers for the SYBR Green system were designed using Primer3 online software http://frodo.wi.mit.edu/cgi-bin/prime-r3/primer3_www.cgi. All results were normalised to the expression of the reference gene, eukaryotic translation initiation factor 4 gamma 2 (EIF4G2), which appeared to be equally transcribed in all tissues analysed by microarrays. Primer specificity was verified by sequencing of selected RT-PCR products for each gene. Sequences of the PCR primer pairs used for each gene are shown in Table 1.
Table 1.
Primers used for quantitative RT-PCR
| Gene | Oligonucleotide | Sequence | product size |
|---|---|---|---|
| SEPHS2 | forward primer reverse primer |
5'-GGAAAGGAGGACCTGCAACCA-3' 5'-ACCAGGAATCTGCCGCAAAAG-3' |
154 bp |
| TOB1 | forward primer reverse primer |
5'-ttgtttctacgacatggtattgcattta-3' 5'-caagtattcgtacattttaattccaccact-3' |
182 bp |
| EIFG2 | forward primer reverse primer |
5'-GCAAGGCTTTGTTCCAGGTGA-3' 5'-AGGCTTTGGCTGGTTCTTTAGTCA-3' |
100 bp |
Results
Unsupervised analysis of obtained data set
We performed Principal Component Analysis (PCA) to determine the major sources of variability in our data (Fig. 1). PCA is an unsupervised algorithm, which, if performed 'on conditions', is able to detect intrinsic similarities and differences in the gene expression profiles of analysed samples. Results may be graphically presented and the distances between the dots visualize the level of similarity/dissimilarity between particular samples. It can be seen that the difference in gene expression profile between breast cancer tissue and normal breast tissue is large and is easily recognized by that unsupervised algorithm (normal-tumour difference was responsible for the sample subdivision by the first component, which accounted for 24.06% of total variance). Using a supervised method of data analysis we found that this precise separation of cancer versus normal tissues in PCA may be ascribed to the differential expression of 8,063 genes (Welch test, Benjamini-Hochberg correction for multiple comparisons, FDR<0.05). In contrast, the difference between hereditary and sporadic breast cancer samples cannot be recognized by PCA, suggesting that the difference in gene expression profile between those two types of breast cancer is not very powerful. Samples obtained from normal glandular breast tissue clustered closely together, while tumours were much more dispersed. By unsupervised analysis we were unable to disclose any differences between hereditary and sporadic tumours; they were not visible also when only cancer samples were subjected to decomposition into principal components (data not shown).
Figure 1.
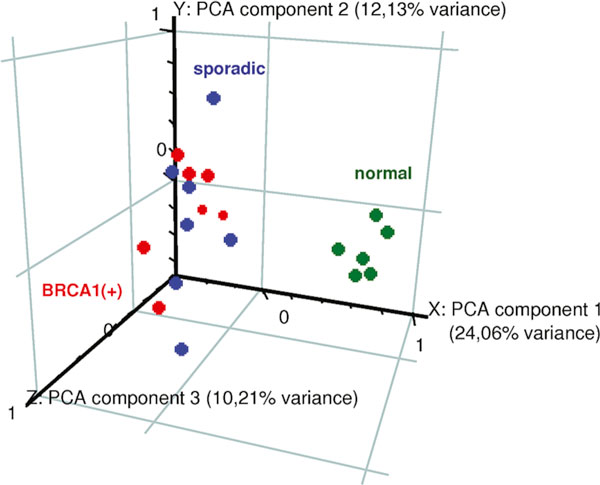
Principal component analysis (PCA) performed on the whole set of 20 samples. The tumor (red and blue) vs. normal (green) difference is clearly visible within the 1st component, which accounted for 24.06% of total variance. PCA is unable to reveal a difference between hereditary (red) and sporadic tumors (blue).
Genes differentiating between hereditary and sporadic breast tumours
As the unsupervised method showed that the distance between hereditary and sporadic tumours is not large and taking into account the limited number of samples in our analysis, we chose a supervised algorithm with a good balance of sensitivity and specificity of analysis. We used the limma package [19], a Bayesian method based on linear modelling with moderated t-statistic.
We selected 100 genes best differentiating between the two analysed groups, ranked according to the statistical power of the expression change (Table 2.). Four times more probe sets were down-regulated in hereditary tumours (78 probesets) in comparison to up-regulated transcripts (22 probe sets). Down-regulated genes showed an average decrease of 1.35 to 5-fold, up-regulated genes were changed by a factor of 1.2-7.8. Among the first hundred genes selected by limma the most prominent classes consisted of genes connected with regulation of transcription (19 genes), metabolism (12 genes), protein synthesis and degradation (10 genes), cellular signalling (8 genes), cell proliferation and death (6 genes) and DNA and RNA replication and processing (5 genes).
