Abstract
The chemokine receptor CCR5 is pivotal in determining an individual’s susceptibility to HIV-1 infection and rate of disease progression. To establish whether population-based differences exist in cell surface expression of CCR5 we evaluated the extent of CCR5 expression across all peripheral blood cell types in individuals from two populations, South African Africans (SAA) and South African Caucasians (SAC). Significant differences in CCR5 expression, both in number of CCR5 molecules per cell (density) and the percentage of CCR5-expressing cells, were observed between the two study groups, within all cell subsets. Most notably, the percentage of all CCR5+ cell subsets was significantly lower in SAC compared with SAA individuals (P < 0·01) among natural killer (NK) -cell subsets (CD56+, CD16+ CD56+ and CD56dim) whereas CCR5 density was significantly higher in SAC compared with SAA individuals in CCR5+ CD8+ T-cell subsets and CCR5+ NK-cell subsets (CD56+, CD16+ CD56+ and CD56dim) (all P < 0·05). These relationships were maintained after exclusion of CCR5Δ32 heterozygous individuals (n = 7) from the SAC dataset. The SAA individuals exhibited significantly higher cell activation levels, as measured by HLA-DR expression, than SAC individuals in CD4+ T-cell subsets (P = 0·002) and CD56+ NK-cell subsets (P < 0·001). This study serves to demonstrate that ethnically divergent populations show marked differences in both cell activation and CCR5 expression, which are likely to impact on both susceptibility to HIV-1 infection and the rate of HIV-1 disease progression.
Keywords: CCR5, HLA-DR, South African populations
Introduction
Shortly after the discovery of CCR5 as an HIV-1 co-receptor, it was identified as one of the host cell proteins that play an important role in the transmission and pathogenesis of HIV infection. Numerous studies have demonstrated the importance of CCR5 receptor density in the context of HIV-1 infection in that the amount of CCR5 expressed on the cell surface can directly influence an individual’s susceptibility to HIV-1.1–3In vitro studies by Platt et al.3 have demonstrated that a co-receptor density threshold of between 7 × 102 and 2 × 103 CCR5 molecules/cell is required for efficient replication of R5 HIV-1 in cell lines expressing CD4 levels similar to those on primary T cells. Furthermore, the density of CCR5 molecules on CD4 T cells correlates positively with the replication of R5 HIV-1.4,5 Increased CCR5 density, determined as the mean number of molecules per cell, in HIV-1 infected individuals correlates with high viral loads,6 faster disease progression,7,8 as well as poorer response to antiretroviral treatment.7,9,10 In addition, CCR5 density is also a determinant of the efficiency of CCR5 in chemotactic response to its ligands.11
The best possible example of the importance of CCR5 in HIV-1 infection has been demonstrated by a 32-bp deletion in the CCR5 open reading frame, CCR5Δ32, which was first discovered in high-risk individuals resisting HIV infection.2,12,13 Individuals heterozygous for the CCR5Δ32 allele have a marked reduction in CCR5 surface expression in comparison to individuals lacking this allele14 and individuals homozygous for this mutant fail to express detectable CCR5 protein on cell surfaces.2 Population studies of CCR5Δ32 show that it is present at an average allele frequency of 10% in Europe; however, it is very rare or absent in Africans15,16 suggesting that this allele is fairly recent in terms of human evolution.15 There is also considerable individual–to-individual variability in surface expression on blood lymphocytes in CCR5 ‘wild-type’ individuals, i.e. individuals lacking polymorphisms in the CCR5 open reading frame.1,14,17
In addition to CCR5 genetic polymorphisms, CCR5 surface expression can also be influenced by its chemokine ligands. For example, inverse associations between gene copy number of the CCR5 ligand, CCL3L, and CCR5 expression levels have been reported.10,18 Similarly, CCL5, the CCR5 ligand most abundant in human plasma, regulates CCR5 density by inducing internalization of the receptor.19 Other molecules, such as interleukin-2, interleukin-12 and interferon-α have also been shown to up-regulate CCR5 expression.20–23 An increase in cell activation levels has also been associated with increased CCR5 expression.14,24–26 Furthermore, HIV-1-infected individuals have significantly greater percentages of CCR5-expressing CD4+ T cells when compared with healthy controls.26,27 No study to date has considered CCR5 expression across different peripheral blood immune cell subsets between ethnically divergent populations, and earlier studies have largely focused on CD4+ T cells and have assessed CCR5 expression mainly in the context of HIV-1 infection. To gain further insight into the many roles that can be attributed to CCR5 in the immune response and its role as an HIV co-receptor, requires an in-depth look at how this molecule is distributed across immune cell types in the absence of any chronic infections or immune disorders, and how this might vary between individuals and ethnic groups. Given that CCR5 expression plays an important role in HIV-1 infection and rate of disease progression, the overall CCR5 expression profile may predispose to these infection/disease outcomes. We therefore performed a cross-sectional study in which we evaluated the expression of the receptor CCR5, as both percentage of CCR5-expressing cells and CCR5 density, on various cell types in whole blood samples taken from healthy, HIV-1-uninfected individuals to evaluate baseline expression in two South African population groups, South African Africans (SAA) and South African Caucasians (SAC).
Materials and methods
Study participants
This study cohort comprised 22 SAA and 31 SAC healthy, HIV-1-uninfected individuals. The SAA cohort had a median age of 33·5 years (range 23–62 years) and comprised 14 (63·6%) women and eight (36·4%) men. The SAC cohort had a median age of 40·0 years (range 25–67 years) and comprised 20 (64·5%) women and 11 (35·5%) men. There were no statistical differences among the median ages or the male : female ratio of the two groups (Mann–Whitney P = 0·129 and Fisher’s exact P = 1, respectively). Since the CCR5Δ32 allele has been shown to impact upon CCR5 expression, participants in this study were genotyped as described previously.28 Seven SAC (22·6%) individuals were heterozygous for the CCR5Δ32 allele. This allele was absent in the SAA population. If CCR5Δ32 allele-bearing individuals were removed from the analysis, the population group remained age matched (P = 0·209: SAA median 33·5 years; SAC median 39·5 years) and gender matched (P = 1; SAA: 14 females and 8 males; SAC: 16 females and 8 males). This study was approved by the University of the Witwatersrand Committee for Research on Human Subjects, and informed written consent was obtained from all participants.
