Abstract
Changes in speciation of copper(II) in reactions with epigallocatechin gallate (EGCG) and gallic acid (GA) as a function of pH have been investigated by multifrequency (X- and S-band) EPR spectroscopy in the fluid and frozen states. The EPR spectra show the formation of three distinct mononuclear species with each of the polyphenols, and these are interpreted in terms of one mono- and two bis-complexes. However, di- or polymeric complexes dominate the Cu(II) speciation in the pH range 4–8, and it is only at alkaline pH values that these mononuclear complexes make appreciable contributions to the metal speciation. Each mononuclear complex displays linewidth anisotropy in fluid solution as a consequence of incomplete averaging of the spin Hamiltonian parameters through molecular motion. Rotational correlation times for the individual complexes have been estimated by analysing the lineshape anisotropy of the fluid solution spectra using parameters determined by simulation of the rigid limit spectra. These show that the molecular masses increase with increasing pH, indicating either coordination of increasing numbers of polyphenol molecules as ligands to the copper or the increasing involvement of polyphenol dimers as ligands in the copper coordination sphere.
Keywords: Copper polyphenol complex, EPR spectroscopy, X-band, S-band, Line shape analysis, Rotational correlation time
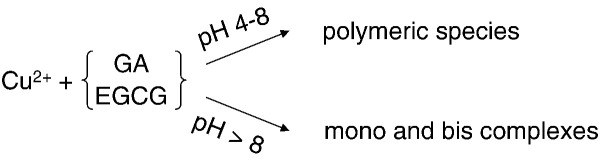
Graphical abstract
Reactions of Cu2 + with GA and EGCG produce complexes at pH > 8, but polymeric species at lower pH.
Highlights
► Cu(II) speciation with two polyphenols (EGCG and GA) at different pH. ► EPR study at two frequencies (X- and S-band). ► Polymerisation dominates at slightly acidic and neutral pH. ► Complexation dominates above neutral pH.
1. Introduction
Interactions between transition metal ions and phenolic compounds are widespread in nature, and can involve complexation of metal ions by the phenols or their oxidation products, polymerisation and redox reactions. Although polymerisation and complexation reactions between Cu(II) and a number of polyphenols have been reported [1,2], it is generally assumed, especially in the biological literature [3–7], that redox is the major reaction process. In redox reactions between Cu(II) and polyphenol molecules, Cu(II) is reduced to Cu(I) and the hydroquinone (H2Q) is oxidised to the semiquinone (HQ). In a second oxidation step, the semiquinone (HQ) is oxidised to the quinone (Q) also by Cu(II) [8].
| Cu(II) + H2Q → Cu(I) + HQ | (1) |
| Cu(II) + HQ· → Cu(I) + Q | (2) |
We have recently investigated the reaction between Cu(II) and gallic acid (GA) over a wide range of pH values, and found no evidence to support either reactions (1) or (2) [9]. The observed oxidation of GA in the alkaline pH region was the result of autoxidation, which was in fact inhibited by Cu(II). In that work, the EPR spectra, which were recorded in fluid solution only, indicated the formation of two, and possibly three, different complexes whose intensities depended on the pH and the Cu:GA ratio, along with the precipitation of a di- or polymeric EPR silent species in the approximate pH range 4–8.
There is extensive epidemiological evidence for the health benefits of green tea (e.g. [10]), and recently there have been proposals to make use of the metal chelating properties of its major polyphenol, epigallocatechin gallate (EGCG), in the treatment of neurodegenerative disorders (e.g. [11–13]). However, there is also evidence that the prion diseases, such as Creuzfeld-Jacob's disease and Alzheimer's, are associated with copper deficiency [14–16]. Thus it is important that the reaction between Cu and EGCG is understood as fully as possible, especially if the chemistry of EGCG mirrors that of GA where precipitation of copper complexes occurs at physiological pH values.
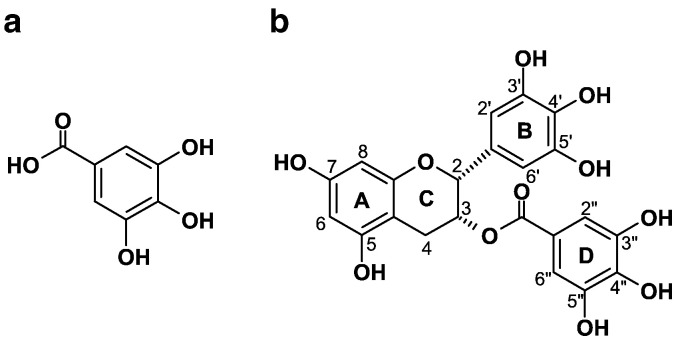
Although both GA and EGCG belong to the same family of polyphenols, there are important differences in their structures. The structure of GA is simple and consists of a carboxyl group attached to a pyrogallol entity (Fig. 1a). The structure of EGCG is more complex with two pyrogallol groups in the molecular structure (one on ring B and one on ring D (Fig. 1b), and one resorcinol group on ring A, but no free carboxyl group. Therefore the principal objective of the present investigation was to determine the extent to which the reactions of GA and EGCG with transition metal ions such as Cu(II) follow similar or different pathways, and to gain information on the complex formation of these polyphenols with Cu(II). For example, the formation of di- or polymeric species involving Cu(II) and the carboxylate group was proposed by Ferreira Severino et al. [9] for the identity of the “EPR silent” species in the reaction of Cu(II) with GA, but since there is no free carboxyl group in EGCG, a similar reaction would not be expected with that polyphenol.
Fig. 1.
Molecular structures of (a) gallic acid (GA) and (b) epigallocatechin gallate (EGCG).
In the previous report of the reactions between Cu(II) and GA, EPR spectra were only obtained from fluid solutions, since the objective of that investigation was simply to distinguish between the relative importance of redox, complexation and polymerisation reactions at different pH values. No anisotropic (rigid limit) spectral parameters were reported, although these could provide additional information on the Cu coordination environment in the mononuclear complexes. Furthermore, the Cu(II) spectra all showed the presence of linewidth anisotropy as a result of incomplete averaging of the anisotropic spectral parameters through molecular motion, but these were not analysed in detail apart from the derivation of approximate values for the isotropic g-values and hyperfine coupling constants. However, if the anisotropic values from the rigid limit spectra are available, it is possible to analyse the fluid solution spectral lineshapes to produce rotational correlation times that are related to the molecular masses of the complexes.