Table 2.
Genes differentiating between hereditary, BRCA1-positive breast cancers and sporadic tumours, selected by limma. Genes are ordered according to the fold change value and grouped into functional classes (according to Gene Ontology annotation, http://www.geneontology.org/)
| Affy_ID | Gene Symbol | Gene Title | Fold change BRCA1(+)vs. sporadic | limma rank |
|---|---|---|---|---|
| Down-regulated in BRCA1(+) | ||||
| Cell proliferation and death | ||||
| 202704_at | TOB1 | transducer of ERBB2, 1 | 0.364 | 75 |
| 243031_at | RTN4 | Reticulon 4 | 0.421 | 71 |
| 1556049_at | RTN4 | reticulon 4 | 0.506 | 46 |
| 215070_x_at | RABGAP1 | RAB GTPase activating protein 1 | 0.731 | 97 |
| Cell signalling | ||||
| 1553986_at | RASEF | RAS and EF-hand domain containing | 0.25 | 49 |
| 244181_at | PIK3R1 | Phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | 0.381 | 80 |
| 238176_at | RAPGEF2 | Rap guanine nucleotide exchange factor (GEF) 2 | 0.461 | 58 |
| 229261_at | SOS1 | Son of sevenless homolog 1 (Drosophila) | 0.565 | 92 |
| 215992_s_at | RAPGEF2 | Rap guanine nucleotide exchange factor (GEF) 2 | 0.583 | 90 |
| 207822_at | FGFR1 | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 0.704 | 82 |
| DNA and RNA | ||||
| 233007_at | XRCC5 | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | 0.667 | 22 |
| 230651_at | THOC2 | THO complex 2 | 0.414 | 45 |
| 243908_at | ZNF638 | Zinc finger protein 638 | 0.531 | 17 |
| 218356_at | FTSJ2 | FtsJ homolog 2 (E. coli) | 0.699 | 57 |
| Metabolism | ||||
| 239545_at | CAS1 | O-acetyltransferase | 0.331 | 26 |
| 238563_at | TPRT | Trans-prenyltransferase | 0.351 | 14 |
| 215316_at | HIBADH | 3-hydroxyisobutyrate dehydrogenase | 0.539 | 30 |
| 200961_at | SEPHS2 | selenophosphate synthetase 2 | 0.576 | 72 |
| 202282_at | HADH2 | hydroxyacyl-Coenzyme A dehydrogenase, type II | 0.585 | 2 |
| 238813_at | ALAS2 | Aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) | 0.607 | 7 |
| 232127_at | CLCN5 | Chloride channel 5 (nephrolithiasis 2, X-linked, Dent disease) | 0.626 | 33 |
| 218124_at | RetSat | all-trans-13,14-dihydroretinol saturase | 0.63 | 67 |
| Protein synthesis and degradation | ||||
| 240146_at | CAPZA2 | Capping protein (actin filament) muscle Z-line, alpha 2 | 0.267 | 28 |
| 235138_at | PUM2 | Vacuolar protein sorting 35 (yeast) | 0.287 | 25 |
| 1560926_at | PPP4R2 | Protein phosphatase 4, regulatory subunit 2 | 0.323 | 88 |
| 1554638_at | ZFYVE16 | zinc finger, FYVE domain containing 16 | 0.43 | 56 |
| 238303_at | SIMP | Source of immunodominant MHC-associated peptides | 0.432 | 35 |
| 232216_at | YME1L1 | YME1-like 1 (S. cerevisiae) | 0.443 | 87 |
| 239175_at | AFTIPHILIN | Aftiphilin protein | 0.486 | 4 |
| 222499_at | MRPS16 | mitochondrial ribosomal protein S16 | 0.537 | 62 |
| 202347_s_at | HIP2 | huntingtin interacting protein 2 | 0.565 | 8 |
| 214843_s_at | USP33 | ubiquitin specific protease 33 | 0.695 | 61 |
| Regulation of transcription | ||||
| 1559949_at | TRPS1 | Trichorhinophalangeal syndrome I | 0.199 | 69 |
