Abstract
AMPA receptor (AMPA-R) complexes consist of channel forming subunits, GluA1–4 and auxiliary proteins including TARPs, CNIHs, synDIG1, and CKAMP44, which can modulate AMPA-R function in specific ways. Combinatorial effects of four GluA subunits binding to various auxiliary subunits amplify the functional diversity of AMPA-Rs. The significance and magnitude of molecular diversity, however, remain elusive. To gain insight into the molecular complexity of AMPA and kainate receptors (KA-Rs), we compared the proteins that co-purify with each receptor type in rat brain. This interactome study identified the majority of known interacting proteins and more importantly, provides novel candidates for further studies. We validate the claudin homologue GSG1L as a novel binding protein and unique modulator of AMPA-R gating, as determined by detailed molecular, cellular, electrophysiological, and biochemical experiments. GSG1L extends the functional variety of AMPA-R complexes and further investigation of other candidates may reveal additional complexity of ionotropic glutamate receptor function.
Introduction
AMPA-R and KA-R are members of the ionotropic glutamate receptor (iGluR) family, functioning as ligand-gated ion channels that mediate excitatory synaptic transmission and plasticity in the brain (Traynelis et al., 2010). Their functions are regulated by the composition of channel forming core subunits, association with auxiliary proteins, phosphorylation, receptor trafficking, and interaction with cytoplasmic scaffolds (Jackson and Nicoll, 2011; Kim and Sheng, 2004; Shepherd and Huganir, 2007). Defining molecules that mediate receptor modulation is critical in understanding basic brain function and disease mechanisms. The molecular composition of AMPA and KA-Rs are diverse and the complete landscape is currently unclear.
The iGluR’s channel core is a tetrameric assembly of receptor subunits, GluA1–4 for AMPA-Rs and GluK1–5 for KA-Rs (Collingridge et al., 2009). Auxiliary transmembrane subunits bind to core iGluR subunits. They are found across species (Wang et al., 2008), and include stargazin (stg)/TARPs (Chen et al., 2000; Tomita et al., 2003), SOL-1 (Zheng et al., 2004), cornichon2/3 (CNIH-2/3) (Schwenk et al., 2009), synDIG1 (Kalashnikova et al., 2010), and CKAMP44 (von Engelhardt et al., 2010) for AMPA-Rs, and Neto1–2 (Zhang et al., 2009) for KARs. The combinatorial effect of various auxiliary subunits binding to channel forming core subunits extends the architectural and functional complexity of iGluRs in the brain (Farrant and Cull-Candy, 2010; Jackson and Nicoll, 2011).
iGluR complexes are extensively studied, yet new binding proteins are continuously reported. Biochemical hurdles in handling intact membrane proteins have been overcome for AMPA and KA-Rs by robust purification protocols (Nakagawa et al., 2005; Zhang et al., 2009). In combination with liquid chromatographic separations in line with tandem mass spectrometers (LC-MS/MS), peptide analysis can identify nearly all proteins present in a low complexity sample (Savas et al., 2011).
In this study, we wished to identify new iGluR interactors that are less abundant or difficult to find. Specifically, we compared the interactomes of native AMPA and KA-Rs and identified a new AMPA-R auxiliary subunit, GSG1L. GSG1L modifies AMPA-R channel function very differently from the known auxiliary modulators, revealing a new functional repertoire of AMPA-Rs. This study provides a proof-of-principal for identifying novel interactors of iGluRs using our interactome data. Our results may further reveal previously unexpected molecular and functional diversity of iGluR complexes.
Results
Identification of candidate proteins that co-purify with AMPA-Rs and KA-Rs in rat brain
We performed immuno-affinity purification of native AMPA and KA-Rs followed by shotgun LC-MS/MS protein analysis (AP-MS/MS). The co-purifying proteins were directly analyzed by multidimensional protein identification technology (MudPIT) (Washburn et al., 2001). As a negative control, we performed a parallel purification with normal rabbit IgG. Any protein binding to IgG was excluded from analysis.
A summary and complete list of the proteins that co-purify with brain AMPA and KA-Rs are shown in Table 1 and S1, respectively. Our purification was highly enriched for the target proteins containing the epitopes of the antibodies used for affinity purification, as demonstrated by numerous spectrum counts (s.c.) and peptides counts (p.c.) for GluA2 (2526 s.c./193 p.c.) and GluK2 (790 s.c./88 p.c.). Nearly all known AMPA-R interacting membrane proteins, such as TARPs (stg/γ-2, γ-3, γ-4, γ-5, γ-7, and γ-8), CNIH2/3, and CKAMP44 were identified in our AMPA-R preparation. While we did not find synDIG1 itself, we identified homologues (Fig 1A and C). Among the known auxiliary subunits, stg/TARPs were most abundantly detected, while fewer s.c.’s and p.c’s were observed for the others. Furthermore, the known KA-R auxiliary subunits Neto-1 and 2 were detected with KA-Rs (Fig 1B and C). These results indicate that our purification was robust and thus, further investigation of the list may identify novel interactors. Our results extend current knowledge on the interactomes of AMPA and KA-Rs.
Table 1. Comparison of AMPA-R and KA-R interactomes by Mass Spectrometry.
Known primary interactors (left) as well as candidate interactors (right) are listed by common name and IPI number. The spectrum count (spec), peptide count (pep) and % coverage (%AA) identified by LC-MS/MS as well as the normalized (norm) abundance of the protein relative to the IPed target protein are listed for proteins in both the GluA2 (A2) and GluK2 (K2) preparations. The current annotated rat protein database does not provide complete representation of the proteins in the rat genome. Thus, to identify Shisa-6, 9, and Neto-1 (shown in italics) we searched against a concatenated database consisting of the humanmouse-rat protein databases. References of known and candidate interactors are provided in the supplementary material.
