Abstract
Background
Esophageal cancer consists of two major histologic types: esophageal squamous cell carcinoma (ESCC) predominant globally and esophageal adenocarcinoma (EAC) with a higher incidence in westernized countries. Five-year overall survival is 15%. Clinical trials frequently combine histologies although they are different diseases with distinct origins. In the evolving era of personalized medicine and targeted therapies, we hypothesized that ESCC and EAC have genomic differences important for developing new therapeutic strategies for esophageal cancer.
Methods
We explored DNA copy number abnormalities (CNAs) in 70 ESCCs with publicly available array data and 189 EAC from our group. All data was from Affymetrix single nucleotide polymorphism (SNP) arrays. Analysis was performed with Nexus 5.0 Copy number software using a SNPRank segmentation algorithm. Log ratio thresholds for copy number gain and loss were set at +/− 0.2 (approximately 2.3 and 1.7 copies respectively).
Results
ESCC and EAC genomes showed some CNAs with similar frequencies (e.g., CDKN2A, EGFR, KRAS, MYC, CDK6, MET) but also many CNAs with different frequencies between histologies, most of which were amplification events. Some of these regions harbor genes to which targeted therapies are currently available (VEGFA, ERBB2) or where agents are in clinical trials (PIK3CA, FGFR1). Other regions contain putative oncogenes that may be targeted in the future.
Conclusions
Using SNP arrays we compared genomic abnormalities in a large cohort of EAC and ESCC. We report here the similar and different frequencies of CNAs in ESCC and EAC. These results may allow development of histology-specific therapeutic agents for esophageal cancer.
Keywords: Esophageal cancer, genetics, genomics
INTRODUCTION
Esophageal cancer, the eighth most common cancer in the world, is composed of two main histologic types: squamous cell carcinoma (ESCC) and adenocarcinoma (EAC). The two tumors have five-year overall survival rates averaging 15% [1], and are often included together in therapeutic and prognostic clinical studies [2;3]. The histologies may share the poor outcome with current therapeutic strategies, but they have distinct differences including causality, cell of origin and epidemiologic distribution [4]. Risk factors for esophageal SCC, the predominant type worldwide, include smoking and alcohol abuse, both inflammatory insults to the esophagus, while EAC is associated with obesity and gastroesophageal reflux disease (GERD) [5;6]. These two malignancies arise from different cell populations: ESCC from squamous epithelium and EAC from intestinal metaplastic epithelial cells or Barrett’s esophagus [7]. They should not necessarily be treated as the same disease in clinical trials and development of therapies.
With a shared poor outcome but distinct biologic, epidemiologic and demographic differences, these tumors are not the same. In the evolving era of personalized medicine and targeted therapies, we hypothesized that ESCC and EAC would have genomic differences important for developing new therapeutic strategies for esophageal cancer. There have been smaller studies using different techniques to compare ESCC and EAC DNA copy number abnormalities but our study is by far the largest and has the highest resolution to date, thus allowing a more precise comparison of similarities and differenced between the genomes.
MATERIALS AND METHODS
Esophageal adenocarcinoma (EAC) copy number data
Seventy-three Affymetrix 250K Sty esophageal adenocarcinoma GeneChip data published as part of Berouhkim et al. [10] were obtained from the authors. Tumor data was normalized to a baseline reference file that was created from matched normals. Additionally, we included our own Affymetrix 6.0 SNP array data from 116 esophageal adenocarcinomas from patients treated at the University of Pittsburgh Medical Center from 2002 – 2008. The baseline reference file for this population was created from normal DNA obtained from the blood of 15 individuals from the same patient population. Genomic DNA was isolated from tumors and blood using QiaAmp DNA Mini Kit (Qiagen, Valencia, CA). DNA concentration and purity was assessed by UV absorbance (NanoDrop 1000, Thermo Fisher Scientific, Waltham, MA) and DNA quality was assessed by gel electrophoresis prior to labeling and array hybridization. All patients provided informed consent and this research was approved by appropriate human research institutional review boards.