In the present paper we report the results of a comprehensive EPR spectroscopic investigation of the EGCG/Cu(II) system along with additional measurements on the GA/Cu(II) reaction to extend those reported by Ferreira Severino et al. [9]. Spectra were recorded with fluid and frozen solutions at X-band (~ 9 GHz) and S-band (~ 3 GHz) frequencies for samples with a wide range of pH values and Cu:polyphenol ratios. The use of S-band measurements for the EGCG/Cu(II) system is important for understanding the fluid solution results, because of the large linewidth anisotropy in the spectra recorded at X-band frequencies [17–19].
2. Materials and methods
2.1. Sources of materials
Epigallocatechin gallate (EGCG, 95% purity) and gallic acid (GA, ≥ 98%) were purchased from Sigma–Aldrich Handels GmbH (Vienna, Austria), and copper sulphate anhydrous (CuSO4) was bought from Merck (VWR International GmbH, Vienna, Austria).
2.2. Sample preparation
Solutions of different concentration ratios of Cu:GA (1:0, 1:0.5, 1:1, 1:2, 1:10 for X-band measurements and 1:5 for S-band measurements) and Cu:EGCG (1:0, 1:0.5, 1:1, 1:2, 1:5 for X-band measurements and 1:5 for S-band measurements) were prepared with pH values ranging between 1 and 13 with a constant Cu(II) concentration of 2 mM. EPR spectra were recorded at room temperature and low temperature (77 K or 160 K) at both X- and S-band frequencies in solutions containing 5% glycerol, which was added to aid glass formation for the frozen solution studies.
2.3. EPR measurements
EPR spectra were acquired as first derivatives of the microwave absorption with either a Bruker EMX CW spectrometer, operating at X-band frequencies (9 GHz) or a Bruker 200D SRC operating at S-band frequencies (3 GHz). For X-band measurements, a high sensitivity cavity was used and microwaves were generated by a Gunn diode; the microwave frequency was recorded continuously with an in-line frequency counter. Low temperature spectra were recorded using a quartz “finger dewar” containing liquid nitrogen inserted into the microwave cavity. S-band EPR spectra were obtained using a S-band bridge (v = 2–4 GHz) SB-1111 Jagmar (Poland), and low temperatures were controlled with a Bruker ER 4111VT variable temperature unit.
The Cu(II) EPR spectra were acquired using 20 mW microwave power (MP) for room temperature and 2 mW MP for low temperature measurements, 100 kHz modulation frequency (MF) and 1 mT modulation amplitude (MA). g-values were determined by reference to the signal of DPPH (g = 2.0036), which was used as an external standard.
2.4. Data analysis
Signal intensities of the fluid solution spectra were determined by double integration (DI) using the Bruker WINEPR software. For determination of the Cu(II) intensity, the DI of the whole Cu(II) spectrum was carried out, followed by subtraction of the DI of the intensity of the free radical signal in the measurements at very high pH.
Easyspin [20] was used for spectral simulation and analysis. Parameters were determined for the frozen solution spectra using the fitting function “pepper”, and these were then used as the basis for simulation of the fluid solution spectra. The Easyspin software assumes the natural abundance ratio of 63Cu and 65Cu isotopes, but returns hyperfine splittings for the 63Cu isotope only; thus the tabulated results apply only to this nucleus (note: the Cu hyperfine parameters for many spectra reported in the literature give a weighted mean from the two isotopes). The frozen solution spectra were all simulated using a model assuming axial symmetry and co-axial g- and hyperfine matrices. For the latter, the spectra provide information on the absolute magnitudes of the A// and A⊥ values, but not on their relative signs. Therefore, simulations to produce the rotational correlation signs were performed initially for situations where these principal values of the hyperfine coupling constant had the same or opposite signs. Fast motion solution spectra (S- and X-band spectra from Complex I, II, and III of GA/Cu and Complex I of EGCG/Cu) were simulated using the “garlic” function, whereas slow motion solution spectra (S- and X-band spectra from Complex II and III of EGCG/Cu) were fitted using the Easyspin function “chili”.
3. Results and interpretation
3.1. Dependence of Cu(II) signals in fluid solutions on pH and polyphenol concentration
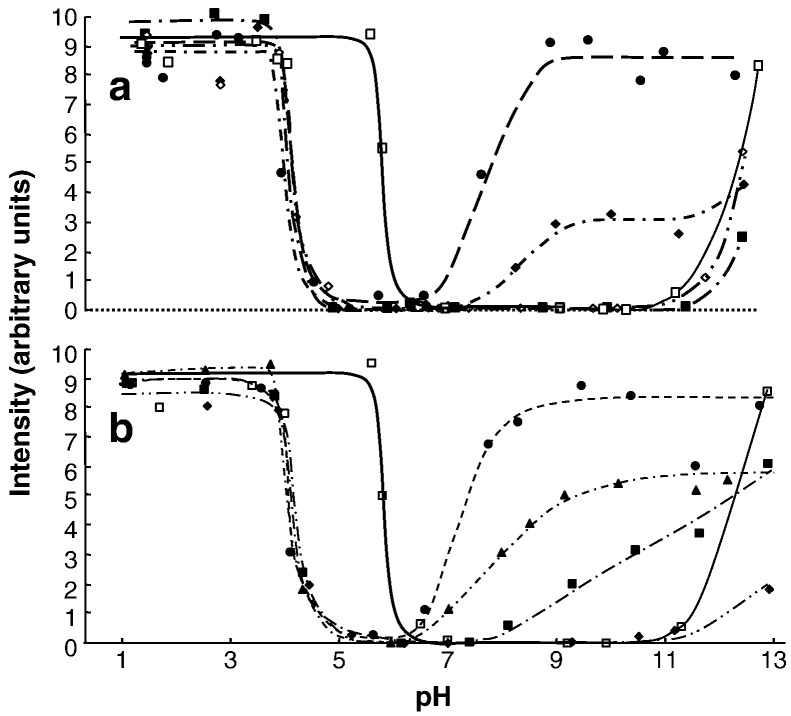
The Cu(II) spectral intensities at X-band frequencies are presented in Fig. 2 as a function of pH for various Cu(II):polyphenol ratios for the Cu/GA and Cu/EGCG reaction systems. Similar curves are observed for both polyphenols; the total signal intensity, and hence the copper speciation, is dependent on both the pH and the Cu(II):polyphenol ratio. The results for the GA system (Fig. 2a) are similar to those reported previously for the Cu/GA system in 1:1 methanol/water [9], except for pH values > 11 and low concentrations of GA. This is because glycerol is able to complex with Cu(II) at high pH when there is deprotonation of the –OH groups [21]. In the absence of polyphenol, the intensity of the Cu(II) signal was constant at pH < 5.5, decreased to zero around pH 6.0, and it remained at zero to pH > 11. In the presence of either EGCG or GA, the decrease in Cu(II) signal intensity occurred around pH 4.0, i.e. ~ 2 pH units lower than in the absence of polyphenol. There was little influence of polyphenol concentration on the spectral intensity at these acidic pH values. However, whereas no signal was observed around pH 6 in the Cu/GA system, except for the 1:10 Cu:GA ratio, a weak signal was observed with the Cu/EGCG solutions in the pH range 4–7. Under alkaline conditions, the intensities of the signals increased with increasing pH and polyphenol concentration, and at high pH and highest polyphenol concentrations approached those observed under acidic conditions.
