Abstract
MCL-1, an anti-apoptotic BCL-2 family member that is essential for the survival of multiple cell lineages, is also among the most highly amplified genes in cancer. Although MCL-1 is known to oppose cell death, precisely how it functions to promote survival of normal and malignant cells is poorly understood. Here, we report that different forms of MCL-1 reside in distinct mitochondrial locations and exhibit separable functions. On the outer mitochondrial membrane, a MCL-1 isoform acts like other anti-apoptotic BCL-2 molecules to antagonize apoptosis, whereas an amino-terminally truncated isoform of MCL-1 that is imported into the mitochondrial matrix is necessary to facilitate normal mitochondrial fusion, ATP production, membrane potential, respiration, cristae ultrastructure, and maintenance of oligomeric ATP synthase. Our results provide insight into how MCL-1's surprisingly diverse salutary functions may control the survival of both normal and cancer cells.
Apoptosis, or programmed cell death, is an essential genetic program required to regulate the proper development and maintenance of tissue homeostasis in metazoans1. However, apoptotic dysregulation can lead to a variety of human pathologies including cancer, neurodegeneration, and autoimmunity2. Members of the BCL-2 family regulate the process of programmed cell death by either promoting or inhibiting apoptosis3. Death signals activate and/or induce BCL-2 homology (BH)3-only molecules (such as BID, BAD, BIM, PUMA, NOXA), promoting the activation of the pro-apoptotic effectors (BAX and BAK) and facilitating the release of apoptogenic factors from the mitochondrial inner membrane space4, 5. This process is antagonized by the anti-apoptotic family members (such as BCL-2, BCL-XL, BCL-W, MCL-1, and A1)6, 7.
Mitochondria, essential for the production of energy by oxidative phosphorylation, possess a selectively-permeable outer mitochondrial membrane (OMM) and a larger surface-area inner mitochondrial membrane (IMM) that folds and forms invaginations called cristae8. Mitochondria are dynamic organelles constantly undergoing fission and fusion to maintain a network important for their function9. Fusion facilitates the exchange of contents, including the mitochondrial genome, while fission is necessary to increase mitochondrial number and allow distribution to daughter cells during cell division10, 11. Imbalances in fission and fusion lead to a variety of cellular and mitochondrial defects12, 13. For example, mitochondrial fusion is required for cell growth, efficient oxidative phosphorylation, and ATP production10. Thus, the integrity of the mitochondrial network is associated with cellular fate and bioenergetics.
MCL-1 is unique among pro-survival BCL-2 family members in that it is essential for embryonic development14 and for the survival of multiple cell lineages in the adult including: lymphocytes15, 16, hematopoietic stem cells17, neutrophils18, 19, and neurons20. Moreover, MCL-1 is frequently amplified in human cancers21 and associated with chemotherapeutic resistance and relapse22, 23. Yet, how MCL-1 functions to promote survival of diverse normal and malignant cells remains poorly understood. Here, we reveal that MCL-1 is localized to distinct mitochondrial sub-compartments with differential functions that affect mitochondrial activity and integrity. On the OMM, MCL-1 exerts its anti-apoptotic activity where it antagonizes BAX and BAK activation to maintain mitochondrial integrity. In contrast, MCL-1 localized to the mitochondrial matrix is unable to inhibit apoptosis, but maintains normal IMM structure, regulates fusion, and promotes the assembly of ATP synthase oligomers; thereby, it facilitates mitochondrial homeostasis and supports mitochondrial bioenergetic function.
RESULTS
Mcl-1-deletion results in mitochondrial morphology defects
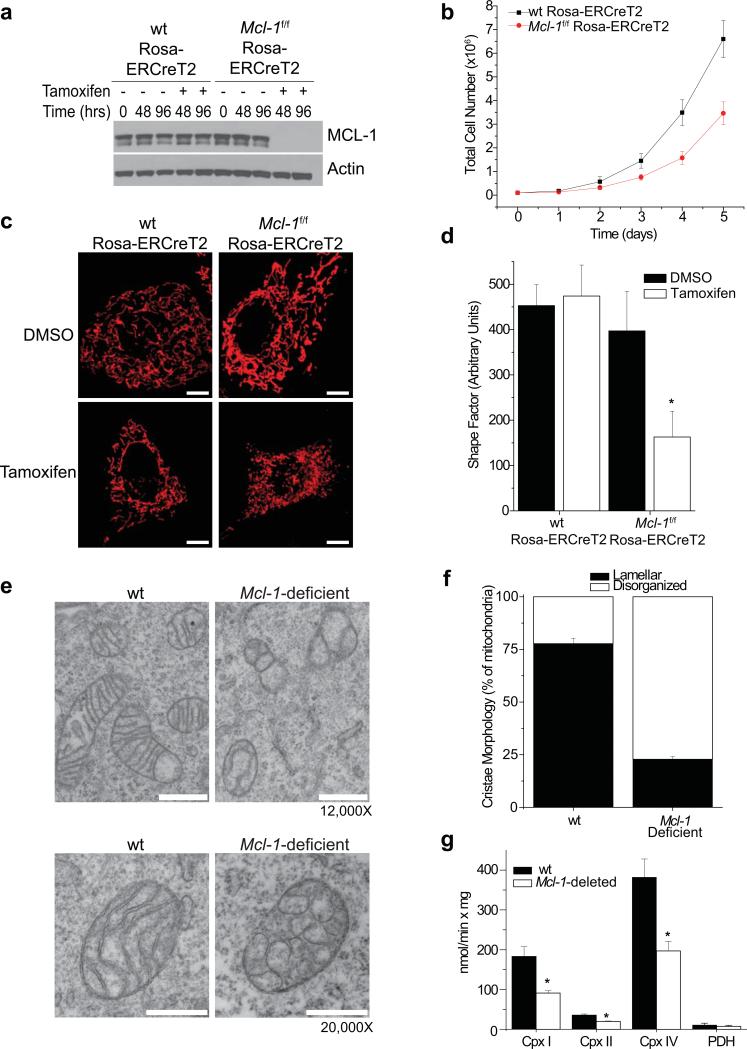
We generated SV40-transformed wild-type (wt) or Mcl-1f/f Rosa-ERCreT2 murine embryonic fibroblasts (MEFs) in which endogenous Mcl-1 can be deleted by tamoxifen-mediated induction of the Cre-recombinase (hereafter referred to as Mcl-1-deletion) (Fig. 1a). Inducible Mcl-1-deletion in MEFs does not trigger spontaneous apoptosis (Sup. Fig. 1a), but results in defective proliferation (Fig. 1b). Notably, Mcl-1-deletion induces the appearance of punctate mitochondria and the loss of the tubular mitochondrial network present in control cells (Fig. 1c,d). Furthermore, ultrastructural defects including defective cristae harboring balloon-like, vesicular structures are apparent within the majority of mitochondria in Mcl-1-deficient MEFs (Fig. 1e,f). These abnormal cristae morphologies are not limited to transformed MEFs as similar defects are observed in primary Mcl-1-deleted liver mitochondria (Sup. Fig. 1b). Proteins involved in oxidative phosphorylation are enriched in the IMM24, 25; therefore, we assessed whether Mcl-1-deletion alters the enzymatic activity of the respiratory chain complexes. Mcl-1-deletion in mouse liver mitochondria decreases the ability of NADH dehydrogenase (complex I), succinate dehydrogenase (complex II), and cytochrome c oxidase (complex IV) to transfer electrons; whereas pyruvate dehydrogenase (PDH) activity is unchanged (Fig. 1g). Thus, loss of MCL-1 expression in MEFs and hepatocytes results in dysregulation of the reticular mitochondrial network, cristae morphological abnormalities, and defects in electron transport chain enzymatic function.
Figure 1. Deletion of Mcl-1 results in mitochondrial morphology defects.
