Abstract
BACKGROUND
Measurement of prostate specific antigen (PSA) in prostate cancer patients following radical prostatectomy (RP) has been limited by the sensitivity of available assays. Because radical prostatectomy removes the tissue responsible for PSA production, post-surgical PSA is typically undetectable with current assay methods. However, evidence suggests that more sensitive determination of PSA status following RP could improve assessment of patient prognosis, response to treatment, and better target secondary therapy to those who may benefit most. We report the development and validation of an investigational digital immunoassay with two logs greater sensitivity than today’s ultrasensitive third-generation PSA assays.
METHODS
Reagents were developed for a paramagnetic bead-based ELISA for use with high-density arrays of femtoliter-volume wells. Anti-PSA capture-beads with immunocomplexes and associated enzyme labels were singulated within the wells of the arrays and interrogated for the presence of enzymatic product. Analytical performance of the assay was characterized, its accuracy compared with a commercially available test, and longitudinal serum samples from a pilot study of 33 RP patients were analyzed.
RESULTS
The assay exhibited a functional sensitivity (20% inter-assay CV) of less than 0.00005 ng/mL (0.05 pg/mL), total imprecision of less than 10% from 1 to 50 pg/mL, and excellent agreement with the comparator method. All RP samples were well within the assay measurement capability. PSA values following surgery were examined in the context of five-year biochemical cancer recurrence.
CONCLUSION
The assay demonstrated a robust two-log advance in measurement sensitivity relative to current ultrasensitive assays, and the analytical performance for accurate assessment of PSA status after RP.
Introduction
Measurement of prostate specific antigen (PSA) following radical prostatectomy (RP) has become standard practice for prostate cancer recurrence monitoring. PSA is typically undetectable by most assay methods following surgery, and it is generally agreed that undetectable post-surgical PSA over time is indicative of a good prognosis (1-3). Selection of individuals for potential post-surgical adjuvant therapy has been informed primarily by pre-surgical clinicopathologic information. Assessment of surgical and secondary treatment effectiveness has relied on monitoring for PSA rise using assays that are unable to measure PSA at very low levels. As long as PSA remains undetectable, the patient can be assured that there is no biochemical evidence of cancer recurrence. Indeed, the less sensitive the assay, the longer this assurance can be offered, while in fact, PSA could be rising significantly below detectable levels. The kinetics of rising PSA after RP can be fast or slow, and the period of undetectable PSA can be brief or on the order of years. Considerations of potential salvage therapies await the emergence of PSA above a reliably quantifiable threshold, generally 0.1 or 0.2 ng/mL. Practitioner response to reemergence of rising PSA after RP has depended on a number of factors, including the definition of biochemical cancer recurrence (BCR), the time to biochemical failure and its potential clinical significance, clinicopathological aspects, and patient life expectancy (4). Limited by the sensitivity of PSA assay methods, BCR is generally defined as a confirmed PSA level of 0.2 ng/mL (200 pg/mL) or greater. Biochemical relapse occurs in up to 40% of patients after RP (5-8), and a third of these patients develop metastatic disease with a 20% probability of dying within 10 years (9, 10). Importantly, clinical data are growing that early adjuvant and salvage radiation therapies following surgery have significantly improved patient outcomes (11-14). However, current assays are unable to measure PSA at levels that could benefit patients with the earliest intervention and prospective selection for adjuvant treatment. A key issue that a greater sensitivity PSA assay could impact is: which patients are most likely to benefit, and for which patients would this represent over-treatment?
Ultrasensitive monitoring of post-surgical PSA was initially examined in the 1990’s with the development of the first ultrasensitive immunoassays capable of measuring down to 0.01 ng/mL. A number of reports established the prognostic significance of serial ultrasensitive PSA measurements (15-18). Evyenia et al. (17) showed an ultrasensitive method could detect PSA rises well before a first-generation assay, providing potentially years of early warning. Ultrasensitive testing also elucidated different categories of recurrence: fast, slow, and apparent non-recurrence (17). Recently, an ultrasensitive radioimmunoassay with a detection cut off of 0.001 ng/mL was used to retrospectively explore the prognostic significance of the lowest post-surgical PSA (nadir PSA) for biochemical recurrence. While PSA was undetectable in approximately half the patients, recurrence risk could nonetheless be stratified depending on nadir PSA level (19). This result mirrored other studies (20-24), indicating a significant relevance of ultra low PSA measurement for predicting long term BCR-free survival.
Despite substantial data on the benefits of ultrasensitive PSA measurement, differences of opinion have persisted on the use and utility of such monitoring. The debate is due in part to differing views on the practical and clinical relevance of BCR, patient anxiety considerations, and both the analytical and biological noise associated with measuring ultra low levels of PSA. It seems clear, however, that inability to measure PSA below 0.1 ng/mL makes it more difficult to objectively assess patient response to surgical and adjuvant therapy in the timeliest manner, and it impedes identification of candidacy for salvage treatment during the critical period when residual cancer is most amenable to such treatment. The ability to accurately quantify PSA values for all RP patients could improve assessment of patient prognosis, response to treatment, and better target secondary therapy to those who may benefit most.
Ultrasensitive PSA assays are referred to as third-generation by being two logs more sensitive than the first generation of PSA radioimmunoassays. It is important to be clear that sensitivity is taken to mean functional day-to-day measurement sensitivity (Limit of Quantification, LoQ), which is commonly defined as the lowest PSA concentration at which measurement variability over time does not exceed 20%. Thus, first-generation PSA assays exhibited LoQs of 0.5 - 1 ng/mL, while third-generation assays are capable of 0.01 ng/mL (5). A fourth-generation assay would represent another 10-fold improvement (</= 0.001 ng/mL), while fifth-generation performance would imply a LoQ of 0.0001 ng/mL or better. Attempts to develop fourth-generation PSA assays have recently been reported. McDermed et al. (25) presented data from an immuno-PCR method with a reported LoQ of < 0.001 ng/mL, but minimal analytical data have been published. Thaxton et al. (26) reported a gold nanoparticle bio-barcode assay with a statistical detection limit of 0.0003 ng/mL. Although the LoQ was not described, the assay was able to measure PSA in most RP specimens tested.