Fig. 2.
Variation of the overall Cu(II) signal intensity as a function of pH for 2 mM Cu(II) in aqueous solution containing 5% glycerol in the presence of various concentrations of (a) gallic acid (GA) and (b) epigallocatechin gallate (EGCG), Cu:polyphenol 1:0 (□), 1:0.5 (♦), 1:1 (■), 1:2 (▲), 1:5 (EGCG) or 1:10 (GA) (●).
Characteristic fluid solution spectra for Cu(II):EGCG in the ratio 1:5 at X- and S-band frequencies are given in Figs. 3 and 4, respectively. The complete set of X-band spectra at different pH values for various Cu(II):EGCG ratios is available as supplementary material (Figures S1–4). Corresponding results for the Cu(II)/GA system at S-band frequencies are presented in Fig. 5, whilst those at X-band frequencies have been published by Ferreira Severino et al. [9].
Fig. 3.
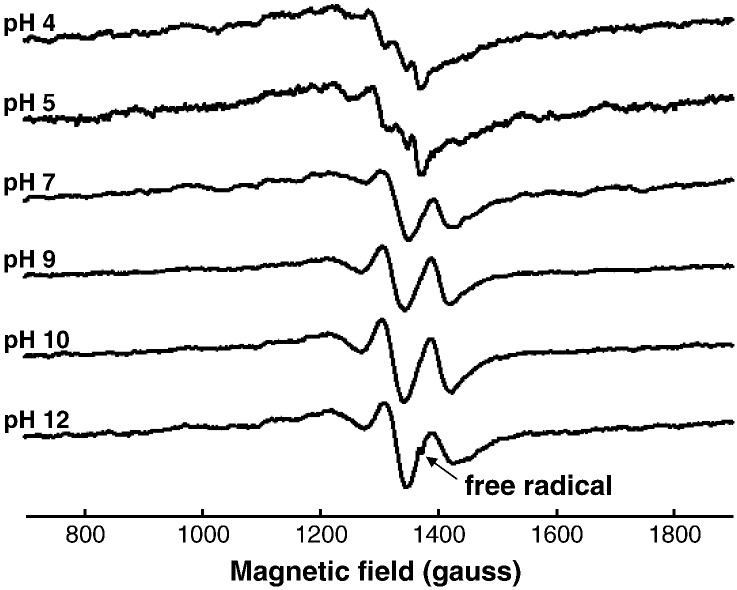
Representative X-band EPR spectra of Cu-EGCG complexes in fluid solution at different pH values; (a) uncomplexed Cu(II), (b) complex I and II, (c) complex II and III, (d) complex III and free radical (which is cut for better clarity). The high field peak of each complex is indicated by an arrow. The concentration ratio of Cu:EGCG was 1:5.
Fig. 4.
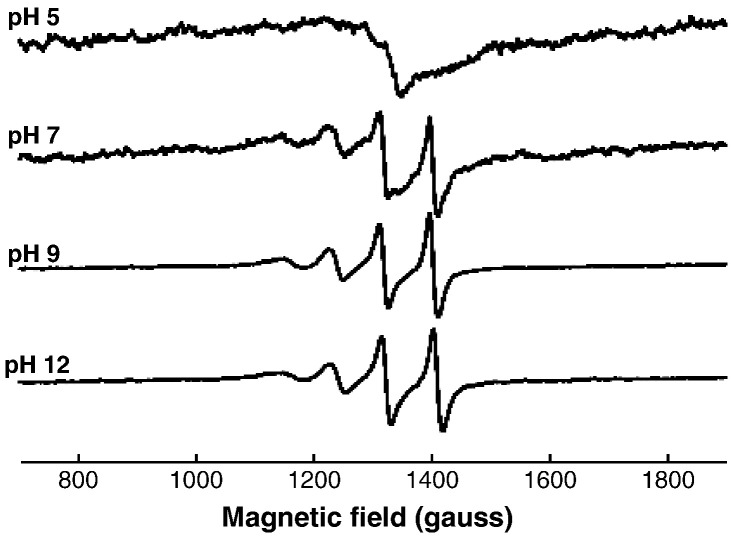
Representative S-band EPR spectra of Cu-EGCG complexes in fluid solution at different pH values. The concentration ratio of Cu:EGCG was 1:5.
Fig. 5.
Representative S-band EPR spectra of Cu-GA complexes in fluid solution at different pH values. The concentration ratio of Cu:GA was 1:5.
