Abstract
Using an original workflow, we have modeled, constructed and characterized two new molecular devices that inducibly activate gene expression in Escherichia coli. The devices, prokaryotic-TetOn and prokaryotic-TetOff, were built by fusing an inducible DNA-binding protein domain to a transcription activation domain and constructing a complementary synthetic promoter sequence through which they could control downstream gene expression. In particular, the transactivators were built using variants of the tetracycline repressor, TetR, and the transactivating domain of the LuxR activator. The complementary promoter sequence included TetR’s operator, tetO, and elements of the lux promoter. These specific protein domains and their operator sites were chosen as they have been thoroughly studied and well characterized. First, our methodology began with optimizing the geometry of the molecular components using molecular modeling. We did so to achieve an unprecedented combination of controllable and transactivating function in bacterial organisms. The devices were then built to activate the expression of green fluorescent protein. Their unique function was found to be robustly tight, and activating many-fold increases of expressed gene levels, as measured by flow cytometry experiments. The devices were further characterized with stochastic kinetic models. The new devices presented herein may become useful additions to the molecular toolboxes used by biologists to control bacterial gene expression. The methodology used may also be a foundation for the design, development and characterization of a library of such devices and more complex gene regulatory networks.
Introduction
Controllable gene regulatory systems are the subject of continued, intense investigations. In addition to being critically important in explaining how phenotypes emerge from genotypes in living organisms, their components are rapidly becoming integral in efforts towards engineered gene expression control (1–10). Two well-studied example regulatory systems are the tetracycline (tet) and luminescence (lux) operons. The tetracycline repressor protein, TetR, and numerous TetR derivatives, afford a remarkably robust function of inducible repression and derepression of gene expression. Thus, they have been employed in numerous synthetic biology and bioengineering applications (11–14). Switching gene expression on and off by TetR and its derivatives simply depends on the presence or absence of the antibiotic tetracycline (Tc). The lux operon’s transcription activator protein, LuxR, is another attractive controllable molecular component for engineering applications. When induced, it activates gene overexpression by recruiting transcriptional machinery to the promoter. In addition, number of variant activators have been identified that upregulate transcription over a range of strengths with varied dependence on inducer molecules (15–18).
Herein, we present two novel synthetic molecular devices that inducibly upregulate bacterial gene expression. We have designed, built and characterized these devices using a unique methodology that is based on both computational and experimental efforts. Both devices are composed of an inducible DNA-binding domain and a transcription activation domain. In particular, TetR and the reverse-TetR (rTetR) derivative were chosen for the inducible domains while the transactivating domain of LuxR was incorporated for transcription upregulation (19–22). TetR protein and its derivative, rTetR, dissociate from and bind to the tetO operator sequence in response to Tc (respectively) (19–21). This behavior made them attractive candidates for controlling our devices. In addition, LuxRΔN(2-162) (LuxRΔN) is the C-terminal, constitutive transactivating domain of the full length LuxR activator. Strong, constitutive transcription activation is achieved with this variant lacking the N-terminal residues 2-162 of the full protein (22). These specific protein components were selected as they have been thoroughly studied and well characterized. Specifically, their protein:DNA-operator crystal structures for both domains as well as the kinetic parameters that govern their protein:protein and protein:DNA interactions were available in the literature.
We designed, constructed and characterized the two new devices using an original workflow that integrates experimental synthetic biology, molecular modeling and stochastic reaction kinetic simulations. This methodology may be directly implemented for the development of other biological devices and larger regulatory networks. The devices we present here, prokaryotic-TetOn and prokaryotic-TetOff (proTeOn and proTeOff), function in an Tc-dependent manner: the proTeOn synthetic protein (PROTEON) activates gene expression in the presence of Tc, and the proTeOff synthetic protein (PROTEOFF) activates expression in the absence of Tc. Orders of magnitude higher expression levels are observed in the activating states compared to the basal expression levels of both systems. With these new devices, there is now a dial of activated expression to complement those of basal and repressed gene expression.
While the proTeOn and proTeOff systems are functionally unique, their designs are conceptually inspired by the widely used TetOn and TetOff systems, which function in eukaryotic systems (23, 24). Gossen and co-workers fused the TetR protein to a mammalian transactivating domain, building molecular devices that activate eukaryotic gene expression in a Tc-dependent manner. To our knowledge, there were no prokaryotic transcription factors homologous to TetOn or TetOff prior to the construction of proTeOn and proTeOff. Additionally, our kind of engineering approach to synthetic biology, using molecular dynamic simulations to guide system design and stochastic simulations to enhance system characterization, has not been previously reported in the literature.
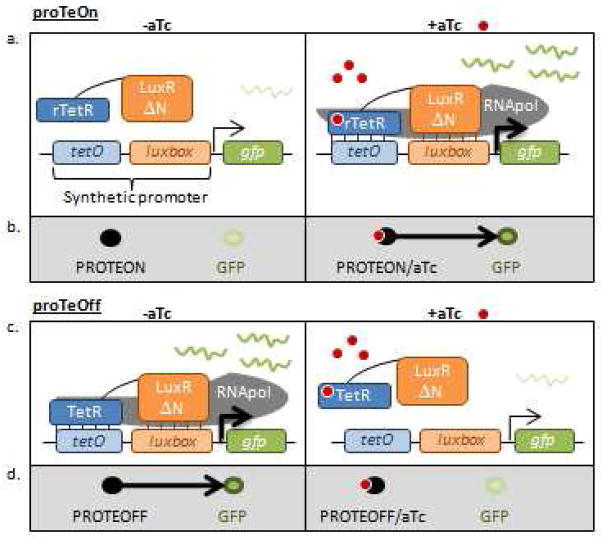
The proposed behaviors of proTeOn and proTeOff are illustrated in Figure 1. As shown, anhydrotetracycline (aTc), a Tc derivative, was used as the inducer in our experiments. For proTeOn, in the absence of aTc, the inducible DNA binding domain, rTetR, does not bind the tetO operator and the PROTEON protein does not upregulate the target gene, green fluorescence protein (gfp) (21). Upon activation with aTc, rTetR binds the inducer, undergoes a conformational change and binds tetO (21). This binding brings LuxRΔN near its operator site, allowing it to bind luxbox and upregulate gfp transcription through RNApol recruitment to the promoter (25–27). For proTeOff, in the absence of aTc, its inducible DNA binding domain, TetR, binds tetO (2, 20, 28, 29). This binding brings LuxRΔN close to its operator site permitting it to bind luxbox and upregulate gfp transcription (25–27). After the addition of aTc, TetR binds the small molecule, undergoes a conformational change and releases tetO (2, 20, 28, 29). Upon dissociation of TetR:tetO, the LuxRΔN:luxbox interaction is destabilized and transcription upregulation is terminated.
