Abstract
The atmospheric dispersion of bacteria over long distances is an important facet of microbial ecology. Certain groups of dispersed bacteria can adapt to their new location and affect established ecosystems. Aeolian dust particles are known to be carriers of microbes but further research is needed to expand our understanding of this field of microbiology. Here we showed the potential of aeolian dust to global migration of bacterial cells. We demonstrated the presence of microbial cells on dust particles directly by bio-imaging. Bacterial abundance on dust particles declined from 105 to less than 103 cells/m3 as the dust event subsided. Taxonomically diverse bacteria were identified by 16S rRNA gene sequencing and some of these bacteria retained growth potential. Our results confirm that bacteria can attach to aeolian dust particles and they have the potential to migrate globally during dust events and thus can contribute to the diversity of downwind ecosystems.
Atmospheric dispersion of bacteria over long distances is an important facet of microbial ecology. Certain species of dispersed bacteria adapt to their new location by reaching preferred environments, with potential effects on established ecosystems. A fundamental paradigm in microbial ecology states that “Everything is everywhere, but the environment selects”1,2; conversely, extreme environmental conditions inhibit wider geographic distribution of dispersed bacteria3,4.
Migration of bacteria is a natural phenomenon promoted by ocean currents and atmospheric events. Aeolian dust, which consists of soil particles originating from deserts or arid regions, is generated from wind erosion of the regolith. Desert dust is transported, sometimes over long distances, on air currents. Microbes present on aeolian dust particles may impact on downwind ecosystems and they are often thought to be detrimental to human health5,6,7,8,9. To demonstrate that aeolian dust particles carry microbial cells, microbial cells on the dust particles should first be visualized. Microbial abundance and community structure on those particles should then be determined.
Major aeolian dust events arise from the Sahara and Sahel deserts (“African dust”), the Australian deserts (“Australian dust”), and the Taklamakan and Gobi deserts and the Loess plateau (“Asian dust”). Over one million tons of Asian dust particles are estimated to fall-out onto Japan each year10, 3,000–5,000 km away from their source regions. It is well-known for Asian desert dust particles to be transported long distances11,12 and to reach the North American Continent (more than 15,000 km distant13,14), and oceanic deposition encourages phytoplankton growth in the North Pacific Ocean by natural iron fertilization15. Asian dust particles can sometimes be transported globally16 in 13 days and have been identified in ice and snow cores of Greenland17 and the French Alps18.
Here we demonstrated that bacteria attach to aeolian dust particles and they have potential to migrate globally during dust events, thus can contribute to the diversity of downwind ecosystems. We first confirmed the presence of microbial cells on dust particles by direct bio-imaging visualization. We then measured the change in bacterial abundance throughout the dust event, and determined the potential for physiological activities of the bacterial cells and their community structure, using culture-independent methods.
Results
Physicochemical characteristics of collected Asian dust particles
Dust particles (Fig. 1a) were collected during a severe dust event (from 12 November 2010, [severe conditions, visibility less than 10 km, supplementary Fig. S1a] through 16 November 2010, [the event was terminated by rain on 15 November 2010, visibility 25 km]) from more than 10,000 liters of air over the Japan Sea (10 km from coasts) at an altitude of 900 meters, using a newly fabricated dust sampler which adsorbs dust particles onto the surface of wet beads. The source of these dust particles was estimated to be the Gobi desert (Supplementary Fig. S1b).
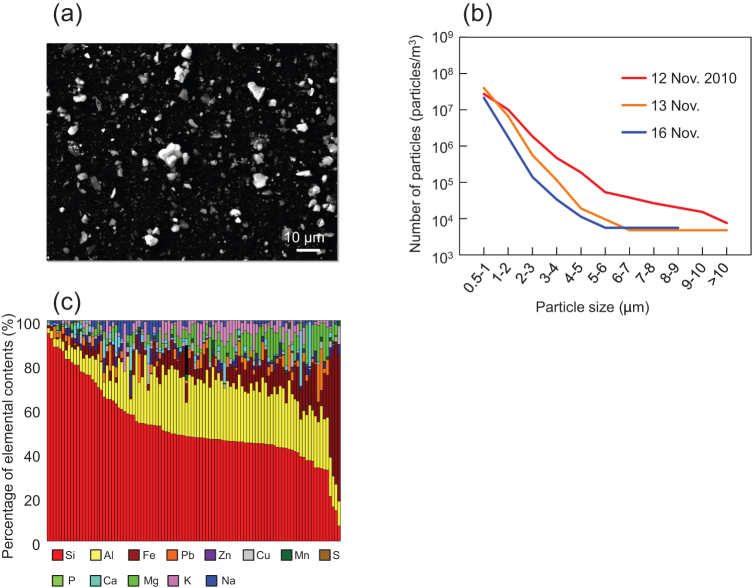
Figure 1.