Esophageal squamous cell carcinoma (ESCC) copy number data acquisition
Raw copy number intensity files (.cel) for 70 ESCC samples from two studies [8;9] were obtained from the Gene Expression Omnibus (GEO). This included 30 samples genotyped with Affymetrix 500K (250K Nsp and 250K Sty) GeneChip data from Hu and colleagues [8]. For this analysis, we only used the data from 250K Sty from Hu in conjunction with another 40 samples (including 11 ESCC cell lines) genotyped with 250K Sty from Bass and colleagues [9]. Each dataset was individually referenced to the respective baseline files. The baseline reference file for the Hu dataset was created using the matched normal data provided. A reference file to normalize the Bass data was created from a subset of 37 non-neoplastic (normal) samples from a larger cohort of 1140 samples that were used in the original work.
Data analysis
All individually normalized data were pooled into a single project and analyzed in Nexus 5.0 Copy number software (Biodiscovery, El Segundo, CA) using the SNPRank segmentation algorithm with a minimum of eight probesets and a significance threshold p-value of 10−6. Log ratio thresholds for copy number gain and loss were set at ± 0.2 (approximately 2.3 and 1.7 copies respectively). Genome positions were mapped to NCBI Build 36.1 (hg18). We then compared the two cancer types using the comparison tool in Nexus 5.0 to guide us in identifying both common and different regions of aberrations. We also manually compared the genomes to identify other regions listed in Tables 1 and 2. Two-tailed Fisher’s exact test was performed in GraphPad software [http://www.graphpad.com/quickcalcs/contingency1.cfm] to determine if there significant difference in the frequencies of copy number changes between the two cancer types.
Table 1.
Comparison of (A) amplified and (B) deleted genes observed at similar frequencies in esophageal squamous cell carcinoma (ESCC, n=70) and esophageal adenocarcinoma (EAC, n=189). Regions previously identified in other studies [11;12]
| (A) | ||||
|---|---|---|---|---|
| Cytoband | Estimated Frequencies | Known target | Other targets | |
| ESCC% (n=70) | EAC% (n=189) | |||
| 1p36.33 | 15 | 7 | Tp73(10) | |
| 1q21.2-q31.2(11) | 20 | 15 | MCL1 | |
| 6p21.1(11;12) | 8 | 19 | VEGFA, BAK (12) | |
| 6p24.1 | 7 | 6 | E2F3(10) | |
| 7p11.2(11) | 27 | 23 | EGFR | |
| 7q21.3(11;12) | 20 | 24 | CDK6 | |
| 7q22.1(11;12) | 23 | 25 | SMURF1 | |
| 7q31.2(11) | 9 | 9 | MET | |
| 8p11.21-p11.1 | 20 | 14 | PLAT(10) | |
| 11q13-q14.1 | 7 | 12 | GAB2(10) | |
| 12p12.1 | 17 | 27 | KRAS | |
| 12p11.21 | 18 | 15 | DDX11(10), FAM60A(10) | |
| 13q34 | 10 | 12 | IRS2(10) | |
| 15q26.3 | 17 | 9 | IGF1R | |
| 17q12(11;;12) | 14 | 22 | ERBB2 | NGFR(11), FGF11(11), C17orf37(10), SCYA3(12) |
| 19q12(11) | 14 | 15 | CCNE1 | |
| 20q(11;12) | 33 | 40 | ZNF217(10), BCL2L1(10), ID1(10), BIRC7(10) | |
| 10q22.2-q22.3 | 4 | 10 | SFTPA1B(10) | |
| (B) | ||||
|---|---|---|---|---|
| Cytoband | Estimated Frequencies | Known target | Other targets | |
| ESCC% (n=70) | EAC% (n=189) | |||
| 3p14.2 | 43 | 50 | FHIT | |
| 4p | 24 | 14 | ZNF141(10) | |
| 4q22.1(11) | 21 | 16 | MGC48628(10), ABCG2(12) | |
| 4q35.2(11) | 24 | 18 | FRG2(10), TUBB4Q(10) | |
| 5q11.2 | 14 | 18 | PDE4D | PLK2(10) |
| 6q26 | 7 | 8 | PARK2 | |
| 8p23.2(12) | 29 | 21 | CSMD1 | DLGAP2(12), MSRA(12) |
| 9p24.1 | 27 | 28 | PTPRD | |
| 11p15.5(12) | 13 | 6 | RASSF7(10) | |
| 12q21-23 | 3 | 9 | ANKS1B | |
| 18q21.2(11;12) | 21 | 33 | SMAD4, DCC | |
| 20p12.1 | 9 | 14 | MACROD2 | FLRT3(10) |
| 21q(12) | 19 | 21 | ||
| 22q11.22 | 4 | 8 | VPREB1(10) | |
Table 2.