We have previously reported preliminary data from a PSA assay based on a novel digital immunoassay technology utilizing high-density arrays of femtoliter-volume wells and single molecule counting (27, 30). Here we report detailed analytical validation data from the assay (brand name AccuPSA®) demonstrating robust fifth-generation performance. The assay has a LoQ of less than 0.00005 ng/mL, and reliably quantified serum PSA in all post radical prostatectomy patients tested. The test can potentially be used to measure PSA in patients following primary and secondary therapy, improve BCR risk stratification, and better inform clinical decisions for use of secondary treatment.
Materials and Methods
SINGLE MOLECULE ARRAYS
Single Molecule Array (SiMoA) technology (27) involves performing a paramagnetic bead-based ELISA, followed by isolation of individual capture beads in arrays of femtoliter-sized reaction wells. Singulation of capture beads within microwells permits buildup of fluorescent product from an enzyme label, such that signal from a single immunocomplex can be readily detected with a CCD camera. At very low PSA concentrations, Poisson statistics predict that bead-containing microwells in the array will contain either a single labeled PSA molecule or no PSA molecules, resulting in a digital signal. With single-molecule sensitivity, concentrations of labeling reagents can be lowered, resulting in reduced non-specific background (27). This effect enables high signal:background ratios at extremely low analyte concentrations.
Arrays of femtoliter-volume wells were prepared as described [27, 30]. In brief, the ends of bundles of 50,000 optical fibers were polished with diamond lapping films. One end of each bundle was etched in mild acid solution. Differential etch rates of the optical fiber core and cladding glass of the bundles causes 4.5 μm diameter, 3.5 μm deep wells to be formed, giving an array of 50,000 microwells across the bundle. Optical fiber arrays were mounted in linear groups of eight within glass holders for bead loading and imaging. Groups of eight arrays were chosen to correspond with microtiter plate columns of eight wells, which were used as rinse troughs for washing array surfaces following bead loading.
ELISA REAGENTS
Three reagents were developed: paramagnetic PSA capture beads, biotinylated detector, and a streptavidin:β-galactosidase (SβG) conjugate. The capture beads were comprised of a monoclonal anti-PSA antibody (BiosPacific) directed to amino acid residues 158-163. The antibody was covalently attached by standard coupling chemistry to 2.7 μm carboxy paramagnetic microbeads (Varian). The antibody-coated beads were diluted to a concentration of 5 × 106 beads/mL in Tris with a surfactant and BSA. Biotinylated detector reagent was comprised of a monoclonal anti PSA antibody (BiosPacific) directed to amino acid residues 3-11. The antibody was biotinylated using standard methods and diluted to a concentration of 0.15 μg/ml in a PBS diluent containing a surfactant and newborn calf serum, NCS (PBS/NCS). SβG was prepared by covalent conjugation of purified streptavidin (Thermo Scientific) and βG (Sigma) using standard coupling chemistry. For assay, aliquots of a concentrated SβG stock were diluted to 15 pM in PBS/NCS with 1 mM MgCl2.
CALIBRATION
The assay was calibrated using WHO 90:10 PSA standards (National Institute for Biological Standards and Control). A stock PSA solution was prepared by dilution to 2 mg/mL in PBS/Tween-20. Assay calibrators were prepared by dilution of the stock solution in 25% NCS/PBS with Tween-20, EDTA and ProClin 300. Calibrators were prepared in a serial series from 0.1 to 100 pg/ml to emphasize quantification accuracy below 100 pg/mL. Recovery studies indicated that use of NCS as a calibrator base gave equivalent accuracy to human serum (not shown).
ELISA ASSAY
Bead-sample incubations and labeling of immunocomplexes in conical 96 well plates (Axygen) were conducted as described (28). In brief, the assay was performed in three steps, starting with analyte capture, incubation with biotinylated detector, and labeling of the immunocomplexes with SβG. Following assay and bead collection with a magnet, beads were loaded onto the arrays for imaging in a loading buffer comprised of PBS and 0.01% Tween-20, MgCl2, and sucrose.
ARRAY IMAGING
Beads from the ELISA were loaded onto the arrays as described (27, 28). Wells containing beads with labeled PSA were visualized by the hydrolysis of enzyme substrate (resorufin β-D-galactopyranoside, RGP, Invitrogen) by βG into fluorescent product. RGP was introduced to the wells during sealing of the arrays with a silicon gasket. Enzyme-containing wells were imaged by fluorescence microscope fitted with a CCD camera. The images were analyzed to determine the average number of label enzymes/bead (AEB) as described (30). At <70% active beads relative to total beads (low PSA), the signal output is a count of active beads corrected for a low statistical probability of multiple enzymes/bead (29). At >70% active beads (higher PSA), the probability of multiple enzymes/bead increases, and average fluorescence of the wells is converted to AEB based on the average intensities of wells containing single enzymes determined at lower concentrations. The AEB unit thus works continuously across the digital and analog realms (30).