(a) Scanning electron micrograph of Asian dust particles collected on 12 Nov. 2010. (b) Size distribution of Asian dust particles collected on 12 Nov. 2010. Size of each dust particle was determined by image analysis of scanning electron micrographs and 20,000 to 100,000 particles were analyzed in each sample. (c) Elemental composition of Asian dust particles (size: 1 to 5 μm) determined by energy dispersive X-ray analysis. 100 dust particles collected on 12 Nov. 2010 were analyzed. Each bar represents one of the 100 dust particles.
Particle size distribution was determined by scanning electron microscopy. The proportion of dust particles exceeding 1 μm increased during the severe Asian dust event, while most particles were less than 1 μm as the event ended (Fig. 1b). The major components of particles 1 to 5 μm in diameter were determined by scanning electron microscopy with energy dispersive X-ray (SEM-EDX) analysis19,20 to confirm that the particles were truly aeolian dust particles and not suspended particulate matter. From the SEM-EDX profile, silicon and aluminum were identified as the major components of collected dust particles. Significant quantities of iron, calcium and magnesium were also present, consistent with soil particle composition19,20 (Fig. 1c).
These results suggest that a portion of Asian dust particles could be sufficiently large to be efficient carriers of bacteria and that those larger dust particles can reach down-wind regions several thousand kilometers away from their source in severe events.
Visualization of microbial cells on Asian dust particles and change in bacterial abundance on the particles during the dust event
Microbial cells on collected Asian dust particles were visualized using a laser scanning microscope equipped with a microspectrophotometer, following fluorescent nucleic acid staining to distinguish microbial cells from dust particles based on their fluorescence21 (Supplementary Fig. S2, S3 and S4, Fig. 2a–c). Microbial cells were attached to particles exceeding 1 μm in size (Fig. 2c). Of 8,500 Asian dust particles collected on 12 Nov. 2010 at the start of the severe event, large particles (> 5 μm) harbored 55% of detected microbial cells, intermediate size particles (2–5 μm) supported 38% of the cells, while particles of 1–2 μm supported only 7% of the cells. No microbial cells were attached to small particles (< 1 μm) (Supplementary Fig. S5) while the detection limit of our laser scanning microscopy is 0.3 μm, and bacteria on the bottom side of particle could not be observed. We also investigated Asian dust particles collected on 16 Nov. 2010, immediately following the dust event, and found few microbial cells on any of the dust particles. The number of microbial cells occupying Asian dust particles was inferred to decrease as the dust event weakened.
Figure 2. Selective detection of microbial cells on Asian dust particles (collected on 12 Nov. 2010) by laser scanning microscopy following fluorescent nucleic acid staining.