| 210282_at | ZNF198 | zinc finger protein 198 | 0.25 | 44 |
| 202600_s_at | NRIP1 | nuclear receptor interacting protein 1 | 0.282 | 70 |
| 243792_x_at | PTPN13 | Protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) | 0.36 | 43 |
| 222320_at | HRPT2 | Hyperparathyroidism 2 (with jaw tumor) | 0.403 | 11 |
| 222544_s_at | WHSC1L1 | Wolf-Hirschhorn syndrome candidate 1-like 1 | 0.408 | 18 |
| 222313_at | CNOT2 | CCR4-NOT transcription complex, subunit 2 | 0.412 | 15 |
| 216022_at | WNK1 | WNK lysine deficient protein kinase 1 | 0.414 | 31 |
| 235409_at | MGA | MAX gene associated | 0.443 | 40 |
| 212881_at | PIAS4 | protein inhibitor of activated STAT, 4 | 0.448 | 55 |
| 227798_at | SMAD1 | SMAD, mothers against DPP homolog 1 (Drosophila) | 0.45 | 16 |
| 213766_x_at | GNA11 | guanine nucleotide binding protein (G protein), alpha 11 (Gq class) | 0.487 | 39 |
| 217550_at | ATF6 | Activating transcription factor 6 | 0.518 | 94 |
| 222180_at | YES1 | V-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | 0.604 | 98 |
| 218955_at | BRF2 | BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like | 0.635 | 83 |
| 212079_s_at | MLL | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | 0.666 | 29 |
| 213944_x_at | GNA11 | guanine nucleotide binding protein (G protein), alpha 11 (Gq class) | 0.693 | 59 |
| Other | ||||
| 1559496_at | --- | --- | 0.284 | 5 |
| 234032_at | --- | PRO1550 | 0.289 | 19 |
| 242343_x_at | ZNF518 | Zinc finger protein 518 | 0.309 | 68 |
| 221543_s_at | SPFH2 | SPFH domain family, member 2 | 0.322 | 6 |
| 204148_s_at | ZP3 /// POMZP3 | zona pellucida glycoprotein 3 (sperm receptor) /// POM (POM121 homolog, rat) and ZP3 fusion | 0.324 | 65 |
| 232489_at | FLJ10287 | hypothetical protein FLJ10287 | 0.33 | 1 |
| 236841_at | FLJ25222 | CXYorf1-related protein | 0.334 | 60 |
| 227931_at | --- | MRNA; cDNA DKFZp686D22106 (from clone DKFZp686D22106) | 0.382 | 54 |
| 238706_at | PAPD4 | PAP associated domain containing 4 | 0.386 | 63 |
| 221542_s_at | SPFH2 | SPFH domain family, member 2 | 0.398 | 24 |
| 231878_at | MVP | Major vault protein | 0.412 | 12 |
| 1564637_a_at | FLJ38426 | hypothetical protein FLJ38426 | 0.47 | 52 |
| 228971_at | --- | --- | 0.476 | 95 |
| 235970_at | MLR1 | transcription factor MLR1 | 0.485 | 73 |
| 230871_at | DHX30 | DEAH (Asp-Glu-Ala-His) box polypeptide 30 | 0.5 | 99 |
| 233228_at | ZNF407 | Zinc finger protein 407 | 0.508 | 48 |
| 237157_at | EVE1 | SH3 domain protein D19 | 0.519 | 9 |
| 215385_at | FTO | Fatso | 0.527 | 41 |
| 222496_s_at | FLJ20273 | RNA-binding protein | 0.528 | 42 |
| 219001_s_at | WDR32 | WD repeat domain 32 | 0.533 | 76 |
| 235927_at | --- | --- | 0.542 | 86 |
| 1569813_at | STRN | striatin, calmodulin binding protein | 0.549 | 66 |
| 240939_x_at | --- | --- | 0.557 | 79 |
| 222642_s_at | TMEM33 | transmembrane protein 33 | 0.558 | 81 |
| 213984_at | SCC-112 | SCC-112 protein | 0.565 | 37 |
| 234488_s_at | GCL /// GMCL1L | germ cell-less homolog 1 (Drosophila) | 0.619 | 100 |
| 201297_s_at | MOBK1B | MOB1, Mps One Binder kinase activator-like 1B (yeast) | 0.626 | 96 |