(a) Mcl-1f/f Rosa-ERCreT2 or wild-type (wt) Rosa-ERCreT2 MEFs (control MEFs) were treated with tamoxifen or DMSO for indicated times to induce deletion of endogenous Mcl-1 then immunoblotted for MCL-1 or Actin (loading control). (b) Mcl-1f/f Rosa-ERCreT2 or wt Rosa-ERCreT2 MEFS were treated with tamoxifen (48 hours), seeded, and counted by hemocytometer at the indicated times. The data represent averages and SEM of 3 independent experiments. (c) Mcl-1f/f Rosa-ERCreT2 or wt Rosa-ERCreT2 MEFs stably-expressing matrix-targeted DsRed-fluorescent protein were treated with tamoxifen or DMSO for 96 hours to induce deletion of endogenous Mcl-1, then imaged to detect mitochondrial morphology. Scale bars represent 10 μm. (d) Average shape factor of mitochondrial matrix for Mcl-1f/f Rosa-ERCreT2 or control MEFs 96 hours after deletion. A shape factor of 1.0 indicates circular mitochondria and the higher the number, the more reticular the network. Error bars represent the standard error of the mean (SEM) from 10 imaged cells (~3000 mitochondria) and statistical significance was determined by unpaired t-test (*p<0.01). (e) Transmission electron micrographs of wt or Mcl-1-deficient MEFs. Scale bars represent 500 nm. (f) Quantification of mitochondrial cristae morphology from wt or Mcl-1-deficient MEFs. Mitochondria were counted from ~100 individual cells and scored for normal (lamellar) or ballooned and swollen cristae (disorganized). Error bars indicated the SEM from 3 independent experiments. (g) Enzymatic assays for indicated electron transport chain components from mouse liver mitochondria prepared from Mcl-1f/f Mx1-Cre or wt Mx1-Cre (wt) mice 14 days after treatment with pI-pC. Three independent animals were assayed in triplicate (n=9) and error bars indicated the SEM. Statistical significance was determined by unpaired t-test (*p<0.01).
MCL-1 localizes to two distinct mitochondrial sub-compartments
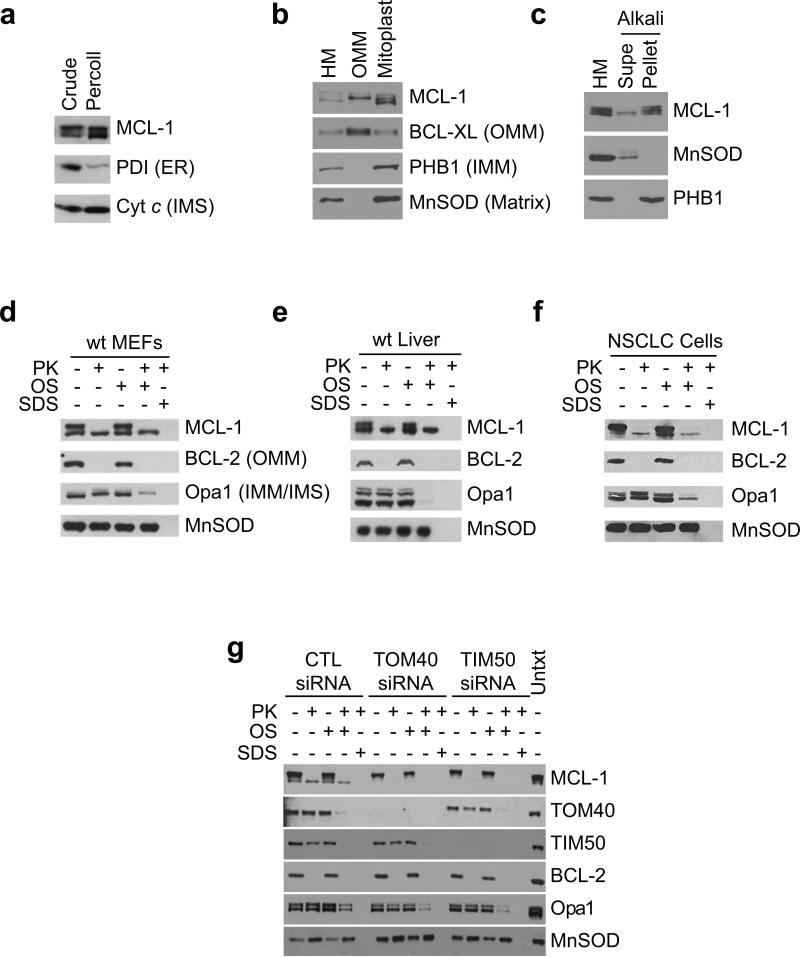
To understand the mechanisms underlying the dependence on MCL-1 for normal cristae morphology, we investigated the localization of MCL-1 within mitochondria. By immunoblot analysis MCL-1 appears as a doublet or triplet in percoll-purified liver mitochondria devoid of contaminating endoplasmic reticulum (ER)(Fig. 2a). MCL-1 is a phosphorylated protein26-28; however, phosphatase-treatment does not abolish the 40 kD species (Sup. Fig. 2a). It has been proposed that non-canonical translational initiation at a non-ATG leads to the production a faster-migrating 36 kD truncated protein29; however, removal of MCL-1's start codon instead produces a 31 kD truncated-protein from a downstream ATG codon (Sup. Fig. 2b). It has also been proposed that non-canonical mRNA splicing generates the 36 kD species30; however, ablation of the putative splice donor and acceptor still produces the doublet in cells (Sup. Fig. 2c). Thus, proteolytic cleavage is the most likely contributor to generating the 36 kD species31, 32. Indeed, Edman sequencing of MCL-1 detects two cleavage sites: between MCL-1's isoleucine-10 and glycine-11 and between leucine-33 and valine-34. Consequently, proteolysis of MCL-1's amino-terminus gives rise to three different MCL-1 species: full-length (40 kD), cleaved at isoleucine-10 (38 kD), and cleaved at leucine-33 (36 kD).
Figure 2. MCL-1 resides in different sub-mitochondrial localizations.
(a) Crude or Percoll-purified mouse liver mitochondria were western blotted for expression of MCL-1, Protein Disulphide Isomerase (PDI, endoplasmic reticulum, ER), or cytochrome c (inner membrane space, IMS). (b) Mouse liver mitochondria (HM) were sub-fractionated into outer mitochondrial membrane or mitoplast and western blotted for MCL-1, BCL-XL (outer mitochondrial membrane, OMM), Prohibitin1 (inner mitochondrial membrane, IMM), or MnSOD (matrix). (c) Mouse liver mitochondria (HM) were swollen and sonicated to disrupt membranes, washed with alkali buffer (pH 11.5) to detach loosely-associated proteins from membranes, and then re-isolated by ultra-centrifugation. Supernatant (Supe) and membrane fractions (Pellet) were western blotted for MCL-1, Prohibitin1, or MnSOD. (d, e, & f) Mitochondria isolated from (d) wt MEFs, (e) wt mouse liver, or (f) human Non-Small Cell Lung Cancer cells were subjected to proteinase K (PK) proteolysis to digest exposed proteins, osmotic shock (OS) was used to disrupt the outer mitochondrial membrane, and detergent (SDS) was used to disrupt both inner and outer mitochondrial membranes. Lysates were resolved and subjected to immunoblot analyses. Submitochondrial markers used are BCL-2 (OMM), Opa1 (IMM/IMS), and MnSOD. (g) Mitochondria isolated from wt MEFs transfected with control or siRNA-oligos specific to TOM40 or TIM50 were subjected to protease treatment as described in d.
The variant forms of MCL-1 exhibit different mitochondrial sub-localization; the 40 kD and 38 kD bands of MCL-1 are enriched in the OMM, while the 36 kD form is only detected in the mitoplast fraction, consisting of IMM and matrix (Fig. 2b). Indeed, while both the 40 kD and 36 kD forms of MCL-1 are tightly-associated with disrupted and alkali-washed mitochondrial membranes, the 38 kD species exhibits a looser association with membranes (Fig. 2c). When mitochondria isolated from MEFs are treated with proteinase K, the 40 kD and 38 kD forms of MCL-1 are sensitive to protease digestion, but the 36 kD species remains resistant to degradation even after osmotic shock-mediated permeabilization of the OMM (Fig. 2d). Moreover, mitochondria from mouse liver and human non-small cell lung cancer cell lines exhibit similar protease sensitivity, suggesting that the sub-mitochondrial localization of MCL-1 is evolutionarily conserved (Fig. 2e,f). Immunoelectron microscopy of isolated liver mitochondria also indicates that endogenous MCL-1 localizes to both the OMM and IMM (Sup. Fig. 3a). Together, these results indicate that the 40 kD and 38 kD forms of MCL-1 reside on the outer leaflet of the OMM, whereas the 36 kD form is associated with the inner membrane and confined within the mitochondrial matrix.
During mitochondrial protein import, nuclear encoded proteins are channeled into the mitochondria by the translocases of outer membrane (TOM) and inner membrane (TIM) proteins33. RNAi-mediated gene silencing of either TOM40 or TIM50 prevents MCL-1's mitochondrial import rendering it completely protease sensitive (Fig. 2g). Accordingly, MCL-1 undergoes TOM- and TIM-dependent mitochondrial import.