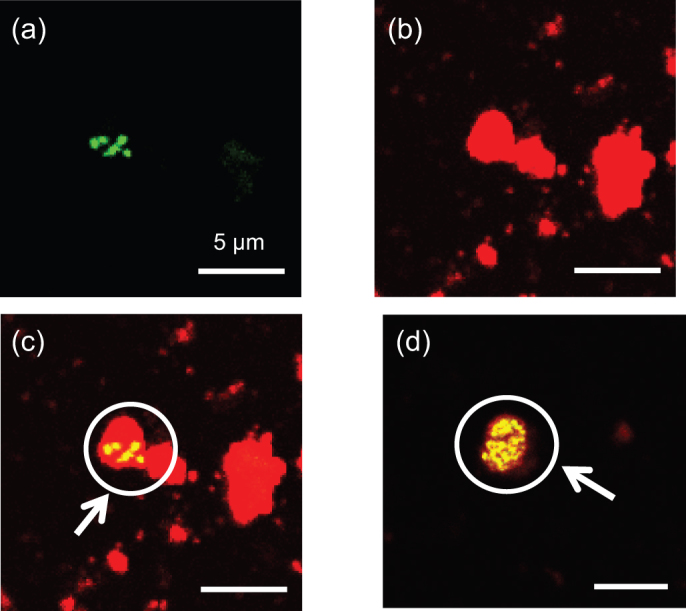
(a) Microbial cells on Asian dust particles. (b) Asian dust particles. (c) Composite image of a) and b). (d) Microcolony of microbial cells formed on an Asian dust particle. Microbial cells fluoresce green/yellow, while particles fluoresce red.
Bacterial abundance on collected dust particles, determined by quantitative real-time PCR targeting eubacterial 16S rRNA gene sequences, declined dramatically to less than 1% of their starting values (from 104 – 105 to less than 103 cells/m3) as the dust event subsided (Supplementary Table S1).
We determined bacterial abundance on Asian dust particles by two different methods: laser scanning microscopy and quantitative PCR, and the results obtained by these methods were similar (Supplementary Fig. S5 and Supplementary Table S1). These data support that Asian dust carries bacterial cells to down-wind areas.
Diversity of bacteria carried with Asian dust
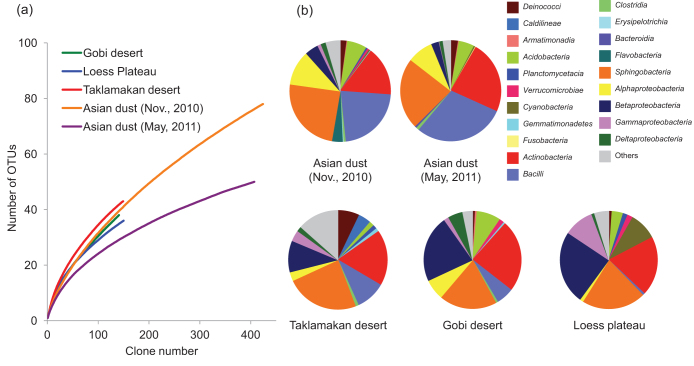
The bacterial community structure on Asian dust particles was analyzed by sequencing nearly the full length of the 16S rRNA gene of 480 clones obtained from Asian dust particles collected on 12 Nov. 2010 and 2 May 2011. Rarefaction curves revealed class richness in the dust source soil and Asian dust (Fig. 3a). Sequences were affiliated with more than 20 bacterial classes, of which Actinobacteria, Bacilli and Sphingobacteria dominated (Fig. 3b). Previous studies reported that these bacteria were often found in African dust5,22 or Asian dust samples23. Since little is known about the source bacterial community structure of Asian dust particles, the bacterial communities of the samples were compared with those of their putative source environments (Taklamakan and Gobi deserts and Loess plateau). A similar diversity of bacterial groups was found in the source regions as in the Asian dust particles (Fig. 3b) and other arid regions24. These results indicate that the diversity of bacterial communities carried by Asian dust reflects the bacterial diversity in dust source regions.
Figure 3. Rarefaction curves of class richness (a) and taxonomic affiliation and relative abundance of the most common bacterial groups (b) in Asian dust and dust source soil samples.