Figure 1. Function of proTeOn and proTeOff.
a. proTeOn behavior. In the absence of aTc, rTetR does not bind tetO and PROTEON does not upregulate gfp. Upon activation with aTc, rTetR binds the inducer, undergoes a conformational change and binds tetO, bringing LuxRΔN near its operator site to bind luxbox and upregulate gfp transcription through RNApol recruitment to the promoter.
b. proTeOn logic. In the absence of aTc, PROTEON does not control GFP expression. Upon induction with aTc, PROTEON upregulates GFP.
c. proTeOff behavior. In the absence of aTc, TetR binds to tetO, bringing LuxRΔN near its operator site to bind luxbox and upregulate gfp transcription through RNApol recruitment. After the addition of aTc, TetR binds to the small molecule, undergoes a conformational change, and releases tetO. Upon dissociation of TetR:tetO, the LuxRΔN:luxbox interaction is destabilized and upregulation by RNApol recruitment terminated.
d. proTeOff logic. In the absence of aTc, PROTEOFF upregulates GFP expression. Upon the addition of aTc, PROTEOFF’s control on GFP expression is terminated.
To achieve these phenotypes, we first used molecular modeling to design the geometry of the synthetic transactivator/promoter pairs of each system. The important feature of proTeOn and proTeOff on which we focused was the optimized interaction between the inducible synthetic transactivator protein that upregulates gene expression and its complementary synthetic promoter. Next, we built and characterized both systems experimentally. Last, implementing stochastic simulations, we explored the system dynamics and quantified unknown kinetic parameters of key interactions. The workflow is detailed in the Methods section. In the Results and Discussion section, we begin by presenting elements of the modeldriven design of the devices.
ProTeOn and proTeOff can be applied to robustly manage prokaryotic gene expression. Variant systems and more complex inducible gene regulatory networks can also be designed and constructed using this workflow and the proTeOn and proTeOff systems as a foundation.
Results and Discussion
proTeOn and proTeOff designs
For proTeOn and proTeOff to perform efficiently, both protein domains (TetR/rTetR and LuxRΔN) must readily bind their operator sequences upon induction, LuxRΔN must recruit RNApol to the promoter and then RNApol must bind to the promoter and begin transcription. Numerous length and space requirements are associated with each of these steps and we systematically accommodated each of them by designing the devices prior to construction. To satisfy the above geometric constraints, we built molecular models of each system using MOE (30) and NAMD (31). With the chosen protein components and DNA operator sites as starting points, the entire DNA promoter sequence and the linker amino acid sequence were designed in tandem.
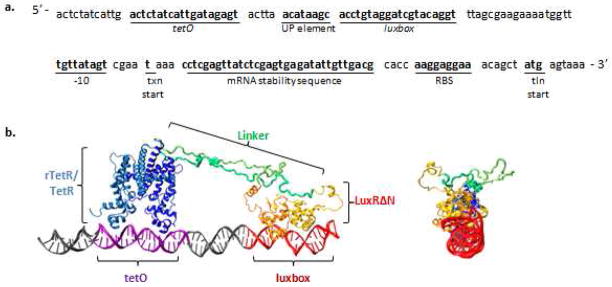
First, the synthetic promoter for both systems was designed, and the optimized sequence is annotated in Figure 2a. The major aim of the promoter design was to minimize the distance between the two DNA-bound protein domains while maintaining favorable binding between the domains and their operators. The interactions between RNApol and the specific residues of the promoter to which it binds (the UP element and the −10 region) had to be accommodated as well. To meet these aims, the operator sites were integrated into the promoter such that the two domains bound along the same face of the DNA (Figure 2b, side view) while maintaining a minimum distance between the two operator sites. The final design was scrutinized to ensure that neither DNA-bound protein domain was encroaching the residues of the UP element or −10 region as this would inhibit efficient RNApol recruitment.
Figure 2. proTeOn and proTeOff systems’ design.
a. proTeOn and proTeOff synthetic promoter sequence. Both PROTEON and PROTEOFF bind and recruit RNApol to this synthetic promoter sequence. Moving from 5′ to 3′: the rTetR/TetR protein domain binds tetO, RNApol binds the UP element and the −10 region, LuxRΔN binds the luxbox, the mRNA stability sequence stabilizes the mRNA transcript, and ribosomes bind the RBS of this resulting mRNA message.
b. proTeOn molecular model. Both proTeOn and proTeOff are designed to assemble as shown. The inducible DNA binding domain (rTetR/TetR, blue) binds the tetO operator (purple), and the transcription activator domain (LuxRΔN, orange) binds the luxbox (red). The two domains bind their operators along the same face of the DNA double helix and are connected (TetR/rTetR’s C-terminus to LuxRΔN’s N-terminus) by a linker peptide (green).