MCL-1 possesses a mitochondrial targeting sequence
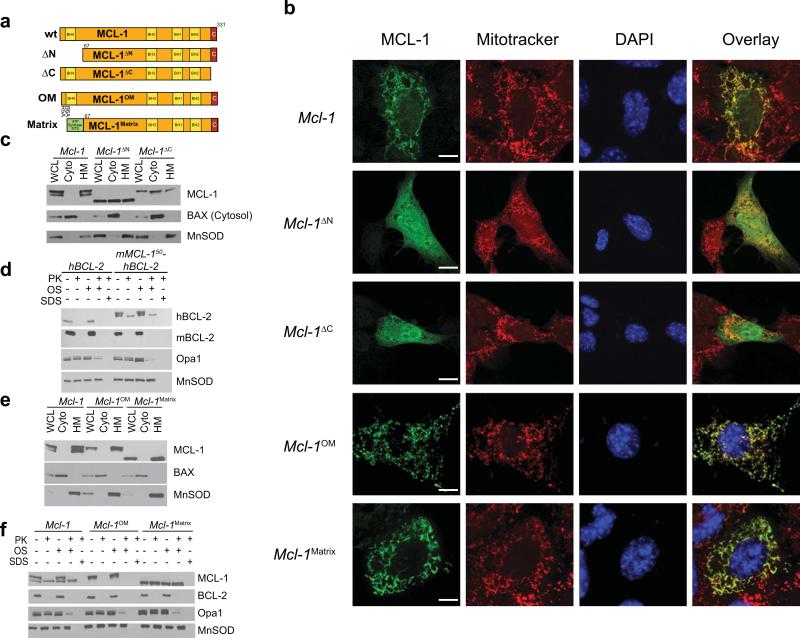
Both MCL-1's amino- and carboxy-terminal domains are necessary for its proper mitochondrial localization and generation of a doublet by immunoblot analysis (Fig. 3a,b,c). Proteins imported into the mitochondrial matrix often contain mitochondrial targeting sequences (MTS)33. To determine if MCL-1's amino-terminus contains a MTS, the first 50 amino-terminal amino acids of MCL-1 were fused to the amino-terminus of human BCL-2 (hBCL-2). Human BCL-2 is normally restricted to the OMM; however, the chimeric protein forms a doublet of which the faster migrating species appears in the matrix, thus validating that MCL-1's amino-terminus possesses an MTS (Fig. 3d). Imported mitochondrial proteins often possess MTS's containing charged residues33. Mutagenesis of the arginine residues at MCL-1's positions 5 and 6 to alanine (hereafter called MCL-1OM) maintains its localization to mitochondria, but abrogates the formation of MCL-1's 36 kD form (Fig. 3a,b,e). Notably, MCL-1OM is completely sensitive to proteolysis indicating that the mutagenesis blocks mitochondrial import (Fig. 3f). To generate a MCL-1 mutant that resides solely within the mitochondrial matrix, we fused the mitochondrial targeting sequence of matrix-localized ATP-synthase34 to amino-truncated MCL-1 (hereafter called MCL-1Matrix) (Fig. 3a). MCL-1Matrix localizes to mitochondria, and only produces a single protein species (Fig. 3b,e). Moreover, MCL-1Matrix is completely resistant to protease attack even under conditions of osmotic shock illustrating that it localizes to the matrix (Fig. 3f).
Figure 3. MCL-1 mutants restrict mitochondrial localization.
(a) Schematic illustration of MCL-1 mutants. Yellow boxes indicate BCL-2 homology (BH) domains and the red box indicates the C-terminal hydrophobic domain. Point mutations are indicated with the original amino acid, site-position and mutated residue. N. crassa ATP-synthase MTS is indicated in green. (b) Confocal immunofluorescence of Mcl-1-deficient MEFs transiently-expressing indicated Mcl-1 constructs were fixed and stained for MCL-1 (green), mitochondria (Mitotracker, in red), and nucleus (DAPI, in blue). Co-localization of green and red channels is shown in yellow overlay. Size bars represent 10 μm. (c & e) Whole cell lysates (WCL), isolated heavy membranes (HM) enriched for mitochondria or cytosolic (cyto) sub-cellular fractions were isolated from Mcl-1-deleted MEFs stably-expressing indicated MCL-1 constructs. Fractions were western blotted for MCL-1, BAX (cytosolic marker) or MnSOD (mitochondrial marker). (d) Mitochondria from Mcl-1-deficient MEFs stably-expressing human BCL-2 (hBCL-2) or a chimeric fusion of the first 50 amino acids of MCL-1 to hBCL-2 (mMCL-150-hBCL-2) were subjected to protease treatment as described in Fig. 2d. Both the fusion protein and hBCL-2 were detected with anti-hBCL-2 antibody. (f) Mitochondria from Mcl-1-deficient MEFs stably-expressing indicated constructs were subjected to protease treatment as described in Fig. 2d.
Inhibition of cell death requires outer membrane-localized MCL-1
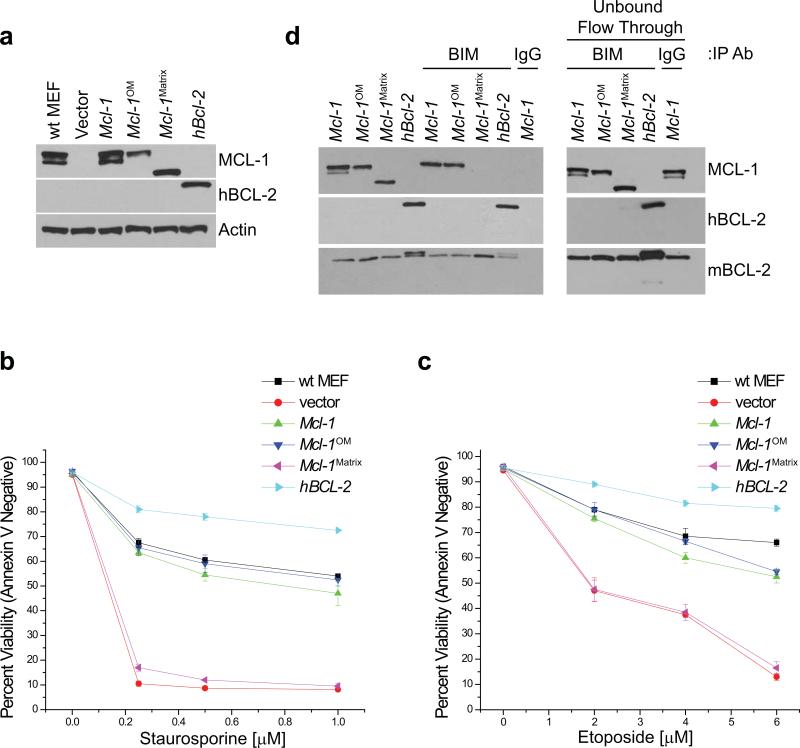
We assessed the anti-apoptotic activities of the different mitochondrial localized mutants of MCL-1. Similar levels of wt MCL-1, MCL-1OM, MCL-1Matrix, or hBCL-2 were stably expressed in Mcl-1f/f Rosa-ERCreT2 MEFs (Fig. 4a). Mcl-1-deletion renders MEFs highly susceptible to cell death stimuli (Fig. 4b,c). Expression of either wt MCL-1 or MCL-1OM protects the MEFs from apoptosis; however, MCL-1Matrix expressing MEFs are completely sensitive to cell death (Fig. 4b,c). Furthermore, the expression of hBCL-2 in the Mcl-1-deleted MEFs renders cells resistant to death stimuli (Fig. 4b,c). MCL-1 binds pro-apoptotic BH3-only molecules such as BIM, preventing OMM permeabilization and activation of the downstream apoptotic cascade15. Both the 40 kD species of wt MCL-1 and MCL-1OM co-immunoprecipitate with pro-apoptotic BIM; however, no interaction is detectable between BIM and the 36 kD species of wt MCL-1 or between BIM and MCL-1Matrix (Fig. 4d). Thus, MCL-1OM can bind pro-apoptotic modulators and prevent cell death similarly to wt MCL-1, but MCL-1Matrix cannot sequester pro-apoptotic molecules or prevent apoptosis.
Figure 4. Anti-apoptotic activity of MCL-1 requires localization to the outer mitochondrial membrane.