Comparison of differentially (A) amplified and (B) deleted genes in esophageal adenocarcinoma (EAC, n=189) and esophageal squamous cell carcinoma (ESCC, n=70). Regions that have been previously identified in other ESCC vs EA studies [11;12] in the similar patterns are indicated along the cytobands. Known targets and other candidate genes listed in the identified regions were obtained from [10–12]. p-values were calculated by two-tailed Fisher’s exact test. All regions with p <0.0001 are in bold.
| (A) | |||||
|---|---|---|---|---|---|
| Cytoband | Estimated Frequencies | Known target | Possible targets | p-value (Fisher’s) | |
| ESCC % (n=70) | EAC % (n=189) | ||||
| 2q31.1-q33.2 | 16 | 5 | ~160 genes | 0.0069 | |
| 3q(11;12) | 60 | 15 | SOX2(9) | pRKCI(10), pIK3CA(10), MME(12) | 0.0001 |
| 5p | 28 | 8 | TERT(10), | 0.0001 | |
| 6q23.3 | 3 | 11 | MYB | hsa-mir-548a-2 | 0.0472 |
| 7p14.1 | 18 | 56 | TARP | 0.0001 | |
| 8p12-11.2(11) | 21 | 9 | FGFR1(11) | 0.0103 | |
| 8p23.1-p22 (11) | 4 | 20 | GATA4(28) | CTSB (11), PDGFRL (11) | 0.0011 |
| 8q (12) | 58 | 38 | MYC | 0.0046 | |
| 9p13.3 | 4 | 13 | CA9 (13;14), TLN1(16) | 0.0429 | |
| 11q13.2(11;12) | 59 | 18 | CCND1 | ORAOV1(10), FGF3(11;12), FGF4, EMS1(11;12), PAK1(11) | 0.0001 |
| *13q12.2(11;12) | 0 | 17 | FOXO1(10), FGF9(11), BRCA2(11), KLF5(24) | 0.0001 | |
| 14q11-32 | 35 | 4 | NKX2-1 | BCL2L2(10), PAX9(33) | 0.0001 |
| 17q11.1-q11.2 | 19 | 9 | ~9 genes | 0.0472 | |
| 17q25.1-q25.3 | 21 | 10 | ~125 genes | 0.0221 | |
| 18q11.2 | 4 | 30 | GATA6(28) | CABLES1(10) | 0.0001 |
| *19p | 11 | 0 | ZNF492(10), ZNF99(10) | 0.0001 | |
| 22q11.1-q23 | 10 | 3 | CRKL(10) | 0.0477 | |
| (B) | |||||
|---|---|---|---|---|---|
| Cytoband | Estimated Frequencies | Known target | Possible targets | p-value (Fisher’s) | |
| ESCC % (n=70) | EAC % (n=189) | ||||
| 1p31.1 | 10 | 3 | NEGR1 | 0.0477 | |
| 1p21.1-p13.2 | 11 | 3 | MAGI3(10), | 0.0248 | |
| 1p36.11-1p35.3(12) | 0 | 8 | SFN(10) | 0.0134 | |
| 2q22.1(12) | 13 | 5 | LRP1B | 0.0297 | |
| 2q37.3(12) | 16 | 3 | TMEM16G(10), ING5(10) | 0.0009 | |
| 3p(11) | 43 | 15 | CHL1(10), CNTN3(10), FRG2C(10) | 0.0001 | |
| 6p25.3 | 4 | 17 | FOXC1(10), GMDS(10) | 0.0072 | |
| 9p21.3(11;12) | 57 | 37 | CDKN2A/B | 0.0046 | |
| 11q23.1 | 24 | 7 | ATM | CADM1(10), OPCML(10), HNT(10) | 0.0003 |
| *13q12.2(11;12) | 20 | 0 | FOXO1(10), FGF9(11), BRCA2(11) , KLF5(24) | 0.0001 | |
| 16q23.1 | 4 | 32 | WWOX | 0.0001 | |
| 17p(11) | 1 | 19 | TP53 | MAP2K4(10), ZNF18(10), ATP1B2(10) | 0.0001 |
| *19p | 1 | 11 | ZNF492(10), ZNF99(10) | 0.0109 | |
RESULTS
Common regions of aberrations