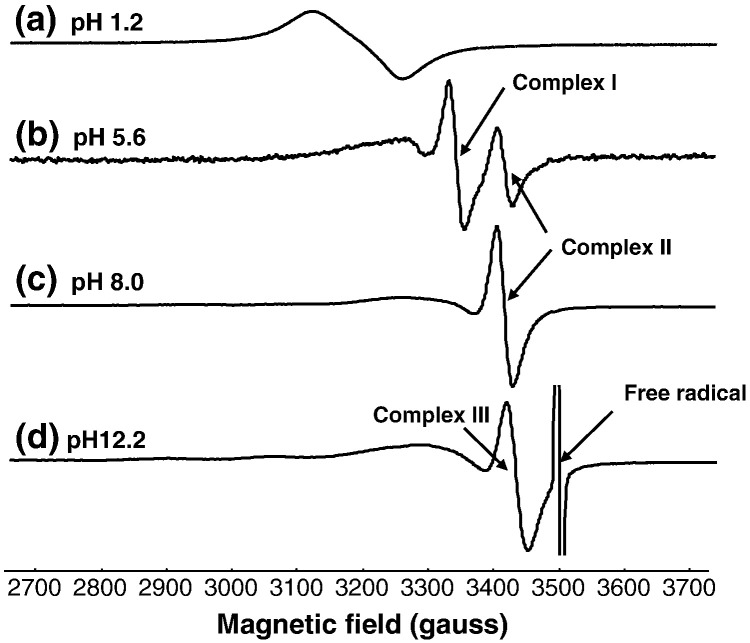
In the low pH-range (pH 1–4) the Cu(II) spectra originate mainly from the uncomplexed [Cu(H2O)6]2 + ion (Figs. 3a, 4a). Around pH 4, the spectral intensity decreased to near zero, but subsequently increased at higher pH values where the spectra were strongly dependent on both the pH and polyphenol concentration. Overall the spectra are consistent with three Cu(II)-EGCG complexes (Figs. 3b–d, 4) (designated Complexes I, II, and III) , and these results are similar to those reported previously with the Cu(II)/GA system [9]. The spectrum from Complex I was always weak, and it was only observed at X-band frequencies; its intensity was too low to produce a spectrum at S-band frequencies. However, it dominated the weak EPR spectra obtained with both the EGCG and GA in the slightly acidic pH range. The contributions from both Complexes II and III increased with increasing pH above pH 7, and above pH 12, only complex III was detected in the solutions which contained more than 2-fold excess of the poyphenols. At this high pH, the spectrum of a Cu(II) glycerol complex was observed from solutions with lower polyphenol concentrations. Thus Complex III might correspond to mixed polyphenol/glycerol complexes of Cu(II), but the formation of a complex between Cu(II) and EGCG with a similar spectrum to that of Complex III in Fig. 3d was observed using pure H2O as the solvent (i.e. without glycerol). All of the spectra from the Cu(II) complexes are complicated by the presence of appreciable linewidth anisotropy; their analysis to produce estimates of rotational correlation times is described below (after consideration of the frozen solution spectra).
3.2. Frozen solution spectra of Cu(II)-GA and Cu(II)-EGCG complexes
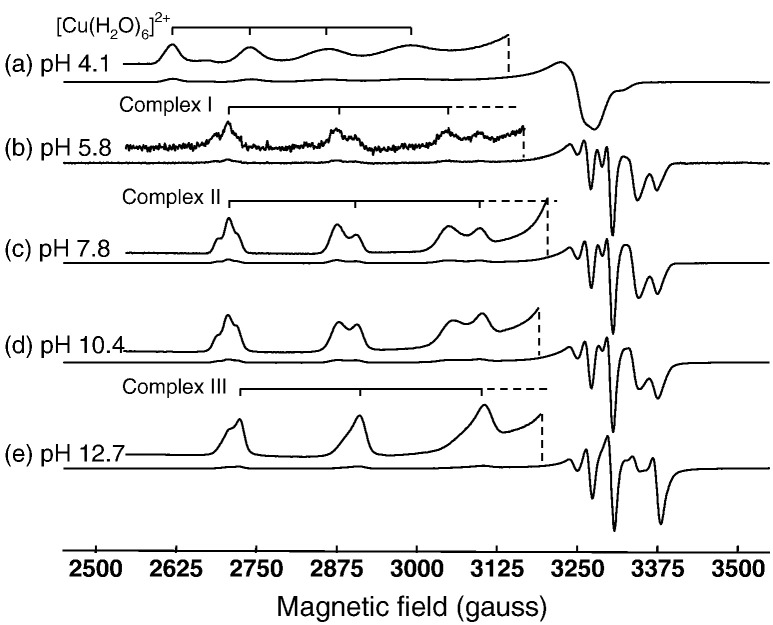
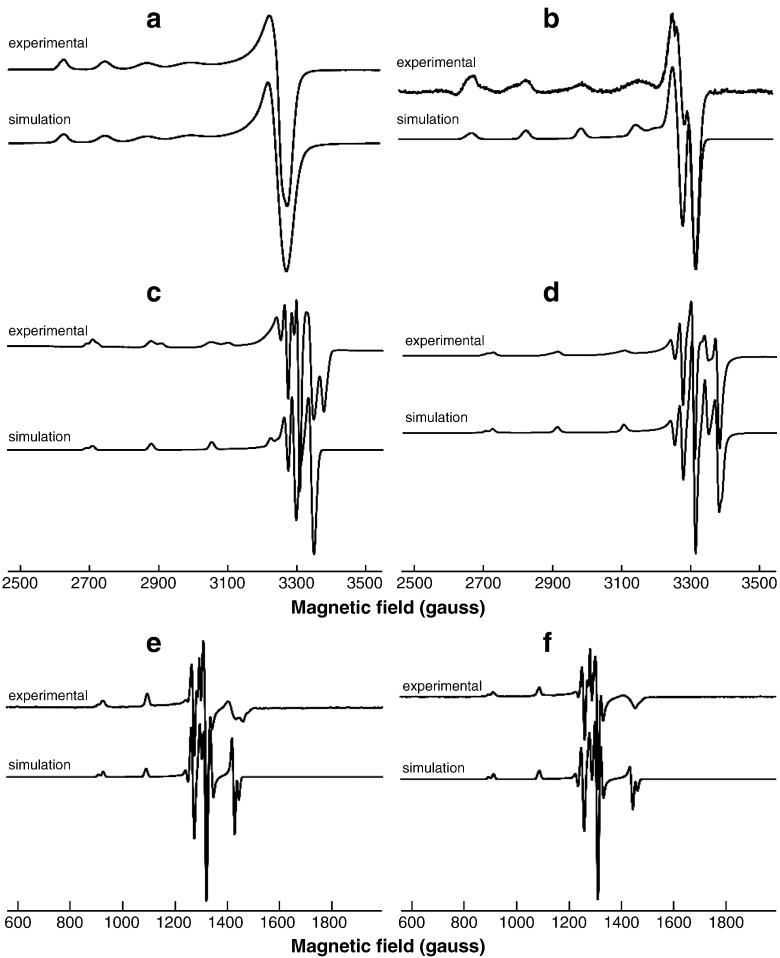
Representative frozen solution spectra from the Cu(II)/EGCG reaction at X-band and S-band frequencies are shown in Figs. 6 and 7 for a Cu:EGCG ratio of 1:5. The g// region in Fig. 6 is expanded to provide better clarity, since this represents the part of the spectrum where different complexes (indicated by stick diagrams) can be discerned. The full range of X-band spectra is available as supplementary information (Figures S5-8). Very similar results were observed with the Cu:GA system and these are also available as supplementary information (Figures S9–12). The spectrum in Fig. 6a corresponds to the uncomplexed [Cu(H2O)6]2 + ion, and that in Fig. 6b belongs primarily to Complex I. Increasing the pH gave results that correspond to mixtures of all three complexes in different ratios (Fig. 6c and d) and at very high pH, Complex III was the major species detected (Fig. 6e). The “pepper” function in the Easyspin software package was used to simulate the spectra of the three mononuclear Cu-EGCG complexes (Fig. 8), and their parameters are summarised in Table 1 along with the corresponding values derived by simulation of the Cu(II)/GA spectra.