| known primary interactors | |||||
|---|---|---|---|---|---|
| IPI | GluA2 spec,pep (%AA) |
GluA2 norm |
GluK2 spec,pep (%AA) |
GluK2 norm |
Common Name |
| IPI00780113.1 | 2526, 193 (71.3)• | 1.0000 | 17, 11 (17.2)• | 0.0215 | GluA2 |
| IPI00324555.2 | 876, 129 (60.4)• | 0.3468 | 6, 3 (5.1) | 0.0076 | GluA1 |
| IPI00231095.1 | 873, 121 (56.5)• | 0.3456 | 6, 4 (4.8) | 0.0076 | GluA3 |
| IPI00195445.1 | 585, 91 (48.7)• | 0.2316 | 3, 2 (2.4) | 0.0038 | GluA4 |
| IPI00207460.1 | 212, 26 (34.0)• | 0.0839 | 0, 0 (0.0) | 0.0000 | TARP gamma-3 |
| IPI00201313.4 | 193, 28 (39.6)• | 0.0764 | 0, 0 (0.0) | 0.0000 | TARP gamma-2 |
| IPI00207426.1 | 162, 28 (36.8)• | 0.0641 | 5, 2 (8.3) | 0.0063 | TARP gamma-8 |
| IPI00207431.1 | 78, 13 (32.4)• | 0.0309 | 0, 0 (0.0) | 0.0000 | TARP gamma-4 |
| IPI00214444.1 | 11, 4 (23.30• | 0.0044 | 0, 0 (0.0) | 0.0000 | TARP gamma-7 |
| IPI00207430.1 | 3, 2 (6.9) | 0.0012 | 0, 0 (0.0) | 0.0000 | TARP gamma-5 |
| IPI00366152.2 | 18, 6 (13.1)• | 0.0071 | 0, 0 (0.0) | 0.0000 | CNIH-2 |
| IPI00358957.3 | 11, 4 (9.0) | 0.0044 | 0, 0 (0.0) | 0.0000 | CNIH-3 |
| IPI00956073.1 | 147, 13 (26.2) | 0.0582 | 0, 0 (0.0) | 0.0000 | Shisa-9/CKAMP-44 |
| IPI00566635.2 | 255, 61 (65.1)• | 0.1010 | 28, 10 (16.0)• | 0.0354 | PSD-95 |
| IPI00777470.1 | 80, 31 (40.7)• | 0.0317 | 208, 62 (62.7)• | 0.2633 | SAP-97 |
| IPI00650099.1 | 53, 21 (27.9)• | 0.0210 | 140, 48 (42.4)• | 0.1772 | PSD-93 |
| IPI00568474.1 | 28, 14 (19.6)• | 0.0111 | 27, 11 (10.0) | 0.0342 | SAP-102 |
| IPI00208830.1 | 2,.2 (3.0) | 0.0008 | 0, 0 (0.0) | 0.0000 | Grip1 |
| IPI00409970.1 | 0, 0 (0.0) | 0.0000 | 2, 2 (6.3) | 0.0025 | Grip2 |
| IPI00204506.1 | 5, 5 (6.7)• | 0.0020 | 42, 20 (22.8)• | 0.0532 | protein4.1 |
| IPI00210635.2 | 16, 13 (19.5)• | 0.0063 | 32, 20 (36.0)• | 0.0405 | NSF |
| IPI00471901.3 | 11, 6 (8.4)• | 0.0044 | 10, 6 (10.2)• | 0.0127 | AP-2 alpha2 |
| IPI00389753 1 | 6, 6 (9.3)• | 0.0024 | 10, 6 (7.6) | 0.0127 | AP-2 beta |
| IPI00203346.4 | 5, 4 (6.3)• | 0 0020 | 8, 6 (10.1)• | 0.0101 | AP-2 alpha1 |
| IPI00196530.1 | 4, 3 (5.7) | 0.0016 | 5, 4 (11.5)• | 0.0063 | AP-2 mu |
| IPI00198371.1 | 2, 2 (14.1) | 0.0008 | 4, 3 (24.6) | 0.0051 | AP-2 sigma |
| IPI00324708.1 | 0, 0 (0.0) | 0.0000 | 790, 88 (47.8)• | 1.0000 | GluK2 |
| IPI00207006.1 | 0, 0 (0.0) | 0.0000 | 190, 52 (48.0)• | 0.2405 | GluK5 |
| IPI00231400.2 | 0, 0 (0.0) | 0.0000 | 187, 36 (29.1)• | 0.2367 | GluK1 |
| IPI00231277.4 | 2, 2 (2.2)• | 0.0006 | 686, 77 (45.4)• | 0.8684 | GluK3 |
| IPI00326553.1 | 0, 0 (0.0) | 0.0000 | 105, 32 (34 5)• | 0.1329 | GluK4 |
| IPI00359373.3 | 0, 0 (0.0) | 0.0000 | 125, 37 (59.7)• | 0.1582 | Neto-2 |
| IPI00367046.2 | 0, 0 (0.0) | 0.0000 | 37, 14 (38.3) | 0 0468 | Neto-1 |
| IPI00370061.1 | 0, 0 (0.0) | 0.0000 | 19, 14 (22.8)• | 0.0241 | Kelch |
| candidate interactors | |||||
|---|---|---|---|---|---|
| IPI | GluA2 spec,pep (%AA) |
GluA2 norm |
GluK2 spec,pep (%AA) |
GluK2 norm |
Common Name |
| IPI00763858.2 | 0, 0 (0.0) | 0.0000 | 9, 5 (13.3) • | 0.0114 | MAGUK p55 |