Next, we optimized the peptide linker required to connect the two DNA-bound protein domains (TetR/rTetR’s C-terminus to LuxRΔN’s N-terminus). We did this by assuming it resembles a polymer in good solution and choosing a length that would minimize the entropic elastic tension effects. The optimal linker length was determined to be 150 Å and the final sequence is the product of 5 repeats of a 9-amino acid subunit, ARTQYSESM. The individual subunits are connected by single glycine residues, and this 49-amino acid peptide is flanked by 3 glycine residues on each side. These were chosen to ensure linker flexibility and promote correct protein domain folding. The final optimized proTeOn system, meeting all geometric requirements, is illustrated in Figure 2b.
Characterization of proTeOn and proTetOff behavior
To investigate the behavior of both systems and determine the optimal conditions and applications of each device, we performed two sets of experiments. First, cells expressing the transactivators were induced with a range of aTc concentrations. Second, cultures were primed with these aTc amounts prior to transactivator expression. In both experiments we monitored the resulting differential expression of the target gene, gfp, over time. Induction experiments were repeated for all inducer concentrations and time points. The reported trends were observed across the replicates.
proTeOn
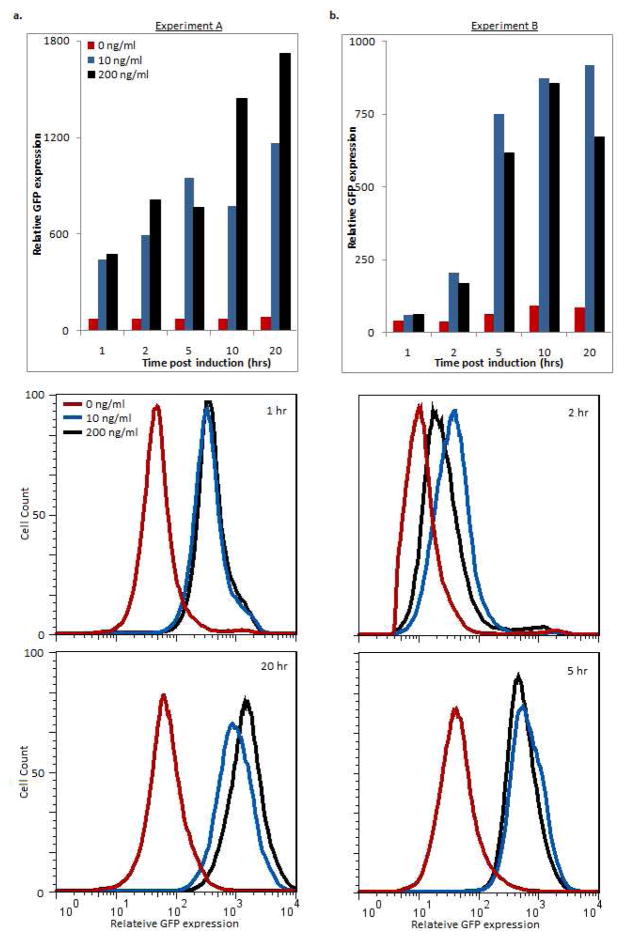
Upon administration of aTc, proTeOn upregulates target genes by one hour post-treatment and achieves 15-fold upregulation through long times. With a low (10 ng/ml) aTc concentration, proTeOn can achieve steady state expression of targets 10-fold over untreated cultures just 5 hours post-treatment. With a high (200 ng/ml) aTc concentration, target gene expression can reach steady state by 10 hours post-treatment at levels 15-fold over untreated cultures. These behaviors are displayed as both expression means and population distributions in Figure 3a. Basal GFP expression levels are observed from the synthetic promoter in the absence of aTc. There is no repressed state for the proTeOn system as presented here.
Figure 3. proTeOn system phenotype analysis by flow cytometry.
Mean GFP expression and expression distribution were analyzed by flow cytometry 1, 2, 5, 10, and 20 hours post-treatment for both experimental set-ups as described. Induction experiments were repeated for all inducer concentrations and time points, and the reported trends were observed across replicates.
a. Cells expressing PROTEON were induced with 0, 10 and 200 ng/ml aTc. By one hour after aTc treatment, proTeOn upregulates GFP expression. Steady state expression is reached by 5 hours and 10 hours with low and high aTc concentrations (respectively) and maintained through 20 hours. Maximum overexpression is 10 and 15-fold above uninduced controls with low and high aTc levels respectively.
b. PROTEON was expressed in cells pre-cultured with 0, 10 and 200 ng/ml aTc. In low and high aTc, significant proTeOn activity is observed 2 hours after PROTEON expression is induced. Steady state activity is achieved by 5 hours and maintained through 20 hours. Maximum upregulation is 10–fold above uninduced controls with both aTc concentrations
PROTEON expressed in the presence of aTc gives target gene upregulation within 2 hours. Steady state upregulation is achieved by 5 hours after the transactivator’s initial expression, with GFP expression 10-fold over uninduced controls. PROTEON is thus rapidly transcribed, translated, folded and becomes functional when under the control of a free, non-repressed, promoter. This is show in Figure 3b. The mean behaviors taken across biological replicates are discussed in the Supporting Information and shown in Supporting Figure 3.