Analyses of copy number data from 70 ESCC and 189 EAC samples resulted in at least 18 regions of gain and 14 regions of loss that occur in both ESCC and EAC at similar frequencies (p>0.05 by Fisher’s exact test) (Table 1). These changes include previously reported cancer loci including gains [VEGFA (6p), EGFR (7p), CDK6 (7q21), MET (7q31), KRAS (12p), ERBB2 (17q12)] and losses [FHIT (3p), CSMDI (8p), and SMAD4 (18q)]. We also observed other regions of gain such as on 1p, 1q, 6p, 7q22, 8p, 10q, 11q, 12p11, 13q, 15q, 19q, 20q, and loss on 4p, 4q, 5q, 6q, 9p, 11p, 12q, 18q, 20p, 21q, and 22q in both ESCC and EA. Some of these regions have been previously identified in a similar pattern (gain or loss) in two studies that compared ESCC and EAC but with smaller cohort sizes and different hybridization techniques [11;12] (Table 1). Various putative target genes within these regions have been previously described by other studies [8–12] (Table 1).
Genomic differences between ESCC and EAC
In addition to regions with copy number aberrations at similar frequencies, we identified 17 regions of copy number (CN) gains and 13 regions of CN losses with a significant difference in their frequencies as determined by p < 0.05 by Fisher’s exact test (Table 2). Eleven of the 17 (65%) regions displayed higher frequencies of gain in ESCC and some of these regions harbored known cancer-associated genes such as SOX2, PIK3CA (ESCC=60% vs EAC=15%, p=0.0001), MYC (58% vs 38%, p=0.0046), and CCND1, ORAOV1 (59% vs 18%, p=0.0001) (Table 2). Similarly, 8/13 (62%) of the copy number loss regions were observed at significantly higher frequencies in ESCC in comparison to EAC. Genes in some of these regions include known cell cycle regulatory genes such as CDKN2A/B (57% vs 37%, p= 0.0046) and ATM (24% vs 7%, p=0.0003). In addition, we observed higher frequency gains targeting 2q (16% vs 5%, p=0.0069), 5p (28% vs 8%, p=0.0001), 8p (21% vs 9%, p=0.01), 14q (35% vs 4%, p=0.0001), 17q11 (19% vs 9%, p=0.047), 17q25 (21% vs 10%, p=0.022), and 22q (10% vs 3%, p=0.04). Higher frequency losses were seen targeting ESCC at 1p (~10% vs 3%, p<0.05), 2q (16% vs 3%, p=0.0009), and 3p (43% vs 15%, p=0.0001. Putative target genes in many of these regions have been previously pointed out in Berouhkim et al. [10] and include cancer susceptibility genes such as TERT (5p gain), FGFR1 (8p12 gain), NKX2-1, BCL2L2, PAX9 (14q gain), CRKL (22q gain), ING5 (2q loss), and CHL1 (3p loss) (Table 2).
On the other hand, the EAC genome displayed six CN gains and five CN losses that were more frequent in EAC in comparison to the ESCC genome. CN gains and losses in EAC were observed to affect cancer-associated loci such as gains at MYB (EA=11% vs ESCC=3%, p=0.047), TARP (56% vs 18%, p=0.0001), GATA4 (20% vs 4%, p=0.001), GATA6 (30% vs 4%, p=0.0001) and loss at 1p targeting SFN (8% vs 0%, p=0.01), GMDS (17% vs 4%, p=0.007), WWOX (32% vs 4%, p=0.0001) and TP53 (19% vs 2%, p=0.0001) (Table 2). We also observed higher frequency gains at 9p (13% vs 4%, p=0.04) containing putative cancer loci such as CA9 [13–15] and TLN1 [16;17].