(a) Mcl-1f/f Rosa-ERCreT2 MEFs stably-expressing indicated constructs were treated for 48 hours with tamoxifen to induce deletion of endogenous Mcl-1 then western blotted for MCL-1, hBCL-2, or Actin (loading control). Vector-expressing, tamoxifen-treated wt Rosa-ERCreT2 MEFs (lane 1) serve as control. (b & c) Rosa-ERCreT2 MEFs stably-expressing indicated constructs were treated for 96 hours with tamoxifen to induce deletion of endogenous Mcl-1 and were administered indicated doses of (b) staurosporine for 16 hours or (c) etoposide for 24 hours after which cell death was determined. Annexin-V and PI double-negative cells were scored as viable. Bars represent the average of 3 independent experiments and the error bars denote the SEM. Vector-expressing, tamoxifen-treated wt Rosa-ERCreT2 MEFs serve as control. (d) Lysates were immunoprecipitated with anti-BIM or anti-rat IgG antibody and immune complexes were resolved and immunoblotted for MCL-1 and hBCL-2. Endogenous murine BCL-2 (mBCL-2) serves as a control for equal BIM immunoprecipitation. 1/10th input of immunoprecipitation is depicted. Post-immunoprecipitated supernatants (Unbound Flow Through) indicate non-precipitated proteins remaining in lysate.
Matrix-localized MCL-1 is required for normal IMM structure and efficient mitochondrial fusion
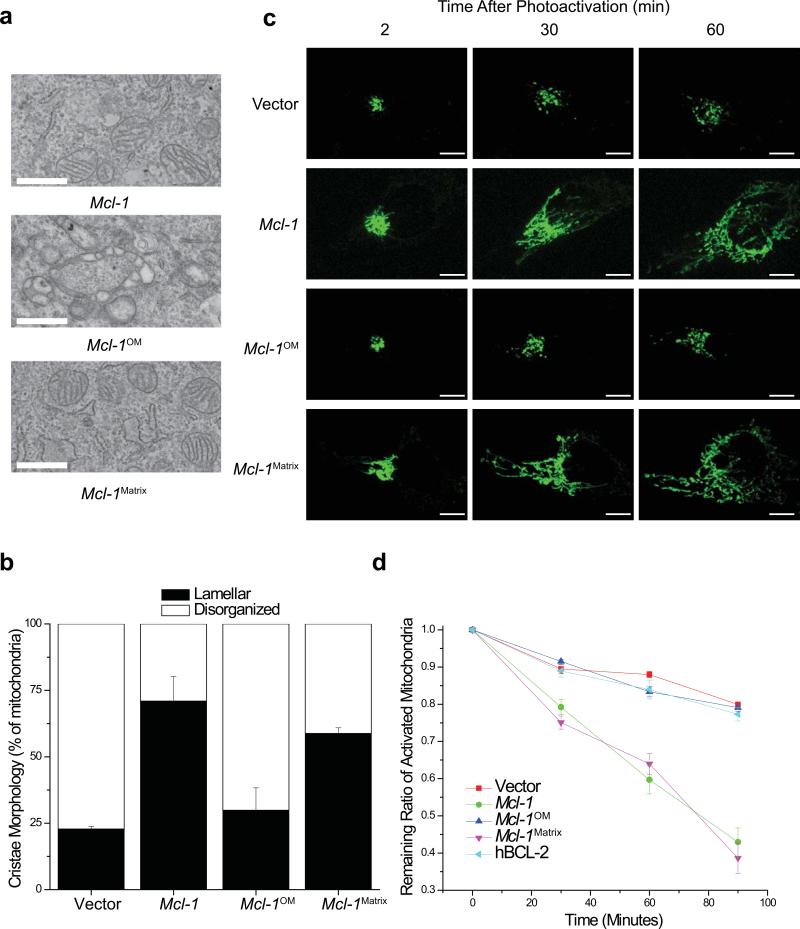
Expression of either wt MCL-1 or MCL-1Matrix in Mcl-1-deleted MEFs largely restores normal IMM structure whereas MCL-1OM does not (Fig. 5a,b). We used photo-activatable-GFP targeted to the matrix35 to assess mitochondrial fusion. Upon Mcl-1-deletion, there is a delay in fusion that can be corrected by expression of either wt MCL-1 or MCL-1Matrix but not by MCL-1OM or hBCL-2 (Fig. 5c,d). Furthermore, expression of human MCL-1 ameliorates the delay in fusion and compensates for loss of mouse MCL-1 implying that this function is evolutionarily conserved (Sup. Fig. 3b). These data demonstrate that matrix-localized MCL-1 is necessary to promote normal IMM structure, mitochondrial fusion and maintenance of the mitochondrial reticular network.
Figure 5. Lacking matrix-localized MCL-1 results in mitochondrial IMM structure and fusion defects.
(a) Transmission electron micrographs (12,000X) of Mcl-1f/f Rosa-ERCreT2 MEFs stably-expressing indicated constructs were treated with tamoxifen to induce deletion of endogenous Mcl-l. Scale bars represent 500 nm. (b) Quantification of mitochondrial cristae morphology from Mcl-1f/f Rosa-ERCreT2 MEFs stably-expressing indicated constructs that were treated with tamoxifen to induce deletion of endogenous Mcl-1. Mitochondria were counted from ~100 individual cells and scored for normal (lamellar) or ballooned and swollen cristae (disorganized). Error bars indicated the SEM from 3 independent experiments. (c) Mitochondrial fusion was measured 96 hours after Mcl-1-deletion in Mcl-1f/f Rosa-ERCreT2 MEFs stably-expressing indicated constructs. Cells were transiently-transfected with photoactivatable-GFP (PA-GFP) targeted to the mitochondrial matrix. Depicted are representative images from 2, 30, and 60 minutes after PA-GFP activation. Scale bars represent 10 μm. (d) Quantitation of the rate of mitochondrial fusion measured 96 hours after Mcl-1-deletion as the ratio of remaining activated mitochondrial area. Data represent average and SEM of fluorescence area over time of 3 independent experiments (n=16 cells). As mitochondria fuse, the PA-GFP signal dilutes decreasing intensity of signal in the area of activation.
Normal mitochondrial bioenergetics requires matrix-localized MCL-1
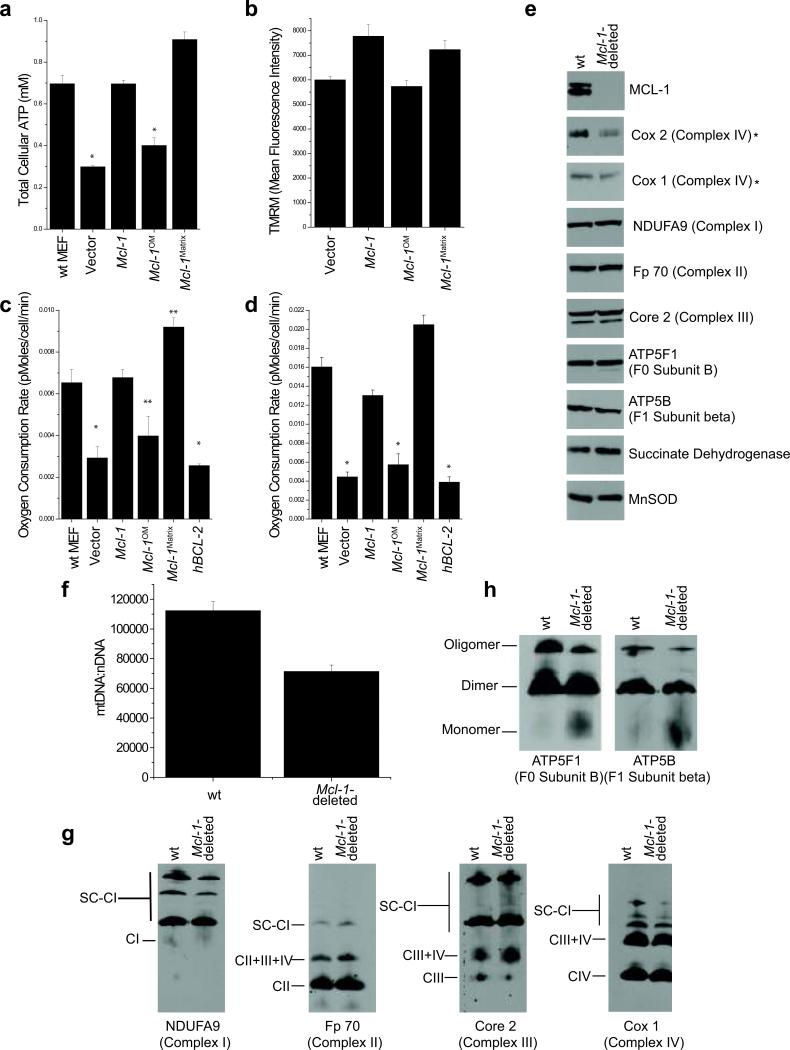
Respiratory complexes pass electrons through the electron transport chain to reduce oxygen, thus generating a proton gradient that drives ATP production by the F1F0-ATP synthase, this process is called oxidative phosphorylation36. To stimulate oxidative phosphorylation, MEFs were cultured in glucose-free media supplemented with galactose37, 38. Under these conditions, Mcl-1-deletion reduces ATP levels, but expression of either wt MCL-1 or MCL-1Matrix restores normal ATP levels (Fig. 6a). In contrast, expression of either MCL-1OM or hBCL-2 in the Mcl-1-deleted MEFs does not rescue ATP levels (Fig. 6a). The proton gradient across the IMM drives ATP synthesis; therefore, we assessed whether the mitochondrial membrane potential was altered in cells lacking MCL-1. Indeed, Mcl-1-deletion decreases mitochondrial membrane potential, which is improved by expression of either wt MCL-1 or MCL-1Matrix, but not MCL-1OM (Fig. 6b & Sup. Fig. 4a). Furthermore, Mcl-1-deleted MEFs also exhibit slower proliferation compared to control cells (Sup. Fig. 4b). Expression of either wt MCL-1 or MCL-1Matrix corrects the proliferative defect, whereas MCL-1OM or hBCL-2 expression is unable to compensate for Mcl-1-deletion (Sup. Fig. 4b). Consequently, MEFs lacking matrix-localized MCL-1 generate less energy, have decreased mitochondrial membrane potential, and grow poorly in galactose-media even in the presence of anti-apoptotic MCL-1 on the OMM.