Applications of proTeOn would be appropriate in systems when quick bursts or long term gene upregulation is desired. Such behavior is governed by rTetR’s responses to induction with aTc. This control is achievable with inducer levels as low as 10 ng/ml aTc. aTc concentrations outside of this range, or scaled throughout an experiment, may be useful depending upon the desired level of activation. Notably, long term target gene upregulation increases with aTc up to 200 ng/ml. Thus, inducing with a range of aTc concentrations, proTeOn can be tuned to robustly upregulate gene expression for both acute and long time scales.
proTeOff
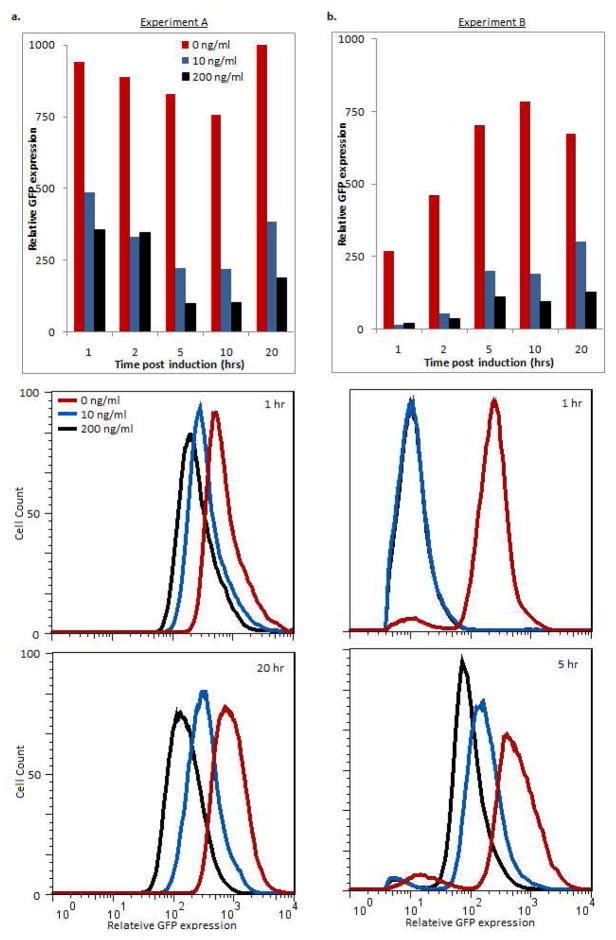
In the absence of aTc, proTeOff activates target gene expression. Upon administration of aTc, upregulation can be reduced by half just one hour post-treatment. Steady state target gene expression is only one-half and one-fifth that of untreated samples with low and high aTc concentrations respectively. This is observed as early as 5 hours and remains through long times. While activation by proTeOff is significantly reduced with low aTc concentrations, a subset of activator proteins still appears to bind the promoter and upregulate transcription. With GFP levels at only one-fifth of the untreated controls, we consider the low GFP expression observed with 200 ng/ml aTc to be the synthetic promoter’s basal expression level. These low basal expression levels are observed through long times. These behaviors are presented also as expression means and population distributions in Figure 4a. Please note that there is also no repressed state for the proTeOff system as presented here.
Figure 4. proTeOff system phenotype analysis by flow cytometry.
Mean GFP expression and expression distribution were analyzed by flow cytometry 1, 2, 5, 10, and 20 hours post-treatment for both experimental set-ups as described. Induction experiments were repeated for all inducer concentrations and time points, and the reported trends were observed across replicates.
a. With 0 ng/ml, proTeOff upregulates GFP expression. With 10 and 200 ng/ml aTc, expression is reduced to half that of untreated samples by one hour. Steady state expression is reached by 5 hours and maintained through 20 hours post-treatment. Minimum expression is one-half and one-fifth that of untreated samples with low and high aTc respectively.
b. PROTEOFF was expressed in cells pre-cultured with 0, 10 and 200 ng/ml aTc. In 0 ng/ml aTc, proTeOff activity is observed (via GFP upregulation) one hour after PROTEOFF expression is induced. In low and high aTc, reduced proTeOff activity is observed across all times and steady state activity is achieved by 5 hours after initial PROTEOFF expression. High aTc levels maintain the reduced device activity, thus basal GFP expression. Low concentrations permit GFP levels to rise to one-third that of untreated cultures. Both of these behaviors are observed through 20 hours.
PROTEOFF expressed in the absence of aTc leads to target gene upregulation within one hour and maintains it through long times. PROTEOFF is therefore quickly transcribed, translated, folded and functional when under the control of a free, non-repressed, promoter. In the presence of low and high aTc concentrations, the device activity is maintained at less than one-fifthof untreated cultures one hour after the PROTEOFF’s initial expression. An aTc concentration of 200 ng/ml maintains this very low basal activity, thus basal gene expression, through long times. Treatment with low aTc allows the target’s expression to rise to onethird of untreated cultures by 5 hours after the transactivator’s initial expression, where it remains through long times. This elevation is due to a subset of free PROTEOFFs binding to the promoter and recruiting RNApol. These data are shown in Figure 4b. The mean behaviors taken across biological replicates are discussed in the Supporting Information and shown in Supporting Figure 3. proTeOff can be applied to upregulate gene expression for both short and long times. Continuous long term upregulation as well as upregulation with intermittent periods of low expression can be achieved. These behaviors are governed by TetR’s response to aTc. aTc concentrations as low as 10 ng/ml effectively achieve periods of low expression. Target gene expression drops with increasing aTc concentrations 10–200 ng/ml. aTc concentrations outside of this range or scaled throughout an experiment, may also achieve a desired phenotype. Thus, proTeOff can be tuned to upregulate gene expression for short, long-continuous and long-intermittent time scales using a range of aTc concentrations.