| 238660_at | WDFY3 | WD repeat and FYVE domain containing 3 | 0.663 | 13 |
| 212602_at | WDFY3 | WD repeat and FYVE domain containing 3 | 0.708 | 47 |
| 212602_at | WDFY3 | WD repeat and FYVE domain containing 3 | 0.708 | 47 |
| Up-regulated in BRCA1(+) | ||||
| Cell proliferation and death | ||||
| 203139_at | DAPK1 | death-associated protein kinase 1 | 1.896 | 74 |
| 209074_s_at | TU3A | TU3A protein | 2.865 | 89 |
| Cell signalling | ||||
| 227125_at | IFNAR2 | Interferon (alpha, beta and omega) receptor 2 | 1.791 | 23 |
| 204613_at | PLCG2 | phospholipase C, gamma 2 (phosphatidylinositol-specific) | 2.083 | 20 |
| DNA and RNA | ||||
| 203805_s_at | FANCA | Fanconi anemia, complementation group A /// Fanconi anemia, complementation group A | 2.056 | 93 |
| Immune response | ||||
| 211530_x_at | HLA-G | HLA-G histocompatibility antigen, class I, G | 1.575 | 38 |
| 205067_at | IL1B | interleukin 1, beta | 1.809 | 78 |
| 205671_s_at | HLA-DOB | major histocompatibility complex, class II, DO beta | 2.37 | 3 |
| 206407_s_at | CCL13 | chemokine (C-C motif) ligand 13 | 2.89 | 10 |
| 234764_x_at | IGLC2 | Ig lambda chain V-region (VL-AIG) /// Immunoglobulin lambda variable 3-21 | 6.297 | 51 |
| Metabolism | ||||
| 222046_at | ARS2 | arsenate resistance protein ARS2 | 1.475 | 77 |
| 204428_s_at | LCAT | lecithin-cholesterol acyltransferase | 1.767 | 27 |
| 208964_s_at | FADS1 | fatty acid desaturase 1 | 1.9 | 64 |
| 1555745_a_at | LYZ | lysozyme (renal amyloidosis) | 7.787 | 36 |
| Regulation of transcription | ||||
| 221010_s_at | SIRT5 | sirtuin (silent mating type information regulation 2 homolog) 5 (S. cerevisiae | 1.378 | 32 |
| 206090_s_at | DISC1 | disrupted in schizophrenia 1 | 1.72 | 85 |
| Other | ||||
| 237883_at | --- | Transcribed locus | 1.316 | 84 |
| 213938_at | CAST | CAZ-associated structural protein | 1.339 | 53 |
| 1561759_at | --- | Homo sapiens, clone IMAGE:5276804, mRNA | 1.369 | 50 |
| 218600_at | MGC10986 | hypothetical protein MGC10986 | 1.46 | 21 |
| 226410_at | LOC348180 | hypothetical protein LOC348180 | 1.694 | 91 |
| 241383_at | LOC201181 | similar to hypothetical protein A930006D11 | 2.053 | 34 |
Figure 2 shows that use of a set of 100 genes for hierarchical clustering results in almost perfect classification of hereditary and sporadic tumour samples. Only one sporadic sample falls into the branch of hereditary cases.
Figure 2.
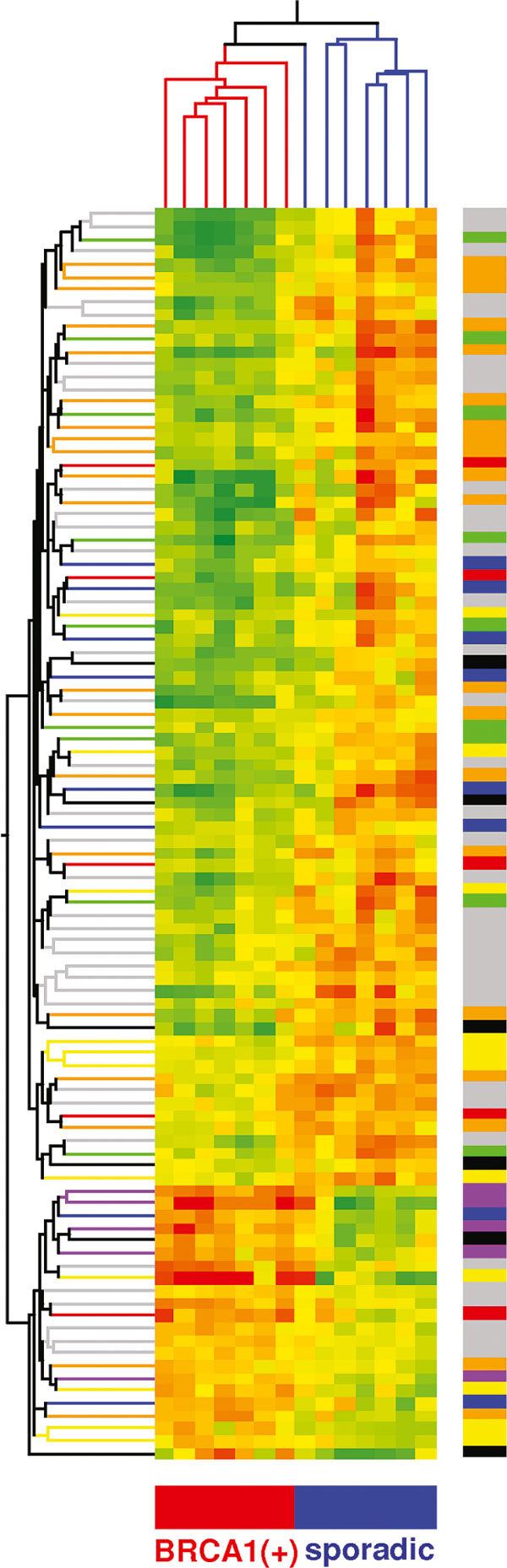
Hierarchical clustering of samples, based on 100 genes differentiating between hereditary and sporadic breast tumors. Colors on the right bar code for: regulation of transcription (orange), cell signalling (blue), cell proliferation and death (black), DNA and RNA replication transcription and processing (red), cellular metabolism (yellow), protein synthesis and degradation (green), immune response (violet) and other (grey). It may be seen, that most prominent cluster of genes upregulated in hereditary tumors consists of genes engaged to immune response, what probably reflects lymphocyte infiltrate of those tumors.