Figure 6. MCL-1 regulates mitochondrial bioenergetics.
(a) Mcl-1f/f Rosa-ERCreT2 MEFs stably-expressing indicated constructs were grown in glucose-free media containing galactose and treated for 96 hours with tamoxifen and total cellular ATP was evaluated. Vector-expressing, tamoxifen-treated wt Rosa-ERCreT2 MEFs are positive control. Data represent the averages of 3 independent experiments and the error bars denote the SEM. Statistical significance was determined by unpaired t-test (*p<0.01). (b) Quantification of mitochondrial membrane potential from Mcl-1f/f Rosa-ERCreT2 MEFs, stably-expressing indicated constructs, were treated for 96 hours with tamoxifen and stained with 10 nM TMRM and imaged. Data represent average mean fluorescence intensities and SEM from 20 cells. (c & d) Oxygen consumption measured 96 hours after Mcl-1-deletion in Mcl-1f/f Rosa-ERCreT2 MEFs stably-expressing indicated constructs under (c) basal and (d) FCCP-uncoupled (maximal) respiration. Vector-expressing, tamoxifen-treated wt Rosa-ERCreT2 MEFs serve as positive control. Experiments were performed in triplicate and the bars represent averages (n=9) and the error bars denote the standard of deviation. Statistical significance was determined by unpaired t-test (*p<0.01, **p<0.05). (e) Mouse liver lysates from Mcl-1f/f Mx1-Cre (Mcl-1-deleted) or wt Mx1-Cre (wt) mice 14 days after treatment with pI-pC were lysed in RIPA and resolved by SDS-PAGE. Immunoblots were probed to determine expression of Complex I (NDUFA9), Complex II (Fp70), Complex III (Core 2), Complex IV (Cox I and Cox 2), ATP Synthase subunit F0 B (5F1), ATP Synthase F1 subunit beta (5B), Succinate dehydrogenase, MnSOD, and MCL-1. Asterix (*) denotes mtDNA-encoded protein (f) Assessment of mtDNA and nDNA by quantitative real-time PCR. Error bars represent the standard error of mean (n=3). (g & h) Mouse liver mitochondria from Mcl-1f/f Mx1-Cre (Mcl-1-deleted) or wt Mx1-Cre (wt) mice 14 days after treatment with pI-pC were lysed in digitonin and resolved by blue native gel polyacrylamide electrophoresis (BN-PAGE) and blotted to determine native complexes and supercomplexes (SC). (g) complex I (CI, detected by NDUFA9), complex II (CII, detected by Fp70), complex III (CIII, detected by Core 2) and complex IV (CIV, detected by Cox I) and (h) ATP Synthase subunit F0 B (5F1), F1 subunit beta (5B). The native migration of monomers, dimers, and oligomers are denoted.
We determined if respiration was altered by measuring oxygen consumption rates (OCR) using the Seahorse XF24 analyzer39. Mcl-1-deletion decreases both basal and maximal OCR compared to non-deleted MEFs (Fig. 6c,d, & Sup. Fig. 5). Expression of either wt MCL-1 or MCL-1Matrix restores OCR; whereas neither MCL-1OM nor hBCL-2 expression can restore normal OCR (Fig. 6c,d, & Sup. Fig. 5). Moreover, Mcl-1-deletion results in elevated mitochondrial superoxide levels, but expression of either wt MCL-1 or MCL-1Matrix prevents the increase in superoxide production consistent with their ability to rescue normal mitochondrial function (Sup. Fig. 4c). In contrast, neither hBCL-2 nor MCL-1OM can negate superoxide generation (Sup. Fig. 4c). Collectively, these data implicate that matrix-localized MCL-1 is required for optimal oxidative phosphorylation and prevention of superoxide in cells, a function that is separate from its anti-apoptotic role.
Mcl-1 is required for assembly of F1F0-ATP synthase oligomers
Structural and functional evidence has revealed that the individual electron transport chain complexes (I, II, III, and IV) organize into inter-complex assemblies known as supercomplexes that are disrupted in the absence of one of their component complexes40. This supramolecular organization of the respiratory complexes has been proposed to increase the efficiency of electron transport and minimize ROS production40, 41. Mcl-1-deletion does not affect the total protein expression of nuclear-encoded respiratory chain subunits; however, the mitochondrial DNA-encoded subunits of complex IV (Cox 1 and Cox 2) protein levels are decreased (Fig. 6e). Therefore, we investigated whether the decreased Cox 1 and Cox 2 expression may be due to alterations in mitochondrial DNA (mtDNA). Indeed, Mcl-1-deletion decreases mtDNA content (Fig. 6f). To investigate the structure of the respiratory complexes and supercomplexes, we used blue native-polyacrylamide gel electrophoresis (BN-PAGE). Consistent with the loss of the Cox 1 and Cox 2 subunits of complex IV, Mcl-1-deleted liver mitochondria also exhibit aberrant assembly of large supercomplexes (composed of complexes I, III, and IV) as detected by Cox I (Fig. 6g). These data are in agreement with the observations that the deletion of complex IV prevents the assembly of large supercomplexes40, 42. In contrast, the smaller respiratory supercomplexes appear relatively unaffected by Mcl-1-deletion (Fig. 6g). Therefore, Mcl-1-deletion results in diminished mtDNA, reduced protein expression of Cox 1 and Cox 2 subunits, and alterations in the large supercomplexes containing complexes I, III, and IV.
The F1F0-ATP synthase also adopts supramolecular structures including the constitutive assembly of ATP synthase homo-dimers and higher-order homo-oligomers43. Furthermore, the assemblage into dimers and oligomers has been proffered as a determinant of the mitochondrial cristae structure and suggested to enhance efficiency of ATP synthesis44-46. As such, mutations that prevent the oligomerization of ATP synthase result in disorganized cristae membranes that form concentric circles and onion-like structures47, 48. By BN-PAGE, Mcl-1-deficient liver mitochondria possess less oligomeric ATP synthase and exhibit a corresponding increase in the monomer (Fig. 6h). The lack of F1F0-ATP synthase oligomers is not due to loss of the individual subunits, implying that the defect is in the oligomerization of ATP synthase (Fig. 6e,h). Accordingly, these data indicate that Mcl-1 is required for efficient assembly of F1F0-ATP synthase oligomers.
DISSCUSION
Gene-deletion studies have demonstrated that MCL-1 expression is critical to the survival of multiple cellular lineages, despite the concomitant expression of other pro-survival BCL-2 proteins17, 49-51. However, the physiological reason why MCL-1 is essential for promoting the survival of so many different cell types remains elusive14-20. In this study, we reveal a role for MCL-1 in regulating mitochondrial inner membrane structure and function that is distinct from its ability to antagonize cell death. We demonstrate that cells express different isoforms of MCL-1 (36 kD, 38 kD, and 40 kD) that perform separate functions and localize to distinct mitochondrial sites. The 40 kD and 38 kD species of MCL-1 reside on the outer mitochondrial membrane where they antagonize pro-apoptotic molecules, thereby preventing cell death similar to other anti-apoptotic BCL-2 family members. During TOM- and TIM-dependent import, MCL-1's amino-terminus undergoes proteolytic processing resulting in a 36 kD species that is tethered to the IMM and exposed to the matrix. In the matrix, MCL-1 maintains normal IMM structure, fusion, and maintenance of mitochondrial bioenergetics, but does not prevent apoptosis. Additionally, Mcl-1 is necessary to maintain normal mtDNA levels and mitochondrial-encoded proteins of complex IV. Depletion of mtDNA may be due to defective mitochondrial fusion since mitochondrial fusion is required for maintenance of mtDNA52. Furthermore, it is possible that loss of the mitochondrial DNA-encoded Cox 1 and Cox 2 subunits of complex IV may lead to the de-stabilization or the aberrant assembly of the large supercomplexes containing complexes I, III, and IV and may result in inefficient respiration. However, the underlying cause of the respiratory dysfunction is still unclear and will require further investigation.