Characterization of proTeOn and proTeOff kinetics
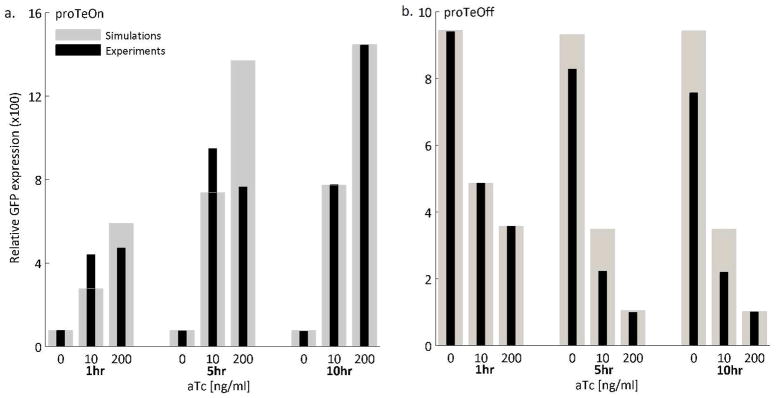
In addition to experimentally testing proTeOn and proTeOff, we have assessed the kinetics of both systems by conducting stochastic simulations. This was done to characterize the systems at a finer resolution than can be achieved in the lab alone. We aimed to quantify the strength of the interactions between the device components by developing a stochastic model that captures the time profiles of measured GFP probability distributions. We modeled the transcription, translation, regulatory and degradation events with stochastic kinetics. We built the models and conducted the simulations as described before (32–41) and discussed in the Supporting Information. Stochastic model parameters that did not exhist in the literature were fit to match the untreated experimental phenotypes (0 ng/ml) and the behaviors upon induction with both low and high aTc concentrations (10 and 200 ng/ml). The mean GFP expression is captured by the model for both proTeOn and proTeOff at short times post-aTc induction (1 and 5 hours) as well as at steady state (10 hours), as shown in Figure 5. Overall, the mean GFP levels achieved by the simulation trajectories agree well with the experimental observations.
Figure 5. proTeOn and proTeOff average GFP by stochastic simulations.
Simulation and experimental results of the average GFP expression at 1, 5, and 10 hours, when 0, 10, and 200 ng/ml aTc are administered. Overall, a good agreement between experimental and computational results is observed.
a. proTeOn average culture GFP.
b. proTeOff average culture GFP.
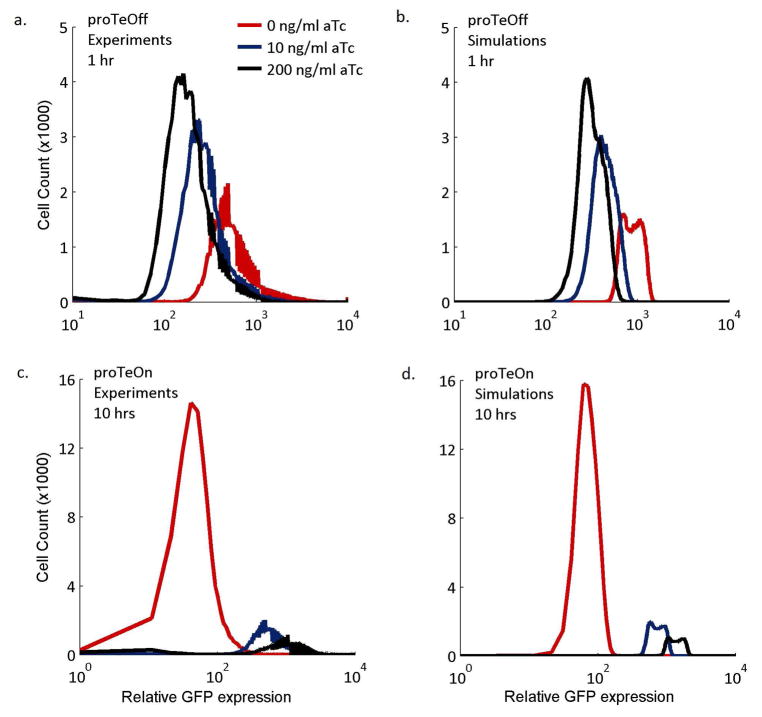
A discrepancy is observed between the simulation and experimental results at 5 hours with 200 ng/ml aTc. This may be attributed to high aTc levels retarding cellular processes, such as protein overexpression (42). By 10 hours, this effect is no longer experimentally significant and a good match is therefore observed between theoretical and experimental results. Due to their stochastic nature, the models can also capture the GFP distributions observed experimentally at short and long times. The GFP distributions at 10 hours for proTeOn and 1 hour for proTeOff, when 0, 10 and 200 ng/ml aTc are administered, are presented in Figure 6. For both systems, and across all aTc concentrations, the distributions generated in silico match those observed in vivo.
Figure 6. proTeOn and proTeOff GFP distribution by stochastic simulations.
Simulation (Figures 6b, 6d) and experimental (Figures 6a, 6c) results of the distribution of GFP expression throughout the cell population when 0, 10 and 200 ng/ml aTc are administered. The simulation results largely agree with the experimental phenotypes.
a, b. proTeOff GFP distribution at 1 hour.
c, d. proTeOn GFP distribution at 10 hours.
The protein-DNA binding strengths in the proTeOn and proTeOff systems are characterized by four kinetic parameters. These values, extracted from the models, are given in Table 1. Binding of aTc to PROTEON increases the affinity of the latter for the promoter 10,000-fold. Binding of PROTEOFF to aTc leads to a decrease in the affinity of PROTEOFF to tetO by 108 times. Both PROTEON and PROTEOFF enhance gfp expression significantly when bound to tetO by recruiting RNApol to the promoter. In our simulations, this is realized by increasing the binding strength of RNApol to DNA. Conforming to the simulation results, binding of PROTEON to tetO increases the binding strength of RNApol to DNA approximately 22 times, whereas binding of PROTEOFF to tetO increases the binding strength around 14 times.
Table 1. Dissociation constants of the key biomolecular interactions underlying proTeOn and proTeOff.