Fig. 6.
Characteristic frozen solution spectra for Cu(II):EGCG in the ratio 1:5 recorded at 77 K at X- band frequencies and different pH values. For each spectrum the low field region is also presented in expanded form, and the positions of the g// peaks for the individual 63Cu complexes are indicated by stick diagrams. The shoulder on the low field side of the 63Cu peak at lowest field in (b–d) corresponds to part of the spectrum from the 65Cu isotope from Complexes I and II. The feature to lowest field in (e) corresponds in part to the 65Cu isotope of Complex III and in part to the 63Cu isotope of Complex II.
Fig. 7.
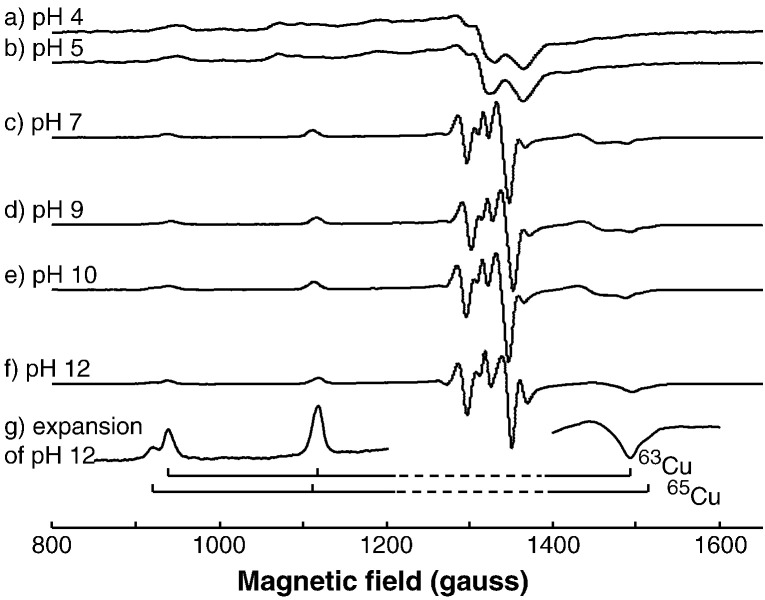
Representative frozen solution S-band spectra for various species formed in the Cu(II): EGCG (1:5) reaction system (a–f), along with an expansion of the peaks associated with the g// feature in (f) showing the contributions from the individual 63Cu and 65Cu isotopes.
Fig. 8.
Simulations of the spectra of the individual components observed with the Cu(II):EGCG (1:5) reaction system; (a) the uncomplexed Cu(II) ion (pH 1), (b) Complex I (pH 4), (c) Complex II (pH 8), and (d) Complex III (pH 13) as examples for X-band and (e) Complex II (pH 9) and (f) Complex III (pH 12) as examples for S-band frequencies.
Table 1.
g- and A-values for frozen solution spectra of Cu(II) with EGCG or GA derived from spectral analysis using the “pepper” subroutine in the Easyspin software [20].
| g// | g⊥ | giso | A//a | A⊥a | Aisoa | A// − Aisoa | ||
|---|---|---|---|---|---|---|---|---|
| Cu-EGCG, LT | ||||||||
| Complex I | X-band | 2.324 | 2.064 | 2.151 | 157 | 4 | 55 | 102 |
| S-band | n.d. | |||||||
| Complex II | X-band | 2.281 | 2.052 | 2.128 | 171 | 28 | 76 | 95 |
| S-band | 2.282 | 2.053 | 2.129 | 171 | 28 | 76 | 95 | |
| Complex III | X-band | 2.251 | 2.052 | 2.119 | 190 | 28 | 82 | 108 |
| S-band | 2.254 | 2.054 | 2.121 | 182 | 28 | 79 | 103 | |
| Cu-EGCG, RT | ||||||||
| Complex I | X-band | 2.324 | 2.064 | 2.151 | 154 | 4 | 54 | 100 |
| S-band | n.d. | |||||||
| Complex II | X-band | 2.274 | 2.05 | 2.124 | 170 | 28 | 75 | 95 |
| S-band | 2.280 | 2.055 | 2.130 | 171 | 28 | 76 | 95 | |
| Complex III | X-band | 2.254 | 2.053 | 2.120 | 177 | 24 | 75 | 102 |
| S-band | 2.254 | 2.054 | 2.121 | 178 | 28 | 78 | 100 | |
| Cu-GA, LT | ||||||||
| Complex I | X-band | 2.32 | 2.055 | 2.143 | 152 | 23 | 66 | 86 |
| S-band | n.d. | |||||||
| Complex II | X-band | 2.28 | 2.052 | 2.128 | 172 | 28 | 76 | 96 |
| S-band | 2.265 | 2.053 | 2.124 | 178 | 28 | 78 | 98 | |
| Complex III | X-band | 2.248 | 2.05 | 2.115 | 187 | 29 | 82 | 105 |
| S-band | 2.253 | 2.053 | 2.120 | 184 | 28 | 80 | 104 | |
| Cu-GA, RT | ||||||||
| Complex I | X-band | 2.32 | 2.068 | 2.152 | 139 | 17 | 58 | 81 |
| S-band | n.d. | |||||||
| Complex II | X-band | 2.27 | 2.052 | 2.124 | 170 | 28 | 75 | 95 |
| S-band | 2.27 | 2.065 | 2.133 | 176 | 26 | 76 | 100 | |
| Complex III | X-band | 2.26 | 2.05 | 2.119 | 174 | 28 | 77 | 97 |
| S-band | 2.27 | 2.06 | 2.130 | 176 | 28 | 77 | 99 | |
| Cu-glycerol complex, LT | X-band | 2.241 | 2.048 | 2.112 | 189 | 30 | 83 | 106 |
In Gauss
The various Cu-complexes are distinguished by a progressive shift in g// to lower values and A// to higher values from the uncomplexed ion through Complex I to Complex III (Table 1). Table 1 also includes the parameters for the Cu-glycerol complex, which can be formed at very high pH. However, since its g- and A-values differ significantly from those of Complex III, it can be concluded that polyphenol complexes dominate the spectra in high pH solutions with a Cu:EGCG ratio of 1:5. The shoulders on the low field side of the low field peak associated with the g// features in Fig. 6b-e correspond to the 65Cu isotope (μ/(μNI) (65Cu) = 1.5877 compared to μ/(μNI) (63Cu) = 1.484897, [22]).