Our data also indicate that MCL-1 is required for the proper assembly of the F1F0-ATP synthase into higher-order oligomers. The formation of F1F0-ATP synthase oligomers has been shown to be required for maintaining the proper organization of the IMM44, 45. Moreover, defects in mitochondrial cristae ultrastructure can result in the breakdown of the mitochondrial network53. Therefore, it is plausible that the loss of F1F0-ATP synthase oligomers in Mcl-1-deleted mitochondria may be responsible for the ultrastructural defects including disorganized cristae as well as the delay in mitochondrial fusion. To that end, identifying how MCL-1 promotes the maintenance of ATP synthase oligomers will be critical to understanding this function.
MCL-1 is a unique anti-apoptotic BCL-2 family member in that its genetic ablation results in cell autonomous deficiencies in a myriad of cellular lineages14-20. However, Mcl-1 gene ablation disables both its anti-apoptotic activity at the OMM as well as its ability to facilitate mitochondrial function within the matrix. For that reason, it will be important to determine the relative contributions of MCL-1's different functional roles for promoting the survival and development of hematopoietic and other cell lineages. MCL-1's critical function during these stages may merely be to support cell survival by antagonizing apoptosis. However, MCL-1 may also facilitate mitochondrial fitness during cellular proliferation and differentiation when metabolic demands on the mitochondria are modulated.
MCL-1 is one of the most highly amplified genes in a variety of human cancers21. As a result, most therapeutic strategies have been directed at antagonizing MCL-1's anti-apoptotic activity attempting to foster malignant cell death. However, these findings raise the possibility that the non-apoptotic function of MCL-1 may also fuel mitochondrial function in cancer cells, thus facilitating the generation of mitochondrially-derived substrates required to maintain the levels of biomolecular synthesis that drives hyper-proliferation54. Therefore, inhibiting MCL-1's ability to promote mitochondrial function may represent a new therapeutic target in cancer cells.
Materials and Methods
Cells and Cell Culture
SV40-transformed wild-type (wt) and Mcl-1-deficient mouse embryonic fibroblasts (MEFs) have been previously described15, 55. Mcl-1f/f Rosa-ERCreT2 MEFs were generated from E12.5 embryos and SV40-transformed. To induce Cre-expression, MEFs were treated with 100 nM (4-hydroxy)-tamoxifen (Sigma) in media. HEK293T cells and human non-small cell lung cancer cells were obtained from American Type Culture Collection. Cells were grown in Dulbecco's-modified Eagle's medium (DMEM, Invitrogen) with 5% fetal bovine serum, 2 mM glutamine (Gibco) and 10 mM HEPES (Gibco). Cells grown in galactose were grown in glucose-free DMEM with 10% fetal bovine serum, 2 mM glutamine, and 10 mM galactose.
Plasmids, Expression Constructs, and Generation of Mutants
Mcl-1 constructs were developed using mouse Mcl-1 cDNA and site-directed mutagenesis (Stratagene) and polymerase chain reaction (PCR). Mcl-1OM replaces arginines 5 and 6 with alanines. The amino-terminal 67 amino acids (aas) of MCL-1 (MCL-1ΔN) and the carboxy-terminal 22 aas (MCL-1ΔC) were deleted by PCR. The MCL-1-hBCL-2 chimera fuses the amino-terminal 50 aas of MCL-1 to full-length human BCL-2. Mcl-1Matrix fuses amino-terminal 58 aas of N. crassa ATP-synthase34 to Mcl-1ΔN. Mitochondrial morphology and fusion were measured using matrix-localized photoactivatable-GFP35 and Mito-DsRed (Clontech). MCL-1ΔATG1 was generated by PCR. The non-canonical mRNA splicing mutant Mcl-1SD;SAmut mutates nucleotide 52 from G to A and nucleotide 189 from G to T30. PCR primer sequences are available by request.
Ecotropic Retroviral Production and Cell Transduction
Retroviruses were produced by FuGene6 (Roche) co-transfection in 293T cells with packaging plasmids.
Western Blotting, Co-Immunoprecipitation, and Antibodies
For immunoblot analysis, cells were lysed in Flag Lysis Buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% TritonX-100, 1 mM EDTA) containing protease inhibitors (Roche) on ice. Whole cell lysates (WCL) were cleared by centrifugation and protein concentrations determined by BCA assay (Pierce). WCLs were pre-cleared and immunoprecipitated with anti-BIM short and anti-BIM long rat monoclonal antibodies (Millipore) or rat IgG (Santa Cruz Biotechnology) and protein-A/G+ agarose (Santa Cruz Biotechnology). Immunocomplexes were resolved using Bis-Tris gels (Invitrogen), transferred to PVDF (Millipore) and developed using Western Lightning (Perkin Elmer). The following antibodies and dilutions were used: anti-MCL-1 (Rockland Immunochemical) 1:10,000, anti-hBCL-2 (Clone 6C8, gift of S. Korsmeyer) and anti-mBCL-2 (Clone 3F11, gift of S. Korsmeyer) 1:3,000, anti-Tim50 (Abcam ab23938) 3μg/ml, anti-Tom40 (Santa Cruz H-300) 4μg/ml. The following antibodies were used at 1:1000 dilution: anti-Prohibitin1 (Abcam ab55618 ), anti-Opa1 (BD Biosciences 612606 clone 18/OPA-1), anti-MnSOD (BD Biosciences 611580 clone 19/MnSOD), anti-PDI (BD Biosciences 610947), anti-hBCL-2 (Clone 6C8, gift of S. Korsmeyer), anti-Complex II SDH (MitoSciences MS204 clone 2E3GC12FB2AE2), anti-Complex IV, anti-Cox-2 (Mitosciences ab110258 clone 12C4F12), anti-Co m p l e x I , a n t i-NDUFA9 (Abcam ab55521), anti-Pyruvate dehydrogenase (Cell Signaling ab67592), anti-Actin mouse (Millipore MAB1501 clone C4), anti-AKT (Cell Signaling 9272), anti-phospho-AKT (Cell Signaling Ser473) Complex II, anti-Fp70 (Molecular Probes 459200 clone 2E3GC12FB2AE2), Complex III, anti-Core 2 (Molecular Probes 459220 clone 13G12AF12BB11); Complex IV, anti-Cox 1 (Abcam ab14705), ATP Synthase, anti-5B (Santa Cruz Biotechnology), ATP Synthase, anti-5F1 (Aviva Systems Biology ARP48188P 050).
Cell Death Experiments
Cell viability was determined by staining with Annexin-V-FITC and propidium iodide (BD Biosciences) and flow cytometry.
RNAi Experiments
MEFs were plated in Opti-MEM media (Invitrogen), transfected with 70nM Stealth siRNAs (Invitrogen) specific for mouse Tom40 and Tim50 using Lipofectamine RNAi-Max (Invitrogen), after 72 hours they were collected, lysed and immunoblotted. Stealth siRNA sequences: TOM40: ACUGAACAACUGGUUGGCUACAGUA, CCCUCUGUAUGAAAUAGUCAUCUUC, GAAGAUGACUAUUUCAUACAGAGGG. TIM50: GCCCUACUACCAGCCACCUUAUACA, UGUAUAAGGUGGCUGGUAGUAGGGC, UCAAGACCAUUGCACUGAACCAAGU.
Measurement of Superoxide Anion
MEFs were treated with 5 μM MitoSOX-red (Invitrogen) for 30 minutes at 37°C and fluorescence measured by flow cytometry.
Isolation and Purification of Mouse Liver Mitochondria
Mitochondria were isolated from mouse livers homogenized in Mito Isolation Buffer (MIB) (200 mM Mannitol, 68 mM Sucrose, 10 mM HEPES-KOH pH 7.4, 10 mM KCl, 1 mM EDTA, 1 mM EGTA. 0.1% BSA) by differential centrifugation and further purified using a 70%/30% Percoll-gradient (Pierce).
Subcellular Fractionation and Isolation of Heavy Membrane (HM) from Cells
Cells were swollen in 220 mM Mannitol, 70 mM Sucrose, 10 mM HEPES-KOH (pH 7.4) and homogenized with a 30-gauge needle. Non-lysed cells were sedimented at 600×g and disposed. The supernatant was centrifuged at 5,500×g and the pellet was the HM fraction. The supernatant was centrifuged at 100,000×g and the soluble fraction is the cytosolic fraction.