| IPI00365736.3 | 14, 11 (12.0)• | 0.0055 | 5, 5 (6.1) | 0.0063 | Liprin alpha 3 |
| IPI00392157.3 | 0, 0 (0.0) | 0.0000 | 14, 13 (13.9)• | 0.0177 | Liprin alpha 4 |
| IPI00388795.3 | 11, 8 (12.6) | 0.0044 | 94, 32 (36.6) | 0.1190 | CASK |
| IPI00214300.1 | 0, 0 (0.0) | 0.0000 | 37, 12 (42.6) | 0.0342 | Lin 7 |
| IPI00367477.1 | 56, 21 (29.8)• | 0.0222 | 0, 0 (0.0) | 0.0000 | NGL-3 (LRRC 4b) |
| IPI00207958.1 | 11, 7 (11.4)• | 0.0044 | 0, 0 (0.0) | 0.0000 | NGL-l (LRRC 4c) |
| IPI00360822.3 | 4, 3 (5.1) | 0.0018 | 0, 0 (0.0) | 0.0000 | LRRTM3 |
| IPI00454354.1 | 3, 3 (4.3) | 0.0012 | 8, 6 (5.3)• | 0.0101 | LRRC 7 |
| IPI00206020.1 | 3, 3 (19.2) | 0.0012 | 5, 3 (11.1) | 0.0063 | LRRC 59 |
| IPI00372074.1 | 2, 2 (4.0) | 0.0008 | 0, 0 (0.0) | 0.0000 | LRRC 8 |
| IPI00359172.2 | 0, 0 (0.0) | 0.0000 | 3, 2 (5.9) | 0.0038 | LRRC 47 |
| IPI00367715.3 | 2, 2 (3.9) | 0.0008 | 0, 0 (0.0) | 0.0000 | FLRT-2 |
| IPI00829463.1 | 10, 8 (7.8)• | 0.0040 | 10, 7 (6.5)• | 0.0127 | Nrxn-1 |
| IPI00195792.3 | 10, 7 (6 8)• | 0.0004 | 6, 6 (7.8)• | 0.0076 | Nrxn-2 |
| IPI00829491.1 | 5, 4 (6.1) | 0.0020 | 4, 2 (2.7)• | 0.0051 | Nrxn-3 |
| IPI00325649.1 | 3, 2 (4.9) | 0.0012 | 0, 0 (0.0) | 0.0000 | Nlgn-2 |
| IPI00325804 1 | 0, 0 (0 0) | 0.0000 | 5, 2 (3.3)• | 0.0063 | Nlgn-3 |
| IPI00764645.1 | 30, 15 (23.2)• | 0.0119 | 0, 0 (0.0) | 0.0000 | EphB2 |
| IPI00189428.1 | 4, 3 (5.5) | 0.0016 | 0, 0 (0.0) | 0.0000 | EphB1 |
| IPI00569433.1 | 3, 3 (6.7)• | 0.0012 | 0, 0 (0.0) | 0.0000 | EphA4 |
| IPI00230960.1 | 2, 2 (4.8)• | 0.0008 | 0, 0 (0.0) | 0.0000 | EphA5 |
| IPI00365395.2 | 2, 2 (13.4) | 0 0008 | 0, 0 (0.0) | 0.0000 | EphrinB2 |
| IPI00411236.1 | 10, 7 (8.1)• | 0.0040 | 13, 8 (9.3)• | 0.0165 | Latrophillin 1 |
| IPI00561212.4 | 9, 8 (9.2)• | 0.0036 | 0, 0 (0.0) | 0.0000 | Latrophillin 3 |
| IPI00568123.2 | 4, 3 (4.3) | 0.0016 | 0, 0 (0.0) | 0.0000 | Latrophillin 2 |
| IPI00568245.2 | 0, 0 (0.0) | 0.0000 | 480, 136• | 0.6076 | myosin 18 |
| IPI00193933.3 | 3, 3 (6.0) | 0.0012 | 0, 0 (0.0) | 0.0000 | DHHC5 |
| IPI00357941.4 | 7, 7 (5.8) | 0.0028 | 0, 0 (0.0) | 0.0000 | RTPT delta |
| IPI00231945.4 | 3, 2 (3.3) | 0.0012 | 0, 0 (0.0) | 0.0000 | RTPT |
| IPI00565098.2 | 30, 13 (25.8)• | 0 0119 | 0, 0 (0.0) | 0.0000 | GSG1L |
| IPI00939232.1 | 2, 2 (5.1) | 0.0006 | 0, 0 (0.0) | 0.0000 | Shisa-6 |
| IPI00214724.3 | 4,20 (12.1)• | 0.0016 | 0, 0 (0.0) | 0.0000 | PPRT 1 |
| IPI00366048.3 | 38, 10 (38.1) | 0.0150 | 0, 0 (0.0) | 0.0000 | PPRT 2 |
| IPI00207495.3 | 0, 0 (0.0) | 0.0000 | 32, 13 (24.5)• | 0.0405 | pentraxin-2 (Narp) |
| IPI00192125.1 | 0, 0 (0 0) | 0.0000 | 58, 19 (29.9)• | 0.0734 | pentraxin-1 |
| IPI00212317.1 | 0, 0 (0.0) | 0.0000 | 69, 16 (36.6)• | 0.0873 | pentraxin receptor |
| IPI00206558.4 | 19, 9 (15.1)• | 0.0075 | 0, 0 (0.0) | 0.0000 | Olfm-1 |
| IPI00337161.1 | 5, 3 (8.7) | 0.0020 | 0, 0 (0.0) | 0.0000 | Olfm-3 |
Black dots represent proteins which were also found in a smaller scale duplication experiment.