The following dissociation constants are estimates from our modeling efforts. The units for the second order reactions (#1 and 2) are M whereas for the third order reactions (# 3 and 4) they are M2.
| Biomolecular Interactions | Binding Affinity | |
|---|---|---|
| proTeON | proTeOFF | |
| PROTET + tetO ↔ PROTET:tetO | 2.5E-5 | 2.5E-10 |
| PROTET:aTc2 + tetO ↔ PROTET:tetO:aTc2 | 2.5E-10 | 2.5E-2 |
| RNApol + pro + tetO ↔ RNApol:pro:tetO | 3.7E-9 | 3.7E-9 |
| RNApol + pro + PROTET:tetO ↔ RNAp:pro:tetO:PROTET | 1.67E-10 | 2.56E-10 |
Conclusions
The engineered proTeOn and proTeOff systems can all be applied to tightly control gene expression with aTc in prokaryotes. Incorporating appropriate tags in the target genes’ transcripts may render proTeOn and proTeOff powerful tools for cytosolic, membrane associated and secreted protein over-expression.
More broadly however, the workflow presented here can be implemented in the design, construction and testing of a library of variant devices and regulatory networks. Such devices include those that respond to sugars, proteins, amino acids, metabolites, toxins and other small molecules. With these, one may efficiently engineer gene expression responses as applications demand. As the collection of well characterized synthetic devices grows, we may also be able to combine them, along with naturally occurring parts, into larger gene regulatory networks to achieve more complex desired phenotypes. Feedback loops, feedforward loops, AND and OR logic gates are a few possible networks. These devices and networks may then be used either as tools for controlling the expression of a single gene, or as interoperable parts of larger regulatory networks that control multiple genes independently. The current proTeOn and proTeOff devices, and the kinetic and structural details that have been identified for each, are a firm stepping stone from which this work can expand. Using our workflow, integrating experimental synthetic biology, molecular modeling, and stochastic reaction kinetic simulations, the required effort and expense that scale with system complexity will also be reduced. Molecular devices that can tunably regulate the expression of a single, or a handful of, genes in response to sugar, protein, amino acid, metabolite, toxin and other small molecule levels may provide synthetic biology research and the bioengineering industry the tools they need for efficient, diverse gene expression control.
Methods
PROTEON and PROTEOFF parts
PROTEON and PROTEOFF are composed of an inducible DNA binding domain and a DNA binding transcription activator, connected by a linker peptide. The reverse tetracycline repressor (rTetR) is the N-terminal, inducible DNA binding domain in PROTEON while the tetracycline repressor (TetR) is in PROTEOFF, rendering both systems responsive to anhyrdorotetracycline (aTc) (2, 19–21, 28, 29). LuxRΔN(2–162) (LuxRΔN) is the DNA-binding activator at the C-terminus of both synthetic proteins (22). LuxRΔN is the C-terminal domain of the full length LuxR transactivator. It lacks N-terminal residues 2-162 of the full length LuxR and possesses strong constitutive transactivator activity (22). In both systems, the two domains are connected by a 150 Å, 55 amino acid, peptide linker (its design is discussed below).
proTeOn and proTeOff synthetic promoter parts
The proTeOn and proTeOff synthetic promoter is composed of sequences from the tet and lux operons’ promoters. Operator sites for TetR/rTetR (2, 20, 29) and LuxRΔN (25, 26, 29), RNApolymerase (RNApol) binding sequences (the UP element and −10 region) (27), transcription and translation start sites, an mRNA stabilizing sequence and a ribosomal binding site (43) are included. These sequences are annotated in Figure 2a. Green fluorescence protein mutant 3 (GFP) is the reporter molecule used to monitor the systems’ behavior and is under the control of the synthetic promoter (44). A second synthetic promoter, containing the RBS of the lux promoter and lacking the mRNA stability sequence was also tested. This promoter provided gfp transcript increase upon induction. However, a parallel increase in GFP protein was not observed.
Molecular modeling
We designed the proTeOn and proTeOff systems using molecular modeling with MOE (30) and with NAMD (31). We built models of the systems utilizing the known structure of the inducible DNA binding domain (rTetR or TetR) bound to the tetO operator (PDB code 1QPI) (19), and the transcription activator domain (LuxRΔN) bound to the luxbox (PDB code 1H0M) (45). The linker size was determined by assuming the scaling of polymer’s end-to-end-vector distance in a good solvent (46). The specific sequence was then determined to minimize proteolysis in bacteria. We thus designed a 55-amino acid, linker peptide, which is expected to be linear, flexible, and hydrophilic (47).
System construction
Both synthetic activator genes were synthesized by GENEART and using standard molecular biology techniques, cloned into the expression vector pT7-FLAG1 (p1118 Sigma), at KPN1 restriction sites, in Top10 Escherichia coli (E. coli) cells (C404010 Invitrogen). The final constructs are illustrated in Supporting Figure 1a. The synthetic promoter and gfp (44) were synthesized by GENEART, on pMK, a pUC19 derived expression vector that is compatible in E. coli, high copy, and kanamycin resistant. This construct is illustrated in Supporting Figure 1b. proTeOn and proTeOff are contained on these two plasmids and were transformed into chemically competent BL21(DE3)-T1 E. coli cells (B2935 Sigma) by heat shock for characterization.
Gene expression control by tetracycline in bacterial cultures
BL21(DE3)-T1 cells containing the proTeOn and proTeOff systems were cultured in selective LB media, at 30° C to facilitate temperature sensitive folding of LuxRΔN, agitating at 200 rpm. Cultures were maintained in mid-log growth. We completed initial experiments to first confirm the solubility and stability of PROTEON and PROTEOFF and then to establish promoter specific gene regulation by aTc (rather than a general soluble protein upregulation).