Within a set of 100 genes selected in our study, we found genes coding for proteins known to be interacting with the BRCA1 pathway, such as Fanconi Anemia complementation group A gene (FANCA, increased 2-fold) and XRCC5 (50% decrease), which are both engaged in double-strand break repair. FANCA protein is a component of the multi-subunit FA complex which takes part in sensing and/or regulation of the DNA damage response. The FA complex activates FANCD2 protein which is further targeted to the BRCA1 nuclear loci. Inactivation of the FA/BRCA pathway leads to chromosomal instability, due to impaired DNA repair [20]. DNA repair protein XRCC5 (80 kDa Ku autoantigen) is the DNA-binding component of the DNA-dependent protein kinase, and functions together with the DNA ligase IV-XRCC4 complex in the repair of DNA double-strand break by non-homologous end joining [21].
The TOB1 gene (transducer of ERBB2 gene, decreased in hereditary tumours) encodes a member of the tob/btg1 family of anti-proliferative proteins that have the potential to regulate cell growth, and is probably engaged in several human tumours (breast, lung, thyroid) [22-25]. This protein inhibits T cell proliferation and transcription of cytokines and cyclins. This is the only gene from Hedenfalk's list [15] that appears within the first 100 genes selected by limma.
Another interesting gene is selenophosphate synthetase 2 (SEPHS2, decreased in hereditary tumours). This protein encodes an enzyme that synthesizes selenophosphate from selenide and ATP. Selenophosphate is the selenium donor used to synthesize selenocysteine, which is co-translationally incorporated into selenoproteins at inframe UGA codons. This protein itself contains a selenocysteine residue in its predicted active site. It has been proposed that the effects of selenium in preventing cancer and neurological disorders may be mediated by selenium-binding proteins [26].
The most prominent among genes up-regulated in hereditary carcinomas is a group of immune response genes (5 genes). On the contrary, no immunological genes are found in the list of down-regulated genes. This may reflect a common feature of BRCA1-linked breast cancer, i.e. the inflammatory state and lymphocyte infiltrate of a tumour. It is especially striking as all our tumour samples were inspected by a pathologist, and only pieces of tumour mass without a visible inflammatory state were taken for microarray experiments. Thus we conclude that this immunological imprint must be very prominent in BRCA1(+) breast cancer.
Validation of microarray results by Q RT-PCR
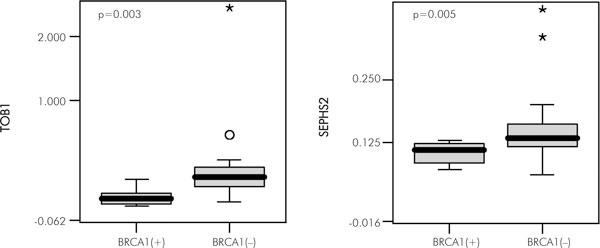
We selected TOB1 and SEPHS2 genes (see Table 1) for further analysis by quantitative RT-PCR. We examined the expression level of these genes in 12 tumours with mutations in BRCA1 and 16 tumours without mutation. The results of this analysis were concordant with the microarray results: both genes seem to be down-regulated in BRCA1-linked tumours, when compared to sporadic ones. The difference in expression level of TOB1 and SEPHS2 genes in BRCA1-linked tumours versus BRCA1 negative tumours was statistically significant (p-value for TOB1: 0.003, for SEPHS2: 0.005, Kolmogorov-Smirnov test, Fig. 3).
Figure 3.