Whole blood surface staining and flow cytometry
Ethylenediaminetetraacetic acid (EDTA) –anti-coagulated whole blood obtained from each of the study participants was stained within 1 hr of blood collection. The following antibody panels were used for each donor: (i) T cells: CD3-allophycocyanin (APC), CD4-FITC, CD8-peridinin chlorophyll protein (PerCP), CCR5-phycoerythrin (PE); (ii) B cells: CD19-APC, CCR5-PE; (iii) natural killer (NK) cells: CD3-PerCP, CD16-FITC, CD56-APC, CCR5-PE; and (iv) granulocytes and monocytes: CD45-APC, CD14-FITC, CCR5-PE. An HLA-DR marker to assess the extent of cell activation (i.e. HLA-DR) was also included in a subset of the cohort in the following panel: (v) CD3-PerCP, CD8-FITC, CD56-APC, HLA-DR-PE. All antibodies were obtained from BD Pharmingen (BD Biosciences, San Jose, CA). The CCR5 antibody used was conjugated to PE at a ratio of 1 : 1, thereby allowing for CCR5 quantification, as the mean number of CCR5 molecules per cell (CCR5 density), in addition to the percentage of CCR5-expressing cells within a cell subset. Quantification was carried out using the QuantiBRITE system (BD Biosciences), which is a set of four pre-calibrated beads to calibrate the fluorescence 2 (FL2) axis in terms of PE molecules.
Fifty microlitres of whole blood was used for each antibody panel described above. After incubating stained samples for 15 min, the red blood cells were lysed with 2 ml FACS® lysing solution (BD Biosciences) for 7 min. The cells were then pelleted by centrifugation at 100 g for 5 min at room temperature and washed with FACS flow (BD Biosciences). The stained cells were suspended in 150 μl paraformaldehyde (Electron Microscopy Services, Pretoria, South Africa) and stored at 4° until flow cytometric acquisition within 6 hr of sample collection. All incubations were performed at room temperature in the dark.
Flow cytometric acquisition and analysis was performed on the FACSCalibur (BD Biosciences). The flow cytometer was set up by running FACSComp 5.2 (four-colour lyse-wash) software (BD Biosciences) with CaliBRITE beads (BD Biosciences). Daily compensation using whole blood stained with a single antibody was conducted to optimize instrument settings for the assay. The lymphocyte population was identified on the basis of forward and side scatter parameters. T cells were defined as CD3-expressing lymphocytes and were further classified as CD4+ or CD8+ T cells. Lymphocytes expressing CD19 were defined as B cells. The NK cells were defined as lymphocytes negative for CD3 and positive for CD56 expression. NK cell subsets analysed were, CD56+, i.e. total CD56-expressing NK cells, CD56dim, CD56bright and CD16+ CD56+. Monocyte and granulocyte populations were identified on the basis of side scatter and CD14 parameters. Monocytes were identified by the presence of CD14 markers, whereas granulocytes are negative for CD14. The mean number of CCR5 receptors per cell was determined for CCR5+ cell subsets. Data were analysed using FloJo 7·6·1 (Tree Star, San Carlos, CA).
Statistical analysis
Mann–Whitney U-tests were conducted to compare CCR5 density between individuals, grouped by population or by the presence or absence of the CCR5Δ32 allele. Correlations between CCR5 density and age of individuals were calculated by bivariate Spearman’s rank coefficients. All statistical analyses were performed using pasw Statistics 18 software (SPSS Inc., Chicago, IL).
Fisher exact tests were performed using the Simple Interactive Statistical Analysis software (Uitenbroek, D. G, Binomial. SISA. 1997. http://www.quantitativeskills.com/sisa/distributions/binomial.htm [1 January 2004]) to test whether there was any significant difference in composition of population groups.
Results
CCR5 expression on lymphocyte populations
Isolation of peripheral blood mononuclear cells by Ficoll purification and a delay in whole blood sample processing is documented to result in acute down-regulation of CCR5 expression on various cell types.29–31 Therefore, flow cytometry was performed on whole blood samples. Data presented as mean/medium fluorescence intensity or as the proportion of positive cells are always relative to controls that are specific for any given experiment. Hence, we used a method for enumerating CCR5 molecules, which allowed better comparison between individuals using a CCR5 antibody conjugated to PE at a ratio of 1 : 1 in combination with antibodies that define different cell types. This allowed for CCR5 quantification as the mean number of CCR5 molecules per cell, i.e. CCR5 density, in addition to the percentage of CCR5-expressing cells within a cell subset.
In agreement with previous studies, a large inter-individual variability on CCR5 expression was observed.1,6 The mean number of CCR5 molecules per cell differed by as much as sixfold between individuals, most notably in the CD8+ T-cell and CD56bright NK-cell subsets.
The potential influence of gender upon expression was examined. Female participants demonstrated a significantly higher percentage of CCR5-expressing cells in the CD56bright NK subset than the men (P = 0·042) when the whole cohort was examined. However, this significance was lost when the population groups were examined separately. No differences in expression between male and female participants were observed in all other cell subsets (data not shown).
It is interesting to note that a high percentage of CCR5-expressing cells within a subset does not necessarily correlate with high CCR5 density, i.e. some individuals have a small percentage of CCR5-expressing cells within a cell subpopulation but express CCR5 at high density on this small proportion of cells and vice versa. This is in agreement with other reports.6
The cell subsets with the highest CCR5 density were T cells and monocytes: T cells (ranging from 1369 to 4820 molecules/cell); CD4+ T cells (ranging from 945 to 3678 molecules/cell); CD8+ T cells (ranging from 1055 to 5953 molecules/cell) and CCR5+ monocytes (ranging from 1628 to 8773 molecules/cell). The subsets with the greatest percentage of cells expressing CCR5 were CD8+ T cells (CD3+ CD8+, 57·8% median) and CD56bright NK cells (64·8% median). Although the CD56bright NK-cell and B-cell populations were observed as high CCR5-expressing subsets, these cell types were present in low numbers, which may affect the accuracy of the CCR5 density values.
CCR5 expression differs between SAA and SAC individuals within all cell subsets
Significant differences in CCR5 expression were noted between SAA and SAC individuals, both in terms of CCR5 density and in the percentage of individual cell types that express CCR5 in the different lymphocyte subpopulations (Table 1). When CCR5Δ32-bearing SAC individuals (n = 7) were removed from the analysis, these differences were maintained or even strengthened (Table 1).
Table 1.