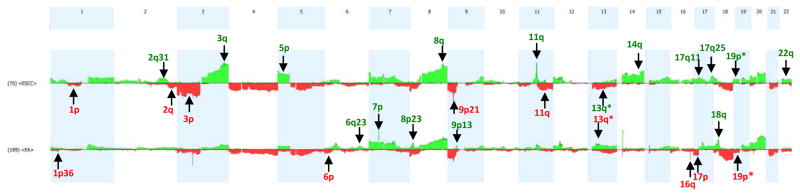
Interestingly, we observed two loci that displayed an opposite pattern of copy number changes in the two cancers (indicated by ‘*’ in Figure 1 and Table 2). The 13q region has been previously identified by Weiss and Rumiato [11;12]. In our analysis, the 13q region displayed a 20% loss in ESCC and harbors the tumor suppressor gene BRCA2 in addition to other candidate tumor suppressor genes including FOXO1 [10], and STARD13/DLC2 [18–20]. However, this region is amplified in 17% of EA and also harbors the putative oncogene, ELF1 [21–23] and KLF5 [24;25]. Similarly, we observed 11% loss in EAC vs 11% gain in ESCC at 19p chromosomal arm. This region contains approximately 600 genes but two genes, ZNF492 and ZNF99, have been proposed as candidate genes within this region [10].
Figure 1.
Genomic copy number differences between esophageal squamous cell carcinoma (ESCC, top pane) and esophageal adenocarcinoma (EAC, bottom panel). Genomic data from 70 ESCC and 189 EAC were analyzed in Nexus 5.0. Figure shows the gains (green) and losses (red) for chromosomes 1–22. High frequency differences occurring in either ESCC or EAC are indicated in the respective histograms. All gains are indicated in green and losses in red. Two regions that show opposite patterns of copy number changes between the two cancer types are indicated by ‘*’.
COMMENT
We have performed a comparative genomic analysis of the largest cohort of EAC and ESCC samples and at the highest resolution to date. We found considerable similarity between these two tumor types but also many focal regions of DNA amplification or loss that are more frequent in one histologic type than in the other. This confirms some findings from smaller studies reported previously [11;12] but our considerably larger sample size allows us to more precisely define regions, accurately determine event frequencies in the two histologies and identify significant differences between histologies. Our results demonstrate that while the genomes of EAC and ESCC are similar in many ways, there are distinct differences that may lead to biomarkers or therapeutic strategies unique to, or more effective in, one tumor type versus the other.
Genes of interest that were amplified in both histologies at similar frequencies include MCL1, EGFR, CDK6, SMURF1, KRAS, ERBB2, CCNE1, VEGFA, MET and IGF1R. Of these, EGFR, ERBB2, VEGFA and MET are the targets of currently available therapeutic agents, some of which have been tested or are in clinical trials for locally advanced and metastatic esophageal cancer for both histologies. For example, studies have been conducted with EGFR inhibitors in esophageal cancer and the results showed 8% partial responses in patients with advanced ESCC and no significant benefit for patients with EAC [26]. Among the other genes, CDK6 and IGFR1 are currently being explored as potential targets for esophageal cancer therapy. CDK6 in particular is interesting as we have recently reported that amplification of this region is associated with poor survival in EAC[27]. We further demonstrated that expression of CDK6 was associated with amplification and was an even stronger prognostic factor than gene amplification [27]. We also showed that small interfering RNA knockdown, or inhibition with the CDK4/6 inhibitor PD-0332991 (Pfizer, New York, NY), resulted in reduced proliferation and anchorage independent growth in EAC cell lines. Thus, CDK6 inhibitors may provide a novel therapeutic target for EAC. Our current study indicates that they should also be assessed in ESCC.