Basal, low, medium and high levels of the synthetic activators were maintained with 0, 0.25, 0.75 and 1 mM IPTG respectively. Each system was induced over a range of aTc concentrations, 1, 10, and 200 ng/ml. The total soluble protein was isolated from cultures with CelLyticB reagent (B7435 Sigma), separated by size on a 10% polyacrylamide gel and transferred to PVDF membrane. PROTEON was detected by primary mouse monoclonal anti-FLAG M2 antibody (F3165 Sigma), GFP by mouse monoclonal anti-GFP antibody [LGB-1] (ab291 Abcam), and loading control RNApol by mouse monoclonal anti-RNApol sigma 70 antibody [2G10] (ab12088). Biotinylated, polyclonal sheep anti-mouse secondary antibody, VECTASTAIN ABC kit (PK-4002 Vector Labs), and Amersham ECL Westernblotting detection reagent (RPN 2109) were used to complete the specific detection of each protein. Relative quantification of each protein was performed using ImageJ software, publically available at http://rsb.info.nih.gov, taking the ratio of protein of interest to RNApol density. PROTEON and PROTEOFF are soluble and stable at high intracellular concentrations, and gene regulation by aTc is specific to genes under the control of the synthetic promoter. This is discussed in Supporting Information and illustrated in Supporting Figure 2.
We characterized each system maintaining PROTEON and PROTEOFF expression with 0.75 mM IPTG and controlling the activity of each device with 0, 10, and 200 ng/ml aTc. Two sets of experiments were performed. In experiment set-up A, PROTEON and PROTEOFF production was induced overnight with 0.75 mM IPTG and 0 ng/ml aTc; at t = 0 hours cultures were treated with 0, 10, and 200 ng/ml aTc. In experiment set-up B cultures were treated overnight with 0, 10, and 200 ng/ml aTc and 0 mM IPTG; at t = 0 PROTEON and PROTEOFF expression was induced with 0.75 mM IPTG. Experiments A investigated the proTeOn and proTeOff system dynamics upon induction with aTc while experiments B provided insight on the PROTEON and PROTEOFF protein production and maturation dynamics.
IPTG and aTc levels were maintained in the cultures at all t > 0 hours. Induction experiments were repeated for all inducer concentrations and time points, and reported trends were observed across replicates.
Cell samples were collected for analysis by flow cytometry at t = 1, 2, 5, 10 and 20 hours, fixed with 4% paraformaldehyde for 30 min at room temperature, washed with 1x phosphate buffered saline (PBS) and stored in 1x PBS at 4° C. Individual cells’ GFP expression was measured by flow cytometry using a FACScalibur (BD Biosciences). 100,000 cells were investigated per sample with excitation at λex = 488 nm and subsequent fluorescence detection at λem = 530 ± 30 nm. The cytometry data was collected using CellQuest (BD Biosciences) and analyzed using FlowJo (Tree Star) software. Each sample’s healthy cell population was selected by first removing erroneous events (due to electronic noise) that fell below a minimum emission at λem = 530 ± 30 nm, then secondly removing events that fell outside of the characteristic side-scatter and forward-scatter range for single E. coli cells. The differential GFP expression of the selected cells was analyzed and compared across samples.
Mean gene expression data analysis
All cytometry data was collected as described above. Prior to evaluating data across replicates, all GFP expression values were normalized to their corresponding (same system and time point) 0 ng/ml aTc samples. Basal expression (achieved with 0 ng/ml) from the proTeOn system is denoted with a value of one. In contrast, maximal expression (also achieved with 0 ng/ml) from the proTeOff system is denoted with a value of one. This basis is consistent for both experimental setups and through all times. Normalized GFP expression averages and standard errors were then calculated for replicates.
Stochastic modeling
We built computer models of proTeOn and proTeOff to further characterize the experimental behavior of each system. A hybrid stochastic-discrete and stochastic-continuous algorithm called Hy3S was used (38, 39). Hy3S couples chemical Langevin equations with discrete kinetic Monte Carlo, modeling a system’s behavior at the resolution of biomolecular interactions in individual cells. Characteristics of numerous natural and synthetic biological systems have previously been described using this approach (32, 37, 38, 40, 41). The simulations were carried out under a number of key assumptions and parameters that are discussed in the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (American Recovery and Reinvestment Act grant GM086865), the National Science Foundation (CBET-0425882 and CBET-0644792), and the University of Minnesota Biotechnology Institute. Computational support from the Minnesota Supercomputing Institute (MSI) is gratefully acknowledged. This work was also supported by the National Computational Science Alliance under TG-MCA04N033. We are thankful to Dr. Emilia Wu for her assistance with the protein structure images.
Footnotes
Author Contributions
All authors contributed extensively to the work presented in this paper.
Competing Financial Interests
The authors declare no competing financial interests.