Differences in CCR5 expression, both as percentage of CCR5-expressing cells and CCR5 density, between two South African population groups, South African Africans (SAA) (n = 22) and South African Caucasians (SAC) (n = 31) across all peripheral blood cell populations*
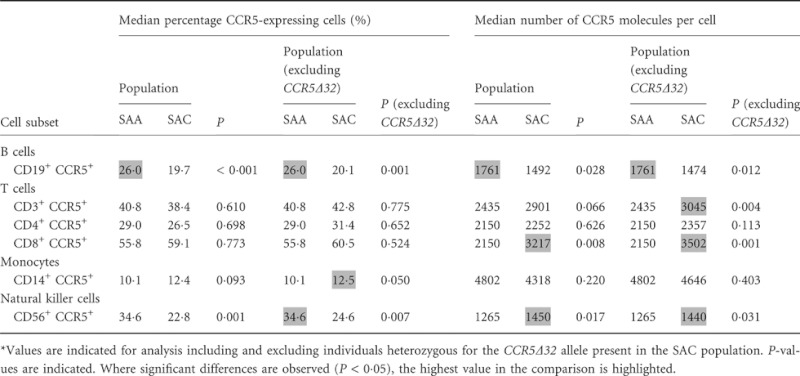 |
As NK cells showed significant differences in CCR5 expression, we examined expression on various NK cell subsets in detail (Fig. 1). Overall, SAC individuals tend to express a greater number of CCR5 receptor molecules per cell, compared with SAA individuals, but on a significantly smaller proportion of CCR5-positive cells.
Figure 1.
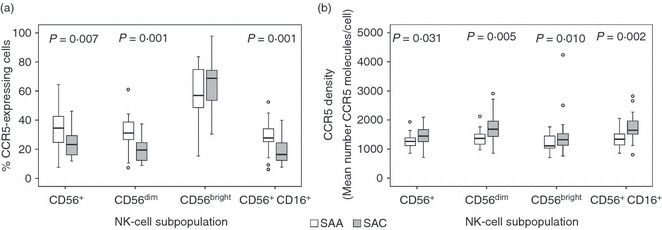
CCR5 expression in South African African (SAA; n = 22) and South African Caucasian (SAC; n = 24) individuals within natural killer (NK) cell subsets. (a) Percentage of CCR5-expressing cells within NK-cell subsets. (b) CCR5 density on CCR5-expressing NK-cell subsets. Heterozygous CCR5Δ32 individuals have been excluded. Box-whisker plots depicting the median (horizontal black line), 25th and 75th centiles (margins of the box) and the 10th and 90th centiles (whiskers). Outliers are indicated with (○). Where significant, P-values have been indicated.
Relationship between CCR5 expression and age of individuals
The age range of participants in our study was broad, ranging between 23 and 67 years of age. Hence, we investigated the relationship between CCR5 expression and age on all peripheral blood mononuclear cell subsets studied. A significant negative correlation with age and the percentage of CCR5-expressing cells was seen in the B-cell subset in SAA individuals (R = −0·478, P = 0·024) but not in CCR5 ‘wild-type’ SAC individuals (R = −0·352, P = 0·092). When grouped together (SAA+SAC), a significant positive correlation was noted on CD8+ T-cell subsets (R = 0·320, P = 0·020). This relationship was lost when the populations were analysed separately, although a trend was still maintained in the SAC population excluding CCR5Δ32 allele-bearing individuals (R = 0·374, P = 0·072). Significant negative correlations were observed in the SAC population in CD56+ NK-cell subsets whether the population was examined as a whole or with the exclusion of CCR5Δ32 heterozygous individuals (R = −0·461, P = 0·009 and R = −0·448, P = 0·028, respectively). The relationship appears to be the result of the CD16+ CD56+ NK-cell subset (R = −0·417, P = 0·043 in SAC CCR5 ‘wild-type’ individuals), because no significant correlations were observed in the other NK-cell subsets. No similar relationships were observed in SAA individuals so the proportion of CCR5-expressing NK cells appear to decrease with age in SAC individuals.
Although correlations between the proportion of CCR5-expressing cell subsets and age were observed in B-cell, T-cell and NK-cell subsets, these relationships were not observed when correlations with CCR5 density and age were analysed in the same cell subsets (data not shown). In monocyte cells, however, a negative correlation between CCR5 density and age only in SAA individuals was observed (R = 0·468, P = 0·028) (Fig. 2).
Figure 2.
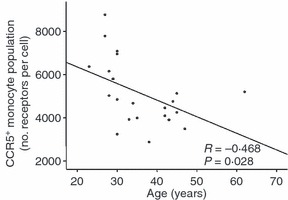
Correlation between CCR5 density (mean number of molecules per cell) on monocyte cells and age of South African African (SAA) study participants (n = 22). P-value is indicated.
SAA and SAC individuals differ in cell activation levels
The levels of activation for a subset of individuals from both population groups, SAA (n = 16) and SAC (n = 24), were measured for T cells and NK cells using HLA-DR as a marker of activation. The SAA individuals exhibited significantly higher expression of HLA-DR, measured as the percentage of HLA-DR-expressing cells, in CD4+ T-cell (P = 0·002) and NK-cell (CD56+, P < 0·001) subsets (Fig. 3). When we investigated the correlations between HLA-DR and CCR5 expression levels, a positive correlation was observed in SAA individuals, between HLA-DR expression (%) and the percentage of CCR5-expressing cells on T-cell (CD3+) and CD4+ T-cell subsets (P = 0·021 and 0·037, respectively, Table 2). However, the proportion of HLA-DR-expressing cells did not correlate with CCR5 density on these same subsets (Table 2). The SAC individuals, on the other hand, demonstrated a positive relationship between proportions of HLA-DR-expressing cells and CCR5 density, i.e. individuals with higher proportions of T-cell subsets expressing HLA-DR had higher numbers of mean CCR5 molecules per cell on CD3+, CD4+ and CD8+ T-cell subsets (Table 2), but no significant correlation was observed when analysing the relationship between the percentage of CCR5-expressing subsets and of HLA-DR-expressing subsets. No significant correlations were observed in any NK-cell subsets (data not shown).
Figure 3.
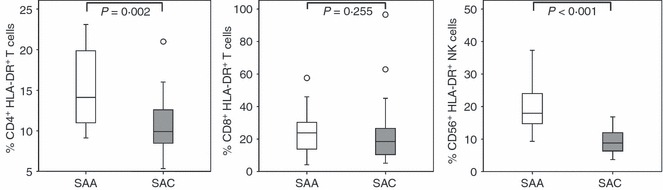
Activation levels, as demonstrated by percentage HLA-DR expression in the two study population groups, South African Africans (SAA; n = 16) and South African Caucasians (SAC; n = 23) within natural killer (NK) -cell and T-cell subsets. Box-whisker plots depicting the median (horizontal black line), 25th and 75th centiles (margins of the box) and the 10th and 90th centiles (whiskers). Outliers are indicated with (○). P-values have been indicated.