Proteolysis of Heavy Membrane (HM) Fraction
HM were treated with the following treatments: proteinase K (0.2 mg/ml), 20 mM KCl (osmotic shock), 1% SDS (membrane solubilization) on ice and precipitated with tricholoroacetic acid (Sigma).
Oxygen Consumption
Respiration was measured 5 hours after 64,000 cells/well were seeded using the XF24 analyzer (Seahorse Bioscience)39. Oxygen consumption rate (OCR) was measured sequentially after addition of 0.5 μM oligomycin, 0.5 μM FCCP, and 0.5 μM rotenone.
Alkali Wash
Mitochondria were sonicated and centrifuged at 10,000×g. The supernatant was centrifuged at 100,000×g and the supernatant is the soluble fraction. The pellet was treated with 0.1 M sodium carbonate (pH 11.5) in MIB buffer on ice and then centrifuged at 100,000×g and supernatant and pellet collected.
Mitochondria Sub-Fractionation
Mitochondria were swollen in 14 ml of 10 mM KH2PO4 (pH 7.4). One-third volume of 1.8 M sucrose, 10 mM MgCl2 was added to shrink the mitochondria. Mitochondria were sonicated and centrifuged at 12,000×g to get supernatant 1 and pellet 1. Supernatant 1 was spun at 100,000×g and the pellet 2 is the OM. Pellet 1 was resuspended in 7 ml of 1.8 M sucrose, 10 mM MgCl2 on ice for 5 minutes and centrifuged at 5,500×g and pellet 3 is the mitoplast.
Cellular ATP
Assays were performed using the ATP Bioluminescence Assay Kit HSII (Roche) following manufacturer's instructions. Luminescence was measured using a Biotek Synergy Luminometer and ATP concentration was determined by comparing values to a standard curve.
Image analysis
Images were captured using a Marianas spinning disk confocal (SDC) imaging system consisting of a Zeiss Axio Observer (Carl Zeiss MicroImaging) inverted microscope with motorized-stage, a CSU-X confocal head (Yokogawa Electric Corporation), LaserStack laser launch and Vector laser scanner head (Intelligent Imaging Innovations-3i) or a Zeiss Plan-Apochromat 63x 1.4 NA oil objective and Evolve 512 EMCCD camera (Photometrics). Fluorescence intensity was measured using Slidebook (Intelligent Imaging Innovations-3i).
Mitochondrial photoactivation fusion assay
Cells were transiently-transfected with a photoactivatable-GFP targeted to the mitochondrial matrix35. A region of interest (~2 μm) of the mitochondria was activated using 405 nm laser and time-lapse images were acquired. Total area of intensity (pixels above threshold) was normalized to the post-activation time point.
Analysis of mitochondrial morphology and shape
Cells were stably-transfected with mito-DsRed (Clontech). A Laplacian 2D-spatial filter was applied to the maximum intensity projection (MIP) to highlight mitochondria and an intensity threshold applied to the images to mask mitochondrial objects. Shape factor consisting of p2/4πA (p=perimeter and A=area) was applied to mitochondria56.
Mitochondrial DNA isolation and quantification
Mcl-1f/f Rosa-ERCreT2 or wt control MEFs were treated with tamoxifen (8 days) to induce deletion and quantification of relative copy number differences was carried out by analyzing the difference in threshold amplification between mtDNA and nuclear DNA as previously described52. DNA was extracted using the DNeasy kit (Qiagen).
Mitochondrial membrane potential and image analysis
Cells were stained with 10 nM tetramethylrhodamine (Invitrogen) in Hank's balanced salt solution (Gibco) at 37°C for 20 minutes. Imaging conditions (laser excitation power at 561 nm, exposure time and camera gain) were identical for all samples. Mean pixel-intensity values of mitochondria in the image were masked by Ridler-Calvard automatic thresholding algorithm.
Transmission electron microscopy
Livers were fixed in 2.5% gluteraldehyde in 0.1 M sodium cacodylate buffer and post-fixed in 2% osmium tetroxide in 0.1 M sodium cacodylate buffer with 0.3% potassium ferrocyanide. Tissue was stained with 4% aqueous uranyl acetate, dehydrated, infiltrated and embedded in epoxy resin. Ultra-thin sections (80 nm) were cut and imaged using a JEOL 1200 electron microscope with an AMT XR 111 camera.
Immuno-electron microscopy
Mitochondrial fractions were fixed for 2 hours using a 4% Paraformaldehyde in 0.1 M Phosphate Buffer, embedded in 12% gelatin, immersed in 2.3 M sucrose, frozen using liquid nitrogen, and cryo-sectioned (85 nm) at temperature of -110°C using a Leica UCT ultramicrotome. The sections were labeled with anti-MCL-1 antibody (Abcam) and 10 nm protein-A gold secondary (Cell Microscopy Center) as described previously57.
Immunofluorescence
Mcl-1-deficient MEFs were transfected with Mcl-1 constructs, labeled with Mitotracker-Red (Invitrogen) for 45 minutes at 37°C, fixed in 4% paraformaldehyde and immunostained with anti-MCL-1 (Rockland) and Alexa488 anti-rabbit IgG (Invitrogen). Images were collected using Leica DM IRBE microscope equipped with a Leica TCS Confocal System using a 63X magnification.
Enzymatic activity of electron transport chain components
The activity of NADH-ubiquinone oxidoreductase (complex I) was assayed as previously described58. Succinate-ubiquinone oxidoreductase (complex II) activity was assayed by measuring succinate dependent reduction in DCIP, mediated by CoQ1. The reaction was followed by recording absorbance at 600 nm following succinate addition59. Cytochrome c oxidase activity (complex IV) was assayed by measuring oxidation of cytochrome c at 550 nm and calculated using the extinction coefficient of 19.6 mM-1cm-1as previously described60. Pyruvate dehydrogenase (PDH) activity was assayed by measuring tpyruvate-dependent reduction of INT, mediated by PMS, as previously described61.
Isolation of respiratory complexes and supercomplexes
Mitochondria were isolated as described previously62. Isolation of the complexes and supercomplexes was completed as previously described40.
Blue native polyacrylamide gel electrophoresis (BN-PAGE)
Mitochondrial lysates (20 μg) were resolved on 3-12% gradient BN-PAGE gel (Invitrogen) as previously described40. The gel was transferred onto PVDF and immunoblotted with indicated antibodies.
λ-Phosphatase Treatment
WCLs were incubated with λ-protein phosphatase (New England Biolabs) for 30 minutes at 30°C. Samples were resolved and immunoblotted with anti-MCL-1 (Rockland), anti-Phospho-AKT(Ser473) (Cell Signaling), anti-AKT (Cell Signaling) and anti-Actin (Millipore).
Edman Sequencing of MCL-1
MCL-1 protein was immunoprecipitated, resolved, and transferred to PVDF membrane. Coomassie-stained membrane was submitted to the W.M. Keck Foundation Biotechnology Resource Laboratory (Yale University) for amino-terminal sequencing.
Statistical Tests
Statistical significance was calculated using the two-tailed student t-test; p-values are denoted in figure legends.
Supplementary Material
Acknowledgments
We thank the St. Jude Cell and Tissue Imaging Facility for assistance on live cell imaging and S. Frase for assistance on electron micrographs; the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University for Edman sequencing; B. Xia, E. Parganas and D. Gable for technical assistance; C. Shaner for animal husbandry; members of the St. Jude Biochemistry Department, S. Oakes, J. Ihle, and C. Sherr for helpful discussions. J.T.O is supported by the Pew Scholars Program in the Biomedical Sciences; the National Institutes of Health HL-102175; the American Cancer Society RSG-10-255-01-LIB; a Cancer Center Support Grant P30CA021765; and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital.
Footnotes
Author Contributions
R.M.P. and J.T.O. conceived the study, designed the experiments, and wrote the manuscript. R.M.P. performed the experiments, analyzed data, and prepared figures. D.P.S., B.K. and M.B generated reagents and performed experiments. J.L. and J.D.S. carried out electron transport chain enzymatic assays. J.T. assisted in imaging data acquisition and quantification of mitochondrial morphology. M.M.C. and R.J.Y. carried out pilot experiments on mitochondrial morphology and provided reagents. S.P. assisted in acquisition of immunofluorescence images. S.M. and D.R.G. carried out experiments on oxygen consumption and provided reagents. J.T.O. supervised the project.