Real-time quantitative PCR validation of microarray results for two selected genes. Box-and-whisker plot represent median and quartile values, with non-outlier range denoted by whiskers and outliers marked by asterisks and circles. Statistical comparison was performed by Kolmogorov-Smirnov test.
Discussion
Our preliminary study was done on a relatively small number of cases; however it may be informative, as the samples were chosen carefully. In all previous microarray studies on breast cancer it was observed that the most striking difference in the gene expression profile is connected with oestrogen receptor status. Initially it was stated that ER(+) and ER(-) breast cancers represent distinct categories of breast tumours [27,28]. More detailed studies performed on a larger set of samples revealed further subtypes, e.g. luminal A and luminal B subtypes among ER(+) tumours and at least two subtypes: ERBB2+ and 'basal' within and ER(-) group [Perou et al., 2000; Sorlie et al., 2001]. Alternatively, another subdivision of ER(-) cases was proposed by Farmer at al., who claim two subgroups: 'basal', characterized by oestrogen and androgen receptors negativity and expression of cytokeratins 5 and 17, and 'molecular apocrine', negative for ER and positive for AR [29]. It was also shown previously that BRCA1 mutation-linked breast cancer tissues cluster within a 'basal' subtype [30]. However, these specific cases account only for the minority of 'basal' tumours; thus the genes that are differentially expressed between basal and other subgroups do not necessarily reflect a difference in the biology of hereditary (BRCA1-linked) and sporadic carcinomas. Moreover, a set of genes differentiating between breast cancer subtypes in the studies of Perou and Sorlie was selected from genes that are stably expressed in the biopsy specimens, taken from the same tumour before and after chemotherapy. This allowed the construction of so-called 'molecular portraits' of each tumour, but may have eliminated some significant genes that could be suppressed by chemotherapy. In our study we analyse only tumours that have not been subjected to neoadjuvant chemotherapy. To reduce, at least partially, the sources of variability that are not linked to BRCA1 status, we decided to analyse only ER(-) tumour samples.
The aim of our study was to reveal a set of genes differentially expressed between BRCA1 mutation-linked and sporadic breast cancer. Similar study have been performed by Hedenfalk et al., who analysed seven sporadic, seven BRCA1- and eight BRCA2-linked breast cancers and published a set of 51 differentially expressed genes. We tried to retrieve information about expression of those 51 genes from HG U133 Plus 2.0 Gene Chip used in our study. However, due to the incompatibility of both types of DNA microarrays, we were able to find expression data only for 40 genes (represented by 88 probesets). This set of genes was unable to discriminate between sporadic and hereditary cases from our cohort (not shown). This may be caused by shortening of the original list of genes and the fact that the set of 51 genes was selected by comparison of BRCA1 and BRCA2 tumours with the sporadic ones, all of them of rather mixed histopathology, grade and ER status. Of the genes from Hedenfalk's list only one (TOB1) is represented, at the 62nd position, in our list of 100 genes selected by limma.
It should be mentioned that Jazaeri et al., who analysed a cohort of hereditary and sporadic ovarian cancers, found by the unsupervised method of data analysis that BRCA1 and BRCA2 tumours localize separately from each other, while sporadic cases were placed by the algorithm between these two groups, some of them being closer to BRCA1 tumours, others to BRCA2 tumours [31]. This result confirms all previous observations that BRCA1 and BRCA2 mutation-linked carcinomas are different entities, and suggests that at least in some sporadic cases BRCA1 or BRCA2 pathways may be truncated, thus accounting for BRCA1-like and BRCA2-like sporadic tumours. It should be verified whether this hypothesis applies also to breast cancer. Interestingly, in our group of samples Jazaeri's set of genes performed better than a set published by Hedenfalk [15] (not shown).
Prediction of BRCA status in patients with breast cancer on the basis of clinical and molecular features would be very useful for genetic counselling and for cost-reduction of genetic screening [32,33]. It was shown recently by Lakhani et al., 2005, that ER status together with immunochemistry for some markers of 'basal' subtype (CK14, CK5/6, CK17 and osteonectin) is better able to predict BRCA1 mutation status in breast cancer patients than previously used criteria [34]. Although it must be experimentally proven, we believe that among 100 genes selected by limma some markers may appear to improve the specificity of prediction of BRCA1 status.