Table 2.
Correlation between cell activation, as measured by HLA-DR percentage expression, and CCR5 expression (percentage CCR5-expressing cells and density)*
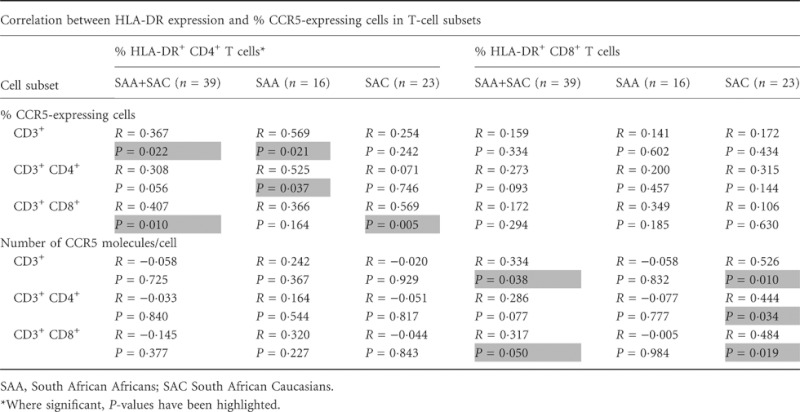 |
To investigate possible relationships between CCR5 density and HLA-DR density on the same cell subsets, we used the geometric mean of HLA-DR+ subsets as a substitute measure for the mean number of molecules per cell as we did not use an HLA-DR antibody conjugated to PE at a 1 : 1 ratio. No significant differences in HLA-DR mean fluorescence intensity between the two populations were observed (data not shown). Furthermore, no correlations between HLA-DR mean fluorescence intensity and CCR5 density were observed.
Influence of CCR5Δ32 allele on CCR5 expression within the SAC population
Individuals bearing the CCR5Δ32 allele tended to have a lower percentage of CCR5-expressing cell subsets in all cell populations examined, yet statistical significance was only observed in CD4+ T cells (P = 0·026) (Fig. 4a). Individuals heterozygous for the CCR5Δ32 allele demonstrated lower CCR5 density on T-cell populations relative to individuals that lack the allele (P < 0·005) (Fig. 4b). This is consistent with other reports.14 However, it is interesting to note that differences in expression in other cell populations are not significant with the exception of CD56bright NK cells (Fig. 4b). In addition, some SAC individuals heterozygous for the CCR5Δ32 allele had comparable CCR5 expression, both in the percentage of CCR5-expressing cells and CCR5 density, compared with homozygous ‘wild-type’ individuals (Fig. 4a,b).
Figure 4.
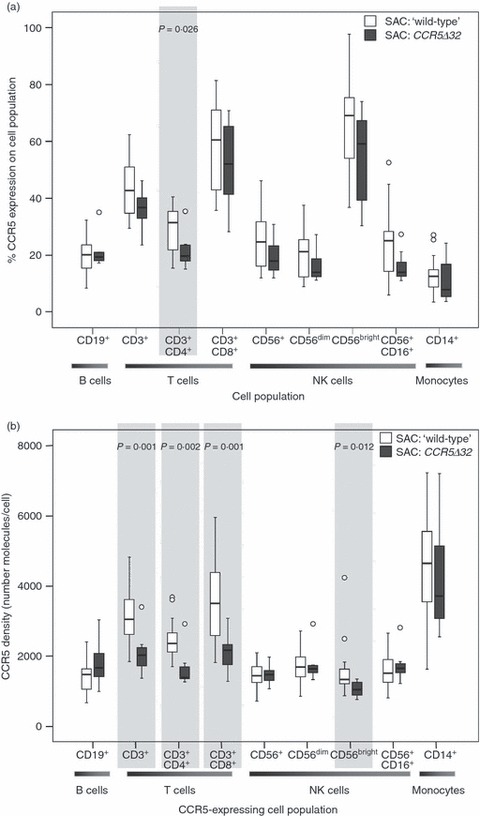
Influence of CCR5Δ32 bearing alleles on CCR5 expression in South African Caucasian (SAC) population within all cell populations examined. (a) Percentage of CCR5-expressing cells in the different cell subsets examined in SAC individuals heterozygous for the CCR5Δ32 allele (n = 7) and CCR5 ‘wild-type’ SAC individuals (n = 24). (b) CCR5 density (mean number of molecules/cell) on the different cell subsets examined in SAC individuals heterozygous for the CCR5Δ32 allele (n = 7) and CCR5 ‘wild-type’ SAC individuals (n = 24). Where significant, P-values have been indicated. Box-whisker plots depicting the median (horizontal black line), 25th and 75th centiles (margins of the box) and the 10th and 90th centiles (whiskers). Outliers are indicated with (○).
Discussion
Extensive variation exists between individuals, in their susceptibility to HIV-1 and the rate of disease progression to AIDS. Cell surface expression of the HIV-1 co-receptor, CCR5, is highly variable, even in individuals homozygous for the wild-type open reading frame region.1,14,17 Although the influence of the CCR5Δ32 mutation on CCR5 expression on T cells has been studied extensively in the context of HIV-1 infection, we lack information on how healthy HIV-1 uninfected individuals may differ in terms of individual CCR5 expression levels. Given that the extent of expression of CCR5 plays a pivotal role in determining HIV-1 susceptibility and that striking ethnic or population differences in SNP frequencies of CCR5 exist, it is surprising that very few studies have questioned how populations may differ in terms of their CCR5 expression levels.
Transsexual men receiving oestrogens and anti-androgens have been reported to have increased CCR5 expression on T cells32 and female mice have higher CCR5 expression on CD4+ T cells compared to their male counterparts.33 Oestrogen therefore appears to influence CCR5 expression, so we looked at potential differences in expression between genders in the current study. On the whole, no differences in expression were observed between male and female study participants, whether examined collectively or separately in the two population groups.