Supporting Information Available: A detailed description of the stochastic kinetic reaction models of protein devices and further details on experimental characterization. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Foster TJ, Davis MA, Roberts DE, Takeshita K, Kleckner N. Genetic organization of transposon Tn10. Cell. 1981;23:201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- 2.Hillen W, Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 3.Hillen W, Gatz C, Altschmied L, Schollmeier K, Meier I. Control of expression of the Tn10-encoded tetracycline resistance genes. Equilibrium and kinetic investigation of the regulatory reactions. J Mol Biol. 1983;169:707–721. doi: 10.1016/s0022-2836(83)80166-1. [DOI] [PubMed] [Google Scholar]
- 4.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan HB, Greenberg EP. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaefer AL, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci U S A. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kritzer JA. Grand challenge commentary: Beyond discovery: probes that see, grab and poke. Nat Chem Biol. 6:868–870. doi: 10.1038/nchembio.469. [DOI] [PubMed] [Google Scholar]
- 8.Vickers CE, Blank LM, Kromer JO. Grand challenge commentary: Chassis cells for industrial biochemical production. Nat Chem Biol. 6:875–877. doi: 10.1038/nchembio.484. [DOI] [PubMed] [Google Scholar]
- 9.Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, Collins JJ. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc Natl Acad Sci U S A. 2010;107:15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaacs FJ, Collins JJ. Plug-and-play with RNA. Nat Biotechnol. 2005;23:306–307. doi: 10.1038/nbt0305-306. [DOI] [PubMed] [Google Scholar]
- 11.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 12.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 13.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 14.Ramalingam KTJ, Maynard J, Kaznessis Y. Forward engineering of synthetic bio-logical AND gates. Biochemical Engineering Journal. 2009:38–47. [Google Scholar]
- 15.Egland KA, Greenberg EP. Quorum sensing in Vibrio fischeri: analysis of the LuxR DNA binding region by alanine-scanning mutagenesis. J Bacteriol. 2001;183:382–386. doi: 10.1128/JB.183.1.382-386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuqua C, Winans SC, Greenberg EP. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 17.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxRLuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sitnikov DM, Schineller JB, Baldwin TO. Transcriptional regulation of bioluminesence genes from Vibrio fischeri. Mol Microbiol. 1995;17:801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- 19.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat Struct Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 20.Saenger W, Orth P, Kisker C, Hillen W, Hinrichs W. The Tetracycline Repressor-A Paradigm for a Biological Switch. Angew Chem Int Ed Engl. 2000;39:2042–2052. doi: 10.1002/1521-3773(20000616)39:12<2042::aid-anie2042>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 21.Scholz O, Henssler EM, Bail J, Schubert P, Bogdanska-Urbaniak J, Sopp S, Reich M, Wisshak S, Kostner M, Bertram R, Hillen W. Activity reversal of Tet repressor caused by single amino acid exchanges. Mol Microbiol. 2004;53:777–789. doi: 10.1111/j.1365-2958.2004.04159.x. [DOI] [PubMed] [Google Scholar]
- 22.Choi SH, Greenberg EP. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc Natl Acad Sci U S A. 1991;88:11115–11119. doi: 10.1073/pnas.88.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 25.Devine JH, Shadel GS, Baldwin TO. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc Natl Acad Sci U S A. 1989;86:5688–5692. doi: 10.1073/pnas.86.15.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egland KA, Greenberg EP. Quorum sensing in Vibrio fischeri: elements of the luxl promoter. Mol Microbiol. 1999;31:1197–1204. doi: 10.1046/j.1365-2958.1999.01261.x. [DOI] [PubMed] [Google Scholar]
- 27.Finney AH, Blick RJ, Murakami K, Ishihama A, Stevens AM. Role of the C-terminal domain of the alpha subunit of RNA polymerase in LuxR-dependent transcriptional activation of the lux operon during quorum sensing. J Bacteriol. 2002;184:4520–4528. doi: 10.1128/JB.184.16.4520-4528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berens C, Hillen W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur J Biochem. 2003;270:3109–3121. doi: 10.1046/j.1432-1033.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- 29.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Group CC. MOE: Molecular Operating Environment. 2005. [Google Scholar]
- 31.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biliouris K, Daoutidis P, Kaznessis YN. Stochastic simulations of the tetracycline operon. BMC Syst Biol. 5:9. doi: 10.1186/1752-0509-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill AD, Tomshine JR, Weeding EM, Sotiropoulos V, Kaznessis YN. SynBioSS: the synthetic biology modeling suite. Bioinformatics. 2008;24:2551–2553. doi: 10.1093/bioinformatics/btn468. [DOI] [PubMed] [Google Scholar]
- 34.Salis H, Kaznessis YN. Numerical simulation of stochastic gene circuits. Comp Chem Eng. 2005;29:577–588. [Google Scholar]
- 35.Kaznessis YN. Computational methods in synthetic biology. Biotechnol J. 2009;4:1392–1405. doi: 10.1002/biot.200900163. [DOI] [PubMed] [Google Scholar]
- 36.Kaznessis YN. Mathematical models in biology: from molecules to life. Wiley Interdiscip Rev Syst Biol Med. 2011;3:314–22. doi: 10.1002/wsbm.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weeding E, Houle J, Kaznessis YN. SynBioSS designer: a web-based tool for the automated generation of kinetic models for synthetic biological constructs. Brief Bioinform. 2010;11:394–402. doi: 10.1093/bib/bbq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salis H, Kaznessis Y. Accurate hybrid stochastic simulation of a system of coupled chemical or biochemical reactions. J Chem Phys. 2005;122:54103. doi: 10.1063/1.1835951. [DOI] [PubMed] [Google Scholar]
- 39.Salis H, Sotiropoulos V, Kaznessis YN. Multiscale Hy3S: hybrid stochastic simulation for supercomputers. BMC Bioinformatics. 2006;7:93. doi: 10.1186/1471-2105-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sotiropoulos V, Kaznessis YN. Synthetic tetracycline-inducible regulatory networks: computer-aided design of dynamic phenotypes. BMC Syst Biol. 2007;1:7. doi: 10.1186/1752-0509-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuttle LM, Salis H, Tomshine J, Kaznessis YN. Model-driven designs of an oscillating gene network. Biophys J. 2005;89:3873–3883. doi: 10.1529/biophysj.105.064204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golan DE, AHT, Armstrong EJ, Armstrong AW. Principles of pharmacology: the pathophysiologic basis of drug therapy. Lippincott Williams and Wilkins; Philadelphia: 2005. [Google Scholar]
- 43.Meynial-Salles I, Cervin MA, Soucaille P. New Tool for Metabolic Pathway Engineering in Escherichia coli: One-Step Method To Modulate Expression of Chromosomal Genes. Appl Environ Microbiol. 2005;71:2140–2144. doi: 10.1128/AEM.71.4.2140-2144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 45.Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Marco SD. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doi M, SFE . The Theory of Polymer Dynamics. Clarendon Press; 1986. [Google Scholar]
- 47.George RA, Heringa J. An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng. 2002;15:871–879. doi: 10.1093/protein/15.11.871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.