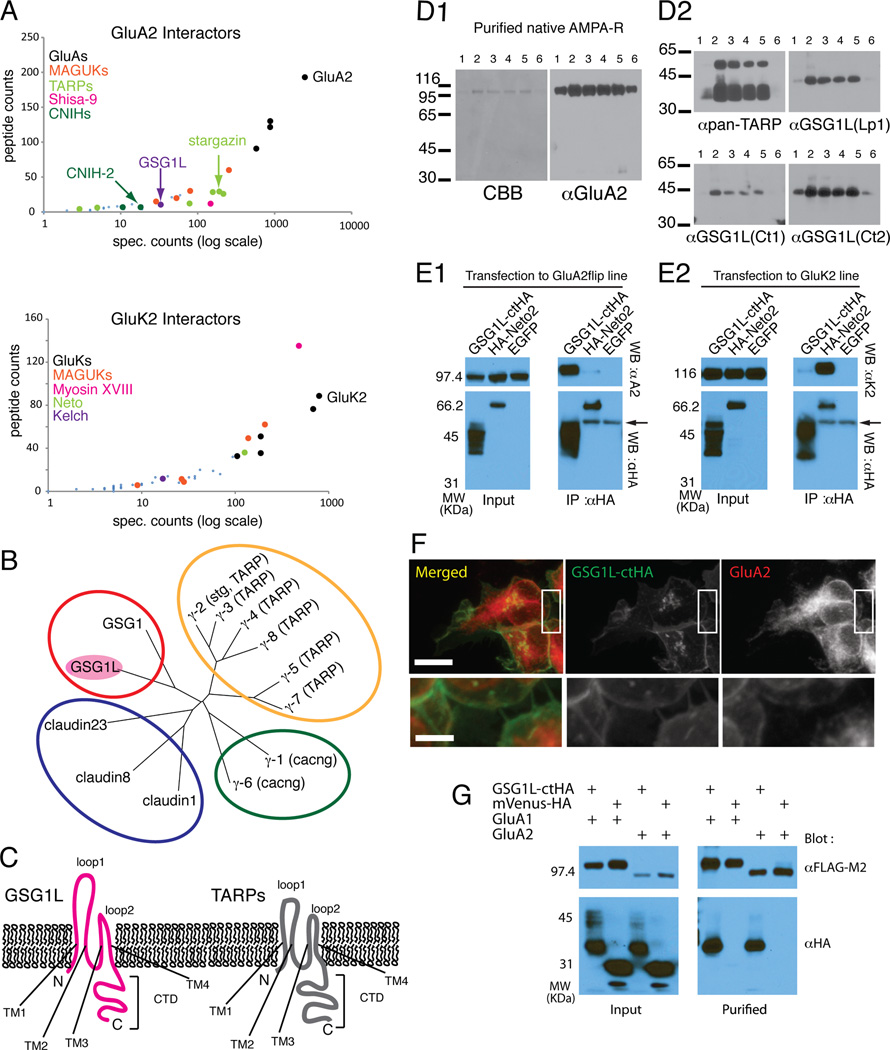
Figure 1. Comparative interactomes of native AMPA-R and KA-R identify GSG1L as AMPA-R interacting protein.
A. Graphical representation of proteins identified as interacting with GluA2 (top) and GluK2 (bottom). Each dot represents a protein identified by mass spectrometry. The-y axis is the number of peptides (log scale) and the x-axis represents the number of spectra in which the identified proteins were found. The black dots are the bait protein receptor subunits. Other known interactor protein families are highlighted by different color dots (see legend). The larger dots indicate known interactors, whereas the smaller dots indicate potential candidates. Note the location of GSG1L between data points for stg and CNIH-2.
B. Phylogenic tree of representative proteins in claudin family constructed using neighbor-joining algorithm in CLUSTALW. The red, yellow, green, and blue circles represents families of GSG, stg/TARPs, gamma subunit of calcium channels, and conventional cluadins.
C. Topology of GSG1L (magenta) and TARPs (gray) relative to the membrane. TM1–4 = transmembrane domain 1–4, loop1–2 = extracellular loop 1–2, CTD = C-terminal domain. GSG1L loop1 is ~50% longer compared to TARPs.
D1. Left: CBB staining of purified native AMPA-Rs. Fractions 1–6 are consecutive elutions from the antibody column using antigen peptide. Right: Western blots of same fractions probed with anti-GluA2CT (αGluA2). Molecular weight markers are on left (kDa).
D2. The duplicated membranes resolving the fractions in D1 were probed with anti-pan-TARP and anti-GSG1L (three different antibodies; Lp1, Ct1, and Ct2, each recognizing different epitopes) antibodies. GSG1L co-purifies with AMPA-Rs from rat brain.
E1. Western blots of the input and immunoprecipitate (IP). Stable HEK cell line expressing GluA2flip was transfected with plasmids expressing GSG1LctHA (ctHA indicates an HA tag at the C-terminus), HA-Neto2, and EGFP. Cellular lysates were IPed using anti-HA antibody. The Western blot was probed with antibodies indicated on right. The arrow indicates the IgG derived from the antibody used for IP.
E2. Similar experiment as E1 was conducted but using stable HEK cell line expressing GluK2.
F. Confocal images of HEK cells cotransfected with GSG1LctHA and GluA2. Scale bar = 10µm (upper) and 2.5 µm (lower).
G. HEK cells were transfected with plasmids expressing the proteins indicated at the top of each lane. FLAG tagged GluA1 and 2 subunits were affinity purified using FLAG beads. The bound protein was eluted using FLAG epitope peptide. Western blots were conducted using the indicated antibodies. mVenus variant of EYFP was used as a negative control.
Predicted protein GSG1L is expressed and binds to AMPA-Rs
Among the candidates, we focused on a predicted protein GSG1L, a membrane protein specifically co-purifying with AMPA-Rs (Fig 1A). It is a distant homologue of stg/TARPs belonging to the extended claudin family (Fig 1B). Furthermore, its peptide counts were comparable to known AMPA-R auxiliary subunits (Fig 1A and Table 1). GSG1L was reproducibly identified from rat brain (Table 1 and S1) and also co-purified with AMPA-Rs from human cortex (Fig S1A–C), indicative of evolutionary conservation of the interactome. Collectively, this evidence provided confidence for further investigation.
While it is in the claudin family, GSG1L is distinct from stg/TARPs, as there is a large evolutionary distances between GSG1L and stg/TARPs. The nearest family member of GSG1L is the product of germ line specific gene 1 (GSG1) whose transcript is specifically expressed in the germ line and whose function is unknown (Tanaka et al., 1994).
Similar to claudins, the predicted topology of GSG1L has a cytoplasmic N-terminus, four transmembrane segments, two extracellular loops, and a cytoplasmic C-terminus (Fig 1C). Loop1 is ~50% longer in GSG1L than in TARPs. The extracellular and cytoplasmic domains of GSG1L are not conserved with stg/TARPs (Fig S1D). These regions are responsible for modulating AMPA-R function in stg/TARPs (Tomita et al., 2005), and thus GSG1L may potentially have unique modulatory function.
GSG1L was annotated as a predicted protein in the rat genome. Its protein existence was unknown and two alternatively spliced transcripts were predicted (Genbank entries XP_002725730.1 and XP_574558.2; predicted molecular weights, 26 and 36 kDa). The shorter variant lacks the first 102 amino acids including the first transmembrane domain. We first created three polyclonal antibodies against different epitopes of the predicted GSG1L protein (Fig S1D). The first epitope Lp1 is only present in the product of longer spliced variant. When we purified native AMPA-Rs from rat brain and examined GSG1L by Western blot, all three antibodies detected a band at the molecular weight of 43 kDa, consistent with the long isoform (Fig 1D1 and D2). These results establish that GSG1L is a protein expressed in rat brain and co-purifies with native AMPARs.