Previous studies have demonstrated CCR5 receptor expression to be stable within individuals over multiple time-points despite the wide range of variability that exists between individuals.6,26,34 However, aging has been associated with increased CCR5 expression on T cells both in mice through gene expression studies35 and in humans using ribonuclease protection assays and Western blots.36 Hence, we investigated the relationship between CCR5 expression and age on all studied cell subsets in the two population groups (age and sex matched). The proportion of CCR5-expressing CD8+ T cells was observed to significantly increase with age in the pooled cohort but no correlations were observed in T cells when the two populations were analysed separately. In a human study in which CCR5 expression on CD4+ T cells was shown to be higher in older individuals, CCR5 expression was compared between two groups, young adults (aged 18–40 years) and older subjects (≥ 60 years, mean age 73 years).36 Only three of our participants were aged ≥ 60 years (one SAA individual, aged 62, and two SAC individuals, aged 64 and 67), so it is likely that our study did not have sufficient ‘older’ individuals to show this relationship. Also, levels of expression were determined differently, where we have looked at cell surface expression by means of flow cytometry, Yung and Mo36 used ribonuclease protection assays and Western blots. However, in SAA individuals, but not SAC individuals, we observed a decrease in CCR5 density on CCR5+ monocytes in older individuals. Further studies with a larger cohort would be necessary to unequivocally establish the effects of age on CCR5 expression in healthy adults.
Although only a few studies have compared CCR5 expression in different population groups, differences have been noted. For example, Ethiopians living in Israel have been shown to express CCR5 at higher levels, both in terms of proportions of CCR5-expressing CD4+ cells and the number of molecules per CD4+ cell, than Israeli Caucasians, despite comparable activation levels as measured by HLA-DR expression.37 However, the prevalence, and hence potential influence upon expression, of the CCR5Δ32 allele in this study was not indicated. In the present study, we show significant expression differences, both in terms of percentage of CCR5-expressing cells and the CCR5 density, between our two study populations, in all cell subsets. The SAA individuals tended to express fewer CCR5 molecules per cell but on a larger proportion of cells than SAC individuals. The most significant differences in CCR5 expression between the two study groups were seen in the NK-cell subsets. The role of NK cells in HIV-1 infection is complex and is not fully understood. Although NK cells are able to directly mediate the killing of HIV-1-infected cells, they are also thought to be involved in the killing of uninfected CD4+ T cells and so contribute towards CD4+ T-cell decline (reviewed by Funke et al.38). Cytotoxic NK-cell subsets, expressing CD4, CCR5 and CXCR4 have been identified.39,40 Furthermore, in vitro studies have shown HIV-1 to infect stimulated CD4+ NK cells expressing HIV-1 co-receptors,39–41 an indication that these cells could serve as HIV-1 reservoirs. It would follow that, similar to T-cell subsets, both the proportion of CCR5-expressing and CCR5 density of NK cells could impact on the progression of HIV-1 infection.
Immune activation levels have previously been reported as environmentally driven.42 Individuals living in Africa, of Italian (Caucasian) and Ugandan origin, were demonstrated to have higher activation levels when compared with individuals from the same population groups living in Italy.42 Interestingly, comparable activation levels, as measured by HLA-DR expression, were seen in both the Italian and Ugandan group living in Uganda,42 contrasting the results in our study where SAA and SAC individuals, largely controlled for environment, differ significantly in HLA-DR expression, i.e. activation levels, in CD4+ T-cell and NK-cell subsets. These differences could be indicative of either the two South African population groups being exposed to differing environmental/activation triggers or that corresponding geographically distinct population groups (SAA versus Ugandans and SAC versus Italians) differ with respect to their genetic background. Similarly, in a study comprising participants of Ethiopian and Israeli origin, Ethiopian individuals that had recently moved to Israel had higher CD4+ HLA-DR+ and CD8+ CD38+ levels compared with Ethiopian individuals who had been living in Israel for > 7 years and Caucasian Israelis.37 Ethiopians who had been living in Israel for > 7 years still had higher activation levels than their Caucasian Israeli counterparts, implying that there must be factors other than environmental, i.e. genetic, to which the higher activation levels can be attributed.37 Furthermore, Kenyan women negative for HIV-1 and other sexually transmitted infections, have increased activated mucosal T cells (CD4+ CD69+ T-cell, CD4+ CD69+ CCR5+ T-cell and CD8+ CD69+ T-cell subsets) compared with women of mixed ethnicity in the USA.43 It was postulated that these differences could be attributed to differences in host genetics or from immune activation caused by chronic systemic infections, which may be more prevalent in Africa.43 Given that the participants in our study were all living in the same environment and had no overt infections or known immune conditions, we postulate that differences in activation levels are more likely to be driven by genetic factors. These studies collectively highlight the complexity of understanding what influences the level of immune activation measurable in a healthy individual and point towards an interplay between the genetics and environment of the individual.
Cell activation results in an increase in CCR5 expression.14,24–26 Activated (HLA-DR+) CD4+ cells have been shown to express CCR5 at higher levels than non-activated (HLA-DR−) CD4+ cells.6,14,37,44 Our results in healthy SAC individuals are in keeping with this; higher proportions of HLA-DR-expressing CD4+ and CD8+ T cells correlated with correspondingly higher density of CCR5 expression. Furthermore, the low expression of activation markers HLA-DR and CD38 on CD4+ T cells is linked to low HIV-1 susceptibility in HIV-1-exposed uninfected individuals.44,45 It is interesting to note that in a report by Reynes et al.,6 the observation is made that although activation, measured by HLA-DR expression on CD4+ T cells, leads to a significant increase in both the number of CCR5 molecules and the proportion of CCR5-expressing cells, the difference in percentage of CCR5-expressing cells between HLA-DR− and HLA-DR+ cell subsets is much greater than differences seen in CCR5 density. A recent report has demonstrated in vivo that activated (HLA-DR+ CD38+) T cells in lymphoid tissues are highly susceptible to infection by R5 tropic HIV-1 viruses and postulated that this is probably because of the observed higher expression of CCR5 on these cells in comparison to other CD4+ T-cell subsets.24
Consistent with the significantly higher activation levels observed in T and NK cells, SAA individuals had greater proportions of CCR5-expressing subsets compared with SAC individuals. However, an inverse relationship was observed in terms of CCR5 density. Several studies have shown that CCR5 density is capable of influencing HIV-1 susceptibility, with higher density associated with increased susceptibility to infection.1–3 Furthermore, it has been reported that CCR5 density on CD4+ T cells of slow progressors versus normal progressors, but not the percentage of CCR5-expressing cells, correlate positively with viral loads.8 Interestingly, CCR5 density was also lower in the slow progressors compared with the uninfected controls.8 However, Meditz et al.24 have recently demonstrated that both CCR5 density and the percentage of CCR5-expressing cells are important determinants of HIV-1 susceptibility in vivo. Hence, both density and the percentage of CCR5-expressing cells appear to be important determinants, coupled with the extent of immune activation, and it is the combinatory effects of these that may predispose to different infection and disease outcome phenotypes. It can be envisaged that larger proportions of CCR5-expressing cells in the periphery would result in larger numbers of cells entering an immune response at the site of infectious encounter; if these are CD4+ cells then more target cells would be available for HIV-1 infection, if among these there are CCR5-expressing NK cells, for example, then those co-expressing CD4 may be vulnerable to infection but others may be more effective killer cells – providing a counter response to control the extent of infection. The overall outcome would be dependent on many factors, including also the concentration of the natural chemokines that bind CCR5, which aside from their role as chemoattractants would also prevent HIV-1 entry by blocking or down-regulating CCR5 expression.