Abbreviations
CK5/6, CK14, CK17: cytokeratines 5/6, 14 i 17; FANCA: Fanconi Anemia complementation group A protein; PCA: Principal Component Analysis; SEPHS2: selenophosphate synthetase 2; TOB1: transducer of ERBB2 gene; XRCC5: X-ray repair complementing defective repair in Chinese hamster cells 5
Acknowledgements
This work was supported by grant PBZ-KBN-040/P04/2001, from the State Committee for Scientific Research. Volha Dudaladava is a fellow of a Fellowship Programme totally supported by the National Cancer Institute - Office for International Affairs, NIH, Bethesda, MD, USA
References
- Perou CM, Sorlie T, Eisen MB, Rijn M van de, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Rijn M van de, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer LJ, Dai H, Vijver MJ van de, He YD, Hart AA, Mao M, Peterse HL, Kooy K van der, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Vijver MJ van de, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, Velde T van der, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Jeffrey SS, Lonning PE, Hillner BE. Genomics-based prognosis and therapeutic prediction in breast cancer. J Natl Compr Canc Netw. 2005;3:291–300. doi: 10.6004/jnccn.2005.0016. [DOI] [PubMed] [Google Scholar]
- Pathology of familial breast cancer: differences between breast cancers in carriers of BRCA1 or BRCA2 mutations and sporadic cases. Breast Cancer Linkage Consortium. Lancet. 1997;349:1505–1510. doi: 10.1016/S0140-6736(96)10109-4. [DOI] [PubMed] [Google Scholar]
- Lakhani SR. The pathology of familial breast cancer: Morphological aspects. Breast Cancer Res. 1999;1:31–35. doi: 10.1186/bcr10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osin PP, Lakhani SR. The pathology of familial breast cancer: Immunohistochemistry and molecular analysis. Breast Cancer Res. 1999;1:36–40. doi: 10.1186/bcr11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijver MJ Van de. The pathology of familial breast cancer: The pre-BRCA1/BRCA2 era: historical perspectives. Breast Cancer Res. 1999;1:27–30. doi: 10.1186/bcr9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappuis PO, Nethercot V, Foulkes WD. Clinico-pathological characteristics of BRCA1- and BRCA2-related breast cancer. Semin Surg Oncol. 2000;18:287–295. doi: 10.1002/(SICI)1098-2388(200006)18:4<287::AID-SSU3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Metcalfe K, Sun P, Hanna WM, Lynch HT, Ghadirian P, Tung N, Olopade OI, Weber BL, McLennan J, Olivotto IA, Begin LR, Narod SA. Estrogen receptor status in BRCA1- and BRCA2-related breast cancer: the influence of age, grade, and histological type. Clin Cancer Res. 2004;10:2029–2034. doi: 10.1158/1078-0432.CCR-03-1061. [DOI] [PubMed] [Google Scholar]
- Eerola H, Heikkila P, Tamminen A, Aittomaki K, Blomqvist C, Nevanlinna H. Relationship of patients' age to histopathological features of breast tumours in BRCA1 and BRCA2 and mutation-negative breast cancer families. Breast Cancer Res. 2005;7:R465–469. doi: 10.1186/bcr1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JN, Watson P, Page DL, Narod SA, Lenoir GM, Tonin P, Linder-Stephenson L, Salerno G, Conway TA, Lynch HT. Hereditary breast cancer: pathobiology, prognosis, and BRCA1 and BRCA2 gene linkage. Cancer. 1996;77:697–709. doi: 10.1002/(SICI)1097-0142(19960215)77:4<697::AID-CNCR16>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Verhoog LC, Brekelmans CT, Seynaeve C, Bosch LM van den, Dahmen G, van Geel AN, Tilanus-Linthorst MM, Bartels CC, Wagner A, Ouweland A van den, Devilee P, Meijers-Heijboer EJ, Klijn JG. Survival and tumour characteristics of breast-cancer patients with germline mutations of BRCA1. Lancet. 1998;351:316–321. doi: 10.1016/S0140-6736(97)07065-7. [DOI] [PubMed] [Google Scholar]