Growth potential of bacterial cells on Asian dust particles
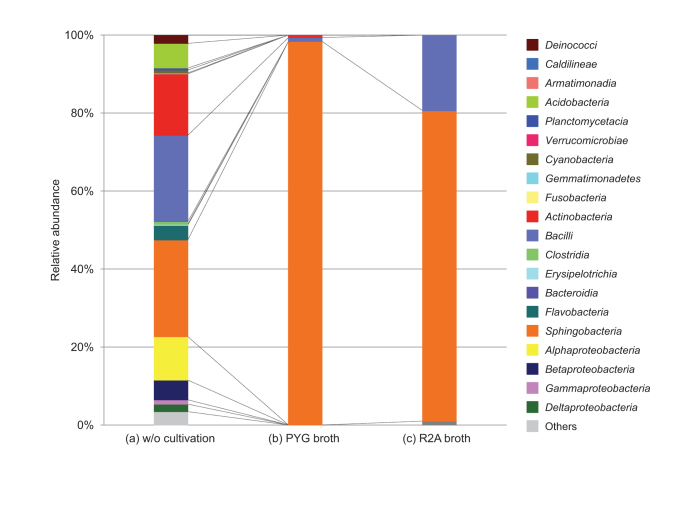
We incubated the dust particles in both nutrient-poor and nutrient-rich liquid media and phylogenetically analyzed the proliferated cells. This experiment revealed that Bacilli and Sphingobacteria maintain the potential for physiological activity during long distance transportation on Asian dust particles (Fig. 4b and 4c). We also plated Asian dust particles on agar media and incubated them for 48 hours. Some microbes on Asian dust particles proliferated into microcolonies (Fig. 2d and Supplementary Fig. S6), suggesting that taxonomically diverse bacteria are not only transported long distances by Asian dust events, but that some groups remain potentially physiologically active.
Figure 4. Bacterial community composition of Asian dust particles collected on 12 Nov. 2010, as determined by bacterial DNA analysis.
(a) following direct extraction. (b) and (c) following cultivation in specified media.
Discussion
In this study, we used bio-imaging with laser scanning microscopy and quantitative PCR of bacterial 16S rRNA genes to demonstrate that bacterial cells can be transported several thousand kilometers on Asian dust particles. We also confirmed the phylogenetic diversity of these bacteria, and that some groups retain growth potential despite such long-distance transportation.
Bacterial populations on aeolian dust particles are considered likely to impact natural environments and human health5,6,7,8,9. The abundance of bacteria moving with dust particles (104 – 105 cells/m3) is equivalent to or less than bacterial abundance in the air of our typical non-dust environment (105 cells/m3)25, and their diversity is similar to that of the soil in the dust source regions (Fig. 3). Dominant bacterial classes found on Asian dust particles are commonly found in natural environments. Actinobacteria inhabit extreme environments such as hypersaline lakes, thermal springs, and arid soils. Bacilli include spore-forming bacterial genera, while Sphingobacteria are commonly found in soil and aquatic environments. We may therefore conclude that the bacteria carried with Asian dust probably rarely affect human health, based on their abundance and phylogenetic composition, although they constitute a possible source of opportunistic infection, and their potential as allergens must also be considered.
On the other hand, these phylogenetically diverse bacteria could affect established ecosystems. Most bacteria transported by aeolian dust will be stressed by atmospheric transport (UV exposure, reduced nutrient availability, etc.). Some of them, however, will adapt favorable environments, where they retain characteristics which may render them highly resilient to competition in their new habitat. Transported bacteria are therefore a potential driving force of bacterial diversity maintenance or enhancement in these habitats.
Bacterial genes are recognized as important resources. Fair and equitable sharing of the benefits arising from international genetic resource use was proposed at the tenth meeting of the Conference of the Parties to the Convention on Biological Diversity (COP 10) held in 201026. Bacteria are transported worldwide during aeolian dust events and the genes of these transported bacteria may be transferred to indigenous bacteria, introducing new traits and expanding gene diversity within bacterial communities. Bacteria found in a given geographic area are likely to be found elsewhere27, and aeolian dust has an important role in global bacterial inoculation.
Asian dust particles have been transported from deserts or arid regions in China and Mongolia to Korea, Japan, and the Pacific islands in geologic time scale, and they have carried phylogenetically and functionally diverse bacterial groups. Aeolian dust events may contribute to global migration of bacterial cells and their genes, and therefore can be important sources of bacterial diversity in the earth's ecosystem.
Methods
Collection of Asian dust particles
The occurrence of atmospheric Asian dust and its drift were confirmed by LIDAR (Light Detection and Ranging) data from the Ministry of the Environment, Japan (http://www-gis5.nies.go.jp/eastasia/DustLider.php), Asian dust information from Japan Meteorological Agency (http://www.jma.go.jp/jp/kosa/index.html) and direct calling to aviation weather service centers. The geographic origin of Asian dust was determined by the backtrajectory analysis (http://ready.arl.noaa.gov/HYSPLIT.php; Supplementary Fig. S1b).
Asian dust particles were collected with a sampler set from a small airplane. Air outside of the airplane was collected in the sampler, which consisted of wet glass beads in a stainless can (volume: 2 liter) and a sterilized Teflon inlet tube. The inlet was inserted into the air intake, which opens at the leading edge of a small airplane, to avoid boundary effects. Asian dust particles in the air were adsorbed on the surface of the wet glass beads. To prepare the sampler, glass beads were washed with particle-free water (commercial distilled water for injection) in an ultrasonic washer and sterilized in dry oven (180°C, 2 hours × twice). The stainless can was washed with particle-free water and sterilized in dry oven (180°C, 2 hours × twice). Teflon tubes were washed with particle-free water and sterilized by autoclaving.
Asian dust particles were collected on 12 November 2010 (severe conditions, visibility: less than 10 km), 13 November 2010 and 16 November 2010 (end of the event, visibility: 25 km). Asian dust particles were also collected on 2 May 2011 when a severe dust event occurred. The airplane took off from Yao Airport, Osaka, Japan and started collection of Asian dust particles in the air over the Japan Sea (10 km from coasts) at an altitude of 900 meters (latitude: N 35° 42.004', longitude: 134° 52.134'). The airplane route was westbound to near Akasaki (latitude: N 35° 34.187', longitude: 133° 41.870'), then eastbound and collected Asian dust particles along this out and inbound flight path. Asian dust particles from a total of more than 10,000 liters of the air were collected for this study.
During sampling, we recorded the time, altitude, longitude and latitude, visibility, aircraft heading and course, airspeed and ground speed, and we confirmed that there was no wind from land during sampling. In addition, the numbers of particles in the atmosphere were measured on-site with a particle counter (KR-12A, Rion, Tokyo, Japan).
Dust particles collected on wet beads were suspended in 2.5 L of particle-free water and used for the following experiments.
Collection of soil in dust source regions
Soil samples were collected in three dust source regions: (i) Gobi desert (site 1: latitude: N 40° 21.259', longitude: E 109° 23.581'; site 2: latitude: N 40° 28.309', longitude: E 108° 39.338'; site 3: latitude: N 40° 24.337', longitude: E 108° 36.287') on 22 July 2011, (ii) Loess Plateau (site 1: latitude: N 35° 35.450', longitude: E 109° 10.074'; site 2: latitude: N 35° 42.324', longitude: E 109° 25.390'; site 3: latitude: N 35° 42.279', E longitude: 109° 25.450') on 30 June 2009, and (iii) Taklamakan desert (site 1: latitude: N 40° 22.598', longitude: E 84° 19374'; site 2: latitude: N 40° 20.230', longitude: E 84° 19285'; site 3: latitude: N 40° 09.290', longitude: E 84° 19537') on 3 March 2008. These samples were analyzed within 72 hours after sampling.
Determination of size distribution and elemental composition of collected Asian dust particles
Dust particles (50 ml of the 2.5 L suspension) were trapped onto a sterilized 0.4 µm-pore size polycarbonate membrane filter and sputter coated with evaporated gold.
Samples were observed under a scanning electron microscope (JSM-5610L; JEOL, Tokyo, Japan) using high vacuum mode. Electron micrographs were obtained at magnifications of × 1,000 using a backscatter electron detector.
Elemental composition of Asian dust particles were analyzed under a scanning electron microscope (JSM-5500LV; JEOL) equipped with EDX (JED-2200; JEOL) operating at 30 kV. For the particle size distribution, image processing was carried out by automated particle size distribution analysis software28.
To determine the size distribution and elemental composition of collected dust particles accurately, particles of more than 0.5 µm were utilized and analyzed.
Visualization of microbial cells on Asian dust particles
Dust particles (100 ml of the 2.5 L suspension) were fixed with 4% (w/v) paraformaldehyde at 4°C for 16 h. After fixation, 10 ml-aliquots were filtered through a sterilized 0.4 µm-pore size polycarbonate membrane filter and rinsed twice with particle-free water. The filters were then transferred to a microtube and dehydrated in an ethanol series (50%, 80% and 100 % ethanol for 3 min each) and dried under vacuum. The filters were stained with nucleic acid-staining dye, SYBR Green I (Invitrogen, Carlsbad, USA; 1/10,000-dilution of the supplied product) which can be used to distinguish cells from non-biological backgrounds21, containing 2% Tween 20 for 10 min. After washing with particle-free water (10 min × three), the filters were mounted in immersion oil for observation by laser scanning microscopy (TCS-SP5, Leica Microsystems, Wetzlar, Germany).
To discriminate microbial cells from dust particles, a microscope with a microspectrophotometer was used. Escherichia coli W3110 cells inoculated in sterilized soil (collected in Loess Plateau, Shaanxi, China) as well as Asian dust particles collected in Beijing, China, were used to determine the protocol to distinguish microbial cells from dust particles based on their fluorescence (Supplementary Fig. S2 and S3).
All particles were observed under blue excitation (wavelength 480 nm) and particles with green fluorescence (wavelength 500 – 540 nm) were enumerated as microbial cells; those with yellow to red fluorescence (wavelength 550 – 680 nm) were enumerated as non-biological particles such as soil particles.
In this analysis, at least 200 different microscopic fields were observed for each sample; more than 6,000 particles were observed.
Visualization of microcolony-forming microbial cells on Asian dust particles
Dust particles (10 ml of the 2.5 L suspension) were filtered through a sterilized 0.4 µm-pore size polycarbonate membrane filter. The filter was then removed immediately from the funnel and placed on standard agar or R2A agar media containing 20 µg/ml pimaricin (fungicide). After 48 h of incubation at 25°C, microbial cells on the filters were fixed on a filter paper soaked with 4% (w/v) paraformaldehyde at 4°C for 16 h. After fixation, the filters were washed with particle-free water, dehydrated in an ethanol series and dried under vacuum. Microbial cells on the filter were then fluorescently stained and observed by a laser scanning microscope equipped with a microspectrophotometer, as described above.
Direct DNA extraction
Dust particles (2.0 L of the 2.5 L suspension) were filtered onto a 0.4 µm-pore size sterilized polycarbonate membrane filter. DNA was extracted and purified by the methods reported by Tsai and Olson29. DNA of bacteria in the soil samples collected from the various dust source regions was extracted by FastDNA SPIN Kit for Soil (MP-Biomedicals, Solon, OH, USA).
Cultivation of bacteria in aeolian dust and DNA extraction
Dust particles (1 ml of the 2.5 L suspension) were inoculated into 9 ml of nutrient rich PYG broth (peptone 2 g, yeast extract 0.2 g, glucose 0.5 g per 100 ml water) or nutrient poor R2A broth. Samples were incubated at 25°C for 7 days with slow shaking. Bacterial DNA was extracted and purified using Wizard Genomic DNA Purification Kit (Promega, Madison, USA) according to the manufacturer's instructions.
Random cloning
Nearly the full-length 16S rRNA gene sequences of the bacterial domain were amplified by PCR using the universal primers 8f (5′-AGAGTTTGATCCTGGCTCAG-3′)30 and 1492r (5′-TACCTTGTTACGACTT-3′)31. PCR amplification was performed with the reagents supplied with TaKaRa LA Taq Hot Start Version (Takara Bio, Shiga, Japan). The PCR mixture, containing 5 U of TaKaRa LA Taq, 0.5 μM of each primer, 16 μl of a 2.5 mM concentration of each deoxyribonucleoside triphosphate, 10 μl of 10×PCR buffer II (containing Mg2+) and 1 µl of 2.5 mg/ml 8-methoxypsoralen (dissolved in dimethyl sulfoxide) was made up to 99 μl with DNA-free water. A DNA suspension was added last in a 1 µl volume after irradiation of the PCR mixture with UV light32. After an initial denaturing step at 94°C for 1 min, 30 cycles were performed as follows: denaturing at 94°C for 1 min, annealing at 42°C for 1 min, and extension at 72°C for 2 min. The final extension step was at 72°C for 10 min. Construction of 16S rRNA gene clone libraries and sequencing were performed at Dragon Genomics Center (Takara Bio, Mie, Japan). Insert 16S rRNA gene fragments of randomly selected 480 clones from directly extracted DNA and 960 clones from extracted DNA of bacteria cultured in PYG and R2A liquid media (480 clones each) were sequenced. Fifty clones obtained from each dust source soil sample were also sequenced (total 150 clones per each dust source region). No clone was obtained when DNA suspension was not added. Sequences were analyzed by the ribosomal database project33. Rarefaction curves were constructed with DOTUR at 85% similarity34.
Quantitative real time PCR
For determination of bacterial abundance, 16S rRNA gene occurrence was quantified by real time PCR with a LightCycler (Roche Diagnostics, Mannheim, Germany). Real time PCR was performed according to the procedure reported by Nishimura et al.35.
To determine the rate of recovery of DNA during extraction, known amounts of PCR products of the luciferase gene (luc) were inoculated into the samples as an internal standard and quantified after DNA extraction according to Nishimura et al.35. The DNA recovery rate was calculated by comparing the copy number of the inoculated luc gene before and after DNA extraction. The copy number of the 16S rRNA gene quantified by real time PCR was calibrated based on the recovery rate.
Author Contributions
N. Y., T. I. and T. B. contributed equally to this work. M. N. conceived the study; N. Y., T. I. and T. B. collected samples; T. I. performed quantitative PCR and phylogenetic analysis; A. S. performed SEM-EDX analysis; T. B. performed scanning electron microscopy and laser scanning microscopy; N. Y. and M. N. wrote the paper; all authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary Figures and Table
Acknowledgments
This research was supported by the Environment Research and Technology Development Fund of the Ministry of the Environment, Japan (B-0902). We thank the following for technical assistance: Katsuji Tani and Takehiko Kenzaka at Osaka Ohtani University; Pin Fang Li at China Agricultural University; Kanami Enoki, Yuka Himezawa and Makoto Saraya at Osaka University. We used the systems of Genome Information Research Center, Osaka University for bacterial community analysis. Collection of Asian dust particles by a small airplane was supported by First Flying Co. Ltd.
Nucleotide sequence accession numbers: Sequences were deposited with DNA Data Bank of Japan (DDBJ) and assigned accession numbers were AB636927 to AB637350 (samples without cultivation), AB637351 to AB637760 (samples incubated with PYG broth) and AB637761 to AB638212 (samples incubated with R2A broth) for samples collected on November 2010. Assigned accession numbers were AB695733 to AB696139 (samples collected on 2 May 2011), AB696140 to AB696280 (Gobi desert samples), AB696281 to AB696430 (Loess Plateau samples) and AB696431 to AB696579 (Taklamakan desert samples).
References
- Beijerinck M. W. De infusies en de ontdekking der backteriën, Jaarboek van de Koninklijke Akademie v. Wetenschappen. Müller, Amsterdam, The Netherlands. (1913).
- Baas-Becking L. G. M. Geobiologie of Inleiding Tot de Milieukunde. W. P. van Stockum & Zoon N. V., The Hague, The Netherlands. (1934).
- Finlay B. J. Global dispersal of free-living microbial eukaryote species. Science 296, 1061–1063 (2002). [DOI] [PubMed] [Google Scholar]
- Fenchel T. & Finlay B. J. The ubiquity of small species: patterns of local and global diversity. Bioscience 54, 777–784 (2004). [Google Scholar]
- Griffin D. W. et al. African desert dust in the Caribbean atmosphere: Microbiology and public health. Aerobiologia 17, 203–213 (2001). [Google Scholar]
- Griffin D. W. et al. Atmospheric microbiology in the northern Caribbean during African dust events. Aerobiologia 19, 143–157 (2003). [Google Scholar]
- Prospero J. M. et al. Interhemispheric transport of viable fungi and bacteria from Africa to the Caribbean with soil dust. Aerobiologia 21, 1–19 (2005). [Google Scholar]
- Griffin D. W. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin Microbiol Rev. 20, 459–477 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas A. et al. Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environ. Microbiol. 11, 1612–1623 (2009). [DOI] [PubMed] [Google Scholar]
- National Institute for Environmental Studies. http://www.nies.go.jp/kanko/kankyogi/08/05.html, in Japanese.
- Iwasaka Y. et al. The transport and spacial scale of Asian dust-storm clouds: a case study of the dust-storm event of April 1979. Tellus 35B, 189–196 (1983). [Google Scholar]
- Iwasaka Y. et al. The transport of Asia dust (KOSA) particles; importance of weak KOSA events on the geochemical cycle of soil particles. Tellus 40B, 494–503 (1988). [Google Scholar]
- Duce R. A. et al. Long-range atmospheric transport of soil dust from Asia to the tropical North Pacific: temporal variability. Science 209, 1522–1524 (1980). [DOI] [PubMed] [Google Scholar]
- Kellogg C. A. & Griffin D. W. Aerobiology and the global transport of desert dust. Trends Ecol. Evol. 21, 638–644 (2006). [DOI] [PubMed] [Google Scholar]
- Bishop J. K. B. et al. Robotic observations of dust storm enhancement of carbon biomass in the North pacific. Science 298, 817–821 (2002). [DOI] [PubMed] [Google Scholar]
- Uno I. et al. Asian dust transported one full circuit around the globe. Nature Geosci. 2, 557–560 (2009). [Google Scholar]
- Bory A. J.-M. et al. Two distinct seasonal Asian source regions for mineral dust deposited in Greenland (NorthGRIP). Geophys. Res. Lett. 30, 1167 (2003). [Google Scholar]
- Grousset F. E. et al. Case study of a Chinese dust plume reaching the French Alps. Geophys. Res. Lett. 30, 1277 (2003). [Google Scholar]
- Mori I. et al. Change in size distribution and chemical composition of kosa (Asian dust) aerosol during long-range transport. Atmos. Environ. 37, 4253–4263 (2003). [Google Scholar]
- Matsuyama S. et al. Microbeam analysis of yellow sand dust particles. X-ray Spectrometry 37, 151–155 (2008). [Google Scholar]
- Morono Y. et al. Discriminative detection and enumeration of microbial life in marine subsurface sediments. ISME J. 3, 503–511 (2009). [DOI] [PubMed] [Google Scholar]
- Griffin D. W. et al. Atmospheric microbiology in the northern Caribbean during African dust events. Aerobiologia 19, 143–157 (2003). [Google Scholar]
- Jeon E.-M. et al. Impact of Asian dust events on airborne bacterial community assessed by molecular analyses. Atmos. Environ. 45, 4313–4321 (2011). [Google Scholar]
- Abed R. M. M. et al. Microbial diversity of eolian dust sources from saline lake sediments and biological soil crusts in arid Southern Australia. FEMS Microbiol. Ecol. 80, 294–304 (2012). [DOI] [PubMed] [Google Scholar]
- Bowers R. M. et al. Sources of bacteria in outdoor air across cities in the Midwestern United States. Appl. Environ. Microbiol. 77, 6350–6456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Foreign Affairs of Japan. http://www.mofa.go.jp/policy/economy/summit/2009/fact7.pdf
- Hua N. -P. et al. Detailed identification of desert-originated bacteria carried by Asian dust storms to Japan. Aerobiologia 23, 291–298 (2007). [Google Scholar]
- Ichijo T. et al. New digital image analysis software for analyzing size distribution of Asian Dust particles. 130th Annual Meeting of the Pharmaceutical Society of Japan (Okayama, Japan), 30P–pm348 (2010), in Japanese.
- Tsai Y. L. & Olson B. H. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57, 1070–1074 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards U. et al. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17, 7843–7853 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. H. et al. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28, 1942–1946 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M. et al. 16S ribosomal DNA-based analysis of bacterial diversity in purified water used in pharmaceutical manufacturing processes by PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 68, 699–704 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R. et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. & Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y. et al. Similarity of bacterial community structure between Asian dust and its sources determined by rRNA gene-targeted approaches. Microbes Environ. 25, 22–27 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Table