RP PATIENTS
Retrospective longitudinal serum samples from 20 non-recurring (BCR-free for five years) and 13 biochemically recurring RP patients were obtained under IRB approval and de-identified from New York University (Urology Associates) and Johns Hopkins University. All subjects had undergone radical retropubic prostatectomy without neo-adjuvant hormonal therapy. Targeted longitudinal sampling was a serum draw between 3 and 6 months after RP (nadir PSA), followed 3-6 months later by two subsequent draws separated by 3-6 months. Patients with positive lymph nodes at the time of surgery were excluded, as were patients who received neo-adjuvant or adjuvant therapy prior to BCR. BCR was defined as two consecutive PSA levels ≥ 0.2 ng/mL (200 pg/mL) after the initial collected sample, or secondary treatment. Table 1 summarizes specific data for each patient.
Table 1.
Patient Data
| Patient | Age | Follow up, years |
Pre-RRP PSA, ng/mL |
Pathologic Gleason score |
Pathologic stage |
Postoperative treatment |
|---|---|---|---|---|---|---|
| 193 | 54 | 7.8 | 4.8 | 3+3=6 | pT2a | No |
| 219 | 66 | 7.0 | 6.5 | 3+3=6 | pT2b | No |
| 120 | 65 | 8.2 | 5.9 | 4+4=8 | pT2a | No |
| 125 | 68 | 8.0 | 5.8 | 3+3=6 | pT2a | No |
| 335 | 67 | 7.2 | 4.9 | 3+3=6 | pT2b | No |
| 393 | 61 | 8.0 | 5.0 | 3+4=7 | pT2b | No |
| 157 | 70 | 6.9 | 5.5 | 3+4=7 | pT2b | No |
| 190 | 70 | 5.1 | 6.3 | 3+4=7 | pT2b | No |
| 394 | 46 | 3.8 | 5.7 | 3+3=6 | pT2a | No |
| 9458 | 61 | 10.0 | 15.0 | 3+2=5 | pT2c | No |
| S569 | 61 | 10.1 | 6.9 | 3+4=7 | pT2c | No |
| S1576 | 56 | 10.2 | 6.7 | 3+3=6 | pT2a | No |
| S2278 | 56 | 10.4 | 4.1 | 5+4=9 | pT2b | No |
| S9956 | 66 | 7.1 | 6.2 | 3+3=6 | pT2c | No |
| A3181 | 63 | 6.4 | 1.5 | 3+3=6 | pT2c | No |
| A6815 | 66 | 5.1 | 9.2 | 3+3=6 | pT2a | No |
| 9082 | 56 | 7.9 | 5.3 | 3+4=7 | pT2c | No |
| 8802 | 44 | 11.1 | 1.7 | 3+3=6 | pT2c | No |
| 5644 | 61 | 5.6 | 7.3 | 3+3=6 | pT2a | No |
| 9569 | 68 | 8.2 | 14.7 | 3+4=7 | pT3a | No |
| 172 | 62 | 0.5 | 10.0 | 3+4=7 | pT3a | Yes |
| 367A | 61 | 2.0 | 4.3 | 4+3=7 | pT2a | Yes |
| 475A | 63 | 1.0 | 6.5 | 4+3=7 | pT3c | Yes |
| 8929 | 61 | 1.9 | 11.2 | 4+5=9 | pT3a | No |
| 9908 | 56 | 3.0 | 13.4 | 3+2=5 | pT2a | No |
| 7288 | 59 | 2.0 | 10.0 | 3+4=7 | pT3a | Yes |
| 0138 | 62 | 2.0 | 8.7 | 3+4=7 | pT3b | Yes |
| 2209 | 57 | 1.2 | 6.6 | 3+4=7 | pT3a | Yes |
| 9645 | 46 | 4.4 | 12.2 | 3+4=7 | pT3b | Yes |
| 3710 | 57 | 5.3 | 27.9 | 4+5=9 | pT3b | No |
| 4789 | 59 | 6.0 | 7.3 | 4+5=9 | pT2c | No |
| 7795 | 67 | 1.1 | 3.5 | 3+3=6 | pT2c | No |
| 2049 | 69 | 0.5 | 5.6 | 4+3=7 | pT3a | Yes |
SAMPLE HANDLING AND MEASUREMENT OF SERUM PSA
Specimens were stored at −70°C until assayed. To limit effects of potential interferences, thawed samples were centrifuged at 9000g for 3-5 minutes and pre-diluted 1:4 in a diluent containing PBS with 0.01% Tween-20, heterophilic blocker, and EDTA prior to assay. Samples and calibrators were assayed in triplicate, and serial patient samples were tested within a single plate. Specimens above the highest calibrator were diluted 100-fold with the zero calibrator and re-assayed.
Results
DOSE-RESPONSE, LINEARITY, AND RECOVERY
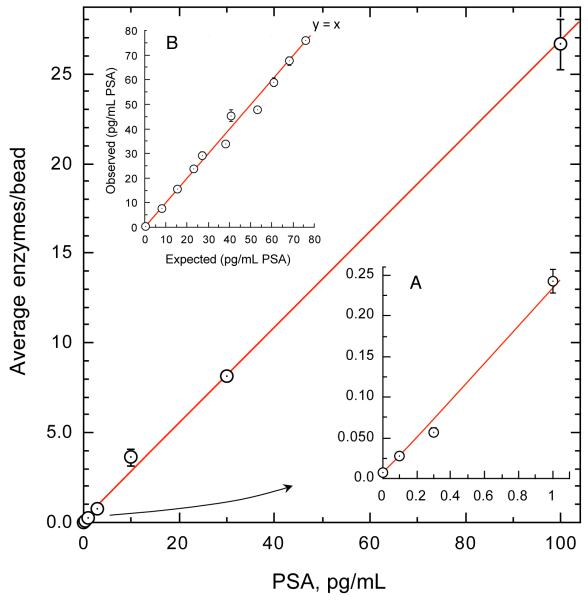
Fig. 1 shows a representative dose-response across three and a half logs of range. The assay demonstrated a highly linear response (R2=0.999). In a study of 20 calibration curves over 10 days, the mean signal to noise ratio at 0.1 pg/mL was 4.33 (SD 0.76). Linearity, conducted with guidance from CLSI protocol EP6-A (31), was evaluated with admixtures of female serum exhibiting relatively high and very low PSA levels (Fig. 1B). Linear (depicted) and 3rd order polynomial fit goodness was virtually identical (R2=0.988 and 0.990 respectively). Percent deviation from linearity between the two models was within 5% across the range. Recovery of spiked PSA from serum in the absence and presence of supplemented high levels of potential endogenous interferences (20 mg/dL bilirubin, 1000 mg/dL triglycerides, 12 g/dL protein, 20 mg/dL hemoglobin) was within 10% of expected.