References
- 1.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 2.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–56. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 3.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 4.Wei MC, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–30. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat Cell Biol. 2000;2:156–62. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- 6.Cheng EH, et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8:705–11. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 7.Zong WX, Lindsten T, Ross AJ, MacGregor GR, Thompson CB. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 2001;15:1481–6. doi: 10.1101/gad.897601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey TG, Mannella CA. The internal structure of mitochondria. Trends Biochem Sci. 2000;25:319–24. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- 9.Chan DC. Mitochondrial dynamics in disease. N Engl J Med. 2007;356:1707–9. doi: 10.1056/NEJMp078040. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet. 2005;14:R283–9. doi: 10.1093/hmg/ddi270. Spec No. 2. [DOI] [PubMed] [Google Scholar]
- 11.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009;106:11960–5. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bleazard W, et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinkenberger JL, Horning S, Klocke B, Roth K, Korsmeyer SJ. Mcl-1 deficiency results in peri-implantation embryonic lethality. Genes Dev. 2000;14:23–7. [PMC free article] [PubMed] [Google Scholar]
- 15.Opferman JT, et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–6. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 16.Dzhagalov I, Dunkle A, He YW. The anti-apoptotic Bcl-2 family member Mcl-1 promotes T lymphocyte survival at multiple stages. J Immunol. 2008;181:521–8. doi: 10.4049/jimmunol.181.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opferman JT, et al. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 2005;307:1101–4. doi: 10.1126/science.1106114. [DOI] [PubMed] [Google Scholar]
- 18.Dzhagalov I, St John A, He YW. The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood. 2007;109:1620–6. doi: 10.1182/blood-2006-03-013771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steimer DA, et al. Selective roles for antiapoptotic MCL-1 during granulocyte development and macrophage effector function. Blood. 2009;113:2805–15. doi: 10.1182/blood-2008-05-159145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arbour N, et al. Mcl-1 is a key regulator of apoptosis during CNS development and after DNA damage. J Neurosci. 2008;28:6068–78. doi: 10.1523/JNEUROSCI.4940-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei G, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–42. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Wuilleme-Toumi S, et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia. 2005;19:1248–52. doi: 10.1038/sj.leu.2403784. [DOI] [PubMed] [Google Scholar]
- 24.Vogel F, Bornhovd C, Neupert W, Reichert AS. Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol. 2006;175:237–47. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilkerson RW, Selker JM, Capaldi RA. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 2003;546:355–8. doi: 10.1016/s0014-5793(03)00633-1. [DOI] [PubMed] [Google Scholar]
- 26.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–60. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene. 2004;23:5301–15. doi: 10.1038/sj.onc.1207692. [DOI] [PubMed] [Google Scholar]
- 28.Morel C, Carlson SM, White FM, Davis RJ. Mcl-1 integrates the opposing actions of signaling pathways that mediate survival and apoptosis. Mol Cell Biol. 2009;29:3845–52. doi: 10.1128/MCB.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warr MR, Shore GC. Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr Mol Med. 2008;8:138–47. doi: 10.2174/156652408783769580. [DOI] [PubMed] [Google Scholar]
- 30.Kojima S, Hyakutake A, Koshikawa N, Nakagawara A, Takenaga K. MCL-1V, a novel mouse antiapoptotic MCL-1 variant, generated by RNA splicing at a non-canonical splicing pair. Biochem Biophys Res Commun. 2010;391:492–7. doi: 10.1016/j.bbrc.2009.11.086. [DOI] [PubMed] [Google Scholar]
- 31.De Biasio A, et al. N-terminal truncation of antiapoptotic MCL1, but not G2/M-induced phosphorylation, is associated with stabilization and abundant expression in tumor cells. J Biol Chem. 2007;282:23919–36. doi: 10.1074/jbc.M700938200. [DOI] [PubMed] [Google Scholar]
- 32.Huang CR, Yang-Yen HF. The fast-mobility isoform of mouse Mcl-1 is a mitochondrial matrix-localized protein with attenuated anti-apoptotic activity. FEBS Lett. 2010;584:3323–30. doi: 10.1016/j.febslet.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–67. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 34.Pfanner N, Muller HK, Harmey MA, Neupert W. Mitochondrial protein import: involvement of the mature part of a cleavable precursor protein in the binding to receptor sites. Embo J. 1987;6:3449–54. doi: 10.1002/j.1460-2075.1987.tb02668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karbowski M, et al. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol. 2004;164:493–9. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–8. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 37.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–76. [PubMed] [Google Scholar]
- 38.Rossignol R, et al. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–93. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 39.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–74. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–39. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 41.Genova ML, et al. Is supercomplex organization of the respiratory chain required for optimal electron transfer activity? Biochim Biophys Acta. 2008;1777:740–6. doi: 10.1016/j.bbabio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Diaz F, Fukui H, Garcia S, Moraes CT. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol Cell Biol. 2006;26:4872–81. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krause F, Reifschneider NH, Goto S, Dencher NA. Active oligomeric ATP synthases in mammalian mitochondria. Biochem Biophys Res Commun. 2005;329:583–90. doi: 10.1016/j.bbrc.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Giraud MF, et al. Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochim Biophys Acta. 2002;1555:174–80. doi: 10.1016/s0005-2728(02)00274-8. [DOI] [PubMed] [Google Scholar]
- 45.Thomas D, et al. Supramolecular organization of the yeast F1Fo-ATP synthase. Biol Cell. 2008;100:591–601. doi: 10.1042/BC20080022. [DOI] [PubMed] [Google Scholar]
- 46.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–98. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paumard P, et al. The ATP synthase is involved in generating mitochondrial cristae morphology. Embo J. 2002;21:221–30. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bornhovd C, Vogel F, Neupert W, Reichert AS. Mitochondrial membrane potential is dependent on the oligomeric state of F1F0-ATP synthase supracomplexes. J Biol Chem. 2006;281:13990–8. doi: 10.1074/jbc.M512334200. [DOI] [PubMed] [Google Scholar]
- 49.Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci U S A. 1993;90:3516–20. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krajewski S, et al. Immunohistochemical analysis of Mcl-1 protein in human tissues. Differential regulation of Mcl-1 and Bcl-2 protein production suggests a unique role for Mcl-1 in control of programmed cell death in vivo. Am J Pathol. 1995;146:1309–19. [PMC free article] [PubMed] [Google Scholar]
- 51.Yang T, Kozopas KM, Craig RW. The intracellular distribution and pattern of expression of Mcl-1 overlap with, but are not identical to, those of Bcl-2. J Cell Biol. 1995;128:1173–84. doi: 10.1083/jcb.128.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–9. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velours J, Dautant A, Salin B, Sagot I, Brethes D. Mitochondrial F1F0- ATP synthase and organellar internal architecture. Int J Biochem Cell Biol. 2009;41:1783–9. doi: 10.1016/j.biocel.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart DP, et al. Ubiquitin-independent degradation of antiapoptotic MCL-1. Mol Cell Biol. 2010;30:3099–110. doi: 10.1128/MCB.01266-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koopman WJ, et al. Inhibition of complex I of the electron transport chain causes O2-. -mediated mitochondrial outgrowth. Am J Physiol Cell Physiol. 2005;288:C1440–50. doi: 10.1152/ajpcell.00607.2004. [DOI] [PubMed] [Google Scholar]
- 57.Peters PJ, Bos E, Griekspoor A. Cryo-immunogold electron microscopy. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0407s30. Chapter 4, Unit 4 7. [DOI] [PubMed] [Google Scholar]
- 58.Frost MT, Wang Q, Moncada S, Singer M. Hypoxia accelerates nitric oxide-dependent inhibition of mitochondrial complex I in activated macrophages. Am J Physiol Regul Integr Comp Physiol. 2005;288:R394–400. doi: 10.1152/ajpregu.00504.2004. [DOI] [PubMed] [Google Scholar]
- 59.Miyadera H, et al. Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proc Natl Acad Sci U S A. 2003;100:473–7. doi: 10.1073/pnas.0237315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miro O, et al. Cytochrome c oxidase assay in minute amounts of human skeletal muscle using single wavelength spectrophotometers. J Neurosci Methods. 1998;80:107–11. doi: 10.1016/s0165-0270(97)00204-5. [DOI] [PubMed] [Google Scholar]
- 61.Hinman LM, Blass JP. An NADH-linked spectrophotometric assay for pyruvate dehydrogenase complex in crude tissue homogenates. J Biol Chem. 1981;256:6583–6. [PubMed] [Google Scholar]
- 62.Fernandez-Vizarra E, Lopez-Perez MJ, Enriquez JA. Isolation of biogenetically competent mitochondria from mammalian tissues and cultured cells. Methods. 2002;26:292–7. doi: 10.1016/S1046-2023(02)00034-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.