Although increased cellular activation in CD4+ T-cell subsets is associated with increased susceptibility to HIV-1 infection, it is not clear what the functional significance of HLA-DR expression on NK cells may be. The expression of class II MHC proteins such as HLA-DR is usually considered to be restricted to professional antigen-presenting cells (reviewed by Chaplin46). However, HLA-DR expression on both NK-cell and T-cell subsets has been documented and in both these subsets, HLA-DR expression is often used as a biomarker of activation.47–49 HLA-expressing NK cells have been shown to present antigen to trigger T-cell proliferation and interleukin-2 production.50,51 In addition, HLA-DR-expressing NK cells have significantly stronger cytolytic activity in comparison to other NK cells.49 Evans et al.49 demonstrate that the magnitude of an individual’s NK-cell interferon-γ response triggered by the model pathogen, Mycobacterium bovis bacillus Calmette–Guérin, was associated with the initial proportion of HLA-DR-expressing NK cells in peripheral blood mononuclear cells. Hence, HLA-DR-expressing NK cells appear to play an important role in differences in individual responses to bacillus Calmette–Guérin and potentially other pathogens. Although SAA individuals appear to have a larger proportion of activated CD4+ T cells in comparison to SAC individuals, and therefore potentially increased risk of acquiring HIV-1 if exposed, we hypothesize that the concomitant higher NK-cell activation may counteract this potential risk either entirely or to some extent through enhanced killing of activated/infected CD4+ T cells.
Across all subsets examined, the influence of CCR5Δ32 on CCR5 expression was most notable in the T-cell subsets. We observed individuals heterozygous for the CCR5Δ32 allele to have significantly lower percentages of CCR5-expressing CD4+ T cells in comparison to CCR5 ‘wild-type’ individuals, an observation which is in agreement with previous studies.14,52,53 SAC individuals bearing this allele had significantly reduced receptor density on T-cell (CD4+ and CD8+ subsets) and CD56bright NK-subsets. Interestingly, the statistical relationships between CCR5 expression of CCR5Δ32 and ‘wild-type’ individuals were stronger when analysing CCR5 density than when analysing the percentage of CCR5-expressing cells. Similar results have been reported by Shalekoff and Tiemessen.53 Although CCR5Δ32 heterozygosity does have an impact on CCR5 expression, this was not observed across all subsets and furthermore, in agreement with previous studies, some ‘wild-type’ individuals lacking CCR5Δ32 had CCR5 densities comparable to individuals bearing this allele.14,52,53 Consequently, although a significant factor, heterozygosity for the CCR5Δ32 allele is not the only determinant of CCR5 cell surface density.
Although several studies have described differences in CCR5 expression levels between individuals, these have been mainly restricted to a single lymphocyte population (eg. CD4+ T cells), and have mainly addressed only the role of CCR5 as co-receptor for entry of HIV-1 into CD4 T cells. Importantly, we should not overlook that CCR5 also plays a role in T-cell immunity.54,55 In this study we analysed CCR5 expression on T cells, NK cells, B cells and monocytes. Although HIV-1 mainly infects CD4+ T lymphocytes, it is known to have a broad cellular host range infecting a number of different cell populations of the brain, bowel, heart, kidney and liver in addition to haematopoietic cells.56 Aside from the ability of HIV-1 to infect cell types other than CD4+ T cells, the role of differential CCR5 expression on other cell types and their role in the immune response on initial encounter with HIV-1, or in existing chronic infection, need to be better understood to establish the overall impact of differing degrees of CCR5 expression in vivo.
We found CCR5 expression to differ substantially between SAA and SAC individuals and that these differences can be attributed to factors other than the CCR5Δ32 allele. In addition, these two groups differ substantially in their activation levels as measured by HLA-DR. Further understanding of what contributes towards the large inter-individual and inter-population variability in CCR5 expression, and understanding the implications thereof, will assist in the design of prophylactic and therapeutic strategies that work in conjunction with the host’s immune response to HIV-1.
Acknowledgments
This study was supported by grants from the Poliomyelitis Research Foundation (PRF) and the Technology Innovation Agency [South African HIV/AIDS Research (and Innovation) Platform (SHARP)]. We would like to thank all our volunteer participants in this study.
Disclosures
All authors declare that there is no conflict of interest.