GSG1L interacts specifically with AMPA-R subunits in vitro
To reconstitute the interaction in non-neuronal cells, we transfected HA-tagged GSG1L into stable HEK cell lines that express either GluA2 or GluK2, and immunoprecipitated (IPed) using an anti-HA antibody. GluA2 co-IPed with GSG1L whereas GluK2 did not (Fig 1E1 and E2). Under the same conditions the known KA-R auxiliary subunit Neto-2 specifically interacted with GluK2 but not GluA2. Conversely, the specific interaction of GSGIL with GluA2 and not GluK2 was also observed when the IP was done using antibodies against each glutamate receptor subunit (Fig S2A1 and 2). Furthermore, GSG1L and GluA2 partially co-localize near the plasma membrane when co-expressed in a stable HEK cell line using a DOX inducible expression system (Fig 1F). Similar results were obtained when the two proteins were co-expressed using transient transfection (Fig S2B). GluA1 also forms a complex with GSG1L, as determined by co-IP experiments (Fig 1G). These observations establish the physical interaction between GSG1L and AMPA-R subunits.
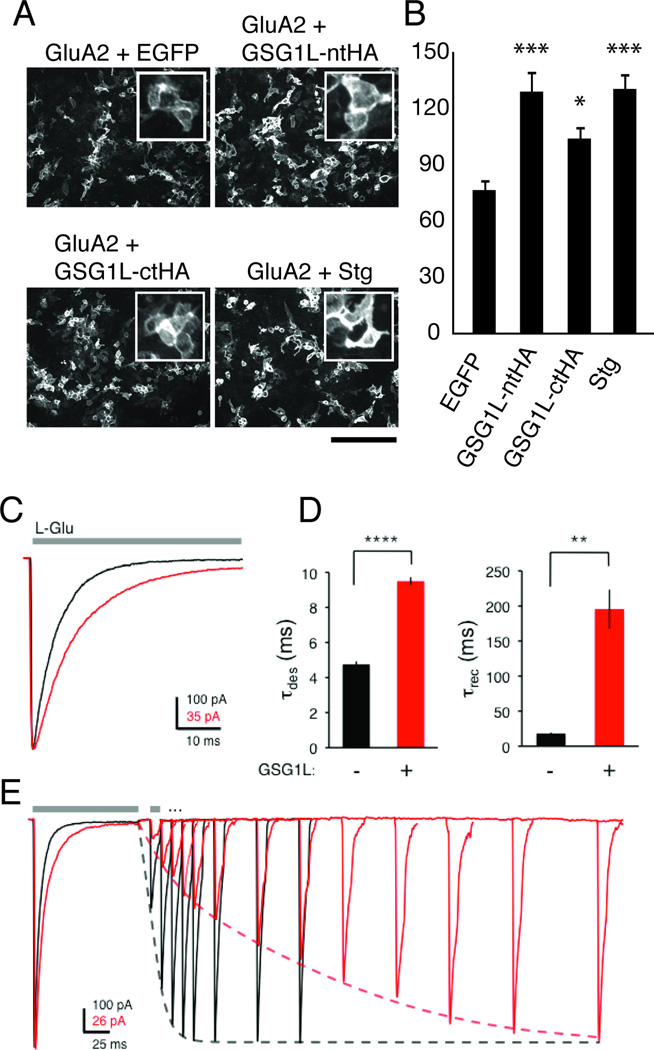
Figure 2. Functional modulation of AMPA-R by GSG1L.
A. Cell surface staining of GluA2 in HEK cells co-transfected with plasmids expressing the proteins indicated above each image. Scale bar = 200µm. Insets are representative enlarged views.
B. Histogram summarizing the quantification obtained from C. *** and * indicate, respectively, p < 0.0003 and p < 0.0166 against control experiments using EGFP according to Bonferroni’s corrected student t-test. The vertical axis represents arbitrary units of fluorescence intensity.
C. Example current responses of outside-out patches from HEK293T cells expressing GluA2-Q(flip) without (black) or with (red) GSG1L to a 100 ms application of 10 mM L-Glu (holding potential −60 mV). Data were fitted with two exponentials. The weighted τdes of the traces presented here is 5.55 ms and 10.70 ms in the absence and presence of GSG1L, respectively.
D. Summary histogram for the time constants of desensitization (left) and recovery from desensitization (right). Data are presented as mean ± SEM. ****p < 0.0001 (t-test); **p < 0.005 (Mann-Whitney U test)
E. Representative current traces of outside-out patches from HEK293T cells expressing GluA2-Q(flip) demonstrating recovery from desensitization in the presence (red) or absence (black) of GSG1L. The paired-pulse protocol consisted of a 100 ms pulse of 10 mM L-Glu followed by a 10 ms pulse in an interval increasing by 10 ms (only selected sweeps are shown). Traces are peak-scaled to the amplitude of the first pulse. Dashed lines indicate the single-exponential fits of the recovery (τrec = 15 ms and 140 ms for GluA2-Q(flip) without and with GSG1L, respectively; summarized in D).
Functional interaction of GSG1L with AMPA-Rs
We next investigated functional interactions between GSG1L and AMPA-Rs. Transfection of GSG1L into a stable HEK cell line that expresses GluA2 increased surface expression of GluA2 compared to EGFP. In fact, GSG1L increased surface GluA2 as efficiently as stg (Fig 2A and B), indicating that surface expression of AMPA-Rs is positively modulated by GSG1L.
A functional interaction was also detected by a cell death assay (Sans et al., 2003; Shanks et al., 2010) (Fig S3). For this purpose, we created stable TetON HEK cell lines that DOX dependently express GluA2 and constitutively express GSG1L or stg (Fig S3A). Cell death was observed after GluA2 expression was induced by DOX in the cell line constitutively expressing stg or GSG1L. Cytotoxicity was blocked by AMPA-R antagonist NBQX and was not detected in the absence of stg or GSG1L (Fig S3C). Glutamate in the media thus triggered the cell death by activating AMPA-Rs whose function was enhanced by stg or GSG1L.