- Veronesi A, de Giacomi C, Magri MD, Lombardi D, Zanetti M, Scuderi C, Dolcetti R, Viel A, Crivellari D, Bidoli E, Boiocchi M. Familial breast cancer: characteristics and outcome of BRCA 1-2 positive and negative cases. BMC Cancer. 2005;5:70–76. doi: 10.1186/1471-2407-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, Wilfond B, Borg A, Trent J. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- Górski B, Jakubowska A, Huzarski T, Byrski T, Gronwald J, Grzybowska E, Mackiewicz A, Stawicka M, Bębenek M, Sorokin D, Fiszer-Maliszewska L, Haus O, Janiszewska H, Niepsuj S, Góźdź S, Zaremba L, Posmyk M, Płużańska M, Kilar E, Czudowska D, Waśko B, Miturski R, Kowalczyk JR, Urbański K, Szwiec M, Koc J, Dębniak B, Rozmiarek A, Dębniak T, Cybulski C, Kowalska E, Tołoczko-Grabarek A, Zajączek S, Menkiszak J, Mędrek K, Masojć B, Mierzejewski M, Narod SA, Lubiński J. A high proportion of founder BRCA1 mutations in Polish breast cancer families. Int J Cancer. 2004;110:683–686. doi: 10.1002/ijc.20162. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3(1) doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- Thacker J, Zdzienicka MZ. The XRCC genes: expanding roles in DNA double-strand break repair. DNA Repair (Amst) 2004;3:1081–1090. doi: 10.1016/j.dnarep.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Nakamura T, Komoda M, Satoh H, Suzuki T, Tsuzuku JK, Miyasaka T, Yoshida EH, Umemori H, Kunisaki RK, Tani K, Ishii S, Mori S, Suganuma M, Noda T, Yamamoto T. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes Dev. 2003;17:1201–1206. doi: 10.1101/gad.1088003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebig C, Seitz S, Korsching E, Kristiansen G, Gustavus D, Scherneck S, Petersen I. Profile of differentially expressed genes after transfer of chromosome 17 into the breast cancer cell line CAL51. Genes Chromosomes Cancer. 2005;44:233–246. doi: 10.1002/gcc.20240. [DOI] [PubMed] [Google Scholar]
- Iwanaga K, Sueoka N, Sato A, Sakuragi T, Sakao Y, Tominaga M, Suzuki T, Yoshida Y, K-Tsuzuku J, Yamamoto T, Hayashi S, Nagasawa K, Sueoka E. Alteration of expression or phosphorylation status of tob, a novel tumor suppressor gene product, is an early event in lung cancer. Cancer Lett. 2003;202:71–79. doi: 10.1016/j.canlet.2003.08.019. [DOI] [PubMed] [Google Scholar]
- Ito Y, Suzuki T, Yoshida H, Tomoda C, Uruno T, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Kuma K, Yamamoto T, Miyauchi A. Phosphorylation and inactivation of Tob contributes to the progression of papillary carcinoma of the thyroid. Cancer Lett. 2005;220:237–242. doi: 10.1016/j.canlet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Dodig S, Cepelak I. The facts and controversies about selenium. Acta Pharm. 2004;54:261–276. [PubMed] [Google Scholar]
- Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, Borg A, Ferno M, Peterson C, Meltzer PS. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61:5979–5984. [PubMed] [Google Scholar]
- Pusztai L, Ayers M, Stec J, Clark E, Hess K, Stivers D, Damokosh A, Sneige N, Buchholz TA, Esteva FJ, Arun B, Cristofanilli M, Booser D, Rosales M, Valero V, Adams C, Hortobagyi GN, Symmans WF. Gene expression profiles obtained from fine-needle aspirations of breast cancer reliably identify routine prognostic markers and reveal large-scale molecular differences between estrogen-negative and estrogen-positive tumors. Clin Cancer Res. 2003;9:2406–2415. [PubMed] [Google Scholar]
- Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, Duss S, Nicoulaz AL, Brisken C, Fiche M, Delorenzi M, Iggo R. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazaeri AA, Yee CJ, Sotiriou C, Brantley KR, Boyd J, Liu ET. Gene expression profiles of BRCA1-linked, BRCA2-linked, and sporadic ovarian cancers. J Natl Cancer Inst. 2002;94:990–1000. doi: 10.1093/jnci/94.13.990. [DOI] [PubMed] [Google Scholar]
- Vahteristo P, Eerola H, Tamminen A, Blomqvist C, Nevanlinna H. A probability model for predicting BRCA1 and BRCA2 mutations in breast and breast-ovarian cancer families. Br J Cancer. 2001;84:704–708. doi: 10.1054/bjoc.2000.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, Gumpper KL, Scholl T, Tavtigian SV, Pruss DR, Critchfield GC. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.20.6.1480. [DOI] [PubMed] [Google Scholar]
- Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, Vijver M van der, Parry S, Bishop T, Benitez J, Rivas C, Bignon YJ, Chang-Claude J, Hamann U, Cornelisse CJ, Devilee P, Beckmann MW, Nestle-Kramling C, Daly PA, Haites N, Varley J, Lalloo F, Evans G, Maugard C, Meijers-Heijboer H, Klijn JG, Olah E, Gusterson BA, Pilotti S, Radice P, Scherneck S, Sobol H, Jacquemier J, Wagner T, Peto J, Stratton MR, McGuffog L, Easton DF. Breast Cancer Linkage Consortium. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]