References
- 1.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci U S A. 1999;96:5215–20. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–77. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 3.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–64. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heredia A, Gilliam B, DeVico A, et al. CCR5 density levels on primary CD4 T cells impact the replication and Enfuvirtide susceptibility of R5 HIV-1. AIDS. 2007;21:1317–22. doi: 10.1097/QAD.0b013e32815278ea. [DOI] [PubMed] [Google Scholar]
- 5.Lin YL, Mettling C, Portales P, Reynes J, Clot J, Corbeau P. Cell surface CCR5 density determines the postentry efficiency of R5 HIV-1 infection. Proc Natl Acad Sci U S A. 2002;99:15590–5. doi: 10.1073/pnas.242134499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynes J, Portales P, Segondy M, et al. CD4+ T cell surface CCR5 density as a determining factor of virus load in persons infected with human immunodeficiency virus type 1. J Infect Dis. 2000;181:927–32. doi: 10.1086/315315. [DOI] [PubMed] [Google Scholar]
- 7.Gervaix A, Nicolas J, Portales P, et al. Response to treatment and disease progression linked to CD4+ T cell surface CC chemokine receptor 5 density in human immunodeficiency virus type 1 vertical infection. J Infect Dis. 2002;185:1055–61. doi: 10.1086/339802. [DOI] [PubMed] [Google Scholar]
- 8.Reynes J, Portales P, Segondy M, et al. CD4 T cell surface CCR5 density as a host factor in HIV-1 disease progression. AIDS. 2001;15:1627–34. doi: 10.1097/00002030-200109070-00004. [DOI] [PubMed] [Google Scholar]
- 9.Heredia A, Latinovic O, Gallo RC, Melikyan G, Reitz M, Le N, Redfield RR. Reduction of CCR5 with low-dose rapamycin enhances the antiviral activity of vicriviroc against both sensitive and drug-resistant HIV-1. Proc Natl Acad Sci U S A. 2008;105:20476–81. doi: 10.1073/pnas.0810843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ketas TJ, Kuhmann SE, Palmer A, Zurita J, He W, Ahuja SK, Klasse PJ, Moore JP. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology. 2007;364:281–90. doi: 10.1016/j.virol.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desmetz C, Lin YL, Mettling C, Portales P, Rabesandratana H, Clot J, Corbeau P. The strength of the chemotactic response to a CCR5 binding chemokine is determined by the level of cell surface CCR5 density. Immunology. 2006;119:551–61. doi: 10.1111/j.1365-2567.2006.02470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–62. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 13.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 14.Wu L, Paxton WA, Kassam N, et al. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–91. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galvani AP, Novembre J. The evolutionary history of the CCR5-Delta32 HIV-resistance mutation. Microbes Infect. 2005;7:302–9. doi: 10.1016/j.micinf.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Petersen DC, Kotze MJ, Zeier MD, Grimwood A, Pretorius D, Vardas E, van Rensburg EJ, Hayes VM. Novel mutations identified using a comprehensive CCR5-denaturing gradient gel electrophoresis assay. AIDS. 2001;15:171–7. doi: 10.1097/00002030-200101260-00005. [DOI] [PubMed] [Google Scholar]
- 17.Moore JP, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–62. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 19.Lin YL, Mettling C, Portales P, Rouzier R, Clot J, Reynes J, Corbeau P. The chemokine CCL5 regulates the in vivo cell surface expression of its receptor, CCR5. AIDS. 2008;22:430–2. doi: 10.1097/QAD.0b013e3282f46a6f. [DOI] [PubMed] [Google Scholar]
- 20.Yang YF, Tomura M, Iwasaki M, et al. IFN-α acts on T-cell receptor-triggered human peripheral leukocytes to up-regulate CCR5 expression on CD4+ and CD8+ T cells. J Clin Immunol. 2001;21:402–9. doi: 10.1023/a:1013173610032. [DOI] [PubMed] [Google Scholar]
- 21.Yang YF, Tomura M, Iwasaki M, et al. IL-12 as well as IL-2 upregulates CCR5 expression on T cell receptor-triggered human CD4+ and CD8+ T cells. J Clin Immunol. 2001;21:116–25. doi: 10.1023/a:1011059906777. [DOI] [PubMed] [Google Scholar]
- 22.Zou W, Foussat A, Houhou S, et al. Acute upregulation of CCR-5 expression by CD4+ T lymphocytes in HIV-infected patients treated with interleukin-2. ANRS 048 IL-2 Study Group. AIDS. 1999;13:455–63. doi: 10.1097/00002030-199903110-00003. [DOI] [PubMed] [Google Scholar]
- 23.Weissman D, Dybul M, Daucher MB, Davey RT, Jr, Walker RE, Kovacs JA. Interleukin-2 up-regulates expression of the human immunodeficiency virus fusion coreceptor CCR5 by CD4+ lymphocytes in vivo. J Infect Dis. 2000;181:933–8. doi: 10.1086/315303. [DOI] [PubMed] [Google Scholar]
- 24.Meditz AL, Haas MK, Folkvord JM, et al. HLA-DR+ CD38+ CD4+ T lymphocytes have elevated CCR5 expression and produce the majority of R5-tropic HIV-1 RNA in vivo. J Virol. 2011;85:10189–200. doi: 10.1128/JVI.02529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moriuchi H, Moriuchi M, Fauci AS. Cloning and analysis of the promoter region of CCR5, a coreceptor for HIV-1 entry. J Immunol. 1997;159:5441–9. [PubMed] [Google Scholar]
- 26.Ostrowski MA, Justement SJ, Catanzaro A, et al. Expression of chemokine receptors CXCR4 and CCR5 in HIV-1-infected and uninfected individuals. J Immunol. 1998;161:3195–201. [PubMed] [Google Scholar]
- 27.Shalekoff S, Pendle S, Johnson D, Martin DJ, Tiemessen CT. Distribution of the human immunodeficiency virus coreceptors CXCR4 and CCR5 on leukocytes of persons with human immunodeficiency virus type 1 infection and pulmonary tuberculosis: implications for pathogenesis. J Clin Immunol. 2001;21:390–401. doi: 10.1023/a:1013121625962. [DOI] [PubMed] [Google Scholar]
- 28.Picton AC, Paximadis M, Tiemessen CT. Genetic variation within the gene encoding the HIV-1 CCR5 coreceptor in two South African populations. Infect Genet Evol. 2010;10:487–94. doi: 10.1016/j.meegid.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berhanu D, Mortari F, De Rosa SC, Roederer M. Optimized lymphocyte isolation methods for analysis of chemokine receptor expression. J Immunol Methods. 2003;279:199–207. doi: 10.1016/s0022-1759(03)00186-8. [DOI] [PubMed] [Google Scholar]
