Abstract
Expression of the human epidermal growth factor receptor 2 (HER2) is amplified in 25 – 30% of breast cancers and has been associated with an unfavorable prognosis. Here we report the construction, purification, and characterization of Affitoxin – a novel class of HER2-specific cytotoxic molecules combining HER2-specific Affibody molecule as a targeting moiety and PE38KDEL, which is a truncated version of Pseudomonas exotoxin A (PE), as a cell killing agent. It is highly soluble and does not require additional refolding, oxidation, or reduction steps during its purification. Using Surface Plasmon Resonance (SPR) technology and competitive binding assays, we have shown that Affitoxin binds specifically to HER2 with nanomolar affinity. We have also observed a high correlation between HER2 receptor expression and retention of Affitoxin bound to the cell surface. Affitoxin binding and internalization is followed by PE activity domain-mediated ADP-ribosylation of translation elongation factor 2 (eEF2) and, consequently, inhibition of protein synthesis as shown by protein expression analysis of HER2-positive cells treated with Affitoxin. Measured IC50 value for HER2-negative cells MDA-MB468 (65±2.63 pM) was more than 20 times higher than the value for low HER2 level-expressing MCF7 cells (2.56±0.1 pM), and almost three orders of magnitude higher for its HER2-overexpressing derivative MCF7/HER2 (62.7±5.9 fM). These studies suggest that Affitoxin is an attractive PE38-based candidate for treatment of HER2-positive tumors.
Keywords: Affibody molecule, HER2, breast cancer, targeted therapy, Pseudomonas exotoxin A
Introduction
Breast cancer is the most common female cancer and the second most common cause of female cancer-related deaths in the United States. Worldwide, more than one million patients are diagnosed with breast cancer annually. Approximately 25–30% of all breast cancer cases are characterized by overexpression of HER2 receptors, which is associated with increased proliferation and survival rate of cancer cells leading to poor therapy outcomes and unfavorable prognosis (1, 2). Thus, HER2 has become an attractive target for breast cancer therapy. Several approaches, including inhibition of HER2 receptor-triggered signal transduction by humanized antibodies and small molecules targeting catalytic activity of receptors, have been tested in clinical trials (2).
In spite of the development of new HER2-targeted therapies, such as trastuzumab (Herceptin®; Genentech, Inc., South San Francisco, CA), which has revolutionized the treatment of HER2-overexpressing breast cancers (3–5), there is a significant number of patients with HER2-positive tumors who do not respond or acquire resistance to these therapies (6). Therefore, there is a need for novel therapeutic approaches using HER2 not only as a target for blocking the EGF signaling pathway but also for receptor-mediated delivery of cytotoxic agents. Recently, Gail et al. presented a group of trastuzumab-maytansinoid conjugates claiming that since HER2 expression remained unchanged in tumors that become resistant to HER2-targeted therapies, trastuzumab-based cytotoxic conjugates may present a promising therapeutic modality (7).
Immunotoxins are hybrid proteins that are composed of a targeting moiety such as an antibody, antibody fragment or ligand directed to an antigen or a receptor on the surface of tumor cells, and a toxic domain derived form plant (ricin) or bacteria (diphtheria toxin or Pseudomonas aeruginosa exotoxin A) (8–10). The targeting moiety directs the toxin to the tumor cell and then, the activity domain induces apoptosis by inhibition of protein synthesis. The PE38, which is a truncated version of Pseudomonas exotoxin A (PE), is widely used for construction of immunotoxin due to its high toxic potential and the fact that its cytotoxic pathways are well described and understood (11).
A large number of PE38-based immunotoxins directed against various surface antigens overexpressed in tumors were constructed and tested in preclinical and clinical trials. For example, immunotoxin therapy based on PE38 delivery is proven to be efficient in treatment of such blood malignancies as CD22-positive lymphomas (12). Similar approaches have been applied to target solid tumors. For example, interleukins IL4 and IL13 were used as targeting moieties for PE38 delivery in treatment of breast (13), pancreas (14), and head and neck cancers (15). Similarly, TGFα fused to PE was shown to be effective in preclinical studies on glioma, prostate or epidermoid cancer expressing EGFR (16, 17). However, according to the literature, the most frequently used targeting molecule fused with PE38 is single chain variable fragment of antibody (scFv) or its disulfide-stabilized derivative (dsFv). These molecules were successfully applied in targeting LeY receptors (18), mesothelin (19), and osteosarcoma antigen (20). Recombinant scFv and dsFv attached to PE38 moiety were already tested in treatment of HER2-overexpressing breast (21–24), ovarian (25), prostate (26, 27), lungs (28), and gastric (29) cancers.
Affibody molecules are a new class of relatively small, ~7-kDa, proteins based on a 58-amino-acid scaffold, derived from the Z domain of Staphylococcus Aureus protein A (30). They are almost 20 times smaller than antibodies and 4 times smaller than scFvs. These very stable molecules can be readily expressed in soluble form in bacterial systems alone or as a fusion protein. Their size, high affinity, and specificity make these proteins an interesting alternative to antibodies or scFvs as targeting agents (31).
We have genetically fused HER2-specific Affibody molecule with a truncated and optimized version of Pseudomonas Exotoxin A (PE38KDEL) (32). The resulting recombinant protein called HER2-Affitoxin combines high HER2 specificity and affinity of Affibody molecules with the tumoricidal potential of PE38KDEL. In the present work we report cloning, overexpression, purification, and in vitro characterization of this novel recombinant anti-cancer agent.
Material and Methods
Affitoxin cloning
The PE38 toxin part of the Affitoxin was amplified by PCR using plasmid pRB98 (kindly provided by Drs. Richard Beers and Ira Pastan, NCI, NIH, Bethesda, MD) and primers P1 and P2 (Supp. Table 1). The PCR product was purified by agarose gel electrophoresis and cloned into pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA). The resulting plasmid containing the PE38 insert was digested with AccI and BlpI enzymes, and recloned into a vector containing the HER2-Affibody molecule sequence (Affibody AB, Sweden, www.affibody.se). The HER2-portion of the resulting plasmid was amplified by PCR using primers P3 and P4 (Supp. Table 1). HA22 plasmid kindly provided by Dr. Ira Pastan was used for PCR with primers P5 and P6 (Supp. Table 1) as a source of PE38 cDNA. Finally, an Affitoxin-expressing construct was built, combining the separately PCR-amplified building blocks by mega-primer PCR using primers P3 and P6. The PCR product was cloned into pET101/D-TOPO® vector (Invitrogen, Carlsbad, CA), and the insert of the resulting plasmid AfTxKDEL31 was sequenced.
Affitoxin overexpression and purification
BL21 Star™ (DE3) E. coli cells (Invitrogen, Carlsbad, CA) transformed with the AfTxKDEL31 plasmid were grown in Super Broth medium supplemented with 100 μg/ml ampicillin until A600 reached value 1.5. Affitoxin expression was induced by addition IPTG to 1 mM final concentration. Two and a half hours after induction, cells were harvested, resuspended in 50 ml of the buffer containing 300 mM NaCl, 50 mM sodium phosphate pH=8.0, and one tablet of Complete EDTA-free protease inhibitor cocktail (Roche Diagnostic GmbH, Basel, Switzerland), followed by two 5-minute cycles of ultrasonication on ice at 20W with Cole-Parmer Ultrasonic Processor Model CP70T. The lysate was clarified by centrifugation at 50,000 g for 20 minutes followed by 0.45 μm syringe filtration. ÄKTAprime plus chromatography system (Amersham Biosciences, Pittsburgh, PA) was used for Ni-affinity chromatography on 20 ml column with PrepEase High Specificity resin (USB Corporation, Cleveland, OH). The “histidine-tagged protein purification gradient elution” program was used with the following buffers: A1 (300 mM NaCl and 50 mM sodium phosphate pH=8.0) and B (400 mM imidazole, 300 mM NaCl and 50 mM sodium phosphate pH=8.0). The eluted fractions were analyzed by 4–12% gradient Bis-Tris PAAG electrophoresis in MOPS-SDS buffer and the gel was stained with MicrowaveBlue® (PROTIGA Inc., Frederick, MD). The fractions containing Affitoxin were combined and diluted 5 times with water to adjust NaCl concentration to 60 mM. ÄKTAprime plus was used for anion-exchange chromatography on 1 ml HiTrap Q HP column (Amersham Biosciences, Pittsburgh, PA). The “Anion Exchange” program was used with the buffers A1 (60 mM NaCl and 10 mM sodium phosphate pH=8.0) and B (1 M NaCl and 50 mM sodium phosphate pH=8.0). The eluted fractions were analyzed by 4–12% gradient Bis-Tris PAAG electrophoresis in MOPS-SDS buffer and the gel was stained with MicrowaveBlue® (PROTIGA Inc., Frederick, MD).
Preparation of Affitoxin sample for physicochemical characterization
Before physicochemical characterization, 0.5 ml of Affitoxin solution eluted from Ni-column was mixed with dry guanidine-hydrochloride to final volume of 0.8 ml and 10 μl of trifluoroacetic acid (TFA). This sample was fractionated on a C-18 column (8 × 100 mm cartridge column, Delta-Pak, Waters Corp., Milford, MA) connected to Waters HPLC system (including 626 Pump system, 6000 S Controller with Millenium-32 software and 996 Photodiode Array Detector). Chromatography was performed at a flow rate of 1 ml/min using aqueous acetonitrile-TFA solvents (buffer A: 0.1% TFA in water; buffer B: acetonitrile containing 0.1% TFA). The column was equilibrated in 5% buffer B. After injection the column was rinsed for 20 min prior to gradient elution using a linear gradient from 5 to 95% of buffer B over 60 min. The column eluate was monitored at 206 nm. 1.25 ml fractions were collected. The aliquots of the fractions were analyzed by MALDI-TOF mass spectrometry. Fractions containing Affitoxin were pooled. 10 μl aliquot was used for N-terminal sequence analysis on automated protein sequencer Procise 494 cLC (Applied Biosystems, Foster City, CA) to assess protein homogeneity and to determine protein concentration. The 0.5 nmol aliquots of Affitoxin were transferred into 1.5 ml tubes and lyophilized.
MALDI-TOF MS analysis of Affitoxin
For MS analysis, 1 μl of Affitoxin solution eluted from C-18 column as described above, was mixed with 0.7 μl of matrix (2 mg/ml of 2,5-dihydroxybenzoic acid (DHB) in 50% acetonitrile/water, 0.05% TFA) directly on the target plate and analyzed using an Applied Biosystems Voyager-DE Pro MALDI-TOF mass spectrometer. The accelerating voltage was 25 kV, guide wire 0.15% and grid voltage 91.5%. The instrument was operated in linear mode under positive ion conditions. A nitrogen laser was used at 337 nm with 250 laser shots averaged per spectrum. Calibration was performed using instrument default settings (mass accuracy 0.1%) and data analysis was carried out using Data Explorer software resident on the instrument.
Characterization of Affitoxin binding
Surface plasmon resonance (SPR) assay was used to assess the Affitoxin binding affinity to the extracellular domain of HER2. The ProteOn GLM sensor chips were preconditioned with one 18-second injection of 10 mM HCl, 50 mM NaOH, 0.1% surfactant p20 and 100 mM phosphoric acid in 1 M NaCl in the horizontal and vertical direction. Following preconditioning the system was equilibrated with PBS-T buffer (20 mM sodium phosphate, 0.15 M NaCl, and 0.005% surfactant p20, pH 7.4). The alginate-based layer was then activated for 5 min. with 0.2M EDAC (1-ethyl-3[3-dimethylaminopropyl] carbodiimide HCl) and 0.05 M sulfo-NHS (N-hydroxysulfosuccinimide) at 30 μl/min. Protein A/G (20 μg/ml) was injected in the horizontal direction over all 6 channels for 5 minutes to obtain a response level of 3000 RU. The reaction was then quenched with an injection of 1 M ethanolamine over all channels for 5 minutes. Different levels of HER2/Fc (R&D Systems Inc, Minneapolis, MN) were achieved by injecting five concentrations (50, 25, 12.5, 6.25 and 3.12 μg/ml) of HER2/Fc, for 5 minutes, in running buffer (10 mM Hepes pH 7.5, 150 mM NaCl, 3 mM EDTA, 0.005% (v/v) surfactant p20 and 0.1% (w/v) BSA) in the vertical direction. A buffer blank was used for channel 4 so this could be used as a reference channel. The HER2/Fc surface was then equilibrated with 1-minute injections of running buffer in both directions.
The kinetics of ZHER2:342 Affibody molecule or Affitoxin binding to the HER2/Fc surface was then measured. Specifically, ZHER2:342 Affibody molecule (5, 2.5, 1.25, 0.63, 0.32, 0 nM) or Affitoxin (500, 166.6, 55.5, 18.5, 6.17, 0 nM) was injected for 120 seconds at 200 μl/min in the horizontal direction at 25°C. Bound Affibody molecules were allowed to dissociate for 2000 seconds, and the surface was regenerated with two 18-second pulses of 100 mM phosphoric acid/0.05 % Surfactant P20. The processed data was fit globally using a simple Langmuir model.
Labeling of Affitoxin with Alexa Fluor® 488
Alexa Fluor® 488 C5–malemide (0.1 mg, Invitrogen, Carlsbad, CA) was dissolved by adding 10 μl of DMSO, then 40 μl of 0.1 M NaHCO3. This solution was immediately transferred to the tube with 0.5 nmol of lyophilized reduced Affitoxin and incubated for 15 minutes at room temperature in the dark. Then 1 μl of 1 M DTT was added and incubated for additional 5 minutes. Two 0.5 nmol aliquots of the modified Affitoxin were combined and desalted on 10×250 mm Sephadex G-50-fine column equilibrated in ammonia solution (pH 8.5). MALDI-TOF MS analysis of the fractions confirmed that Affitoxin was mostly modified (data not shown). The combined fractions of the modified Affitoxin were lyophilized.
Cell lines
The human breast ductal carcinoma BT474 and the human breast adenocarinomas: MCF7, MDA-MB468 and MDA-MB361 cell lines were obtained from American Type Tissue Collection (Manassas, VA). MCF7 cell line stably transfected with HER2 (MCF7/HER2), was kindly provided by Drs. John W. Park and Byron Hann (University of California, San Francisco, CA, USA). Cells were maintained in cell culture media at 37°C in 5% CO2 and humidified atmosphere. MCF7, MCF7/HER2, and MDA-MB361 were grown in DMEM, BT474 in RPMI, and MDA-MB468 in DMEM F12. All media were supplemented with 10% FBS, 100 units/ml of penicillin and 0.1_mg/ml streptomycin. When necessary, cells were detached from the culture plates with 0.05% trypsin and 0.02% EDTA in PBS. All media, FBS, antibiotics and trypsin were purchased from Invitrogen (Carlsbad, CA).
Flow cytometry analysis
Cells were incubated with 5 μg/ml of Affitoxin labeled with Alexa Fluor® 488 alone or in the presence of variable concentrations of Affibody molecules. After one hour incubation at 37°C live cells were analyzed by flow cytometry using a FACS Calibur instrument (BD Biosciences, San Jose, CA) equipped with a 15-mW argon-ion laser (500 counts per second). CellQuest Pro (BD Biosciences, San Jose, CA) software was used for data acquisition and FlowJo (Tree Star Inc, Ashland, OR) for data analysis.
Confocal microscopy
BT474, MCF7/HER2, MCF7 and MDA-MB468 cells were plated on 35 mm confocal dishes at densities 2×105 cells/dish. After an overnight attachment, Alexa Fluor® 488-labeled Affitoxin was added to a final concentration of 5 μg/ml in regular growth medium containing 10% FBS, and the cells were incubated at standard conditions for additional 6 hours. For competition studies, 50 μg/ml HER2-Affibody (Affibody AB, Sweden) was added simultaneously with labeled Affitoxin. One hour incubation with 10 μg/ml Hoechst 33342 (Invitrogen) was used for nuclei contrasting. Cells were rinsed thrice with PBS and observed using a Zeiss LSM 510 Confocal Microscope (Carl Zeiss Inc, Thornwood, NY) with an Axiovert 100M inverted microscope, operating with a 80 mW argon UV laser tuned to 364 nm and a 25 mW argon laser tuned to 488 nm. A 63×1.3 NA Plan-Apochromat oil immersion objective and a multi-track configuration were applied. Alexa Fluor® 488 and Hoechst signals were collected with a BP 505–550 filter and a BP 385–470 filter after excitation with the 488 nm and 364 nm laser lines, respectively, while the DIC image was collected by a transmitted light detector. Images (8 bit, 512 × 512 pixels) were acquired with a scan zoom of 2.5 and a line average of 8 using the Zeiss AIM software.
Protein expression assay
MCF7 and MCF7/HER2 cells were plated in a 96-well plate followed by transfection with 30 ng/well of Gaussia Luciferase plasmid pCMV-GLuc (New England Biolabs Ipswich, MA). Fugene6 transfection reagent (Roche, Basel, Switzerland) was used according to the manufacturer’s protocol. One hour after transfection, the cells were treated with Affitoxin in concentration ranging from 0.02 pM to 0.02 nM for 24 hours. Then, 25 μl of medium from each well was transferred to white non-transparent 96-well plate and combined with 50 μl Gaussia Luciferase assay solution (New England Biolabs, Ipswich, MA). Signal integration over 10 s was performed using FluoStar Optima plate reader (BMG Labtech, Chicago, IL) equipped with injection system. IC50 was calculated using GraphPad Prism software (GraphPad Software, Inc, San Diego, CA).
Cell viability assay
Cell viability was assessed by measurement of intracellular ATP levels. MDA-MB468, BT474, MDA-MB361, MCF7 and MCF7/HER2 cells were seeded in 96-well plate to yield 25–30% confluency at conditions described above. Immediately after plating, the cells were treated with Affitoxin at indicated concentrations. Intracellular ATP level measurement was performed using CellTiter-Glo (Promega, Madison, WI) according to the provided protocol. Signals were integrated after substrate addition, using FluoStar Optima plate reader. IC50 was calculated using GraphPad Prism.
Determination of HER2 concentration
Concentration of HER2 in BT474, MC7/HER2, MDA-MB361, MDA-MB468, and MCF7 was determined by ErbB2/c-Neu Rapid Format ELISA Kit (EMD Chemicals, Inc., Gibbstown, NJ) ELISA according to the manufacturer’s protocol.
Results
Affitoxin cloning and purification
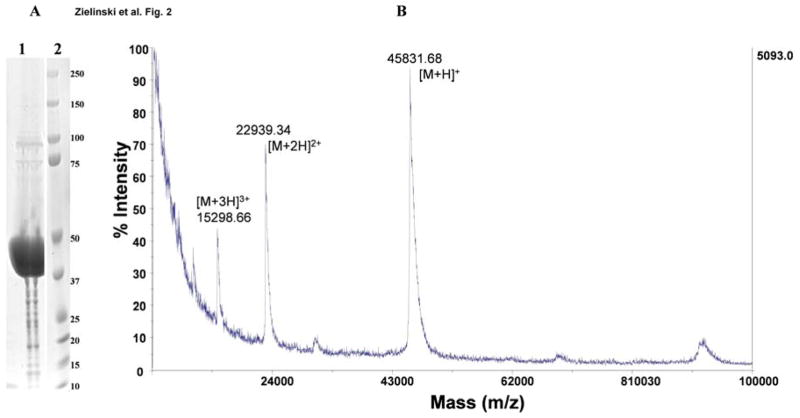
Applying standard cloning procedures, several DNA fragments encoding distinct domains of Affitoxin (Fig. 1, Supp. Fig. 1) were combined and cloned in plasmid pET101/D-TOPO. The construct contains hexahistidine-tagged HER2-specific Affibody moiety fused to the translocation and activity domains of PE38 toxin. The C-terminal part contains optimized KDEL sorting signal. The resulting plasmid was used for bacterial overexpression of Affitoxin. The soluble fraction of bacterial protein extract was subjected to two-step purification protocol including nickel affinity-followed by an anion exchange-chromatography. As a result, we obtained almost homogenous protein fraction as determined by SDS-PAGE analysis (Fig. 2A). Ion-exchange chromatography-purified Affitoxin was used for reverse phase HPLC followed by MALDI-TOF MS analysis (Fig. 2B). Fractions containing protein with molecular mass of 45.8 kDa were combined and an aliquot was subjected to N-terminal sequence analysis. The obtained sequence GSSHHHHHHLQVDNK confirmed the presence of highly purified Affitoxin.
Figure 1.
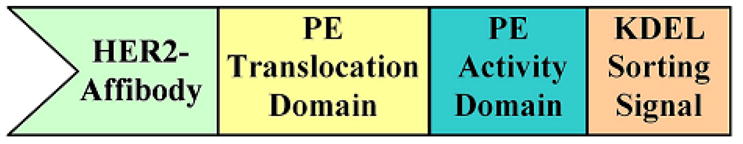
Schematic structure of Affitoxin.
Figure 2. Physicochemical characterization of purified Affitoxin.
A. 50 μg of purified Affitoxin was resolved in 4–12% Bis-Tris polyacrylamide gel along with pre-stained protein weight standards. B. MALDI-TOF MS of purified Affitoxin.
HER2-binding affinity of Affitoxin
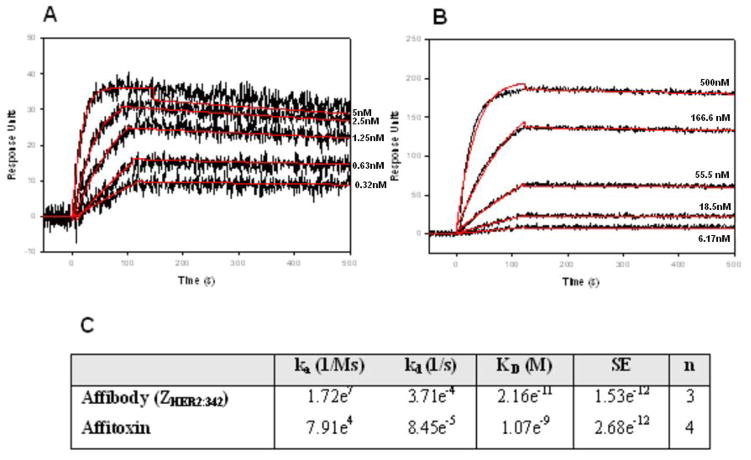
In order to measure the binding affinity of the Affitoxin to HER2, we used SPR-based binding assay. Recombinant HER2/Fc (R&D Systems Inc, Minneapolis, MN) was captured on the chip surface by amine-linked protein A/G. Various concentrations of the Affitoxin were passed over the HER2/Fc-covered surface. The unmodified Affibody ZHER2:342 molecule was used as a positive control. The data were fit globally using a simple Langmuir binding model (Fig. 3A and B). The association and dissociation rates along with KD values are summarized in Fig. 3C.
Figure 3. Kinetic analysis of ZHER2:342 Affibody molecule (A) or Affitoxin (B) binding to HER2/Fc-covered surface.
Binding of 0.32, 0.63, 1.25, 2.5, 5 nM wild type ZHER2:342 Affibody molecule (A) and 6.17, 18.5, 55.5, 166.6, 500 nM of Affitoxin (B) to HER2/Fc on the sensor chip was tested using SPR-based binding assay. The red lines represent a global analysis of data using a Langmuir binding model. The Affibody molecule-HER2/Fc complex was allowed to dissociate for 2000 seconds in order to observe a measurable decayKinetic analysis of ZHER2:342 Affibody molecule (A) or Affitoxin (B) binding to HER2/Fc-covered surface. Kinetic data are summarized in C.
HER2-specificity of Affitoxin
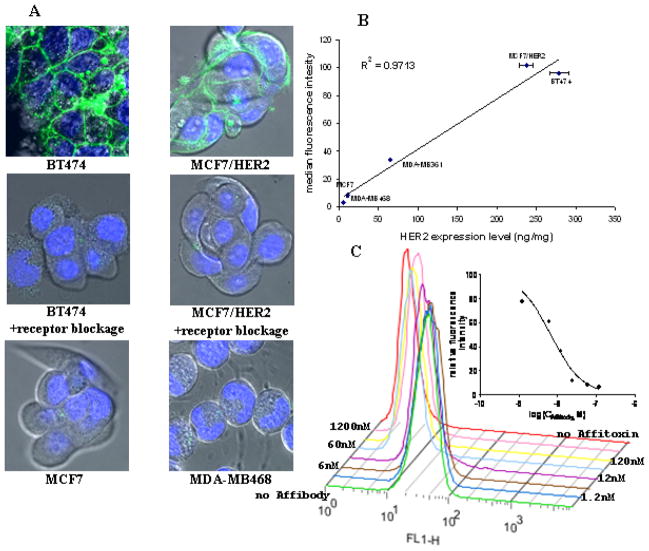
Specificity of Affitoxin binding to HER2 receptor was tested by three different methods. First, several cancer cell lines expressing different levels of HER2 were incubated with Alexa Fluor® 488-labeled Affitoxin and analyzed by flow cytometry (Supp. Fig. 2). The highest binding was recorded for BT474 and MCF7/HER2 cell lines, moderate for MDA-MB361 and low for MDA-MB468 and MCF7. The binding was well correlated with HER2 expression level as determined by ELISA (Fig. 4B).
Figure 4. Specificity of Affitoxin binding confirmed by confocal imaging and flow cytometry.
A. Cells were incubated with 0.1 μM of Alexa Fluor® 488-labeled Affitoxin for 6 hours with and without Affibody molecules. B. HER2 expression level in five different breast cancer cell lines was determined by ELISA and is expressed in nanograms of HER2 per milligram of protein lysate. Cells were incubated with 5 μg/ml of Alexa Fluor® 488-labeled Affitoxin. Binding efficacy was determined by flow cytometry. C. BT474 cells were incubated with 5 μg/ml of Alexa Fluor® 488-labeled Affitoxin and increasing concentrations of Affibody. Binding efficacy was determined by flow cytometry and quantified by GraphPad software.
Next, the binding specificity was confirmed by competition assay. BT474 cells were incubated with 0.1 μM fluorescently labeled Affitoxin in presence of increasing concentration of unlabeled Affibody molecules. We show that addition of Affibody molecules can interfere with the Affitoxin binding in a dose dependent manner (Fig. 4C). The estimated IC50 is 6.5 nM and is approximately 15 times lower than the applied Affitoxin concentration.
In order to visualize the binding specificity four different cell lines were incubated with 0.1 μM fluorescently labeled Affitoxin, followed by confocal microscopy imaging. Both HER2 positive cell lines: BT474 and MCF7/HER2 showed clear accumulation of fluorescent dye on the cell membrane (Fig. 4A), while cells with low receptor expression: MCF7 and MDA-MB468 did not show any probe retention (Fig. 4A). Affitoxin binding to HER2 receptors was blocked by excess of non-labeled Affibody molecules (Fig. 4A).
Cytotoxic effect of Affitoxin on cells expressing different amount of HER2 antigen
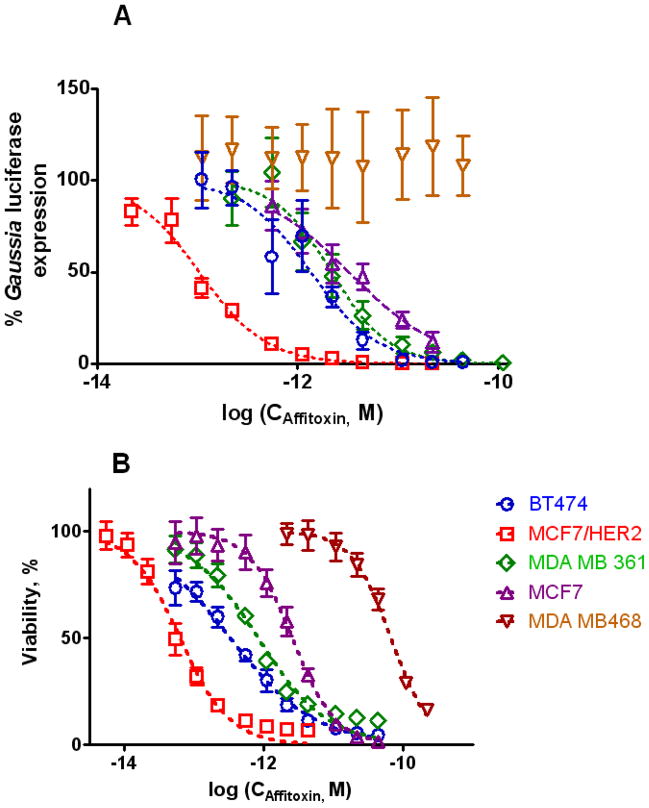
We have shown that inhibition of translation by Affitoxin is not only dose-dependent but it depends also on HER2 expression level (Fig. 5A). As listed in Table 1, the estimated IC50 value for MCF7/HER2 cells was approximately 30 times lower than that observed for non-transfected parental MCF7 cell line (p<0.006). In addition, assessment of the intracellular levels of ATP (correlated with cell viability) after 72-hour treatment with different concentrations of Affitoxin (Fig. 5B) indicated HER2-dependent toxicity. Again, the lowest calculated IC50 value was recorded for MCF7/HER2 and was approximately forty times lower than corresponding value observed for MCF7 (p<0.0003). BT474 was the second cell line in terms of Affitoxin sensitivity (IC50= 0.33 pM). MDA-MB361 (moderate HER2 expression) was around 13 times more resistant to Affitoxin than MCF7/HER2 (p<0.003) but on the other hand, approximately 80 times more sensitive than HER2-negative MDA-MB468 (p<0.0003) (Table 1). These results agree with the decrease of protein production described above.
Figure 5. Efficacy of Affitoxin measured by inhibition of protein synthesis (A) and intracellular ATP level (B).
Five different breast cancer cells lines were transfected with pCMV-GLuc plasmid containing secreted Gaussia luciferase gene under control of strong CMV promoter. Gaussia luciferase activity was measured in the medium 24 hours after treatment with Affitoxin. The same cell lines were assessed for intracellular ATP level 72 hours following treatment (CellTiter-Glo, Promega).
Table 1.
Comparison of HER2 expression and Affitoxin efficacy of different breast cancer cell lines.
| Cell line | HER2 expression* ng/mg | Inhibition of protein synthesis** IC50, pM | Depletion of ATP *** IC50, pM |
|---|---|---|---|
| MCF7/HER2 | 238±8.4 | 0.1 (0.08–0.12) | 0.063 (0.05–0.078) |
| BT474 | 279±12 | 1.32 (0.89–1.97) | 0.33 (0.28–0.38) |
| MDA-MB361 | 65±0.1 | 2.14 (1.61–2.85) | 0.83 (0.63–1.1) |
| MCF7 | 11±1.27 | 3.17 (2.52–3.99) | 2.56 (2.33–2.82) |
| MDA-MB468 | 5.6±0.3 | >43.47 | 65.3 (31.34–73.8) |
HER2 concentration in cell lysates was measured by ELISA expressed in nanograms of HER2 per milligram of cell lysate.
IC50 for inhibition of protein synthesis was estimated based on reduction of Gaussia luciferase signal in transfected cell lines.
IC50 was calculated based on intracellular ATP measurement after 72 hours of cell treatment with Affitoxin.
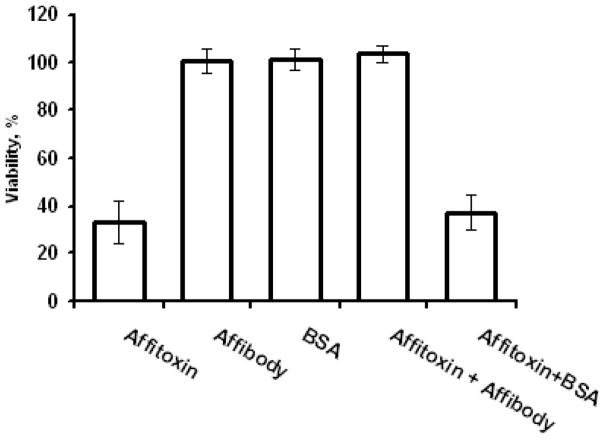
In addition, HER2-dependce of Affitoxin effectiveness was confirmed when the BT474 cells were incubated with Affitoxin in presence of Affibody molecules. As shown in Figure 7, excess of Affibody molecules prevents Affitoxin-induced toxicity. To exclude potentially sequestration of Affitoxin by non-specific protein-protein interaction, we have proved that BSA applied in equivalent concentration does not influence Affitoxin. Moreover, Affibody and BSA applied alone did not affect cells viability (Fig. 6).
Figure 7. Affitoxin efficacy in the presence of Affibody excess.
BT474 cells were treated with 2pM Affioxin in presence of 100nM Affibody and 100nM BSA. Cells were incubated for 72 hours and cell viability was measured using CellTiter-Glo. x
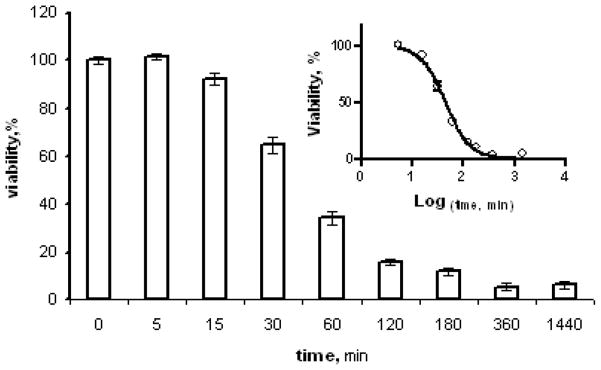
Figure 6. Effect of exposure time on Affitoxin efficacy.
BT474 cells were treated with 43.5 pM Affitoxin for indicated time periods. After that medium containing Affitoxin was changed and incubation was continued up to 72 hours and cell viability was measured using CellTiter-Glo.
We have also compared the effect of Affitoxin exposure time on cell viability. BT474 cells were treated with Affitoxin, and after the indicated time intervals medium containing Affitoxin was replaced with normal growth medium and the incubation was continued for 72 hours. As shown in Fig. 7, the time required to reduce the viability by 50% was forty five minutes and the maximum effect was reached after three hours of exposure time.
Discussion
Affitoxin is a multi-domain protein designed for efficient delivery of PE activity to cells overexpressing HER2 receptors. The Affibody molecule module, smaller than antibody or antibody fragments, is located on the N-terminus of Affitoxin and provides HER2-targeting function (Fig. 1). It has been previously shown that after receptor binding the Pseudomonas exotoxin A uses retrograde pathways from the cell surface to early endosomes (33). In the acidic endosomal environment, it dissociates from the receptor followed by conformational changes and proteolytic cleavage within translocation domain by proteinase furin (Supp. Fig. 1) (34). The released 37 kDa C-terminal fragment containing the translocation and activity domains is transported via late endosomes to the trans-Golgi network (35) where it binds in a pH-dependent manner to KDEL receptor, enabling its translocation to the endoplasmic reticulum (36). The translocation domain induces the transport of the 37 kDa fragment from lumen of the endoplasmic reticulum to the cytosol (37, 38). Once the activity domain of PE has reached cytosol, it catalyzes the ADP-ribosylation of its target protein: eukaryotic translation elongation factor 2 (eEF-2) (39). This results in inhibition of protein synthesis that ultimately leads to cell death (40–42).
In order to lower the probability of PE38 interference with Affibody binding, we incorporated a flexible GGGGSGG spacer between Affibody moiety and the translocation domain of Affitoxin. A hexahistidine tag was introduced on the N-terminal part of the protein for affinity purification of Affitoxin by Ni-NTA chromatography. Finally, since it was shown that mutation of the naturally occurring REDLK to KDEL within the sorting signal leads to about 100-fold increase in specific toxicity (43), we include this modification in our molecule.
The important property of Affitoxin is its high solubility. After two chromatographic steps we obtained almost homogenous protein. The production does not require complicated steps such as refolding and reduction/oxidation steps. Its yield is high and costs are relatively low.
As shown by the SPR-based binding assay, the KD of Affitoxin binding to HER2, although higher than that of Affibody molecule, is still in low nM range (Fig. 3), which is slightly lower than the values reported for stabilized immunotoxins (44). Approximately 50-fold lower affinity of Affitoxin to HER2 as compared with the original ZHER2:342 Affibody molecule is due to a slower association of the much larger chimeric molecule to the immobilized receptor. This difference seems to be less prominent when the binding of Affibody and Affitoxin to HER2 on the cell surface was compared by competition binding assay (Fig. 4C), which also indicated HER2 binding specificity. The high HER2 binding specificity of Affitoxin was independently confirmed by strong correlation between HER2 expression level on different tumor cell lines as determined by ELISA and binding of Affitoxin to these cells as measured by flow cytometry (Fig. 4B). Furthermore, confocal imaging of Alexa Fluor® 488-labeled Affitoxin (Fig. 4A) showed that Affitoxin binds to HER2-expressing cells only and that this binding can be blocked by excess of Affibody molecules.
We have shown that high concentrations of Affibody molecule specifically rescued cells from Affitoxin-induced cell death (Fig. 7). However, the applied Affibody concentrations had to be much higher than observed in competition studies. This observation is in agreement with relatively high KD value when compared to IC50 obtained for HER2-positive cell lines.
HER2-specific cytotoxicity of Affitoxin was confirmed by HER2-dependent inhibition of protein synthesis (Fig. 5A) and the intracellular ATP depletion (Fig. 5B). Differences in IC50 observed for cells expressing low and high levels of HER2 (Table 1), may result in therapeutic advantage allowing for effective treatment of HER2-overexpressing tumor cells using Affitoxin doses harmless for normal tissues that might express low levels of HER2 (45).
It has to be mentioned here that we did not observe linear correlation between HER2 expression and IC50 (R=0.26). MDA-MB468 cells have a huge impact for lack of linearity (R=0.72 for the remaining four cell lines). This phenomenon might be due to the fact that HER2 receptors are responsible only for binding and delivery of Affitoxin into endosomes, which is followed by integration with the trans-golgi network, rentention in the endoplasmic reticulum (ER) via the KDEL receptor, translocation across the ER membrane and release into the cytosol. Although HER2 expression affects the initial events downstream elements of the pathway are HER2-independent. This seems to be value for BT474 is several confirmed by Batra et al. who shown that the estimated IC50 fold lower than recorded for SKOV3 cells, even though both cell lines express high level of HER2 receptors (21). The Affitoxin IC50 values obtained using BT474 cells are approximately 6-fold lower then those reported for e23(scFv)PE38KDEL and comparable to those of disulfide-stabilized e23(dsFv)PE38KDEL immunotoxin when protein inhibition is considered. The difference is more pronounced when ATP depletion is compared (23). It is noteworthy that a 3-hour exposure was enough to reach the maximum effect on HER2-positive tumor cells (Fig. 7), which seems to be compatible with the expected circulation time of Affitoxin.
In spite of the success of currently used HER2-targeted therapies, such as trastuzumab, there are a significant number of patients with HER2-positive tumors who do not benefit from treatment because their tumors do not respond or acquire resistance to these therapies (6). Therefore, new strategies to treat HER2-positive tumors are being developed (46). Typically, commercially available antibodies, or their fragments are conjugated with tumoricidal effectors such as drugs (7), toxins (8–10), or therapeutic radionuclides (47–49). The unique characteristics of Affitoxin presented in this work, makes it an attractive alternative to treat not only patient whose tumors do not respond to current therapies but also in combination with current therapies. As shown in our recent publication (50), Affibody molecules do not interfere with binding or efficacy of trastuzumab. Therefore, Affitoxin and trastuzumab might be applied together to combine obstruction of HER2 signaling pathway by trastuzumab with the blocking of protein production by PE38KDEL.
In this work, we have developed and characterized in vitro a new recombinant protein, Affitoxin, combining the unique HER2 binding characteristics of Affibody molecules with cytotoxic potential of PE38. This new construct presents a potentially attractive therapeutic modality which might complement current antibody-based HER2-directed therapies.
Supplementary Material
Acknowledgments
The authors thank Affibody AB and Dr. Ira Pastan for providing constructs used as templates for cloning of Affitoxin. We also appreciate the support obtained from experts form Affibody AB and SAIC Frederick, Inc., Barbara J Taylor of CCR FACS Core Facility and Susan Garfield of CCR Confocal Microscopy Core Facility. Shreyus Kulnarkis, Monika Kuban, Kandis Stubblefield, and Lakshman Bindu contributed with technical help.
This research was supported in part by the Center for Cancer Research, an Intramural Research Program of the National Cancer Institute, and by Breast Cancer Research Stamp Fund awarded through competitive peer review, and was funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contracts N01-CO-12400 and N01-CO-12401.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government.
References
- 1.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Witton CJ, Reeves JR, Going JJ, et al. Expression of the HER1–4 family of receptor tyrosine kinases in breast cancer. J Pathol. 2003;200:290–297. doi: 10.1002/path.1370. [DOI] [PubMed] [Google Scholar]
- 3.Cobleigh MA, Vogel CL, Tripathy D, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 4.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 5.Pegram MD, Lipton A, Hayes DF, et al. Phase II study of receptor-enhanced chemosensitivity using recombinant humanized anti-p185HER2/neu monoclonal antibody plus cisplatin in patients with HER2/neu-overexpressing metastatic breast cancer refractory to chemotherapy treatment. J Clin Oncol. 1998;16:2659–2671. doi: 10.1200/JCO.1998.16.8.2659. [DOI] [PubMed] [Google Scholar]
- 6.Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 7.Lewis Phillips GD, Li G, Dugger DL, et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008;68:9280–9290. doi: 10.1158/0008-5472.CAN-08-1776. [DOI] [PubMed] [Google Scholar]
- 8.Wolf P, Elsasser-Beile U. Pseudomonas exotoxin A: From virulence factor to anti-cancer agent. Int J Med Microbiol. 2008 doi: 10.1016/j.ijmm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Pastan I, Hassan R, Fitzgerald DJ, et al. Immunotoxin therapy of cancer. Nat Rev Cancer. 2006;6:559–565. doi: 10.1038/nrc1891. [DOI] [PubMed] [Google Scholar]
- 10.Pastan I, Hassan R, FitzGerald DJ, et al. Immunotoxin treatment of cancer. Annu Rev Med. 2007;58:221–237. doi: 10.1146/annurev.med.58.070605.115320. [DOI] [PubMed] [Google Scholar]
- 11.Iglewski BH, Kabat D. NAD-dependent inhibition of protein synthesis by Pseudomonas aeruginosa toxin. Proc Natl Acad Sci U S A. 1975;72:2284–2288. doi: 10.1073/pnas.72.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arons E, Sorbara L, Raffeld M, et al. Characterization of T-cell repertoire in hairy cell leukemia patients before and after recombinant immunotoxin BL22 therapy. Cancer Immunol Immunother. 2006;55:1100–1110. doi: 10.1007/s00262-005-0099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leland P, Taguchi J, Husain SR, et al. Human breast carcinoma cells express type II IL-4 receptors and are sensitive to antitumor activity of a chimeric IL-4-Pseudomonas exotoxin fusion protein in vitro and in vivo. Mol Med. 2000;6:165–178. [PMC free article] [PubMed] [Google Scholar]
- 14.Kawakami K, Kawakami M, Husain SR, et al. Targeting interleukin-4 receptors for effective pancreatic cancer therapy. Cancer Res. 2002;62:3575–3580. [PubMed] [Google Scholar]
- 15.Kawakami K, Kawakami M, Joshi BH, et al. Interleukin-13 receptor-targeted cancer therapy in an immunodeficient animal model of human head and neck cancer. Cancer Res. 2001;61:6194–6200. [PubMed] [Google Scholar]
- 16.Sarosdy MF, Hutzler DH, Yee D, et al. In vitro sensitivity testing of human bladder cancers and cell lines to TP-40, a hybrid protein with selective targeting and cytotoxicity. J Urol. 1993;150:1950–1955. doi: 10.1016/s0022-5347(17)35944-x. [DOI] [PubMed] [Google Scholar]
- 17.Pai LH, Gallo MG, FitzGerald DJ, et al. Antitumor activity of a transforming growth factor alpha-Pseudomonas exotoxin fusion protein (TGF-alpha-PE40) Cancer Res. 1991;51:2808–2812. [PubMed] [Google Scholar]
- 18.Friedman PN, McAndrew SJ, Gawlak SL, et al. BR96 sFv-PE40, a potent single-chain immunotoxin that selectively kills carcinoma cells. Cancer Res. 1993;53:334–339. [PubMed] [Google Scholar]
- 19.Hassan R, Lerner MR, Benbrook D, et al. Antitumor activity of SS(dsFv)PE38 and SS1(dsFv)PE38, recombinant antimesothelin immunotoxins against human gynecologic cancers grown in organotypic culture in vitro. Clin Cancer Res. 2002;8:3520–3526. [PubMed] [Google Scholar]
- 20.Onda M, Wang QC, Guo HF, et al. In vitro and in vivo cytotoxic activities of recombinant immunotoxin 8H9(Fv)-PE38 against breast cancer, osteosarcoma, and neuroblastoma. Cancer Res. 2004;64:1419–1424. doi: 10.1158/0008-5472.can-03-0570. [DOI] [PubMed] [Google Scholar]
- 21.Batra JK, Kasprzyk PG, Bird RE, et al. Recombinant anti-erbB2 immunotoxins containing Pseudomonas exotoxin. Proc Natl Acad Sci U S A. 1992;89:5867–5871. doi: 10.1073/pnas.89.13.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wels W, Harwerth IM, Mueller M, et al. Selective inhibition of tumor cell growth by a recombinant single-chain antibody-toxin specific for the erbB-2 receptor. Cancer Res. 1992;52:6310–6317. [PubMed] [Google Scholar]
- 23.Reiter Y, Brinkmann U, Jung SH, et al. Improved binding and antitumor activity of a recombinant anti-erbB2 immunotoxin by disulfide stabilization of the Fv fragment. J Biol Chem. 1994;269:18327–18331. [PubMed] [Google Scholar]
- 24.Bera TK, Viner J, Brinkmann E, et al. Pharmacokinetics and antitumor activity of a bivalent disulfide-stabilized Fv immunotoxin with improved antigen binding to erbB2. Cancer Res. 1999;59:4018–4022. [PubMed] [Google Scholar]
- 25.Schmidt M, McWatters A, White RA, et al. Synergistic interaction between an anti-p185HER-2 pseudomonas exotoxin fusion protein [scFv(FRP5)-ETA] and ionizing radiation for inhibiting growth of ovarian cancer cells that overexpress HER-2. Gynecol Oncol. 2001;80:145–155. doi: 10.1006/gyno.2000.6040. [DOI] [PubMed] [Google Scholar]
- 26.Skrepnik N, Zieske AW, Bravo JC, et al. Recombinant oncotoxin AR209 (anti-P185erbB-2) diminishes human prostate carcinoma xenografts. J Urol. 1999;161:984–989. [PubMed] [Google Scholar]
- 27.Wang L, Liu B, Schmidt M, et al. Antitumor effect of an HER2-specific antibody-toxin fusion protein on human prostate cancer cells. Prostate. 2001;47:21–28. doi: 10.1002/pros.1043. [DOI] [PubMed] [Google Scholar]
- 28.Skrepnik N, Araya JC, Qian Z, et al. Effects of anti-erbB-2 (HER-2/neu) recombinant oncotoxin AR209 on human non-small cell lung carcinoma grown orthotopically in athymic nude mice. Clin Cancer Res. 1996;2:1851–1857. [PubMed] [Google Scholar]
- 29.Shinohara H, Morita S, Kawai M, et al. Expression of HER2 in human gastric cancer cells directly correlates with antitumor activity of a recombinant disulfide-stabilized anti-HER2 immunotoxin. J Surg Res. 2002;102:169–177. doi: 10.1006/jsre.2001.6305. [DOI] [PubMed] [Google Scholar]
- 30.Nord K, Gunneriusson E, Uhlen M, et al. Ligands selected from combinatorial libraries of protein A for use in affinity capture of apolipoprotein A-1M and taq DNA polymerase. J Biotechnol. 2000;80:45–54. doi: 10.1016/s0168-1656(00)00232-7. [DOI] [PubMed] [Google Scholar]
- 31.Tolmachev V, Orlova A, Nilsson FY, et al. Affibody molecules: potential for in vivo imaging of molecular targets for cancer therapy. Expert Opin Biol Ther. 2007;7:555–568. doi: 10.1517/14712598.7.4.555. [DOI] [PubMed] [Google Scholar]
- 32.Siegall CB, Chaudhary VK, FitzGerald DJ, et al. Functional analysis of domains II, Ib, and III of Pseudomonas exotoxin. J Biol Chem. 1989;264:14256–14261. [PubMed] [Google Scholar]
- 33.Smith DC, Spooner RA, Watson PD, et al. Internalized Pseudomonas exotoxin A can exploit multiple pathways to reach the endoplasmic reticulum. Traffic. 2006;7:379–393. doi: 10.1111/j.1600-0854.2006.00391.x. [DOI] [PubMed] [Google Scholar]
- 34.Ogata M, Fryling CM, Pastan I, et al. Cell-mediated cleavage of Pseudomonas exotoxin between Arg279 and Gly280 generates the enzymatically active fragment which translocates to the cytosol. J Biol Chem. 1992;267:25396–25401. [PubMed] [Google Scholar]
- 35.Lombardi D, Soldati T, Riederer MA, et al. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreitman RJ, Pastan I. Targeting Pseudomonas exotoxin to hematologic malignancies. Semin Cancer Biol. 1995;6:297–306. doi: 10.1006/scbi.1995.0038. [DOI] [PubMed] [Google Scholar]
- 37.Ogata M, Chaudhary VK, Pastan I, et al. Processing of Pseudomonas exotoxin by a cellular protease results in the generation of a 37,000-Da toxin fragment that is translocated to the cytosol. J Biol Chem. 1990;265:20678–20685. [PubMed] [Google Scholar]
- 38.Theuer CP, Buchner J, FitzGerald D, et al. The N-terminal region of the 37-kDa translocated fragment of Pseudomonas exotoxin A aborts translocation by promoting its own export after microsomal membrane insertion. Proc Natl Acad Sci U S A. 1993;90:7774–7778. doi: 10.1073/pnas.90.16.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iglewski BH, Liu PV, Kabat D. Mechanism of action of Pseudomonas aeruginosa exotoxin Aiadenosine diphosphate-ribosylation of mammalian elongation factor 2 in vitro and in vivo. Infect Immun. 1977;15:138–144. doi: 10.1128/iai.15.1.138-144.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsu N, Oda T, Muramatsu T. Involvement of both caspase-like proteases and serine proteases in apoptotic cell death induced by ricin, modeccin, diphtheria toxin, and pseudomonas toxin. J Biochem. 1998;124:1038–1044. doi: 10.1093/oxfordjournals.jbchem.a022197. [DOI] [PubMed] [Google Scholar]
- 41.Chang JH, Kwon HY. Expression of 14-3-3delta, cdc2 and cyclin B proteins related to exotoxin A-induced apoptosis in HeLa S3 cells. Int Immunopharmacol. 2007;7:1185–1191. doi: 10.1016/j.intimp.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Jenkins CE, Swiatoniowski A, Issekutz AC, et al. Pseudomonas aeruginosa exotoxin A induces human mast cell apoptosis by a caspase-8 and -3-dependent mechanism. J Biol Chem. 2004;279:37201–37207. doi: 10.1074/jbc.M405594200. [DOI] [PubMed] [Google Scholar]
- 43.Du X, Ho M, Pastan I. New immunotoxins targeting CD123, a stem cell antigen on acute myeloid leukemia cells. J Immunother. 2007;30:607–613. doi: 10.1097/CJI.0b013e318053ed8e. [DOI] [PubMed] [Google Scholar]
- 44.Bera TK, Onda M, Brinkmann U, et al. A bivalent disulfide-stabilized Fv with improved antigen binding to erbB2. J Mol Biol. 1998;281:475–483. doi: 10.1006/jmbi.1998.1948. [DOI] [PubMed] [Google Scholar]
- 45.Su AI, Cooke MP, Ching KA, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen DL, Andersson M, Kamby C. HER2-targeted therapy in breast cancer. Monoclonal antibodies and tyrosine kinase inhibitors. Cancer Treat Rev. 2008 doi: 10.1016/j.ctrv.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Akabani G, Carlin S, Welsh P, et al. In vitro cytotoxicity of 211At-labeled trastuzumab in human breast cancer cell lines: effect of specific activity and HER2 receptor heterogeneity on survival fraction. Nucl Med Biol. 2006;33:333–347. doi: 10.1016/j.nucmedbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Robinson MK, Shaller C, Garmestani K, et al. Effective treatment of established human breast tumor xenografts in immunodeficient mice with a single dose of the alpha-emitting radioisotope astatine-211 conjugated to anti-HER2/neu diabodies. Clin Cancer Res. 2008;14:875–882. doi: 10.1158/1078-0432.CCR-07-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milenic DE, Garmestani K, Brady ED, et al. Potentiation of high-LET radiation by gemcitabine: targeting HER2 with trastuzumab to treat disseminated peritoneal disease. Clin Cancer Res. 2007;13:1926–1935. doi: 10.1158/1078-0432.CCR-06-2300. [DOI] [PubMed] [Google Scholar]
- 50.Lee SB, Hassan M, Fisher R, et al. Affibody molecules for in vivo characterization of HER2-positive tumors by near-infrared imaging. Clin Cancer Res. 2008;14:3840–3849. doi: 10.1158/1078-0432.CCR-07-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.