GSG1L profoundly slows AMPA-R recovery from the desensitized state
TARPs, which are distantly related to GSG1L (Fig 1B and C), alter AMPA-R gating kinetics (Tomita et al., 2005). Specifically, deactivation and desensitization rates are slowed by both Type I and II TARPs (with the exception of γ-5; (Jackson and Nicoll, 2011) and recovery from desensitization is accelerated (Priel et al., 2005).
To examine its potential function, GSG1L was co-expressed with GluA2-Q (flip) in HEK293T cells. Channel kinetics were assessed by ultrafast agonist application to outside-out membrane patches. In response to a sustained L-glutamate pulse (10 mM for 100 ms), the GSG1L AMPA-R complex desensitized approximately 2-fold slower (data were fitted with two exponentials, weighted τdes : 4.76 ± 0.16 ms, n = 27; versus 9.50 ± 0.21 ms, n = 10; p < 0.0001; t-test) (Fig 2C, D left). This difference is largely due to an increase in the relative amplitude of the slow component of the decay (Aslow = 10 ± 2 % and 47 ± 5 % without and with GSG1L, respectively) and, to a lesser extent, to an increase in the time constants of the individual components (τfast and τslow shift from 4.09 ± 0.13 ms and 11.58 ± 0.85 ms to 4.86 ± 0.40 ms and 15.18 ± 0.82, respectively). In addition, the 20–80% rise time of these responses was also slightly slower with GSG1L (0.23 ± 0.02 ms vs. 0.19 ± 0.01 ms; p < 0.05; t-test).
A more dramatic effect surfaced when analyzing recovery from desensitization via a two-pulse protocol. Whereas GluA2 recovered with a time constant of 18 ± 1 ms (n = 10), the presence of GSG1L slowed recovery by ~10-fold (τrec = 196 ± 28 ms, n = 6; p < 0.005, Mann-Whitney U test) (Fig 2D right, E). Interestingly, despite their structural similarity (Fig 1C and S1D), this recovery phenotype is in fact opposite to what has been described for TARPs but parallels the effect of CKAMP44, a structurally unrelated Cys-knot protein (von Engelhardt et al., 2010). However, GSG1L and CKAMP44 have opposite effects on modulating desensitization. Therefore, GSG1L is an auxiliary factor which confers novel gating properties, further increasing the AMPA-R functional repertoire. Collectively, these data establish the existence of functional interaction between GSG1L and AMPA-Rs.
Localization of GSG1L in neurons
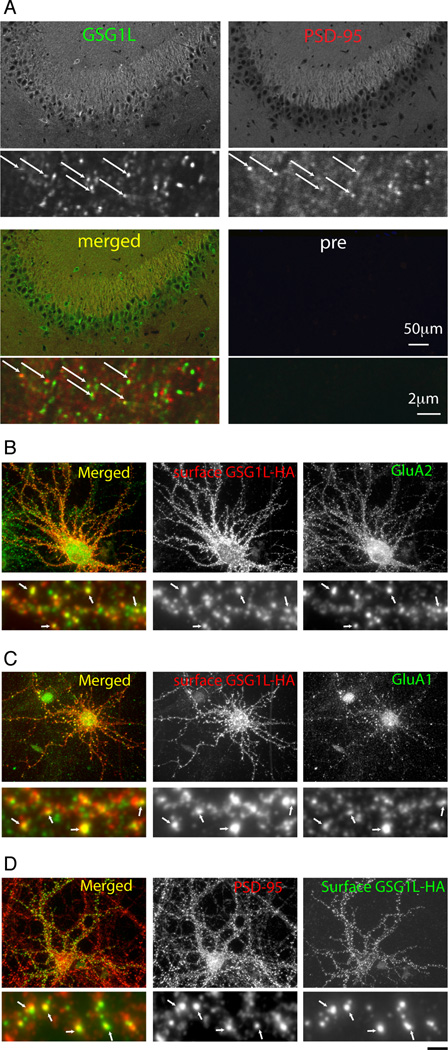
The in situ hybridization data in Allen Brain Atlas indicates GSG1L RNA signal in the hippocampus, striatum, and cortex (Lein et al., 2007). Consistently, GSG1L immunoreactivity was detected in CA3 pyramidal neurons, and partially co-localized with excitatory synaptic marker PSD-95 (Fig 3A). Despite our efforts none of the antibodies generated could detect endogenous GSG1L in dissociated cultured cortical nor hippocampal neurons. However, our antibodies could detect GSG1L when it was moderately overexpressed in cultured neurons. Taken together, we speculate that our antibodies do not have high enough affinity to detect the endogenous proteins in cultured neurons and/or the expression level of GSG1L in culture is lower than in brain tissue.
Figure 3. Localization of GSG1L in neurons.
A. Confocal images of sections of rat hippocampus stained with anti-GSG1L antibody Lp1 and preimmune serum control (pre). Sections were double stained with PSD-95, Scale bar = 50 µm (upper) and 2 µm (lower). Arrows indicate co-localizing puncta.
B. Confocal images of dissociated cortical neurons overexpressing HA tagged GSG1L. The HA tag is in the extracellular loop enabling surface labeling. GSG1L expressed at the cell surface (red) and colocalizes with GluA2 (green). Upper panels; low magnification. Lower panels; enlarged view of the dendrite. The single scale bar corresponds to 20 µm for the upper and 2 µm for the lower panels. Arrows indicate co-localizing puncta.
C. A similar experiment as B was conducted using anti-GluA1 antibody. GSG1L (red) expressed at the cell surface co-localizes with GluA1 (green).
D. A similar experiment as B was conducted using anti-PSD-95 antibody. GSG1L (green) expressed at the cell surface partially co-localizes with PSD-95 (red).
To gain insight into the distribution of GSG1L in neurons, we analyzed the subcellular localization of GSG1L transfected into cortical neurons. To detect GSG1L at the neuronal cell surface, we used a GSG1L construct with an HA epitope tag in the extracellular loop1 (see methods). Consistent with the physical and functional interactions described above, surface GSG1L co-localized with endogenous AMPA-R subunits GluA1 and 2 (Fig 3B and C). The punctate subcellular distribution of surface GSG1L also co-localized with the excitatory synaptic marker PSD-95 (Fig 3D). These results suggest that GSG1L exists at the excitatory synapses in neurons where AMPA-Rs are present.
Discussion
Interactome data identifies novel candidates forming the iGluR complex
By searching through the dataset for membrane proteins that specifically copurify with AMPA-Rs and homologues of known interactors, we reduced the list of candidates significantly. After taking into account the s.c.’s and p.c.’s, we thought it likely that GSG1L is a primary AMPA-R interactor (Fig 1A). Validation of the interaction is the rate-limiting step, requiring multiple experimental approaches. Further investigations of other candidates from our data are expected to validate novel components of AMPA and KA-R complexes (see supplementary material for further discussions).
GSG1L is a new AMPA-R specific auxiliary subunit
The GSG1L gene is implicated to play roles in the nervous system. Its transcript level increases during synapse formation (Bruses, 2010; Lai et al., 2011), and decreases in Huntington’s disease (Becanovic et al., 2010).
Both GSG1L and TARPs are members of the tetraspanin superfamily, with GSG1L belonging to the evolutionarily distant claudin family. The extracellular loop1 of GSG1L is least conserved (19% homology and 6.25% identity) when compared with stg/TARPs and is substantially longer (~50%) (Fig 1C and S1D). Because this loop is essential for ion channel modulation by stg/TARPs (Menuz et al., 2008; Tomita et al., 2005), divergence in channel modulation may be due to mechanistic differences in how the loop interacts with AMPA-Rs. Indeed, whereas TARPs largely speed recovery from the desensitized state, GSG1L slows this parameter, mimicking the structurally unrelated Cys-knot protein CKAMP44 (von Engelhardt et al., 2010). Since desensitization and recovery from the desensitized state impact on high-frequency transmission (Arai and Lynch, 1998), synaptic AMPA-Rs associated with GSG1L are not expected to follow high-frequency trains with great fidelity. Further experiments are necessary to define the mechanisms of binding and functional modulation between GSG1L and TARPs with AMPA-Rs.
Despite that stg/TARPs increase surface expression of AMPA-Rs in HEK cells, there was no change in the amplitude of the AMPA-R mediated current in neurons overexpressing stg (Kessels et al., 2009). Increased surface expression of AMPA-Rs by GSG1L in HEK cells may not warrant that such modulation occurs in neurons. Further experiments are needed to investigate the differences and similarities between GSG1L and stg/TARPs in modulating synaptic physiology.
GSG1L is structurally related to stg/TARPs yet confers completely different function to AMPA-Rs; therefore investigating homologues of known interactors may reveal novel functional repertoire of AMPA-Rs. In fact, we identified many related proteins of known interactors (Table 1 and S1). For example, the LRRC and Shisa family of proteins are related to known AMPA-R interactors, LRRTM2 and CKAMP44 (de Wit et al., 2009; Pei and Grishin, 2012). Similarly, PRRT 1 (NG5 and synDIG4), and pancortin-3 (Olfm1) are shown to co-purify with AMPA-Rs (von Engelhardt et al., 2010). Our study extends the interactome by identifying homologues such as PRRT 2 and Olfm-3.
Given the large number of auxiliary subunits identified for AMPA-Rs, questions regarding their distribution in brain and their stoichiometry remain to be addressed. Different auxiliary subunits simultaneously interact with a single tetramer of GluA subunits (Kato et al., 2010). AMPA-R complexes with different molecular composition may be used during spatio-temporal regulation in specific neurons and synapses. Exactly how this extensive diversity contributes to the activity of neural circuits and behavior remains unclear and is an important question that still needs to be solved.
Methods (The detailed methods are described in the supplementary materials)
Purification of AMPA and KA-Rs from brain
Purification of AMPA and KA-Rs from rat brain follows previous protocols used for single particle EM study of native AMPA-Rs (Nakagawa et al., 2005).
Mass spectrometry
Tryptic digests of resuspended TCA precipitates were subject to multidimensional Protein Identification Technology (MudPIT) (Washburn et al., 2001) and LTQ and LTQ Orbitrap Mass Spectromtery. The RAW files and parameter files will be publically available at http://fields.scripps.edu/published/iGluR upon publication.
Plasmid DNA Construction
Rat GSG1L cDNA was synthesized (Genscript) based on genbank entry XP_574558.2 and tagged with HA or FLAG. The following expression plasmids were used: pTREt, pIRESmcherry (Clontech), and pBOSS (Shanks et al., 2010).
Co-IP experiments
HEK cells were transfected with various expression plasmids and the cellular lysates were prepared from detergent extracts. Anti-HA, FLAG, GluA2CT, and GluK2CT antibodies were used for IP.
Generation of stable TetON HEK cell lines
Cell lines were generated using the previously described methods (Farina et al., 2011; Shanks et al., 2010).
Surface labeling of GluA2 in HEK cells
TetON HEK cells (Clontech) were transfected with appropriate pTREt plasmids in order to co-express GluA2 together with GSG1L or stg or EGFP. Surface GluA2 was live labeled using anti-GluA2-NTD antibody (1:100, Chemicon MAB397).
Neuron transfection and surface labeling
Embryonic day 18 cortical culture and surface labeling were conducted as previously described (Shanks et al., 2010, Sala et al., 2003) with slight modifications.
Electrophysiology
Voltage clamp recordings were performed on outside-out patches from HEK293T cells as described previously (Rossmann et al., 2011).
Immunohistochemistry
Six week old rat (male) was anesthetized and fixed by perfusion using 4% paraformaldehyde in normal rat Ringer solution. 40 µm cryostat brain sections were used.
Supplementary Material
Highlights.
-
-
Mass spectrometry identified proteins co-purifying with AMPA and kainate receptors
-
-
GSG1L is a novel AMPA receptor binding transmembrane protein
-
-
GSG1L uniquely modulates AMPA receptor trafficking and channel gating
-
-
GSG1L co-localizes with AMPA-Rs at synapses
Acknowledgements
This work was supported by John Merck Fund and NIH grant R01 HD061543 to TN. JNS is supported by NRSA fellowship 1F32AG039127. JRY is supported by R01 MH067880 and P41 RR011823. Human brain cortex was obtained through the National Disease Research Interchange (NDRI), Researcher: Yates (code YAJ2), TSRI: IRB-11-5719.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arai A, Lynch G. AMPA receptor desensitization modulates synaptic responses induced by repetitive afferent stimulation in hippocampal slices. Brain Res. 1998;799:235–242. doi: 10.1016/s0006-8993(98)00447-8. [DOI] [PubMed] [Google Scholar]
- Becanovic K, Pouladi MA, Lim RS, Kuhn A, Pavlidis P, Luthi-Carter R, Hayden MR, Leavitt BR. Transcriptional changes in Huntington disease identified using genome-wide expression profiling and cross-platform analysis. Hum Mol Genet. 2010;19:1438–1452. doi: 10.1093/hmg/ddq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruses JL. Identification of gene transcripts expressed by postsynaptic neurons during synapse formation encoding cell surface proteins with presumptive synaptogenic activity. Synapse. 2010;64:47–60. doi: 10.1002/syn.20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Olsen RW, Peters J, Spedding M. A nomenclature for ligand-gated ion channels. Neuropharmacology. 2009;56:2–5. doi: 10.1016/j.neuropharm.2008.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, Sylwestrak E, O'Sullivan ML, Otto S, Tiglio K, Savas JN, Yates JR, 3rd, Comoletti D, Taylor P, Ghosh A. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina AN, Blain KY, Maruo T, Kwiatkowski W, Choe S, Nakagawa T. Separation of Domain Contacts Is Required for Heterotetrameric Assembly of Functional NMDA Receptors. J Neurosci. 2011;31:3565–3579. doi: 10.1523/JNEUROSCI.6041-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrant M, Cull-Candy SG. Neuroscience. AMPA receptors--another twist? Science. 2010;327:1463–1465. doi: 10.1126/science.1187920. [DOI] [PubMed] [Google Scholar]
- Jackson AC, Nicoll RA. The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron. 2011;70:178–199. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalashnikova E, Lorca RA, Kaur I, Barisone GA, Li B, Ishimaru T, Trimmer JS, Mohapatra DP, Diaz E. SynDIG1: an activity-regulated, AMPA- receptor-interacting transmembrane protein that regulates excitatory synapse development. Neuron. 2010;65:80–93. doi: 10.1016/j.neuron.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Ho MT, Yu H, Tu Y, Siuda ER, Wang H, Qian YW, Nisenbaum ES, Tomita S, Bredt DS. Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron. 2010;68:1082–1096. doi: 10.1016/j.neuron.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels HW, Kopec CD, Klein ME, Malinow R. Roles of stargazin and phosphorylation in the control of AMPA receptor subcellular distribution. Nat Neurosci. 2009;12:888–896. doi: 10.1038/nn.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Lai HC, Klisch TJ, Roberts R, Zoghbi HY, Johnson JE. In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J Neurosci. 2011;31:10859–10871. doi: 10.1523/JNEUROSCI.0445-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Menuz K, O'Brien JL, Karmizadegan S, Bredt DS, Nicoll RA. TARP redundancy is critical for maintaining AMPA receptor function. J Neurosci. 2008;28:8740–8746. doi: 10.1523/JNEUROSCI.1319-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. Unexpected diversity in Shisa-like proteins suggests the importance of their roles as transmembrane adaptors. Cell Signal. 2012;24:758–769. doi: 10.1016/j.cellsig.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A, Kolleker A, Ayalon G, Gillor M, Osten P, Stern-Bach Y. Stargazin reduces desensitization and slows deactivation of the AMPA-type glutamate receptors. J Neurosci. 2005;25:2682–2686. doi: 10.1523/JNEUROSCI.4834-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M, Sukumaran M, Penn AC, Veprintsev DB, Babu MM, Greger IH. Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers. Embo J. 2011;30:959–971. doi: 10.1038/emboj.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Prybylowski K, Petralia RS, Chang K, Wang YX, Racca C, Vicini S, Wenthold RJ. NMDA receptor trafficking through an interaction between PDZ proteins and the exocyst complex. Nat Cell Biol. 2003;5:520–530. doi: 10.1038/ncb990. [DOI] [PubMed] [Google Scholar]
- Savas JN, Stein BD, Wu CC, Yates JR., 3rd Mass spectrometry accelerates membrane protein analysis. Trends Biochem Sci. 2011;36:388–396. doi: 10.1016/j.tibs.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, Klocker N. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- Shanks NF, Maruo T, Farina AN, Ellisman MH, Nakagawa T. Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J Neurosci. 2010;30:2728–2740. doi: 10.1523/JNEUROSCI.5146-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yoshimura Y, Nishina Y, Nozaki M, Nojima H, Nishimune Y. Isolation and characterization of cDNA clones specifically expressed in testicular germ cells. FEBS Lett. 1994;355:4–10. doi: 10.1016/0014-5793(94)01155-9. [DOI] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA, Bredt DS. Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature. 2005;435:1052–1058. doi: 10.1038/nature03624. [DOI] [PubMed] [Google Scholar]
- Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, Bredt DS. Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J Cell Biol. 2003;161:805–816. doi: 10.1083/jcb.200212116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R, Sibley D. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Engelhardt J, Mack V, Sprengel R, Kavenstock N, Li KW, Stern-Bach Y, Smit AB, Seeburg PH, Monyer H. CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science. 2010;327:1518–1522. doi: 10.1126/science.1184178. [DOI] [PubMed] [Google Scholar]
- Wang R, Walker CS, Brockie PJ, Francis MM, Mellem JE, Madsen DM, Maricq AV. Evolutionary conserved role for TARPs in the gating of glutamate receptors and tuning of synaptic function. Neuron. 2008;59:997–1008. doi: 10.1016/j.neuron.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Zhang W, St-Gelais F, Grabner CP, Trinidad JC, Sumioka A, Morimoto-Tomita M, Kim KS, Straub C, Burlingame AL, Howe JR, Tomita S. A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron. 2009;61:385–396. doi: 10.1016/j.neuron.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Mellem JE, Brockie PJ, Madsen DM, Maricq AV. SOL-1 is a CUB-domain protein required for GLR-1 glutamate receptor function in C. elegans. Nature. 2004;427:451–457. doi: 10.1038/nature02244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.