- 30.Naranbhai V, Bartman P, Ndlovu D, Ramkalawon P, Ndung’u T, Wilson D, Altfeld M, Carr WH. Impact of blood processing variations on natural killer cell frequency, activation, chemokine receptor expression and function. J Immunol Methods. 2011;366:28–35. doi: 10.1016/j.jim.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shalekoff S, Tiemessen CT. Duration of sample storage dramatically alters expression of the human immunodeficiency virus coreceptors CXCR4 and CCR5. Clin Diagn Lab Immunol. 2001;8:432–6. doi: 10.1128/CDLI.8.2.432-436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giltay EJ, Fonk JC, von Blomberg BM, Drexhage HA, Schalkwijk C, Gooren LJ. In vivo effects of sex steroids on lymphocyte responsiveness and immunoglobulin levels in humans. J Clin Endocrinol Metab. 2000;85:1648–57. doi: 10.1210/jcem.85.4.6562. [DOI] [PubMed] [Google Scholar]
- 33.Mo R, Chen J, Grolleau-Julius A, Murphy HS, Richardson BC, Yung RL. Estrogen regulates CCR gene expression and function in T lymphocytes. J Immunol. 2005;174:6023–9. doi: 10.4049/jimmunol.174.10.6023. [DOI] [PubMed] [Google Scholar]
- 34.Kivisakk P, Trebst C, Lee JC, Tucky BH, Rudick RA, Campbell JJ, Ransohoff RM. Expression of CCR2, CCR5, and CXCR3 by CD4+ T cells is stable during a 2-year longitudinal study but varies widely between individuals. J Neurovirol. 2003;9:291–9. doi: 10.1080/13550280390201001. [DOI] [PubMed] [Google Scholar]
- 35.Mo R, Chen J, Han Y, Bueno-Cannizares C, Misek DE, Lescure PA, Hanash S, Yung RL. T cell chemokine receptor expression in aging. J Immunol. 2003;170:895–904. doi: 10.4049/jimmunol.170.2.895. [DOI] [PubMed] [Google Scholar]
- 36.Yung RL, Mo R. Aging is associated with increased human T cell CC chemokine receptor gene expression. J Interferon Cytokine Res. 2003;23:575–82. doi: 10.1089/107999003322485071. [DOI] [PubMed] [Google Scholar]
- 37.Kalinkovich A, Borkow G, Weisman Z, Tsimanis A, Stein M, Bentwich Z. Increased CCR5 and CXCR4 expression in Ethiopians living in Israel: environmental and constitutive factors. Clin Immunol. 2001;100:107–17. doi: 10.1006/clim.2001.5040. [DOI] [PubMed] [Google Scholar]
- 38.Funke J, Durr R, Dietrich U, Koch J. Natural killer cells in HIV-1 infection: a double-edged sword. AIDS Rev. 2011;13:67–76. [PubMed] [Google Scholar]
- 39.Valentin A, Rosati M, Patenaude DJ, et al. Persistent HIV-1 infection of natural killer cells in patients receiving highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 2002;99:7015–20. doi: 10.1073/pnas.102672999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernstein HB, Wang G, Plasterer MC, Zack JA, Ramasastry P, Mumenthaler SM, Kitchen CM. CD4+ NK cells can be productively infected with HIV, leading to downregulation of CD4 expression and changes in function. Virology. 2009;387:59–66. doi: 10.1016/j.virol.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harada H, Goto Y, Ohno T, Suzu S, Okada S. Proliferative activation up-regulates expression of CD4 and HIV-1 co-receptors on NK cells and induces their infection with HIV-1. Eur J Immunol. 2007;37:2148–55. doi: 10.1002/eji.200737217. [DOI] [PubMed] [Google Scholar]
- 42.Clerici M, Butto S, Lukwiya M, et al. Immune activation in africa is environmentally-driven and is associated with upregulation of CCR5. Italian-Ugandan AIDS Project. AIDS. 2000;14:2083–92. doi: 10.1097/00002030-200009290-00003. [DOI] [PubMed] [Google Scholar]
- 43.Cohen CR, Moscicki AB, Scott ME, et al. Increased levels of immune activation in the genital tract of healthy young women from sub-Saharan Africa. AIDS. 2010;24:2069–74. doi: 10.1097/QAD.0b013e32833c323b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Begaud E, Chartier L, Marechal V, et al. Reduced CD4 T cell activation and in vitro susceptibility to HIV-1 infection in exposed uninfected Central Africans. Retrovirology. 2006;3:35. doi: 10.1186/1742-4690-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koning FA, Otto SA, Hazenberg MD, Dekker L, Prins M, Miedema F, Schuitemaker H. Low-level CD4+ T cell activation is associated with low susceptibility to HIV-1 infection. J Immunol. 2005;175:6117–22. doi: 10.4049/jimmunol.175.9.6117. [DOI] [PubMed] [Google Scholar]
- 46.Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125(2 Suppl 2):S3–23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans RL, Faldetta TJ, Humphreys RE, Pratt DM, Yunis EJ, Schlossman SF. Peripheral human T cells sensitized in mixed leukocyte culture synthesize and express Ia-like antigens. J Exp Med. 1978;148:1440–5. doi: 10.1084/jem.148.5.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ko HS, Fu SM, Winchester RJ, Yu DT, Kunkel HG. Ia determinants on stimulated human T lymphocytes. Occurrence on mitogen- and antigen-activated T cells. J Exp Med. 1979;150:246–55. doi: 10.1084/jem.150.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans JH, Horowitz A, Mehrabi M, Wise EL, Pease JE, Riley EM, Davis DM. A distinct subset of human NK cells expressing HLA-DR expand in response to IL-2 and can aid immune responses to BCG. Eur J Immunol. 2011;41:1924–33. doi: 10.1002/eji.201041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roncarolo MG, Bigler M, Haanen JB, Yssel H, Bacchetta R, de Vries JE, Spits H. Natural killer cell clones can efficiently process and present protein antigens. J Immunol. 1991;147:781–7. [PubMed] [Google Scholar]
- 51.Hanna J, Gonen-Gross T, Fitchett J, et al. Novel APC-like properties of human NK cells directly regulate T cell activation. J Clin Invest. 2004;114:1612–23. doi: 10.1172/JCI22787. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.de Roda Husman AM, Blaak H, Brouwer M, Schuitemaker H. CC chemokine receptor 5 cell-surface expression in relation to CC chemokine receptor 5 genotype and the clinical course of HIV-1 infection. J Immunol. 1999;163:4597–603. [PubMed] [Google Scholar]
- 53.Shalekoff S, Tiemessen CT. CCR5 delta32 heterozygosity is associated with an increase in CXCR4 cell surface expression. AIDS Res Hum Retroviruses. 2003;19:531–3. doi: 10.1089/088922203766774595. [DOI] [PubMed] [Google Scholar]
- 54.Camargo JF, Quinones MP, Mummidi S, et al. CCR5 expression levels influence NFAT translocation, IL-2 production, and subsequent signaling events during T lymphocyte activation. J Immunol. 2009;182:171–82. doi: 10.4049/jimmunol.182.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dolan MJ, Kulkarni H, Camargo JF, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8:1324–36. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 56.Levy JA. HIV pathogenesis: knowledge gained after two decades of research. Adv Dent Res. 2006;19:10–6. doi: 10.1177/154407370601900104. [DOI] [PubMed] [Google Scholar]