Representative spectra from Cu/EGCG at S-band frequencies are presented in Fig. 7, The spectra from the individual Cu isotopes are seen more clearly at this frequency, and are illustrated in the expanded spectrum in Fig. 7g. The corresponding results for the Cu/GA system are available as supplementary material (Figure S13).
3.3. Detailed analysis of the fluid solution spectra
Because of incomplete averaging of the spectral anisotropy, only one of the four Cu(II) hyperfine peaks (that at the highest field) is well resolved in the solution spectra of each of the Cu(II) EGCG complexes in fluid solution at X-band frequencies (Fig. 3). The high field peak of Complex I is clearly visible in Fig. 3b, but the spectra of Complexes II and III strongly overlap (Fig. 3c), and their individual components are not resolved from one another. However, the position of the high field peak from Complex III was determined from the spectrum recorded at very high pH where the contribution from Complex II was weak (Fig. 3d). Somewhat better resolution of the component peaks was observed in the fluid solution spectra from the Cu/GA system at X-band frequencies by Ferreira Severino et al. [9] (see also Figure S14 for a full set of data), but even with this smaller ligand the resolution was not good.
There are a number of reasons for the lack of resolution of the component peaks in the spectra. Firstly, the widths of the four individual hyperfine peaks are unequal because of incomplete averaging of the spectral anisotropy through molecular motion, and in addition the anisotropic data from the frozen solution spectra show that most samples contain more than one type of complex. Furthermore, the peaks from the individual 63,65Cu isotopes are not resolved from one another. Thus there are considerable uncertainties in deriving isotropic parameters from the X-band fluid solution spectra, and these spectra were only able to be analysed by using the parameters obtained from the frozen solution spectra (Table 1) with partial motional averaging. Since the frozen solution spectra provide no information on the relative signs of A// and A⊥, simulations were performed with the A// and A⊥ values having the same and opposite signs. However, only the use of the same signs reproduced the experimental spectra.
The copper hyperfine peaks are much better resolved in the fluid solution spectra recorded at S-band frequencies (Figs. 4 and 5). The magnitudes of the Aiso values derived from these spectra (Table 1) show clearly that A// and A⊥ must have the same signs in Complexes II and III with both the Cu/GA and Cu/EGCG systems, thus providing support for the X-band analyses. Simulated parameters for the spectra of all Cu complexes detected in fluid and frozen solutions are reported in Table 1, the values of Aiso being assumed to be equal to (A// + 2A⊥)/3 for the X-band spectra. In addition, the results for the calculated rotational correlation times are summarised in Table 2. These show that the mobility of the complexes decreased in the order Complex I > Complex II > Complex III for both polyphenols, and that the mobility of the EGCG complexes was considerably less than for the corresponding GA complexes.
Table 2.
Rotational correlation times (s) for the Cu(II)-EGCG complexes derived from spectral analysis using the Easyspin software [20].
| Rotational correlation time (s) |
||
|---|---|---|
| X-band | S-band | |
| Cu-GA, RT | ||
| Complex I | 9.0e − 11a | n.d. |
| Complex II | 1.3e − 10a | 1.0e-10a |
| Complex III | 1.7e − 10a | 1.1e-10a |
| Cu-EGCG, RT | ||
| Complex I | 1.9e − 10a | n.d. |
| Complex II | 3.9e − 10b | 9.0e-10b |
| Complex III | 8.0e − 10b | 9.4e-10b |
calculated using subroutine “garlic”.
calculated using subroutine “chilli”.
4. Discussion
The presence of three distinct mononuclear Cu(II) complexes was identified from the frozen solution spectra of the products of reactions with Cu(II) with both EGCG and GA, and the corresponding complexes from each polyphenol had similar values for their g- and hyperfine parameters. These results are consistent with the unpaired electron residing primarily in the 3dx2-y2 orbital in all of the complexes, and the similarities in the results from the two polyphenols suggests that the binding with Cu is similar with both, and hence that both involve chelation with a pyrogallol entity.
The values for the spectral parameters observed in the present measurements are similar to those reported by Oess et al. [1,2] for the Cu(II)-GA system. Based on the reported trends in g- and A(Cu)-values with coordination environment for Cu(II) amino acid complexes [23–26], Complexes I and II can be assigned respectively to mono- and bis- Cu(II) polyphenol complexes in both the EGCG and GA systems. The spectral parameters for Complex III are similar to those of Complex II, although Complex III has slightly larger A// and Aiso and slightly smaller g//- and giso-values with each polyphenol. The value of (A//-Aiso) is proportional to the 3dx2-y2 electron density and the fact that its magnitude changes in the same direction as that of Aiso is consistent with core polarization of inner shell s-orbitals being the main source of Aiso (e.g. [27]) in these complexes. The fact that similar numbers are obtained for Complexes II and III for both GA and EGCG (Table 1) strongly suggests that they all have similar Cu coordination environments, and that there is no major change in symmetery between Complexes II and III. Since it is well known that dimeric and polymeric species are formed as a result of autoxidation of polyphenols at high pH values [28], it is possible that Complex III involves one or more dimers of GA or EGCG attached to the Cu, although it is also possible that the differences between Complexes II and III simply represent a change in the phenolic groups coordinated to the copper. We do not consider that Complex III corresponds to the coordination of a third bidentate ligand to the Cu-atom as suggested by Oess et al. [1,2]. Such a complex should have some population of the Cu 4 s orbital, and hence a much reduced value of Aiso (since polarization of inner shell orbitals give the opposite sign to population of the 4 s orbital [27]). Finally, we cannot exclude the possibility that Complex III corresponds to a mixed polyphenol/glycerol complex, but in the absence of further evidence any assignment must be regarded as speculative.