Fig. 1. Dose-response and linearity of SiMoA PSA assay.
Y-axis refers to the average number of label enzymes per individual microbead captured in the array. Fitting for optimal read-back utilized four-parameter logistical regression. Fig. 1A highlights the low background obtained with digital quantification. 20 calibration curves gave a mean signal:background ratio at 0.1 pg/mL of 4.33. Fig. 1B depicts linearity obtained from admixtures of high and low female serum samples.
SENSITIVITY
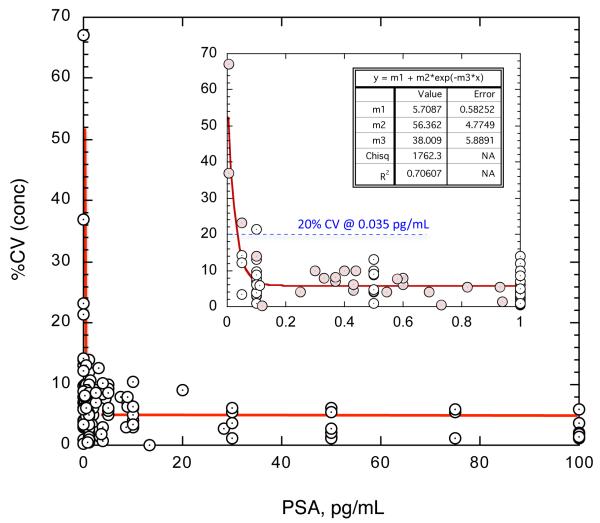
Analytical Limit of Detection (LoD) was estimated as three standard deviations above background. LoD was calculated for each of 20 calibration runs from triplicate measurements of the zero calibrator and the lowest PSA-containing calibrator (0.1 pg/mL). The mean LoD was 0.028 pg/mL (SD 0.039 pg/mL). The LoQ was estimated from sample replicate CVs (n=3) obtained across the assay range over six weeks. The resulting CV profile is depicted in Fig. 2. The replicates were obtained from repeated measurement of assay calibrators, controls, and female serum. CVs for the different sample types were not statistically different. The estimated LoQ was the concentration of PSA corresponding to a 20% CV. From the equation of the power fit, the LoQ was calculated as 0.0352 pg/mL (standard error 0.0340 – 0.0387 pg/mL).
Fig. 2. Limit of Quantification (LoQ) of SiMoA PSA assay.
LoQ was defined as the concentration of PSA at which measurement variation over time reached 20%. LoQ was estimated by non-linear power fit of sample replicate CVs across six weeks of testing. The equation of the fit gave a LoQ of 0.0352 pg/mL (standard error 0.0340 – 0.0387 pg/mL). Female serum samples are highlighted in pink.
REPRODUCIBILITY
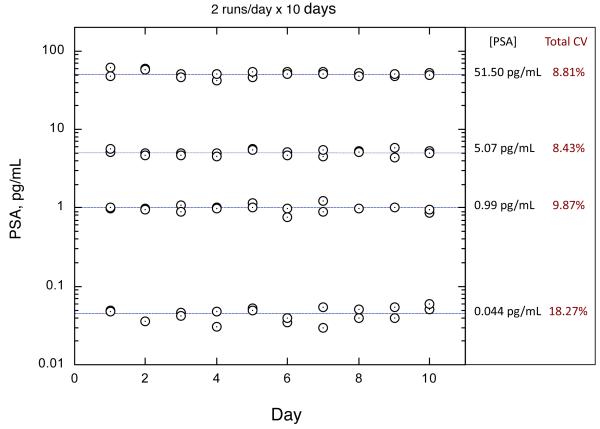
Reproducibility was assessed with guidance from CLSI EP5-A2 (32). Four samples, consisting of 90:10 PSA spiked into 25% NCS, were assayed in triplicate in each of two separate runs per day for 10 days (n=60 for each sample). The lowest sample was prepared near the estimated LoQ (0.035 pg/mL). Because each reportable result is based on triplicate measurements, this protocol gave two results/day for each sample. The plate map was configured such that each PSA result spanned multiple columns, which meant that replicates included variation from different groups of arrays. PSA results were calculated from within-plate calibration curves. Thus, the overall study comprehended array processing variation, calibration variation, and within-run, between-run, and day-to-day variation. The results of the study are depicted in Fig. 3. Total CVs across all variation sources were less than 10% from 1 to 52 pg/mL PSA. The total CV for the 0.04 pg/mL sample was 18.27%, consistent with the LoQ estimate (20% CV at 0.035 pg/mL).
Fig. 3. Reproducibility of SiMoA PSA assay.
Total imprecision was estimated by repeated measurement of a panel of prepared PSA samples over a 10-day period with two runs/day. Variation sources included fiber strips and processing, inter-calibration, and day-to-day reproducibility. The lowest sample was prepared to approximate the LoQ, and the total imprecision obtained was consistent with the LoQ estimate (20% CV at 0.035 pg/mL).
ACCURACY
Accuracy was assessed by comparison to a commercially available equimolar PSA method standardized with WHO reference material (33). 40 serum samples from normal males and eight serum samples from RP patients with PSA levels high enough for measurement in the comparator method (ADVIA Centaur, Siemens; LoD 0.1 ng/mL) were assayed with both methods (see online Supplemental Fig. 1). All samples were diluted 100-fold prior to testing with the SiMoA assay. The assays exhibited excellent agreement (R2=0.970) with no significant bias throughout the range of results (0.17 to >13 ng/mL, mean bias 0.024 ng/mL).
CLINICAL SAMPLES
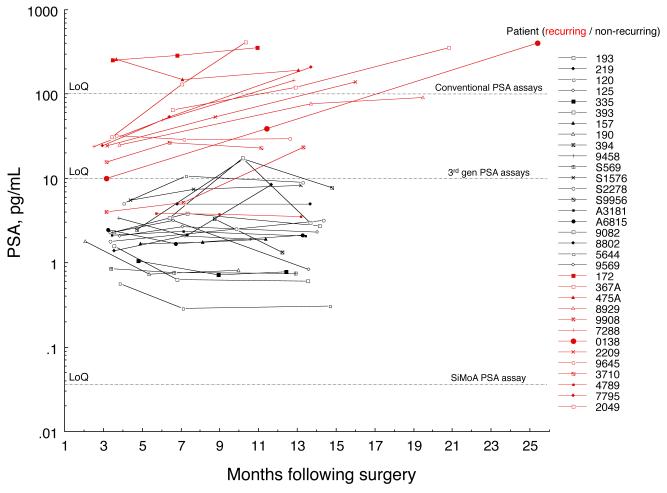
PSA results from all pilot study samples are shown in Fig. 4. Patient demographics, Gleason Scores, and pathology stages were similar for recurrent (n=20) and non-recurrent (n=13) groups (Table 1). Mean ages were 61.0 and 61.5 years for recurrent and non-recurrent men respectively. Gleason Scores were 6-7 for the large majority of men (100% and 80% of recurrent and non-recurring respectively), while pathology stage was fairly evenly distributed among pT2a, pT2b, and pT2c for both cohorts. Clinical stage was T1c for approximately half the recurring men, and three-quarters of non-recurring men. The remaining recurring men were evenly distributed across T2a and T2b, while all but one of the remaining non-recurring patients was T2a, with one non-recurring patient classified T2b. The large majority of both groups exhibited negative margins (91% and 95% of recurring and non-recurring men respectively.) Across all patients, approximately half of the initial PSA values were below the LoQ for commercially available third-generation assays (~10 pg/mL). All samples were at least 10-fold above the LoQ of the SiMoA assay. Replicate CVs were consistent with the 10-day precision study.
Fig. 4. Post RP PSA results.
Longitudinal samples were tested from 13 recurring (red) and 20 non-recurring (black) RP patients. All samples were well above the LoQ of the assay and were measured with good precision. Horizontal lines depict LoQs. The initial PSA value (nadir PSA) was a significant predictor of 5-year biochemical recurrence-free survival (p<0.01), while PSA slope was not a significant predictor in this study (p>0.05).
Fig. 4 highlights the relationship between the nadir PSA and BCR: all patients with a nadir above 10 pg/mL experienced biochemical relapse (red), while all patients with a nadir below 1 pg/mL remained BCR-free for at least five years (black). Bifurcation of the data at 3.0 pg/mL resulted in a sensitivity and specificity of 100% and 75% respectively. This cut point is below the measurement capability of ultrasensitive PSA assays, therefore improvement in clinical sensitivity may be possible from reliable PSA quantification in the formally “undetectable” category. Reciever operating characteristic analysis gave an area under the curve of 0.991, suggesting excellent discrimination between recurring and non-recurring groups (see online Supplemental Fig. 2). Slopes of longitudinal PSA increases were also calculated as the median pairwise slope for each patient. Using a multivariate Cox proportional hazards model comprehending demographic, clinicopathologic, and PSA covariates, PSA nadir was a significant predictor of BCR-free survival (p<0.01), while PSA slope was not a significant predictor in this study (p>0.05). The assay’s clinical performance is described in additional detail elsewhere (34).
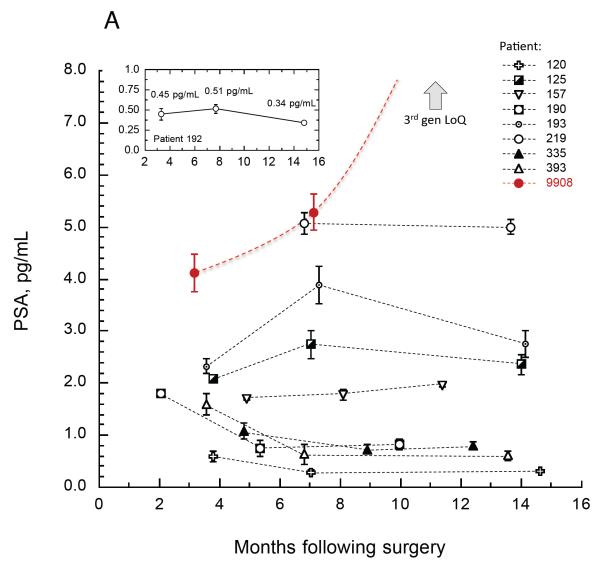
Fig. 5A highlights longitudinal data from five year BCR-free survivors from one of the clinical sites. All patients exhibited extremely low, stable PSA levels over the first year following surgery. Biological noise was minimal; for example, PSA values for patient 192 were 0.45, 0.51, and 0.34 pg/mL, a difference of only 0.17 pg/mL across 12 months (Fig. 5A inset). In contrast, there were other examples of non-recurrent patients (patients S9956, 9082, Fig. 4) exhibiting transient elevations to over 10 pg/mL, followed by PSA reduction back toward the nadir level. A similar phenomenon of lessor magnitude was noted in patients 193 and 125 (Fig. 5A). In contrast, patient 9908 (Fig. 5A red) exhibited a rapid upturn in PSA toward BCR from a similarly ultra low PSA level. Since these patients exhibited similar pre-surgical clinicopathologies (T1c, Gleason Score 5-6, negative margins), factors contributing to successful remission in one patient in contrast to another at these ultra low PSA levels may include surgical, biological, and immunological variables.
Fig. 5. Select longitudinal PSA trends.
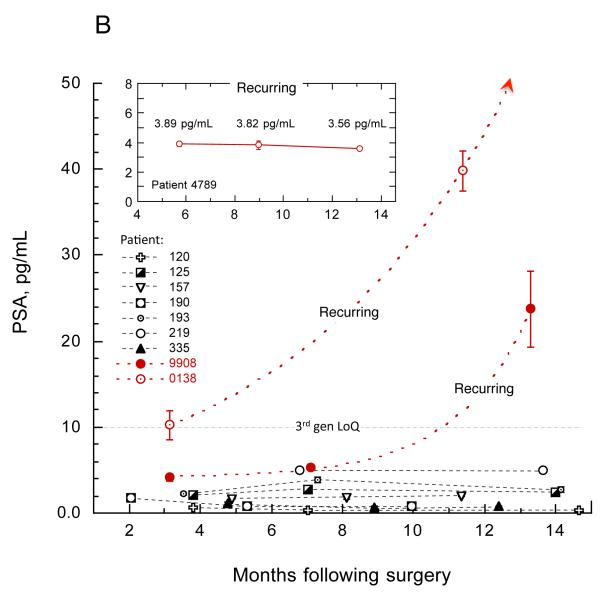
Fig. 5A depicts PSA results from non-recurring patients from one of the clinical sites. Most patients exhibited extremely low, stable PSA levels over the first year following surgery. The early stages of BCR for patient 9908 (red) are also depicted. The LoQ of ultrasenstive PSA methods is off the scale (arrow). Fig. 5B compares the same non-recurring patients with three examples of recurring patients (red) on a broader scale. Exponential projections for the appearance of 200 pg/mL PSA for patients 9908 and 0138 (curved fits, R2 0.999) were consistent with actual BCR. Inset depicts PSA results from a patient in remission who later recurred.
Fig. 5B contrasts the non-recurring patients of Fig. 5A with three examples of recurring patients. While patient 9908 exhibited pre-surgical clinicopathology consistent with many non-recurrers, patient 0138 exhibited less favorable pathology (T2b, pT3b with seminal vescicle invasion, Gleason 7). Perhaps not surprisingly, the more aggressive recurrers (see Fig. 4) tended to have less favorable pre-surgical clinicopathologies.
Consistent with earlier reports (17), the longitudinal profiles suggest three general categories of PSA kinetics following RP: fast rising, slow rising, and apparent non-rising. However, as shown here, very low unrising PSA over a one-year interval is no assurance of non-recurrence. Patient 4789 exhibited highly stable, very low PSA values for 13 months following surgery (Fig. 5B inset), yet was diagnosed with BCR five years later. Unpredictable remission and kinetic characteristics may complicate use of PSA velocity following RP for prediction of long-term recurrence. The only safe generalization that can be made continues to be that the lower the nadir PSA, the more likely the patient will enjoy long term BCR-free survival.
Discussion
The data presented here indicate that the SiMoA PSA assay defines a new analytical standard for extremely sensitive and reproducible PSA testing. Historically, acceptance of ultrasensitive PSA measurement has been inhibited by analytical variability, which has reduced the reliability of the information obtained from these assays. Monitoring PSA after RP is analytically demanding because it requires both sensitivity and day-to-day reproducibility. Compounding the difficulty has been confusion over “analytical sensitivity” (LoD) and true quantification sensitivity (LoQ). Assessing day-to-day reproducibility of results from ultra-low test samples is the most rigorous means of understanding an assay’s quantification sensitivity. This report demonstrates analytically acceptable day-to-day reproducibility in the sub-picogram range, low enough for reliable quantification of PSA in RP patients.
Robust fifth-generation measurement of PSA in all RP patients has the potential to impact management of prostate cancer in a number of significant ways. Reports showing the prognostic value of nadir PSA suggest a category of patients may be identified that represent an extremely low likelihood for BCR. Data from the pilot study reported here and elsewhere (34) indicate that a subgroup of patients below the detection limit of current methods were recurrence-free after five years. As reflected in Fig. 5, PSA levels appear biochemically stable for non-recurrent men. Current practice of hoping that PSA remains undetectable with less sensitive detection methods could be supplanted with reliable data indicating a highly favorable status. Positively discerning these patients with precise measurement of their PSA levels could improve delineation of an ultra low risk category with statistically powered follow-up studies.
PSA trends measured with fifth-generation sensitivity could provide the earliest possible indicator of potential aggressive BCR, with significant potential improvement in early warning time relative to current PSA methods, including third-generation methods. As shown in Fig. 5B, an exponential rise in PSA would not have been measured by a third-generation assay in patient 9908 for 11 months following surgery. Salvage radiation therapy (SRT) is more effective if administered earlier rather than later in the cancer recurrence (13, 14). Stephenson et al. (14) found that post surgical PSA levels prior to SRT were a highly significant predictor of disease progression, with more favorable outcomes observed at lower PSA levels, indicating that intervention with lower cancer burden prior to systemic dissemination leads to improved outcome. With this therapy model, intervention at the earliest sign of recurrence is most likely to lead to the most favorable outcome.
Reliably measuring PSA in every RP patient with fifth-generation sensitivity could also provide additional guidance on who may benefit most from adjuvant radiation treatment (ART). Evidence is growing of significant increases in overall and cancer-specific survival after ART (35-36). However, only about a third of patients who have had RP develop BCR, and about a third of this subset develop metastases (35). Which patients would benefit from ART and which patients would be over-treated remains unclear. Lower risk pathology with nadir PSA in an ultra low risk group might represent a cohort for whom ART represents over treatment. Higher risk pathology with high nadir could be a group most likely to benefit from ART. Treatment decisions for patients between these two groups could be better informed by highly reliable post surgical PSA data.
Clinical studies are needed to further examine the utility of high-resolution post-surgical PSA status. Fifth-generation sensitivity could provide more timely and reliable data around primary treatment effectiveness, monitoring intervals, selection for secondary treatment, and additional therapies based on effectiveness monitoring. Clinical analysis of the pilot study on the utility of the SiMoA PSA assay in recurrence prognosis will be published elsewhere (34).
Supplementary Material
Acknowledgements
This work was supported in part by Award Number R43CA133987 from the National Cancer Institute.
REFERENCES
- 1.Oesterling JE. Prostate-specific antigen: A critical assessment of the most useful tumor marker for adenocarcinoma of the prostate. J Urol. 1991;145:907. doi: 10.1016/s0022-5347(17)38491-4. [DOI] [PubMed] [Google Scholar]
- 2.Diamandis EP. Prostate specific antigen—its usefulness in clinical medicine. Trends Endocrinol Metab. 1998;9:310–316. doi: 10.1016/s1043-2760(98)00082-4. [DOI] [PubMed] [Google Scholar]
- 3.Sia M, Pickles T, Morton G, et al. Salvage radiotherapy following biochemical relapse after radical prostatectomy: proceedings of the Genito-Urinary Radiation Oncologists of Canada consensus meeting. Consensus Statement. Can Urol Assoc J. 2008;2:500–507. doi: 10.5489/cuaj.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunnar A. Second-Line Therapy after Radical Prostatectomy Failure: For Whom? When? How? Eur Urol. 2007;51:1155–58. doi: 10.1016/j.eururo.2007.01.069. [DOI] [PubMed] [Google Scholar]
- 5.Bock JL, Klee GG. How sensitive is a prostate-specific antigen measurement? How sensitive does it need to be? Arch Pathol Lab Med. 2004;128:341–343. doi: 10.5858/2004-128-341-HSIAPA. [DOI] [PubMed] [Google Scholar]
- 6.Freedland SJ, Moul JW. Prostate specific antigen recurrence after definitive therapy. J Urol. 2007;177:1985–1991. doi: 10.1016/j.juro.2007.01.137. [DOI] [PubMed] [Google Scholar]
- 7.Han M, Partin AW, Pound CR, et al. Long-term biochemical disease-free and cancer-specific survival following anatomic radical retropubic prostatectomy: The 15-year Johns Hopkins experience. Urol Clin North Am. 2001;28:555–565. doi: 10.1016/s0094-0143(05)70163-4. [DOI] [PubMed] [Google Scholar]
- 8.Catalona WJ, Smith DS. 5-year tumor recurrence rates after anatomical radical retropubic prostatectomy for prostate cancer. J Urol. 1994;152:1837–1842. doi: 10.1016/s0022-5347(17)32397-2. [DOI] [PubMed] [Google Scholar]
- 9.Bianco FJ, Jr, Scardino PT, Eastham JA. Radical prostatectomy: Long-term cancer control and recovery of sexual and urinary function. Urology. 2005;66:83–94. doi: 10.1016/j.urology.2005.06.116. [DOI] [PubMed] [Google Scholar]
- 10.Pound CR, Partin AW, Eisenberger MA, et al. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 11.Thompson IM, Tangen CM, Paradelo J, et al. Adjuvant radiotherapy for pathological T3N0M0 prostate cancer significantly reduces risk of metastases and improves survival: long-term followup of a randomized clinical trial. J Urol. 2009;181:956–962. doi: 10.1016/j.juro.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trock BJ, Han M, Freedland SJ, et al. Prostate cancer-specific survival following salvage radiotherapy vs observation in men with biochemical recurrence after radical prostatectomy. JAMA. 2008;299:2760–2769. doi: 10.1001/jama.299.23.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephenson AJ, Shariat SF, Zelefsky MJ, et al. Salvage radiotherapy for recurrent prostate cancer after radical prostatectomy. JAMA. 2004;291:1325–1332. doi: 10.1001/jama.291.11.1325. [DOI] [PubMed] [Google Scholar]
- 14.Stephenson AJ, Scardino PT, Kattan MW, et al. Predicting the outcome of salvage radiation therapy for recurrent prostate cancer after radical prostatectomy. J Clin Oncol. 2007;25:2035–2041. doi: 10.1200/JCO.2006.08.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H, Diamandis EP. Ultrasensitive time-resolved immunofluorometric assay of prostate-specific antigen in serum and preliminary clinical studies. Clin. Chem. 1993;39:2108. [PubMed] [Google Scholar]
- 16.Diamandis EP, Wong PY, et al. Detection of prostate cancer relapse with prostate specific antigen monitoring at levels of 0.001 to 0.1 ug/L. J Urol. 1997;157:913–918. [PubMed] [Google Scholar]
- 17.Evyenia JK, Vassilikos JK, Yu H, et al. Relapse and cure rates of prostate cancer patients after radical prostatectomy and 5 years of follow-up. Clin Biochem. 2000;33:115–123. doi: 10.1016/s0009-9120(99)00099-5. [DOI] [PubMed] [Google Scholar]
- 18.Haese A, Huland E, Graefen M, et al. Ultrasensitive detection of prostate specific antigen in the followup of 422 patients after radical prostatectomy. J Urol. 1999;161:1206–1211. [PubMed] [Google Scholar]
- 19.Hong SK, Park HZ, Lee WK, et al. Prognostic significance of undetectable ultrasensitive prostate-specific antigen nadir after radical prostatectomy. Urology. 2010;76(3):723–727. doi: 10.1016/j.urology.2010.03.081. [DOI] [PubMed] [Google Scholar]
- 20.Doherty AP, Bower M, Smith GL, et al. Undetectable ultrasensitive PSA after radical prostatectomy for prostate cancer predicts relapse-free survival. Brit J Cancer. 2000;83:1432–1436. doi: 10.1054/bjoc.2000.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen S, Lepor H, Yaffee R, et al. Ultrasensitive serum prostate specific antigen nadir accurately predicts the risk of early relapse after radical prostatectomy. J Urol. 2005;173:777–780. doi: 10.1097/01.ju.0000153619.33446.60. [DOI] [PubMed] [Google Scholar]
- 22.Sakai I, Harada K, Kurahashi T, et al. Usefulness of the nadir value of serum prostate specific antigen measured by an ultrasensitive assay as a predictor of biochemical recurrence after radical prostatectomy for clinically localized prostate cancer. Urol Int. 2006;76:227–231. doi: 10.1159/000091624. [DOI] [PubMed] [Google Scholar]
- 23.Eisenberg ML, Davies BJ, Cooperberg MR, et al. Prognostic implications of an undetectable ultrasensitive prostate-specific antigen level after radical prostatectomy. Eur Urol. 2010;57(4):622–629. doi: 10.1016/j.eururo.2009.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreira DM, Presti JC, Jr, Aronson WJ, et al. Postoperative prostate-specific antigen nadir improves accuracy for predicting biochemical recurrence after radical prostatectomy: Results from the Shared Equal Access Regional Cancer Hospital (SEARCH) and Duke Prostate Center databases. Int J Urol. 2010 Nov;17(11):914–22. doi: 10.1111/j.1442-2042.2010.02631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDermed JE, Adams TH, Klem RE, Diamandis EP. Ultrasensitive PSA measurements using NADiA technology in men following radical prostatectomy (RP) [Abstract]. Genitourinary Cancers Symposium; 2009. Abstract 121. [Google Scholar]
- 26.Thaxtona CS, Elghanianc R, Thomasa AD, Stoevac SI, Leec JS, Smith ND, et al. Nanoparticle-based bio-barcode assay redefines “undetectable” PSA and biochemical recurrence after radical prostatectomy. PNAS. 2009;106(44):18437–42. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nature Biotech. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song L, Randall JD, Chang L, Rissin DM, Kan CW, Campbell TG, et al. Ultra-sensitive digital immunoassay for amyloid β42 using single molecule arrays. Clin Chem. (submitted) [Google Scholar]
- 29.Rissin D, Walt D. Digital concentration readout of single enzyme molecules using femtoliter arrays and Poisson statistics. Nano Letters. 2006;6:520–523. doi: 10.1021/nl060227d. [DOI] [PubMed] [Google Scholar]
- 30.Rissin DM, Fournier DR, Piech T, Kan CW, Campbell TG, Song L, et al. Simultaneous detection of single molecules and singulated ensembles of molecules enables immunoassays with broad dynamic range. Anal Chem. 2011 doi: 10.1021/ac103161b. ASAP article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical Laboratory Standards Institute Evaluation of the Linearity of Quantitative Measurement Procedure: A Statistical Approach. Approved Guideline - Second Edition. 2003. CLSI Document EP6-A.
- 32.Clinical Laboratory Standards Institute Evaluation of Precision Performance of Quantitative Measurement Methods. Approved Guideline - Second Edition. 2004. CLSI Document EP5-A2.
- 33. [Accessed February 2011];ADVIA Centaur PSA Assay Specifications. http://www.medical.siemens.com/siemens/en_GLOBAL/gg_diag_FBAs/files/Assays/hisstory/PSA_Assay.pdf.
- 34.Lepor H, Cheli CD, Thiel RP, Taneja SS, Laze J, Chan DW, et al. Clinical evaluation of a novel method for measurement of PSA, AccuPSATM, as a predictor of 5-year biochemical recurrence-free survival post radical prostatectomy: results of a pilot study. Brit Jour Urol Intl. doi: 10.1111/j.1464-410X.2011.10568.x. (in press) [DOI] [PubMed] [Google Scholar]
- 35.Lepor1 H, Cheli2 CD, Thiel3 RP, Taneja1 SS, Laze1 J, Chan4 DW, Sokoll4 LJ, Mangold4 L, Partin4 AW. Clinical evaluation of a novel method for measurement of PSA, AccuPSATM, as a predictor of 5-year biochemical recurrence-free survival post radical prostatectomy: results of a pilot study. doi: 10.1111/j.1464-410X.2011.10568.x. (In preparation) [DOI] [PubMed] [Google Scholar]
- 36.Thompson IM, Jr, Tangen CM, Paradelo J, et al. Adjuvant radiotherapy for pathologically advanced prostate cancer: a randomized clinical trial. JAMA. 2006;296:2329–2335. doi: 10.1001/jama.296.19.2329. [DOI] [PubMed] [Google Scholar]
- 37.Van der Kwast TH, Bolla M, Van Poppel H, et al. Identification of patients with prostate cancer who benefit from immediate postoperative radiotherapy: EORTC 22911. J Clin Oncol. 2007;25:4178–4186. doi: 10.1200/JCO.2006.10.4067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.