Genes with different amplification frequencies between ESCC and EAC include SOX2, PIK3CA, MYC, CCND1, FGFR1, GATA4 and GATA6. One of the most striking differences observed is amplification of 3q (60% of ESCC versus 15% of EAC) which contains both SOX2 and PIK3CA. SOX2 has clearly been implicated as a lineage-specific oncogene for squamous cell tumors [9] but the close proximity and possible overexpression of PIK3CA in ESCC should not be ignored, particularly as this pathway is the target of many new therapeutic agents. Also striking is the differential amplification of the transcription factors GATA4 at 8p23.1 and GATA6 at 18q11.2. These very focal amplification events have been identified previously in EAC, and Lin et al, also demonstrated that GATA4 is over-expressed in association with amplification in EAC [28]. The GATA factors are zinc finger DNA binding proteins that control the development of diverse tissues by activating or repressing transcription. Specifically, the GATA transcription factors coordinate cellular maturation with proliferation arrest and cell survival and it is not surprising that they have been implicated in cancer. Targeting of transcription factors is not currently a practical therapeutic approach but this may change in the future if microRNA-based therapeutics live up to their considerable promise [29;30]. In this case, the GATA transcription factors would clearly be a strong potential target for EAC but less likely for ESCC because of the low frequency of amplification.
In addition to regions with well-defined tumor-associated genes, our study also identified several genomic events that occur at different frequencies but for which no known target genes have been confirmed. These include 13q12.2 which is deleted in 20% of ESCC samples but amplified in 17% of the EAC tumors. 13q12.2 contains several interesting genes that could act as putative oncogenes including FGF9 and KLF5 but also contains the tumor suppressor BRCA2 and FOX01, a member of the FOXO tumor suppressor family. Among the putative oncogenes, Kruppel-like factor (KLF5) may be important in esophageal cancer because KLF5 is a cell growth mediator in various epithelial cells and has been implicated in intestinal tumorigenesis where KLF5 was critical in modulating intestinal tumor initiation and progression [25]. Also, expression of KLF5 correlates with cell proliferation in breast cancer and is a poor prognostic factor with increased expression linked with shorter disease-free and overall survival [24].
Another dramatic difference between EAC and ESCC is the amplification of chromosome 14 which occurred in 35% of ESCC cases but only 4% of EAC. While many of these events are whole chromosome gains, there does appear to be a focus on the region containing the gene PAX9. This gene is a member of the paired box transcription factor family, which regulates the expression of the genes involved in cell proliferation, apoptotic resistance, and cell migration [31]. PAX9 has also been implicated in the development of stratified squamous epithelia [32]. PAX9 has been shown to be a lung cell lineage oncogene [33] but its role in esophageal cancer development and prognosis are not yet established [32]. Given the huge disparity in amplification frequency observed in our study, it is possible that PAX9 represents an oncogene specific to the squamous histologic type of esophageal cancer. Finally, other areas of differential amplification include 19p, which was amplified 11% of ESCC but lost in 11% of EAC19p and contains ZNF492, another putative cancer gene. In addition amplification of 2q31-q33 is more prevalent in ESCC but contains approximately 160 genes with no clear candidate as the drivers. In summary, we have used high resolution SNP array analysis to determine the frequencies of DNA copy number gains and losses across the entire genome of 70 ESCC and 189 EAC samples. We have found known targets previously reported to be amplified in smaller cohorts thus corroborating other results and we have identified new regions which represent potential targets for therapy, prognostic biomarkers and aberrations for esophageal cancer studies. To our knowledge, this is the first study to directly compare the genomes of ESCC and EAC at this high resolution and with a large enough sample size to enable meaningful comparisons. Our study has identified several specific changes that differ between histologies including some where therapeutic agents are already available for the putative driver genes. Moving forward, this data shows that EAC and ESCC should not be grouped together when identifying, developing and testing novel therapeutic targets for esophageal cancer. Rather, we conclude that genomic analysis of ESCC and EAC allows for differential selection and potential development of histology-specific targeted therapies for this poor prognosis disease. Similar studies comparing same tumor histologies across different organ sites could also be informative in identification of common or site-specific therapeutic strategies.
Footnotes
Presented at the Fifty-eighth Annual Meeting of the Southern Thoracic Surgical Association, San Antonio, TX, November 9-12, 2011.
References
- 1.Portale G, Hagen JA, Peters JH, et al. Modern 5-year survival of resectable esophageal adenocarcinoma: single institution experience with 263 patients. J Am Coll Surg. 2006;202:588–596. doi: 10.1016/j.jamcollsurg.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 2.Pennathur A, Luketich JD, Landreneau RJ, et al. Long-term results of a phase II trial of neoadjuvant chemotherapy followed by esophagectomy for locally advanced esophageal neoplasm. Ann Thor Surg. 2008;85:1930–1936. doi: 10.1016/j.athoracsur.2008.01.097. [DOI] [PubMed] [Google Scholar]
- 3.Kountourakis P, Correa AM, Hofstetter WL, et al. Combined modality therapy of cT2N0M0 esophageal cancer: the University of Texas M. D. Anderson Cancer Center experience. Cancer. 2011;117:925–930. doi: 10.1002/cncr.25651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pera M, Manterola C, Vidal O, et al. Epidemiology of esophageal adenocarcinoma. J Surg Oncol. 2005;92:151–159. doi: 10.1002/jso.20357. [DOI] [PubMed] [Google Scholar]
- 5.DeMeester SR. Adenocarcinoma of the esophagus and cardia: a review of the disease and its treatment. Ann Surg Oncol. 2006;13:12–30. doi: 10.1245/ASO.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 6.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 7.Chang JT, Katzka DA. Gastroesophageal reflux disease, Barrett esophagus, and esophageal adenocarcinoma. Arch Intern Med. 2004;164:1482–1488. doi: 10.1001/archinte.164.14.1482. [DOI] [PubMed] [Google Scholar]
- 8.Hu N, Wang C, Ng D, et al. Genomic characterization of esophageal squamous cell carcinoma from a high-risk population in China. Cancer Res. 2009;69:5908–5917. doi: 10.1158/0008-5472.CAN-08-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature Genetics. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss MM, Kuipers EJ, Hermsen MAJA, et al. Barrett’s adenocarcinomas resemble adenocarcinomas of the gastric cardia in terms of chromosomal copy number changes, but relate to squamous cell carcinomas of the distal oesophagus with respect to the presence of high-level amplifications. J Pathol. 2003;199:157–165. doi: 10.1002/path.1260. [DOI] [PubMed] [Google Scholar]
- 12.Rumiato E, Pasello G, Montagna M, et al. DNA copy number profile disriminates between esophageal adenocarcinoma and squamous cell carcinoma and represents an independent prognostic parameter in esophageal adenocarcinoma. Cancer Letters. 2011;310:84–93. doi: 10.1016/j.canlet.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Murakami Y, Kanda K, Tsuji M, et al. MN/CA9 gene expression as a potential biomarker in renal cell carcinoma. BJU International. 1999;83:743–747. doi: 10.1046/j.1464-410x.1999.00007.x. [DOI] [PubMed] [Google Scholar]
- 14.Span PN, Bussink J, Manders P, et al. Carbonic anhydrase-9 expression levels and prognosis in human breast cancer:association with treatment outcome. British Journal of Cancer. 2003;89:271–276. doi: 10.1038/sj.bjc.6601122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shafee N, Kaluz S, Ru N, et al. PI3K/Akt activity has variable cell-specific effects on expression of HIF target genes, CA9 and VEGF, in human cancer cell lines. Cancer Letters. 2009;282:109–118. doi: 10.1016/j.canlet.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snijders AM, Schmidt BL, Fridlyand J, et al. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- 17.Lai MT, Hua CH, Tsai MH, et al. Talin-1 overexpression defines high risk for aggressive oral squamous cell carcinoma and promotes cancer metastasis. J Pathol. 2011;224:367–376. doi: 10.1002/path.2867. [DOI] [PubMed] [Google Scholar]
- 18.Ullmannova V, Popescu NC. Expression profile of the tumor suppressor genes DLC-1 and DLC-2 in solid tumors. International Journal of Oncology. 2006;29:1127–1132. [PubMed] [Google Scholar]
- 19.Xiaorong L, Wei W, Liyuan Q, et al. Underexpression of Deleted in liver cancer 2 (DLC2) is associated with overexpression of RhoA and poor prognosis in hepatocellular carcinoma. BMC Cancer. 2008;8:205. doi: 10.1186/1471-2407-8-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kompass KS, Witte JS. Co-regulatory expression quantitative trait loci mapping: method and application to endometrial cancer. BMC Medical Genomics. 2011;4:6. doi: 10.1186/1755-8794-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, Brown C, Ni W, et al. Critical role for the Ets transcription factor ELF-1 in the development of tumor angiogenesis. Blood. 2006;107:3153–3160. doi: 10.1182/blood-2005-08-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang DX, Li NE, Ma Y, et al. Expression of Elf-1 and survivin in non-small cell lung cancer and their relationship to intratumoral microvessel density. Chinese Journal of Cancer. 2010;29:396–402. doi: 10.5732/cjc.009.10547. [DOI] [PubMed] [Google Scholar]
- 23.Andrews PGP, Kennedy MW, Popadiuk CM, et al. Oncogenic Activation of the Human Pygopus2 Promoter by E74-Like Factor-1. Mol Cancer Res. 2008;6:259–266. doi: 10.1158/1541-7786.MCR-07-0068. [DOI] [PubMed] [Google Scholar]
- 24.Tong D, Czerwenka K, Heinze G, et al. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res. 2006;12:2442–2448. doi: 10.1158/1078-0432.CCR-05-0964. [DOI] [PubMed] [Google Scholar]
- 25.Nandan MO, Ghaleb AM, McConnell BB, et al. Krüppel-like factor 5 is a crucial mediator of intestinal tumorigenesis in mice harboring combined ApcMin and KRASV12 mutations. Mol Cancer. 2010;9:63. doi: 10.1186/1476-4598-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilson DH, Kelsen D, Shah M, et al. A phase 2 trial of erlotinib in patients with previously treated squamous cell and adenocarcinoma of the esophagus. Cancer. 2011;117:1409–1414. doi: 10.1002/cncr.25602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ismail A, Bandla S, Reveiller, et al. Early G1 Cyclin-Dependent Kinases as Prognostic Markers and Potential Therapeutic Targets in Esophageal Adenocarcinoma. Clin Cancer Res. 2011;17:4513–4522. doi: 10.1158/1078-0432.CCR-11-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin L, Aggarwal S, Glover TW, et al. A minimal critical region of the 8p22–23 amplicon in esophageal adenocarcinomas defined using sequence tagged site amplification mapping and quantitative polymerase chain reaction includes the GATA-4 gene. Cancer Res. 2000;60:1341–1347. [PubMed] [Google Scholar]
- 29.Wang X, Han L, Zhang A, et al. Adenovirus-mediated shRNAs for co-repression of miR-221 and miR-222 expression and function in glioblastoma cells. Oncol Rep. 2011;25:97–105. [PubMed] [Google Scholar]
- 30.Zhang X, Wang X, Zhu H, et al. Synergistic effects of the GATA-4-mediated miR-144/451 cluster in protection against simulated ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell Cardiol. 2010;49:841–850. doi: 10.1016/j.yjmcc.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JC, Sharma M, Lee NH, et al. Pax9 mediated cell survival in oral squamous carcinoma cell enhanced by c-myb. Cell Biochem Funct. 2008;26:892–899. doi: 10.1002/cbf.1522. [DOI] [PubMed] [Google Scholar]
- 32.Richter T, Kremmer E, Gerber J, et al. Progressive loss of PAX9 expression correlates with increasing malignancy of dysplastic and cancerous epithelium of the human oesophagus. J Pathol. 2002;197:293–297. doi: 10.1002/path.1115. [DOI] [PubMed] [Google Scholar]
- 33.Kendall J, Liu Q, Bakleh, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. PNAS. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]