The anisotropy in the linewidths in the fluid solution spectra provides information about the mobility of the Cu-complexes, which can be expressed as rotational correlation times. Rotational correlation times are influenced by molecular size and shape and by solvent viscosity, although the last of these can be ignored in the present work, because the same solvent composition was used for all measurements. In mononuclear Cu(II) complexes, the major factors affecting the correlation times are, therefore, the size and number of ligand molecules that are coordinated to the copper. The rotational correlation times increase in the order Complex I < Complex II < Complex III for each of the polyphenols, and are consistent with a progressive increase in molecular mass, as proposed from analysis of the spectral parameters in the previous paragraph. The values for the Cu/EGCG system are also appreciably greater than the corresponding values for Cu/GA, as expected for the larger size of the EGCG ligand. Although the trend is the same for X- and S-band results – the rotational correlation times are higher with Complex III than with Complex II – the absolute values differ between the two spectrometer frequencies (Table 2). This result is puzzling, but it may be the consequence of the difficulty in precisely analysing the spectra when the solutions contain a mixture of species. With both polyphenols, there is a mixture of complexes at most alkaline pH values, and with EGCG there is the further complication of two resorcinol groups in the polyphenol. Finally, there is the potential problem that the axial symmetry model may not be precisely correct for all of such components. Thus it was not considered appropriate to attempt to further refine the values reported in Table 2.
Since the effect of molecular rotational correlation time on the shape of an EPR spectrum is dependent on the spectrometer operating frequency, measurements at lower frequency (S-band) [17,18] provided better resolution of fluid solution spectra than those at X-band frequencies. Thus the isotropic spectral parameters for Complexes II and III were able to be determined directly from the S-band spectra, and these results confirmed that the anisotropic hyperfine coupling constants have the same sign, and thus provide agreement with the restricted motion analysis of the X-band spectra.
With each complex, there are small differences between the parameters from the simulations of the frozen and fluid solution spectra, the biggest deviation being observed for Complex III. There are a number of possible explanations for these discrepancies. Firstly, the axial symmetry model assumed for the low temperature simulations may not be strictly correct, and the g- and A-matrices may not be co-axial; in addition there could also be a quadrupolar interaction as a result of the appreciable electric field gradient that can exist at the Cu atom in tetragonal symmetry. Secondly, the process of freezing could favour a certain conformation for a complex, whereas in fluid solutions a number of conformational states could be in equilibrium. Thirdly, at high pH values (i.e. Complex III), dimers of GA or EGCG might be involved in the complexes. However, many of the spectra are derived from more than one complex, the spectra of which are not resolved from one another at RT, so it is inappropriate to discuss these details further.
Characterisation of the EPR silent species that are formed at weakly acidic pH values is problematic. With the Cu/GA system, Ferreira Severino et al. [9] showed that the loss of signal was not the result of reduction of Cu(II) to Cu(I), and proposed that the EPR silent species involved the formation of di- or polymeric complexes with coordination of the carboxyl group, as seen with simple carboxylic acids. However, coordination of carboxyl groups is not an option with EGCG, and any extended structure with this polyphenol must be based on coordination of pyrogallol groups. However, the similarity of the results with GA and EGCG indicates that the chemistry of the reactions with Cu(II) of both phenols is similar, and suggests that the EPR silent species involve extended structures in which the Cu is coordinated to the pyrogallol moiety.
The complexation chemistry of EGCG is further complicated by the presence of two pyrogallol groups in the same molecule. In previous work on the oxidation of EGCG [29], it was shown that the site of oxidation is dependent on the experimental conditions, and that the relative reactivities of the B and D rings is not always the same. The pH of the solution could be a factor in determining this, since it also determines the degree of proton dissociation. Thus it is possible that the products of reaction between Cu(II) and EGCG are not discrete molecules, but a group of closely related complexes. Nevertheless, the EPR results are consistent with those from the Cu(II)/GA system, and are consistent with the formation of extended structures at acidic pH values, with the formation of mononuclear Cu(II) complexes gaining in importance at higher pH values and EGCG concentrations. Furthermore, as stated by Ferreira Severino et al. [9] for the Cu(II)/GA system, there is no convincing evidence for any redox reaction between Cu(II) and either of the polyphenols.
5. Conclusions
The chemistry of the reactions of Cu(II) with the polyphenols EGCG and GA is similar, although the molecular mass of EGCG is four times that of GA with several more phenolic groups, but lacking any carboxyl group. With both polyphenols, EPR silent species are formed at weakly acidic pH values, and strong evidence is presented for these having extended structures rather than being the consequence of reduction of Cu(II) to Cu(I). The polyphenols, therefore, result in the removal of Cu from solution at an appreciably lower pH than is observed in the absence of the polyphenol (by ~ 2 pH units). Such a reaction may, therefore, have significance for understanding the biological activity of these polyphenols, since GA is often used as a food additive, and EGCG, the main polyphenol in green tea, is also widely consumed. Furthermore, EGCG has been proposed as a medicine for the treatment of neurological disorders on the basis of its metal complexing ability. However, the present work shows that the formation of mononuclear Cu(II) chelates is only important at alkaline pH values, and these are not likely, therefore, to feature strongly in biological systems.
Acknowledgements
This work was funded primarily by the Austrian Ministry of Traffic, Innovation and Technology (BMVIT) and the Austrian Science Fund (FWF). In addition, KP is thankful to COST P15 Action for a STMS to visit Prof. Riccardo Basosi's laboratory and MCB was funded by PAR 2007, University of Siena and CSGI (Consorzio Interuniversitario per lo Sviluppo dei Sistemi a Grande Interfase), Italy.
Footnotes
Supplementary data to this article can be found online at doi:10.1016/j.jinorgbio.2011.12.010.
Contributor Information
Katharina F. Pirker, Email: katharina.pirker@ait.ac.at.
Maria Camilla Baratto, Email: baratto@unisi.it.
Riccardo Basosi, Email: basosi@unisi.it.
Bernard A. Goodman, Email: bernard_a_goodman@yahoo.com.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Oess A., Cheshire M.V., McPhail D.B., Vedy J.-C. In: Effect of Mineral-Organic-Microorganism Interactions on Soil and freshwater Environments. Berthelin J., Huang P.M., Bollag J.-M., Andreux F., editors. Kluwer Academic/Plenum; New York: 1999. pp. 151–158. [Google Scholar]
- 2.Oess A., Cheshire M.V., McPhail D.B., Stoll S., Alaili M.E., Vedy J.-C. Sci. Total. Environ. 1999;228:49–58. [Google Scholar]
- 3.Yoshino M., Haneda M., Naruse M., Htay H.H., Iwata S., Tsubouchi R., Murakami K. Toxicol. in Vitro. 2002;16:705–709. doi: 10.1016/s0887-2333(02)00061-9. [DOI] [PubMed] [Google Scholar]
- 4.Aguiar A., Ferraz A. Chemosphere. 2007;66:947–954. doi: 10.1016/j.chemosphere.2006.05.067. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi H., Oikawa S., Hirakawa K., Kawanishi S. Mutat. Res. 2004;558:111–120. doi: 10.1016/j.mrgentox.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Satoh K., Sakagami H. Anticancer. Res. 1997;17:2181–2184. [PubMed] [Google Scholar]
- 7.Labieniec M., Gabryelak T. Cell Biol. Int. 2006;30:761–768. doi: 10.1016/j.cellbi.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Chen C.-H., Liu T.-Z., Chen C.-H., Wong C.H., Chen C.-H., Lu F.-J., Chen S.C. Mol. Nutr. Food Res. 2007;51:962–968. doi: 10.1002/mnfr.200600230. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira Severino J., Goodman B.A., Reichenauer T.G., Pirker K.F. Free Radical Res. 2011;45:123–132. doi: 10.3109/10715762.2010.515220. [DOI] [PubMed] [Google Scholar]
- 10.McKay D.L., Blumberg J.B. J. Am. Coll. Nutr. 2002;21:1–13. doi: 10.1080/07315724.2002.10719187. [DOI] [PubMed] [Google Scholar]
- 11.Mandel S., Amit T., Reznichenko L., Weinreb O., Youdim M.B.H. Mol. Nutr. Food Res. 2006;50:229–234. doi: 10.1002/mnfr.200500156. [DOI] [PubMed] [Google Scholar]
- 12.Mandel S.A., Amit T., Kalfon L., Reznichenko L., Youdim M.B.H. J. Nutr. 2008;138:1578S–1583S. doi: 10.1093/jn/138.8.1578S. [DOI] [PubMed] [Google Scholar]
- 13.Hou R.-R., Chen J.-Z., Chen H., Kang X.-G., Li M.-G., Wang B.-R. Cell Biol. Int. 2008;32:22–30. doi: 10.1016/j.cellbi.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Shiraishi N. Biomed. Res. Trace Elem. 2001;12:231–232. [Google Scholar]
- 15.Brown D. Woodhead Publishing Ltd.; Cambridge: 2002. Prion diseases and copper metabolism: BSE, scrapie and CJD research. [Google Scholar]
- 16.Sigurdsson E.M., Brown D.R., Alim M.A., Scholtzova H., Carp R., Meeker H.C., Prelli F., Frangione B., Wisniewski T. J. Biol. Chem. 2003;278:46199–46202. doi: 10.1074/jbc.C300303200. [DOI] [PubMed] [Google Scholar]
- 17.Basosi R., Antholine W.E., Hyde J.S. In: Biological Magnetic Resonance, Vol. 13: EMR of Parmangetic Molecules. Berliner L.J., Reuben J., editors. Plenum Press; New York: 1993. pp. 103–150. [Google Scholar]
- 18.Basosi R., Della Lunga G., Pogni R. In: Biomedical EPR-Part A: Free Radicals, Metals, Medicine and Physiology. Eaton S.R., editor. Kluwer Acdemic/Plenum Publishers; New York: 2005. pp. 385–416. [Google Scholar]
- 19.Della Lunga G., Pogni R., Basosi R. J. Phys. Chem. A. 1994;98:3937–3942. [Google Scholar]
- 20.Stoll S., Schweiger A. J. Magn. Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Hazimah A.H., Badri M., Crouse K.A., Manas A.R. Palm Oil Dev. 2002;35:8–10. [Google Scholar]
- 22.Almanac 2010. http://www.bruker.com/fileadmin/be_user/news/Almanac/Almanac2010.pdf
- 23.Goodman B.A., McPhail D.B., Powell H.K.J. J. Chem. Soc. Dalton. 1981:822–827. [Google Scholar]
- 24.Pasenkiewicz-Gierula M., Froncisz W., Basosi R., Antholine W.E., Hyde J.S. Inorg. Chem. 1987;26:801–805. [Google Scholar]
- 25.Basosi R., Valensin G., Gaggelli E., Froncisz W., Pasenkiewicz-Gierula M., Antholine W.E., Hyde J.S. Inorg. Chem. 1986;25:3006–3010. [Google Scholar]
- 26.Pogni R., Della Lunga G., Basosi R. J. Am. Chem. Soc. 1993;115:1546–1550. [Google Scholar]
- 27.Goodman B.A., Raynor J.B. Adv. Inorg. Chem. Rad. 1970;13:135–362. [Google Scholar]
- 28.Oniki T., Takahama U. J. Wood Sci. 2004;50:545–547. [Google Scholar]
- 29.Ferreira Severino J., Goodman B.A., Kay C.W.M., Stolze K., Tunega D., Reichenauer T.G., Pirker K.F. Free Radical Biol. Med. 2009;46:1076–1088. doi: 10.1016/j.freeradbiomed.2009.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures